Abstract
The cell wall of Mycobacterium tuberculosis (Mtb) is a complex structure that protects the pathogen in hostile environments. Peptidoglycan (PG), which helps determine the morphology of the cell envelope, undergoes substantial remodeling under stress. This meshwork of linear chains of sugars, cross-linked through attached peptides, is generated through the sequential action of enzymes termed transglycosylases and transpeptidases. The Mtb genome encodes two classical transglycosylases and four transpeptidases, the functions of which are not fully elucidated. Here, we present work on the yet uncharacterized transpeptidase PbpA and a nonclassical transglycosylase RodA. We elucidate their roles in regulating in vitro growth and in vivo survival of pathogenic mycobacteria. We find that RodA and PbpA are required for regulating cell length, but do not affect mycobacterial growth. Biochemical analyses show PbpA to be a classical transpeptidase, whereas RodA is identified to be a member of an emerging class of noncanonical transglycosylases. Phosphorylation of RodA at Thr-463 modulates its biological function. In a guinea pig infection model, RodA and PbpA are found to be required for both bacterial survival and formation of granuloma structures, thus underscoring the importance of these proteins in mediating mycobacterial virulence in the host. Our results emphasize the fact that whereas redundant enzymes probably compensate for the absence of RodA or PbpA during in vitro growth, the two proteins play critical roles for the survival of the pathogen inside its host.
Keywords: bacterial pathogenesis, cell division, cell wall, Mycobacterium tuberculosis, peptidoglycan, Lipid II, PbpA, RodA, transglycosylase, transpeptidase
Introduction
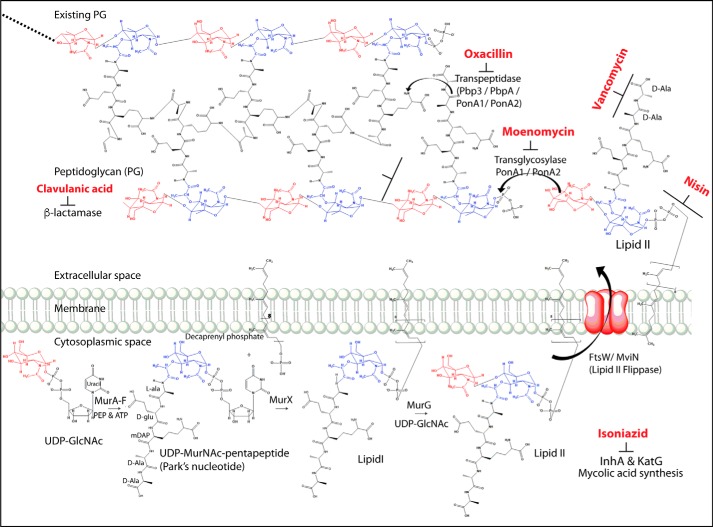
The Mycobacterium tuberculosis (Mtb)2 cell wall is a complex structure that provides osmotic stability, drug resistance, and enhanced virulence (1–3) and protects it from stress conditions in the host, such as reactive oxygen species, starvation, and hypoxia (4–9). Peptidoglycan (PG), a primary morphological determinant of the cell envelope, is a covalently linked network of glycan chains bridged through peptide bonds (10). PG synthesis is critically regulated to rheostat the cell growth and division for optimal bacterial survival. The initial steps of PG biosynthesis from UDP-GlcNAc to Lipid II are performed in the bacterial cytoplasm by Mur family of enzymes (Fig. 1). Subsequently, Lipid II anchored to the intracellular membrane is flipped into the periplasmic space by flippase followed by transglycosylation, wherein the sugar moieties are linked to the existing chain through glycosidic bonds, via transglycosylases. Subsequent cross-linking of peptides through transpeptidation by penicillin-binding proteins (PBPs) completes the cross-linked protective PG structure (Fig. 1). Due to the limited pool of available Lipid II molecules (11, 12), the enzymes involved in its synthesis, transport, and utilization are excellent targets for therapeutic intervention (13–15). Recently, a new class of antibiotic, teixobactin, a specific inhibitor of Lipid II, has been shown to effectively kill multiple Gram-positive bacilli, including drug-resistant Mtb (16).
Figure 1.
Schematic diagram depicting the enzymes catalyzing multiple steps of the peptidoglycan biosynthesis pathway and the stages at which PG biosynthesis inhibitors act. The pathway was drawn with the help of ChemDraw Ultra version 12.0 software.
Lipid II amounts are regulated in the periplasmic space through the enzymes involved in its flipping (Lipid II flippase) and utilization (transglycosylase). The identity of Lipid II flippases involved in regulating its levels in the periplasmic space varies among different classes of bacteria. FtsW in Escherichia coli, RodA in Corynebacterium glutamicum, MurJ from multiple bacterial species, Wzk from Helicobacter pylori, and AmiJ from Bacillus subtilis (17, 18) have been demonstrated to function as Lipid II flippases (19–22). The presence of multiple candidates capable of performing Lipid II flippase activity suggests functional redundancy and possible spatiotemporal regulation. MviN (a homolog of MurJ), FtsW, and RodA are hypothesized to function as possible Lipid II flippase in Mtb. Conditional depletion of MviN (an essential protein for in vitro growth) in Mycobacterium smegmatis (Msm) leads to the accumulation of PG precursors in the cytosol, thus suggesting a possible role for MviN as a flippase (23). RodA, FtsW, and SpoVE are members of the shape, elongation, division, and sporulation (SEDS) family of proteins, with as yet ill-defined roles in cell wall biosynthesis during growth, division, and sporulation (24). Interestingly, recent studies have uncovered a novel role for RodA as an unconventional transglycosylase in B. subtilis and E. coli (25–27). However, the role of FtsW or RodA remains uncharacterized in mycobacteria to date.
The mycobacterial genome encodes for 10 PBPs, which can be broadly categorized into three classes based on their functions (28). Class I consists of two subclasses, A and B; Class A comprises bifunctional enzymes that possess both transglycosylase and transpeptidase activities, and Class B enzymes are monofunctional with only the transpeptidase activity. In Mtb, there are four Class I high-molecular weight PBPs, namely PonA1, PonA2, PbpA, and Pbp3. Whereas PonA1 and PonA2 can perform both transglycosylase and transpeptidase activities, PbpA and Pbp3 can only carry out a transpeptidation reaction. There are six enzymes belonging to Class II and III PBPs, which function as carboxypeptidases and β-lactamases involved in the maintenance of PG. With the exception of Pbp3, all of the remaining PBPs are nonessential for in vitro growth. The genes encoding SEDS members are found in proximity to the genes of Class B PBPs, suggesting functional association among them (25). One such example is FtsW and Pbp3, which are shown to work as a pair in PG biosynthesis in E. coli and Mycobacterium species (26, 29, 30). RodA and PbpA, which are located next to each other, are also hypothesized to work as a pair; however, their roles in cell division and PG biosynthesis remain to be characterized.
Genes encoding for mycobacterial rodA and pbpA are located in the same operon that carries serine/threonine phosphatase pstP and two serine/threonine protein kinases (STPKs), pknA and pknB (31) (Fig. 2a). We have previously reported that PknA, PknB, and PstP are independently essential for in vitro growth as well as in vivo survival of Mtb (32–34). Previous studies by multiple groups have suggested important roles for STPKs in regulating cell division and cell wall synthesis processes (35–37). Due to the presence of rodA and pbpA in the pknA and pknB operon (Fig. 2a), these genes are also speculated to play roles in modulating cell division and cell wall synthesis. The data presented in this study provide the first insight into the roles of rodA and pbpA genes in mycobacterial morphology, growth, and survival in vitro and in vivo.
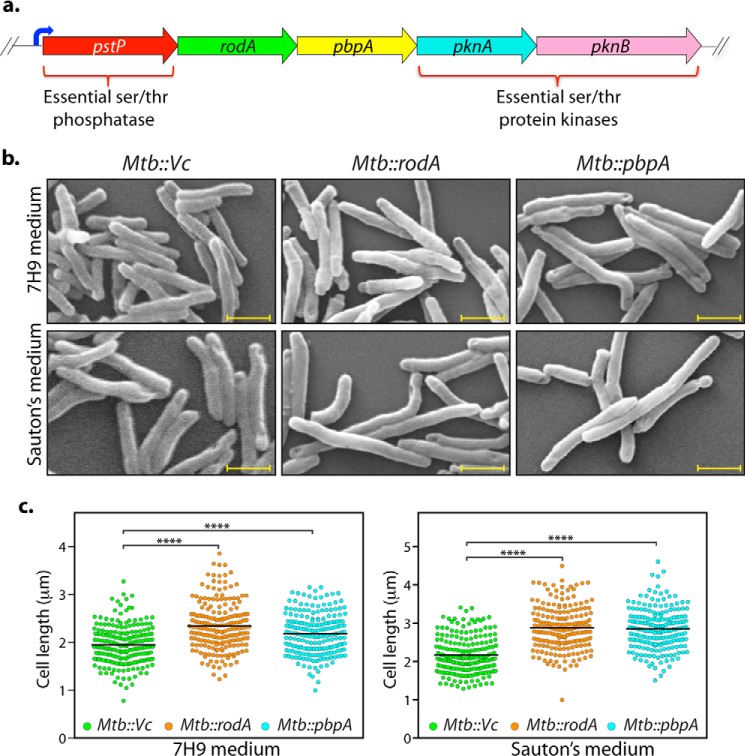
Figure 2.
Overexpression of RodA and PbpA leads to cell elongation in Mtb. a, pictorial representation of the operon encoding rodA and pbpA. Upstream serine/threonine phosphatase (pstp) and downstream serine/threonine kinase genes (pknA and pknB) are indicated. b, fresh cultures of Mtb::Vc (vector control), Mtb::rodA, or Mtb::pbpA were seeded at an initial A600 of 0.1 in either 7H9 (top) or Sauton's medium (bottom) in the presence of 100 ng/ml anhydrotetracycline, and cells were allowed to grow for 6 days at 37 °C at 100 rpm and fixed. Morphology of Mtb::Vc, Mtb::rodA, and Mtb::pbpA was observed through scanning EM at ×20,000. Scale bar, 1.0 μm. c, quantification of cell lengths in Mtb::Vc, Mtb::rodA, and Mtb::pbpA strains (n ∼200) from cells grown in 7H9 medium (left) or Sauton's medium (right). Mean cell lengths obtained were 1.9 μm (Mtb::Vc), 2.34 μm (Mtb::rodA), and 2.18 μm (Mtb::pbpA) in 7H9 medium and 2.1 μm (Mtb::Vc), 2.8 μm (Mtb::rodA), and 2.8 μm (Mtb::pbpA) in Sauton's medium. Cell lengths were measured independently using Smart Tiff software and plotted as a scattered dot plot with mean values using GraphPad Prism version 6. The experiments were biologically and technically repeated three times. Statistical analysis was performed with the help of a one-way ANOVA test. ****, p < 0.0001.
Results
Overexpression of RodA and PbpA leads to cell length elongation in Mtb
To delineate the role of RodA and PbpA on mycobacterial cell morphology regulation, we sought to determine the impact of overexpression of these proteins in Mtb. Toward this, rodA and pbpA genes were cloned downstream of the tetracycline-inducible promoter in pST-KT vector (38), and the plasmids were electroporated into Mtb. Transformants were fixed 6 days postinduction, followed by scanning EM (SEM) analysis. A distinct increase in the cell length of the transformant bacteria was observed, with average cell length of Mtb::rodA and Mtb::pbpA in nutrient-rich 7H9 medium increasing from ∼1.9 μm to 2.34 and 2.18 μm, respectively (Fig. 2, b and c). The increase in the average cell length was more noteworthy in nutrient-limiting Sauton's medium, wherein it increased from ∼2.1 to ∼2.8 μm in the case of both transformant types (Fig. 2, b and c). Similar results were obtained in Msm transformants (Fig. S1, a and b). It is possible that the cell length elongation phenotype observed is due to their probable roles in PG biosynthesis. Overexpression may have resulted in uncoordinated PG biosynthesis and, consequently, increase in bacterial cell lengths.
RodA and PbpA play independent roles in modulating bacterial cell length
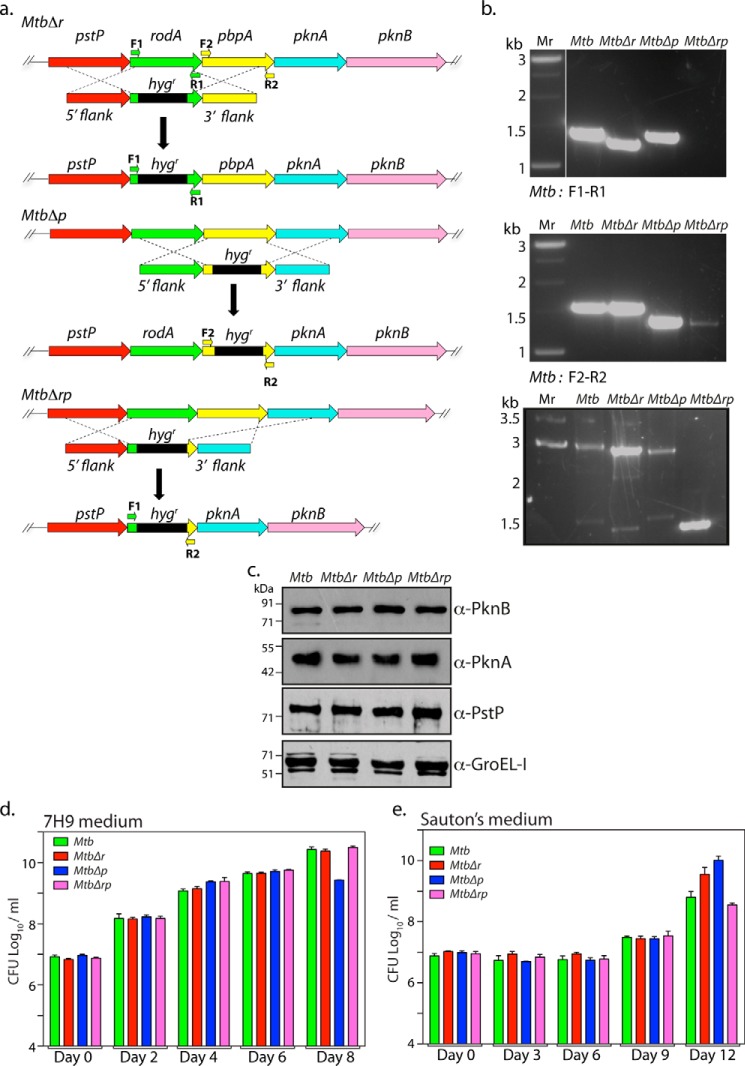
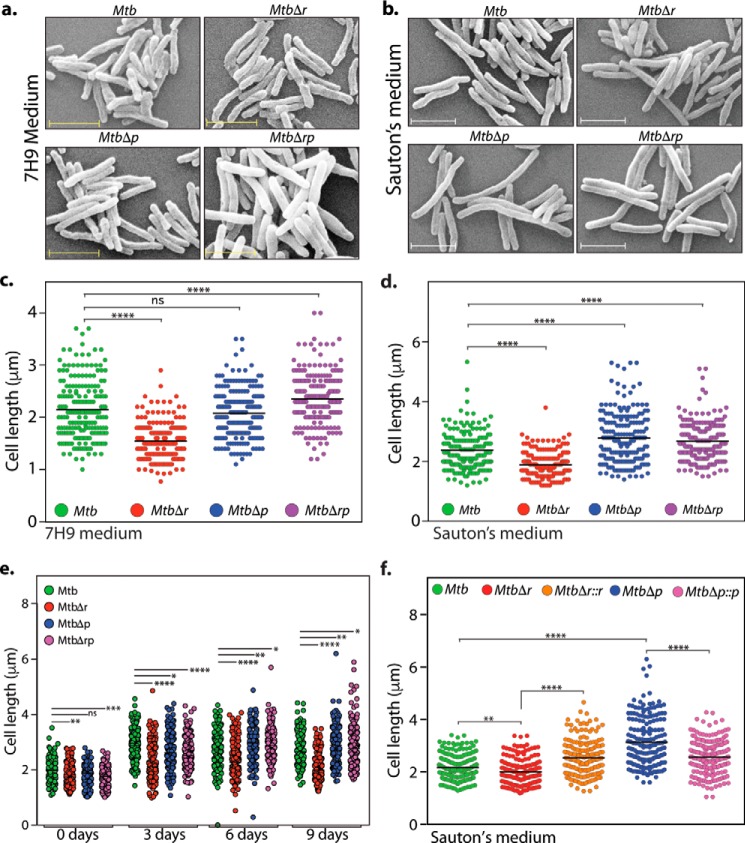
Although E. coli rodA and pbpA orthologs are essential genes, they are not essential in mycobacteria (39, 40). To examine the functions of RodA and PbpA in modulating growth and morphology in vitro and survival in vivo (both independently and combinatorially), rodA, pbpA, and rodA-pbpA gene replacement mutants in Mtb and Msm strains were made using a recombineering method (41) (Fig. 3a and Fig. S1c). PCR analysis with specific primer pairs confirmed the replacement of the genes with an hygr marker at the native locus (Fig. 3b and Fig. S1 (d and e)). Because rodA and pbpA are located upstream of essential kinases pknA and pknB and downstream of essential serine/threonine phosphatase, pstP, it is necessary to ascertain any polarity effects in the deletion mutants. Western blot analysis of lysates isolated from WT and mutant strains showed comparable expression of PstP, PknA, PknB, and GroEL-I (control), indicating that gene replacement mutants are devoid of any polarity effects both in Mtb (Fig. 3c) and Msm (Fig. S1f). We evaluated the impact of deleting rodA or pbpA, or both rodA and pbpA, on mycobacterial survival by enumerating cfu on different days in growth kinetics for both 7H9 and Sauton's medium. No significant differences were observed (Fig. 3, d and e), leading us to conclude that deletion of rodA or pbpA or both does not impact in vitro growth of Mtb. Unlike in E. coli, where conditional depletion of RodA or PbpA alters the cellular morphology from rod to round shape (42), no drastic changes in the morphology of mycobacterial deletion mutants were observed in our SEM studies (Fig. 4, a and b). However, whereas MtbΔr cells shortened significantly in both 7H9 and Sauton's medium, MtbΔp cells showed no significant difference in 7H9 medium but minor and significant changes in bacterial cell lengths in nutrient-limiting Sauton's medium (Fig. 4, c and d). More apparent defects in nutrient-limiting Sauton's medium compared with rich 7H9 medium upon overexpression or deletion could be due to pertinent roles played by these proteins under stress conditions. To analyze whether the cell length phenotype depended on growth phase, we grew all of the strains in Sauton's medium for different periods of time (0, 3, 6, and 9 days) before analyzing the cell lengths with the help of SEM. Whereas the shorter-cell length phenotype observed with MtbΔr was observed at every time point, the cell length phenotype altered across the growth phases in MtbΔp and MtbΔrp strains (Fig. 4e). In the case of MtbΔp and MtbΔrp, the cells were initially shorter. However, at the later phase of growth (days 6 and 9), the cells were more elongated, consistent with observations in Fig. 4d. These differences could be due to nutrient limitations as the growth progresses, leading to the necessity of higher levels of redundant enzymes, such as PBPs, involved in peptidoglycan synthesis. To confirm that the observed aberrations in the cell length were due to the absence of RodA or PbpA, we performed complementation studies to rescue the mutant phenotypes by episomal expression of the respective proteins. The anomalous cell lengths observed in MtbΔr and MtbΔp cells were successfully reversed upon complementation (Fig. 4f).
Figure 3.
Generation of rodA, pbpA, and rodA-pbpA gene replacement mutants in Mtb. a, schematic depiction of the strategy used for the generation of gene deletion mutants. Primers used for the PCR confirmation are indicated. b, genomic DNA was isolated from log cultures of WT and mutants, and PCRs were performed with a defined set of primers (as indicated). Lane 1, Mr 1-kb ladder; lane 2, Mtb; lane 3, MtbΔr; lane 4, MtbΔp; lane 5, MtbΔrp. The expected sizes for the F1 and R1 pair were as follows: Mtb, 1410 bp; MtbΔr, 1300 bp; MtbΔp, 1410 bp; MtbΔrp, 0. The expected sizes for the F2 and R2 pairs were as follows: Mtb, 1476 bp; MtbΔr, 1476 bp; MtbΔp, 1300 bp; MtbΔrp, 0. The expected sizes for the F1 and R2 pairs were as follows: Mtb, 2886 bp; MtbΔr, 2726 bp; MtbΔp, 2660 bp; MtbΔrp, 1300 bp. c, Western blot analysis to detect polarity effects in deletion mutants Mtb, MtbΔr, MtbΔp, and MtbΔrp strains. Strains were grown to an A600 of 0.8–1.0, and WCLs were resolved and probed with α-PstP, α-PknA, α-PknB, and α-GroEL-I antibodies. d and e, Mtb, MtbΔr, MtbΔp, and MtbΔrp strains were inoculated at A600 ∼0.1 in 7H9 or Sauton's medium, and growth was monitored as bacterial survival by cfu enumeration at the indicated time points for 7H9 (d) and Sauton's medium (e). The experiment was performed in triplicate, and the results were plotted using GraphPad Prism version 6. Error bars, S.D.
Figure 4.
RodA and PbpA play independent roles in modulating bacterial cell length. a and b, fresh cultures of Mtb, MtbΔr, MtbΔp, and MtbΔrp were seeded at an initial A600 of 0.1 and grown for 6 days at 37 °C at 100 rpm in 7H9 or Sauton's medium followed by fixation. Morphology of the cells was observed through scanning EM at ×20,000 for 7H9 (a) and Sauton's medium (b). Scale bar, 2.0 μm. c, quantification of cell lengths (n ∼200) of Mtb, MtbΔr, MtbΔp, and MtbΔrp strain cells grown in 7H9 medium was performed. Cell lengths were measured independently using Smart Tiff software and plotted by a scatter dot plot with mean values using GraphPad Prism version 6. Mean cell lengths obtained were as follows: Mtb, 2.1 μm; MtbΔr, 1.5 μm; MtbΔp, 2.0 μm; MtbΔrp, 2.3 μm. The experiments were biologically and technically repeated twice. Cell lengths were analyzed using GraphPad Prism version 6, and statistical analysis was performed using a one-way ANOVA test. ****, p < 0.0001; ***, p < 0.001; ns, not significant. d, quantification of cell lengths in Mtb, MtbΔr, MtbΔp, and MtbΔrp strain (n ∼200) cells grown in Sauton's medium was performed, and mean cell lengths obtained were as follows: Mtb, 2.3 μm; MtbΔr, 1.8 μm; MtbΔp, 2.7 μm; MtbΔrp, 2.6 μm. The experiments were biologically and technically repeated twice. Cell lengths were measured independently using Smart Tiff software and plotted by a scattered dot plot with mean values using GraphPad Prism version 6, and the statistical analysis was performed with a one-way ANOVA test. ****, p < 0.0001; ***, p < 0.001. e, fresh cultures of Mtb, MtbΔr, MtbΔp, and MtbΔrp were seeded at an initial A600 of 0.1 and grown at 37 °C at 100 rpm in Sauton's medium followed by fixation at different time points (days 0, 3, 6, and 9) of growth, and cell lengths were measured as described above. Mean cell lengths obtained for different time points were as follows: day 0: Mtb, 1.98 μm; MtbΔr, 1.80 μm; MtbΔp, 1.84 μm; and MtbΔrp, 1.76 μm; day 3: Mtb, 2.87 μm; MtbΔr, 2.26 μm; MtbΔp, 2.69 μm; and MtbΔrp, 2.62 μm; day 6: Mtb, 2.72 μm; MtbΔr, 2.32 μm; MtbΔp, 2.92 μm; and MtbΔrp, 2.90 μm; and day 9: Mtb, 2.71 μm; MtbΔr, 2.0 μm; MtbΔp, 2.91 μm; and MtbΔrp, 2.88 μm. Cell lengths were analyzed using GraphPad Prism version 6, and statistical analysis was performed using a one-way ANOVA test. ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; ns, not significant. f, fresh cultures of Mtb, MtbΔr, MtbΔr::r, MtbΔp, or MtbΔp::p were seeded at an initial A600 of 0.1 in Sauton's medium and continued to grow for 6 days in the presence of 0.1 μm IVN, and cells were fixed and processed for SEM. Cell lengths were observed through SEM at ×20,000. Cell lengths were measured as described above. Obtained mean cell lengths were as follows: Mtb, 2.1 μm; MtbΔr, 1.9 μm; MtbΔr::r, 2.5 μm; MtbΔp, 3.1 μm; MtbΔp::p, 2.5 μm. Statistical analysis was performed using a two-way ANOVA test; ****, p < 0.0001; **, p < 0.01.
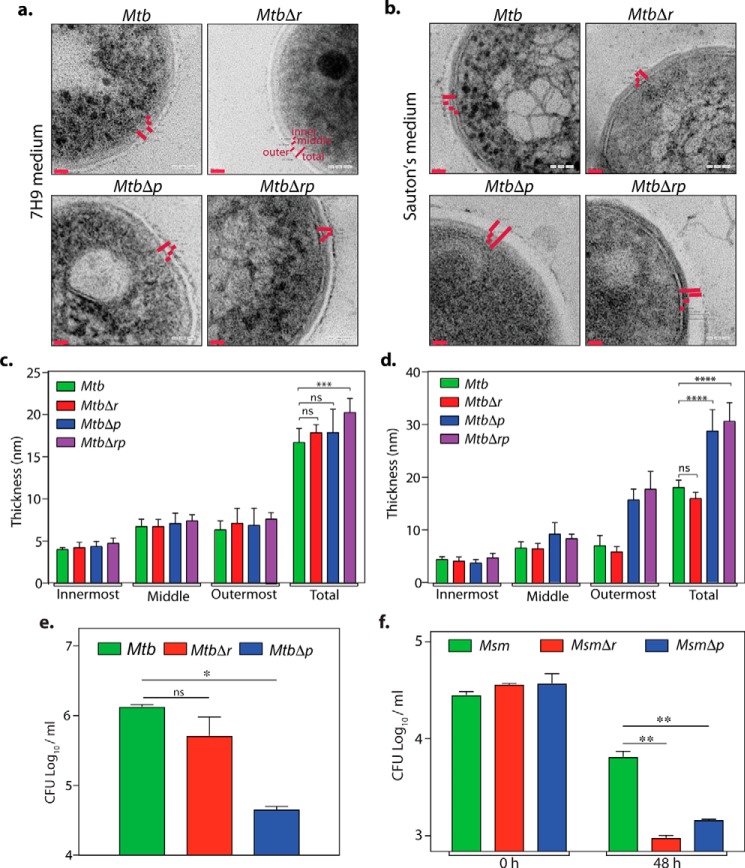
WT and gene replacement mutant strains of Mtb were subjected to transmission EM (TEM) analysis to evaluate the impact of gene deletions on cell wall ultrastructure (Fig. 5, a–d). Whereas the WT and deletion mutants grown in 7H9 medium showed no significant difference in cell wall ultrastructure (Fig. 5, a and c), we observed conspicuous changes in the cell wall architecture of MtbΔp and MtbΔrp in Sauton's medium as compared with the Mtb and MtbΔr strains (Fig. 5, b and d). The outermost electron-dense opaque layer of MtbΔp and MtbΔrp was strikingly thicker (∼30 nm) as compared with Mtb (∼18 nm) and MtbΔr (∼15.9 nm). We speculate that the observed phenotype for MtbΔp and MtbΔrp mutants could be an adaptive response to the compromised cellular fitness under nutrient-limiting conditions. Thus, we evaluated the possible impact of deletion of rodA or pbpA on survival under hypoxia and persistence (Fig. 5, e and f). In line with the results above, we observed decreased survival only with the MtbΔp mutant in the Wayne model of hypoxia (43) (Fig. 5e). However, in persisters analysis, wherein Msm WT and mutant cultures were exposed to 10 μg/ml isoniazid, both MsmΔr and MsmΔp mutants showed a 10-fold decline in cfu (Fig. 5f). Taken together, these data suggest that both RodA and PbpA may play a role in combating survival under different stress conditions.
Figure 5.
Deciphering the roles of rodA and pbpA in mycobacterial cell fitness. a–d, TEM analysis of Mtb rodA and pbpA deletion mutants in 7H9 (a and c) or Sauton's medium (b and d). Fresh cultures of Mtb, MtbΔr, MtbΔp, and MtbΔrp were seeded at an initial A600 of 0.1 and grown for 6 days at 37 °C at 100 rpm in 7H9 or Sauton's medium followed by fixation. Fixed cells were processed for TEM, and cell wall architecture was observed at 200 kV, ×50,000 in 7H9 (a) and Sauton's medium (b). c and d, quantification of cell wall thickness in MtbΔr, MtbΔp, MtbΔrp strains (n = 6–7) grown in 7H9 medium (mean cell wall thickness: Mtb, MtbΔr, and MtbΔp, 17 nm; MtbΔrp, 20.2 nm) (c) or Sauton's medium (mean cell wall thickness: Mtb, 18 nm; MtbΔr, 15.9 nm; MtbΔp, 28.7 nm; MtbΔrp, 30.6 nm) (d). Cell wall thickness was measured independently using SmartTiff software and analyzed using GraphPad Prism version 6, and statistical analysis was performed using a two-way ANOVA test. ****, p < 0.0001; ***, p < 0.001. Red bars, cell wall thickness. e and f, hypoxia and persisters analysis for Mtb and Msm deletion mutants. e, bacterial survival analysis of day 42 posthypoxia (Wayne model) for Mtb, MtbΔr, and MtbΔp. Results were plotted using GraphPad prism version 6. Error bar, S.D. Statistical analysis was performed using a two-tailed test. *, p < 0.05. Similar results were obtained in two independent experiments performed in duplicates. f, persisters analysis of Msm, MsmΔr, and MsmΔp. Cultures were grown until exponential phase and treated with 10 μg/ml isoniazid for 48 h, and cfu obtained before and after isoniazid treatment were plotted. Statistical analysis was performed using a two-tailed test. **, p < 0.005. Error bars, S.D. Similar results were obtained in two independent experiments performed in triplicates.
PbpA functions as a transpeptidase
RodA and PbpA function at different stages of PG biosynthesis (outlined in Fig. 1). RodA has been shown to function either as a Lipid II flippase in E. coli and C. glutamicum or, more recently, as a noncanonical transglycosylase in B. subtilis (25). Based on sequence homology and crystal structure, PbpA is annotated as a transpeptidase (44). To identify the stages of the PG biosynthesis pathway at which mycobacterial RodA and PbpA function, we began with determining the sensitivity of Msm, MsmΔr, and MsmΔp strains to various inhibitors known to act at different steps of the PG biosynthesis pathway (Fig. 1). As expected, Msm, MsmΔr, and MsmΔp strains showed similar sensitivity to isoniazid, an inhibitor of InhA, an enzyme involved in mycolic acid synthesis. However, MsmΔp showed much higher sensitivity to oxacillin + clavulanic acid (oxacillin, a pan-inhibitor of transpeptidases; clavulanic acid, a potent inhibitor of β-lactamases, which enhances the inhibitory potential of oxacillin) compared with MsmΔr or Msm (Fig. 6a), which is consistent with its predicted role as a transpeptidase. In trans complementation of PbpA restored the sensitivity values closer to Msm (Fig. 6b).
Figure 6.
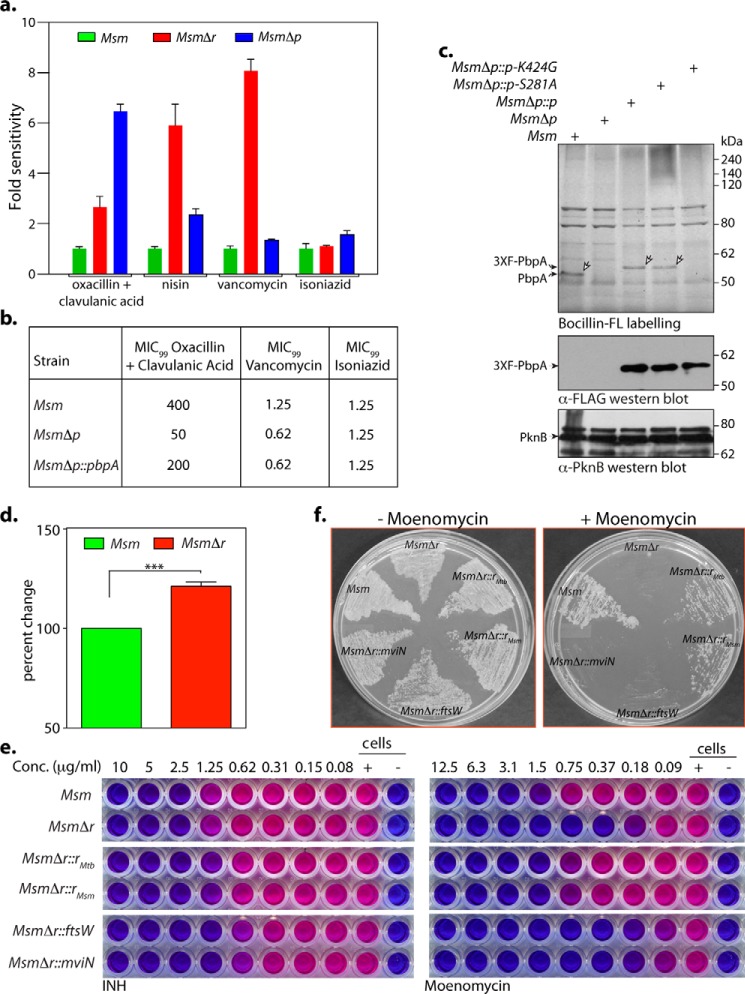
RodA functions as a noncanonical transglycosylase and PbpA as a classical transpeptidase. a, MIC analysis of Msm, MsmΔr, and MsmΔp was performed as described under “Experimental procedures.” The graph presents the -fold difference with respect to Msm in (cfu × MIC) values obtained for MsmΔr and MsmΔp. The experiment was performed in triplicate. b, MIC99 analysis for MsmΔp complementation studies. Shown is a tabular representation of MIC99 values obtained upon a REMA performed with Msm, MsmΔp, and MsmΔp::p strains in Dubo's medium for oxacillin + clavulanic acid, vancomycin, and isoniazid. c, membrane fractions were prepared from cultures of Msm, MsmΔp, MsmΔp::p MsmΔp::pS281A, and MsmΔp::pK424G induced with 5 μm IVN. ∼20 μg of membrane was incubated with Bocillin-FL and resolved on 8% SDS-PAGE, and Bocillin-labeled proteins were detected using a Typhoon scanner. A similar amount of membrane extracts were processed for immunoblot analysis. Bands corresponding to endogenous PbpA and episomal 3X-FLAG-PbpAwt/mut are indicated. d, Msm and MsmΔr strains grown in Sauton's medium until A600 ∼0.2–0.3 were pulsed with [3H]mDAP, and equal amounts of cultures were processed for small-scale Lipid II accumulation analysis. Radiolabeled [3H]Lipid II in the samples was quantified. The reading obtained for Msm in each independent experiment was normalized to 100%, and the counts obtained for MsmΔr are represented as percentage change with respect to counts obtained for Msm. The experiment was performed in biological triplicates. Statistical analysis was performed using a two-tailed test. ***, p < 0.0005. e, REMA for Msm, MsmΔr, MsmΔr::rmtb, MsmΔr::rmsm, MsmΔr::ftsW, and MsmΔr::mviN for isoniazid and moenomycin. The experiment was performed in triplicate, and a representative data set is shown. f, growth analysis of Msm, MsmΔr, MsmΔr::rmtb, MsmΔr::rmsm, MsmΔr::ftsW, and MsmΔr::mviN, streaked on 7H11 in the presence or absence of 1 μg/ml moenomycin. The experiment was performed in triplicate, and a representative data set is shown.
To evaluate PbpA transpeptidase activity, Bocillin-FL labeling assays were performed as described earlier (45). Bocillin-FL is essentially penicillin tagged with a fluorescent probe, which forms a stable covalent intermediate with the catalytically active transpeptidase enzymes. Membrane fractions prepared from Msm, MsmΔp, MsmΔp complemented strains were incubated with Bocillin-FL, reactions were resolved by gel electrophoresis, and the gels were analyzed by scanning them. Although we could detect a band of the appropriate molecular mass corresponding to PbpA (∼50 kDa) in the case of the Msm membrane fraction, the same was absent in the MsmΔp membrane fraction, suggesting that PbpA is indeed a transpeptidase (Fig. 6c). Structural analysis of PbpAMtb suggested Ser-281 and Lys-424 residues to be a part of the catalytic site (44). We generated PbpA-S281A and PbpA-K424G mutants and investigated their ability to rescue the depleted transpeptidase activity of MsmΔp. Whereas WT and PbpA-S281A mutants could rescue the lost transpeptidase activity (Fig. 6c, shown by arrows), PbpA-K424G complementation did not, suggesting that the Lys-424 residue plays an important role in mediating the transpeptidase activity of PbpA.
RodA functions as a noncanonical transglycosylase
Next, we evaluated the role of RodA by assessing the sensitivity of rodA deletion mutant toward nisin and vancomycin. Nisin is an inhibitor, which binds to the pyrophosphate group of Lipid II, forms pores in the membrane, and eventually leads to cell death. If RodA were to function as a Lipid II flippase, the absence of RodA would result in reduced levels of Lipid II in the periplasmic space. In C. glutamicum, where RodA functions as a Lipid II flippase, deletion of rodA resulted in resistance toward nisin (20). Previous data have established that enhanced Lipid II content leads to higher sensitivity to nisin (46). We observed that RodA deletion (but not deletion of PbpA) resulted in higher sensitivity of Msm toward nisin (Fig. 6a and Tables 1 and 2). Vancomycin is another sensor of Lipid II, which binds to the d-Ala-d-Ala terminal amino acids of the pentapeptide in Lipid II, inhibiting nascent PG biosynthesis (13). Approximately 8-fold higher sensitivity of MsmΔr to vancomycin compared with Msm was noted, thus clearly omitting the possibility of RodA functioning as a flippase in mycobacteria (Fig. 6a and Tables 1 and 2). The enhanced sensitivity of MsmΔr to nisin and vancomycin suggests the possibility of higher accumulation of Lipid II molecules in the periplasmic space of MsmΔr mutant, which could be due to hampered noncanonical transglycosylase activity of RodA. To investigate whether higher levels of Lipid II molecules are accumulated in MsmΔr cells compared with WT Msm cells, we pulsed actively growing Msm and MsmΔr cultures with [3H]meso-diaminopimelic acid ([3H]mDAP) for ∼4–5 h to label the lipid-linked peptidoglycan precursors, including Lipid II (Fig. 6d). Equal quantities of these cells were processed for small-scale Lipid II accumulation analysis (47), and the amount of [3H]Lipid II was quantitated. We observed a consistent ∼20% increase in the counts, suggesting that the increased sensitivity to nisin and vancomycin is indeed due to higher accumulation of Lipid II.
Table 1.
MIC99 values
| Strain | Vancomycin | Nisin | Oxacillin + clavulanic acid | Moenomycin | Isoniazid |
|---|---|---|---|---|---|
| μg/ml | |||||
| Msm | 2.5–1.25 | 100–50 | 250–125 | 3.1 | 5.0–2.50 |
| MsmΔr | 0.162–0.15 | 25–12.5 | ≤31.25 | 0.18 | 2.5–1.25 |
| MsmΔp | 0.625 | 50–25 | ≤31.25 | 0.75 | 2.5–1.25 |
| MsmΔrp | 0.3125 | 25–12.5 | ≤31.25 | 0.37 | 2.5–1.25 |
Table 2.
MIC99 values Msm, MsmΔr MsmΔr::r complementation strains
| Strain | Moenomycin | Vancomycin | Nisin | Isoniazid |
|---|---|---|---|---|
| μg/ml | ||||
| Msm | 1.5 | 0.62 | 100 | 2.5 |
| MsmΔr | 0.18 | 0.15 | 50 | 2.5 |
| MsmΔr::rmtb | 0.75 | 0.31 | 50 | 2.5 |
| MsmΔr::rmsm | 0.75 | 0.31 | 100 | 5.0 |
| MsmΔr::ftsW | 0.18 | 0.15 | 50 | 2.5 |
| MsmΔr::mviN | 0.18 | 0.15 | 50 | 2.5 |
| MsmΔr::rmtb-W175R | 0.18 | 0.15 | 50 | 2.5 |
| MsmΔr::rmtb-D343R | 0.37 | 0.15 | 50 | 2.5 |
| MsmΔr::rmtb-DM | 0.18 | 0.15 | 50 | 2.5 |
| MsmΔr::rmsm-W176R | 0.18 | 0.15 | 50 | 2.5 |
| MsmΔr::rmsm-D344R | 0.18 | 0.15 | 50 | 2.5 |
| MsmΔr::rmsm-DM | 0.18 | 0.15 | 50 | 2.5 |
The possibility of RodA functioning as a noncanonical transglycosylase was evaluated by investigating the sensitivity of WT and mutants to moenomycin. Moenomycin specifically targets the active site of canonical PG glycosyltransferases (transglycosylases), leading to compromised cell wall, resulting in cell content leakage and eventual death (48). If mycobacterial RodA were to function as a noncanonical transglycosylase, the MsmΔr deletion mutant would exhibit hypersensitivity to moenomycin compared with Msm. In line with this hypothesis, MsmΔr strain was ∼6-fold more sensitive compared with Msm (Fig. 6e and Table 1). Notably, whereas in trans expression of both Msm and Mtb RodA efficiently restored the moenomycin sensitivity defect, in trans expression of FtsW or MviN failed do so (Fig. 6e and Table 2). Streaking Msm, MsmΔr, and complemented strains on the plates in the presence or absence of 1 μg/ml moenomycin substantiated the above data (Fig. 6f). Based on these data, we suggest mycobacterial RodA to be part of an emerging class of noncanonical transglycosylases.
Amino acid residues critical for transglycosylase function are conserved in mycobacterial RodA
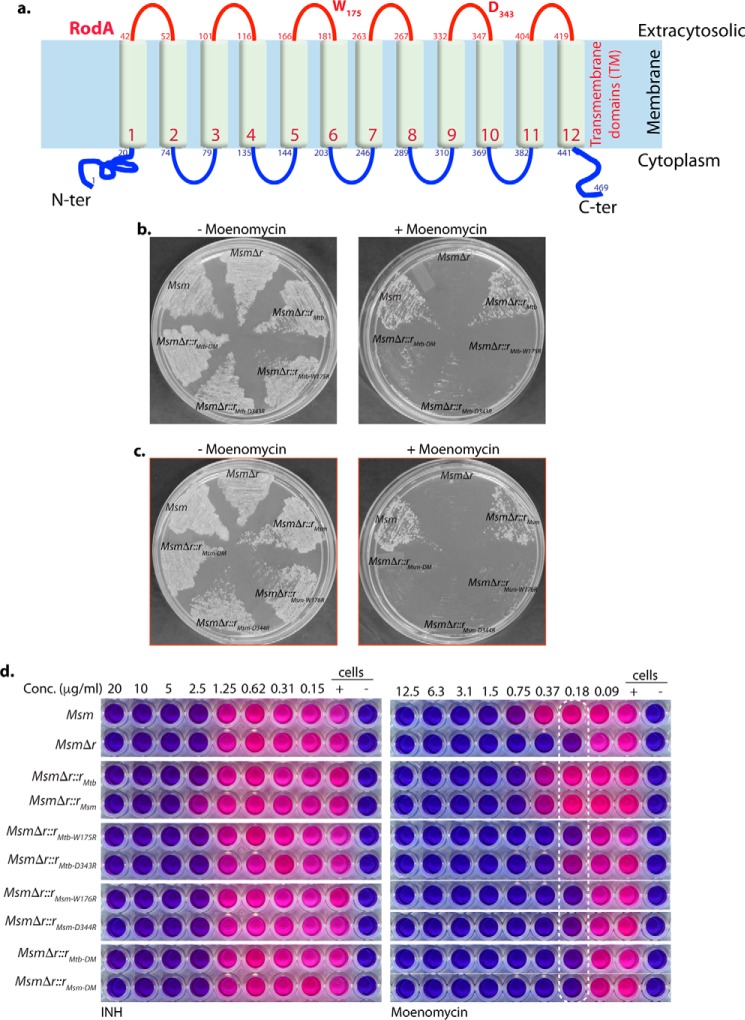
RodA is a well-conserved protein across the bacterial species. Random mutagenesis of B. subtilis RodA has identified a number of residues that are indisputably critical for its function (25). Upon analysis of the primary sequence of mycobacterial RodA, we found ∼80% of these residues to be conserved (data not shown). The amino acid residues Asp-105 and Trp-280 present in the periplasmic loop region of B. subtilis RodA have been biochemically demonstrated to mediate transglycosylase activity in B. subtilis (25). These residues were found to be positionally conserved in the putative periplasmic loop regions of Mtb and Msm RodA at Asp-343/344 and Trp-175/176, respectively (Fig. 7a). To determine the roles of these conserved Asp and Trp residues in mycobacteria, we generated Msm rodA deletion strains complemented with point mutants of these residues in Mtb and Msm RodA. Similar to our previous observations (Fig. 6e), we found MsmΔr mutant to be ∼4–6-fold sensitive to moenomycin compared with Msm. However, whereas in trans expression of WT RodA from Mtb or Msm could rescue the sensitivity phenotype, expression of either single or combinatorial mutants of Asp and Trp residues of RodA failed to complement this moenomycin sensitivity defect. Growth of MsmΔr and MsmΔr complemented with Asp and Trp point mutants from both Mtb and Msm rodA was also specifically attenuated in the presence of moenomycin on solid media on plates (Fig. 7, b and c). In addition, MIC analysis of these mutants in liquid media also yielded similar defect profiles (Fig. 7d and Table 2). Thus, collectively, we can say that conserved catalytic residues Asp-343/344 and Trp-175/176 are critical for the noncanonical transglycosylase activity of mycobacterial RodA.
Figure 7.
Amino acid residues critical for noncanonical transglycosylase function are conserved in mycobacterial RodA. a, schematic depicting the organization of RodA across the membrane. Both the amino and carboxyl termini are predicted to be inside the cytoplasm with 12 transmembrane domains by TMHMM. Residues suggested to be critical for transglycosylase activity, Trp-175 and Asp-343, are marked. b, growth analysis of Msm, MsmΔr, MsmΔr::rMtb, MsmΔr::rMtb-W175R, MsmΔr::rMtb-D343R, and MsmΔr::rMtb-DM, streaked on 7H11 plates in the presence or absence of 1 μg/ml moenomycin. c, growth analysis of Msm, MsmΔr, MsmΔr::rMsm, MsmΔr::rMsm-W176R, MsmΔr::rMsm-D344R, and MsmΔr::rMsm-DM, streaked on 7H11 plates in the presence or absence of 1 μg/ml moenomycin. d, REMA assay for Msm, MsmΔr, MsmΔr::rMtb, MsmΔr::rMsm, MsmΔr::rMtb-W175R, MsmΔr::rMtb-D343R, MsmΔr::rMsm-W176R, MsmΔr::rMsm-D344R, and MsmΔr::rMtb-DM, and MsmΔr::rMsm-DM for isoniazid and moenomycin. The experiment was performed in triplicates, and a representative data set is shown.
RodA is phosphorylated on Thr-463 residue
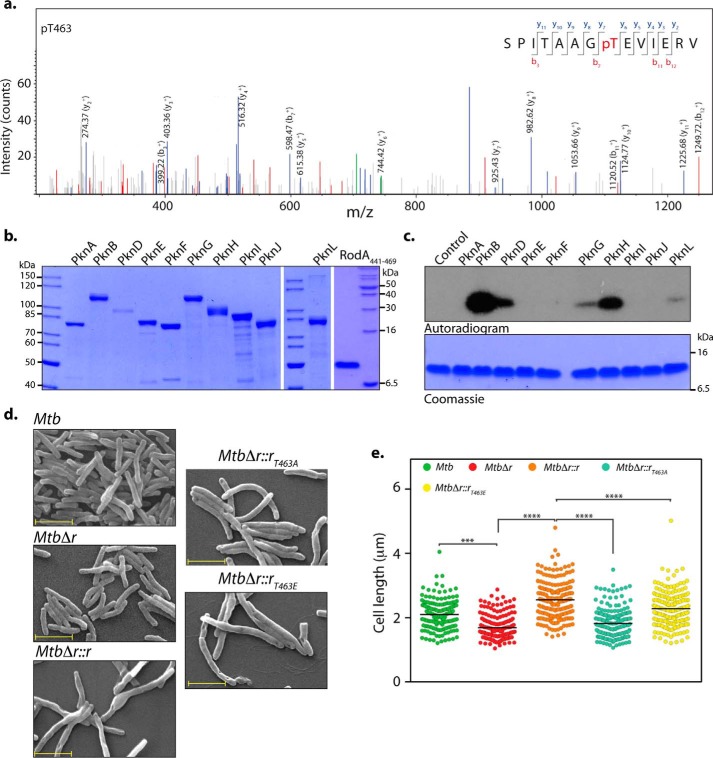
In mycobacteria, phosphorylation of proteins are shown to regulate multiple processes, including mycolic acid synthesis, peptidoglycan synthesis, cell division, and cellular localization (32, 49, 50). PG biosynthesis is a spatiotemporally coordinated event requiring synchronized interactions between numerous proteins involved in cell division and cell wall synthesis. High-throughput analysis of phosphoenriched lysates from Mtb led to the reproducible identification (two of three biological replicates) of a phosphorylation event at Thr-463, a residue belonging to the carboxyl-terminal cytosolic tail region of RodA (Fig. 8a). We sought to identify the STPKs involved in mediating the phosphorylation of the Thr-463 residue of RodA. We were unable to express and purify full-length recombinant RodA due to its poor expression and solubility, perhaps due to the presence of 12 transmembrane domains in the protein (51, 52). Hence, we cloned and purified the carboxyl-terminal RodA(441–469) fragment with amino-terminal His tag (Fig. 8b, right). We next purified 10 MBP-tagged STPKs from a system we have developed previously (49) (Fig. 8b). In vitro kinase assays performed with purified STPKs showed that both PknB and PknH robustly phosphorylate RodA(441–469) (Fig. 8c). In addition to the above kinases, PknG, PknD, and to an extent PknL also mediate RodA(441–469) phosphorylation (Fig. 8c). Together, the data suggest that PknB and PknH are the likely kinases involved in phosphorylating RodA in vivo.
Figure 8.
RodA is phosphorylated on Thr-463 residue. a, MS/MS spectrum of precursor m/z: 761.88025 (+2) and MH+: 1522.75322 Da, of the phosphopeptide SPITAAGpTEVIERV (where pT represents phosphothreonine). The location of Thr-463 was evident by the presence of ion series containing y6, y7, b3, b7, and y8–11 in the spectra. b, Coomassie-stained purified MBP-STPKs and His-FLAG-RodA(411–469). c, an in vitro kinase assay was performed with 10 pmol of MBP-STPKs and 312 pmol of His-FLAG-RodA(411–469). Samples were resolved on 15% SDS-PAGE, stained with Coomassie (bottom), and autoradiographed (top). d, fresh cultures of Mtb, MtbΔr, MtbΔr::r, MtbΔr::rT463A, or MtbΔr::rT463E were seeded at an initial A600 of 0.1 in 7H9 medium and continued to grow for 6 days in the presence of 100 ng of anhydrotetracycline, followed by fixation. SEM was performed to analyze the morphology of the cells (×20,000). Scale bar, 2 μm. e, cell lengths of ∼200 individual cells for each sample were quantified. Mean cell lengths for the samples were 2.1 μm (Mtb), 1.6 μm (MtbΔr), 2.5 μm (MtbΔr::r), 1.8 μm (MtbΔr::rT463A), and 2.2 μm (MtbΔr::rT463E). Similar results were obtained in a biological replicate. Statistical analysis was performed using a one-way ANOVA test; ****, p < 0.0001; ***, p < 0.001.
To investigate the functional significance of phosphorylation at the Thr-463 residue of RodA, we mutated the Thr-463 residue to phosphoablative (T463A) or phosphomimetic (T463E) residues. Toward assessing the role of phosphorylation in modulating catalytic activity of RodA, we evaluated the sensitivity of MsmΔr cells to moenomycin when complemented with WT as well as phosphoablative and phosphomimetic mutants. Both WT and phosphomutants rescued the moenomycin sensitivity phenotype (data not shown), suggesting that phosphorylation does not play any role in modulating noncanonical transglycosylase activity of RodA. Next, we analyzed the impact of phosphorylation on restoring cell length defects observed in the mutant. SEM analysis of Mtb, MtbΔr, and MtbΔr::r complemented strains was consistent with the results in Fig. 4, wherein we observed a decrease in the cell length upon deletion of rodA that was rescued by episomal expression of RodA. The aberrant short-length phenotype could be rescued upon in trans expression RodAT463E; however, the phosphoablative mutant RodAT463A failed to rescue the phenotype, suggesting that phosphorylation of Thr-463 residue is important for the function of RodA (Fig. 8, d and e). The carboxyl terminus of C. glutamicum RodA modulates its interaction with DivIVA (homolog of Wag31Mtb). We speculate that phosphorylation may play an important role in modulating such interactions, thus explaining the inability of phosphomimetic mutant to rescue the observed cell length–defective phenotypes.
RodA and PbpA mutants show compromised bacterial virulence in the host
To investigate the roles of RodA and PbpA in the ability of Mtb to establish infection and survive in the host, we initiated studies using the mouse infection model. We challenged BALB/c mice aerosolically with Mtb, MtbΔr, MtbΔp and MtbΔrp strains, and cfu were enumerated 1 day postinfection to determine the initial bacillary deposition. This was found to be similar in the case of all of the strains (Fig. 9a). Disease progression as assessed by the gross evaluation of lungs and spleen (data not shown) and lung bacillary load 4 weeks postinfection revealed marginal differences between infection by Mtb versus the mutant strains, suggesting that the absence of rodA and pbpA had no impact on mycobacterial survival in the mice model as host (Fig. 9a). We have taken the mouse infection experiment to 12 weeks (data not shown) and have not found any significant differences in the bacterial survival between WT and mutant strains.
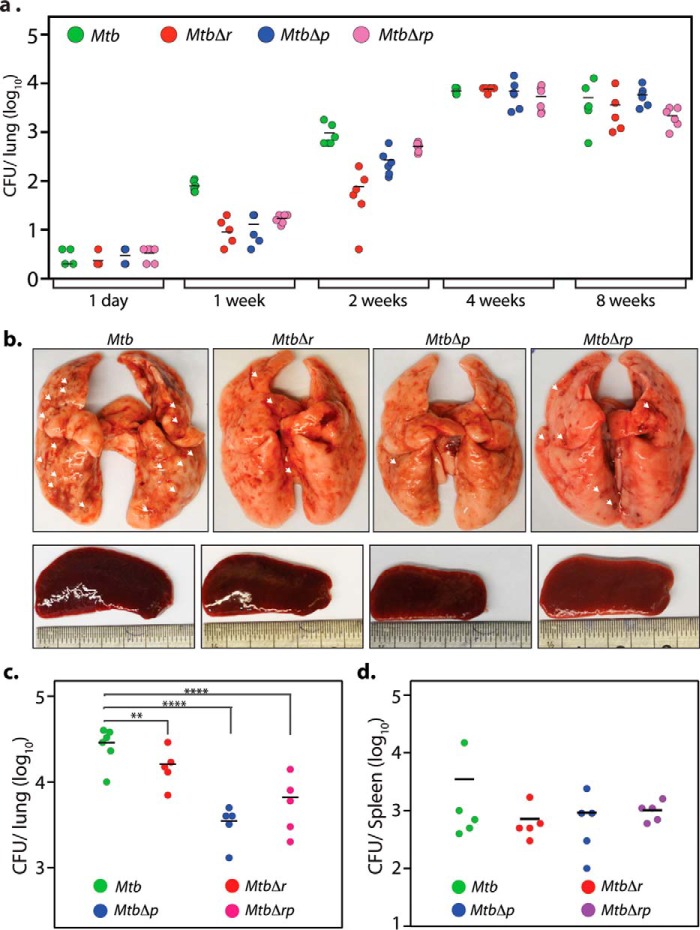
Figure 9.
RodA and PbpA are important for pathogen survival in the host. a, 5 mice/group/time point were aerosolically infected with 200 cfu/lung of Mtb, MtbΔr, MtbΔp, and MtbΔrp strains. cfu were enumerated in the lungs of infected mice after day 1 (4 mice/group), 1, 2, 4, and 8 weeks (5 mice/group) postinfection. b–d, 5 guinea pigs/group/time point were infected with Mtb, MtbΔr, MtbΔp, and MtbΔrp strains, and animals were sacrificed at 4 weeks postinfection. b, representative images for gross assessment of lungs and spleen from infected guinea pigs 4 weeks postinfection. Discrete tubercles in the lungs are shown with white arrows. c and d, cfu enumeration from the infected lungs (c) and spleen (d). Mean cfu values in the lungs of guinea pigs (c) infected with Mtb, MtbΔr, MtbΔp, and Mtbrp were 4.51, 4.2, 3.54, and 3.81 on the log10 scale, respectively. Statistical analysis was performed using a one-way ANOVA test. ****, p < 0.0001; **, p < 0.01. Mean cfu values in the spleens of guinea pigs (d) infected with Mtb, MtbΔr, MtbΔp, and Mtb were 3.44, 3.41, 3.39, and 3.54 on the log10 scale, respectively.
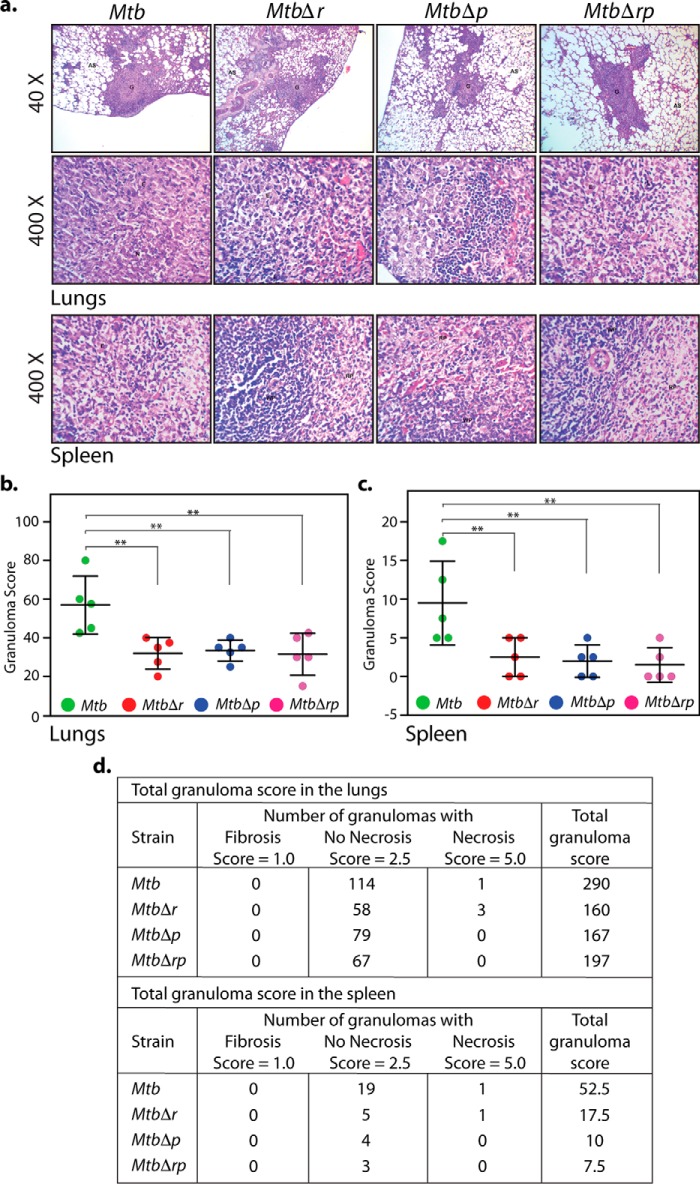
Because granulomas, the hallmark of human pulmonary tuberculosis, are absent in the mouse infection model (53), we used the guinea pig model to study this aspect (54). Accordingly, guinea pigs were infected aerosolically to evaluate the roles of RodA and PbpA in the maintenance of recalcitrant nonreplicating bacilli in such structures. Whereas the lungs of guinea pigs infected with Mtb showed the presence of discrete tubercles, we observed reduction in such tubercles in the lungs of MtbΔr- and MtbΔp-infected guinea pigs (Fig. 9b, white arrows). Bacillary survival in lungs evaluated 4 weeks postinfection revealed the attenuated survival of MtbΔr, MtbΔp, and MtbΔrp, exhibiting 2-, 10-, and 5-fold lower bacillary load, respectively, as compared with Mtb (Fig. 9c). However, the splenic loads for WT and the mutant strains were found to be similar (Fig. 9d). Interestingly, histopathological analysis revealed that irrespective of the bacillary load in lungs or spleen, the granuloma scores were consistently lower in the case of all of the mutants both in lungs and spleen when compared with the WT (Fig. 10). The lower granuloma scores are indicative of a compromised niche that would affect long-term survival. Thus, based on compromised survival studies and lower histopathological scores, obtained in the case of all of the deletion mutants (Fig 10), we suggest that RodA and PbpA play crucial roles in imparting mycobacterial virulence in the host.
Figure 10.
RodA and PbpA mutants show compromised granulomatous pathology in the host. a, representative ×40 and ×400 images (top) of H&E-stained lung sections obtained from guinea pigs infected with Mtb, MtbΔr, MtbΔp, or MtbΔrp at 4 weeks postinfection. Shown are ×400 images (bottom) of H&E-stained spleen sections obtained from guinea pigs infected with Mtb, MtbΔr, MtbΔp, or MtbΔrp at 4 weeks postinfection. G, granuloma; AS, alveolar spaces; FC, foamy histiocytic cells. b–d, granuloma score analysis; scatter plot of the granuloma scores obtained (calculated as described under “Experimental procedures”) of all animals for H&E-stained lung (b) and spleen (c) sections of guinea pigs. Each data point represents the score of an individual animal (n = 5). Statistical analysis was performed using a one-way ANOVA test. **, p < 0.01. d, tabular presentation of granuloma scores of H&E-stained lung and spleen sections for each group showing types of granuloma observed, calculation of granuloma score, and the total granuloma score obtained 4 weeks postinfection.
Discussion
The mycobacterial cell wall is the primary barrier protecting the cell from host-mediated stress and undergoes substantial remodeling in the host (55). Maintenance of cell integrity and shape requires the coordinated regulation of cell wall biosynthesis and cell division processes. Bacterial cell growth broadly has two distinct stages: elongation and subsequent division of the cell. Synthesis of PG, an integral part of the cell wall, takes place at both poles and septum and is critically regulated to sustain optimal bacillary growth and survival. The cellular machinery associated with PG biosynthesis at the poles and septum are termed the elongasome and divisome, respectively (56). These complexes are thought to function independently, yet in a coordinated manner, to preserve the integrity of cell growth and division processes. The SEDS and PBP family proteins function in pairs and modulate PG biosynthesis within these elongasome and divisome complexes; for example, based on interaction and localization studies, the FtsW-Pbp3 pair from E. coli and mycobacteria is shown to function in the divisome (26, 29, 30). In most cases, SEDS-PBP pairs are genetically linked and exhibit high phylogenetic conservation. RodA and pbpA, located next to each other in the same operon (Fig. 2), are members of the SEDS and PBP family, respectively, and are thus speculated to be functionally correlated (25). In S. pneumoniae (57) and E. coli, RodA directly interacts with Pbp2B (homolog of Mtb PbpA) (58) and in C. glutamicum with DivIVA (homolog of Mtb Wag31) (59), a known determinant of apical/polar growth, and these interactions are critical for appropriate localization of the elongasome. The present study aimed to examine the functions of mycobacterial RodA and PbpA by overexpressing and generating gene replacement mutants and analyzing their phenotypes obtained in vitro and in vivo (Figs. 2 and 3).
If RodA and PbpA were to function as a pair, one would expect that the cellular morphology would be similar when either of them is overexpressed or deleted. Whereas the overexpression of either RodA or PbpA resulted in a similar phenotype of elongated cells, deletion of either of them gave contrasting phenotypes (Figs. 2–4). Whereas the cells were elongated upon pbpA deletion, we observed the cells to be shorter upon rodA deletion (Fig. 4). PbpA is known to interact with FhaA, an FHA domain–containing protein, and localize to both poles and septum (60). Whereas mycobacterial PbpA protein interacts with CrgA, a probable scaffold recruiter protein at the divisome, RodA fails to do so. Thus, we hypothesize that RodA and PbpA are most likely participating in different complexes (61). We observed significant changes in the cell wall architecture upon PbpA deletion (both in single and combinatorial mutants) in limiting medium conditions (Fig. 5). Such thickened cell wall architecture is typically observed in microaerobically or anaerobically grown Mtb and is believed to serve the purpose of protecting the bacteria in hypoxic conditions (62). Based on the above findings and the phenotypes observed, we suggest that PbpA and RodA may function in different complexes, with PbpA playing an important role in stress adaptation.
PbpA is predicted to be a transpeptidase involved in cross-linking of stem peptides attached to NAM in the PG monolayer (44). In line with this prediction, the absence of pbpA resulted in higher susceptibility to β-lactam antibiotics (Fig. 6). Furthermore, biochemical assays revealed PbpA to be classical transpeptidase with a role for Lys-424 in the catalysis (Fig. 6). Unlike in mycobacteria, rodA is indispensable in a number of other bacteria, and its conditional depletion results in conversion of bacterial rods to spheres (20, 42, 63, 64). Based on sequence and structural conservation with FtsW, RodA has been proposed to function as lipid II flippase (24, 65). In contrast, recent studies performed in B. subtilis suggested a novel role for RodA as a noncanonical transglycosylase, which upon overexpression can compensate for the absence of all classical PBPs with transglycosylase function (66). If mycobacterial RodA were to function as a Lipid II flippase, one would expect lower levels of Lipid II in the periplasmic space upon its deletion (20). On the other hand, if RodA were to function as a transglycosylase, its absence would result in accumulation of Lipid II in the periplasmic region. We observed increased sensitivity of mycobacterial rodA deletion mutant to vancomycin and nisin (Fig. 6), indicative of Lipid II accumulation (46) in the periplasmic space, strongly suggesting that RodA is likely to function as a transglycosylase rather than a Lipid II flippase. In addition, small-scale Lipid II accumulation analysis performed with [3H]mDAP (an exclusive component of PG precursor) also suggested ∼20% higher accumulation of Lipid II (Fig. 6). B. subtilis conditional rodA mutant is hypersensitive to moenomycin, an inhibitor of canonical transglycosylases (27). In line with this, we found the mycobacterial rodA mutant to be hypersensitive to moenomycin (Fig. 6). Importantly, whereas in trans expression of Msm and Mtb rodA could rescue moenomycin sensitivity defects, expression of ftsW or mviN failed do so. Meeske et al. (25) have identified immutable residues in B. subtilis RodA that are necessary for its function. Furthermore, biochemical experiments revealed a definitive role for Trp-105 and Asp-280 residues in modulating transglycosylase activity (25). In agreement with this, in trans expression of mycobacterial RodA point mutants, wherein residues corresponding to conserved Trp and Asp have been mutated, failed to rescue the moenomycin sensitivity defect of rodA deletion mutant (Fig. 7). Taken together, these are the first data demonstrating RodA function as a noncanonical transglycosylase in mycobacteria.
Phosphorylation events have been shown to regulate various cellular processes, including cell wall biosynthesis and cell division (36, 37). STPKs PknA and PknB have been shown to phosphorylate many target proteins, including those involved in PG biosynthesis, such as Wag31, FtsZ, MviN, MurD, and GlmU (23, 50, 67–69). Phosphorylation of these proteins has been implicated in modulating their enzymatic activities, cellular localization, or protein-protein interactions (50, 67, 70). Mass spectrometry analysis of phosphoenriched Mtb lysates identified phosphorylation at Thr-463 of Mtb RodA (Fig. 8). Our results are in agreement with an independent study wherein RodA was identified to be phosphorylated at Thr-463 in Mycobacterium bovis BCG (71). In vitro kinase assays with purified STPKs showed that PknB and PknH are the most likely STPKs to mediate phosphorylation of RodA (Fig. 8). Interestingly, whereas in trans complementation of MtbΔr with phosphoablative mutant failed to rescue the short-length phenotype, complementation with phosphomimetic mutant fully complemented the mutant phenotype (Fig. 8). Whereas RodA is a structurally conserved protein, so far, no post-translational modifications (and their role in modulating RodA activity) have been identified in other bacterial systems. Interestingly, in C. glutamicum, the negative charges on the carboxyl-terminal residues of RodA have been shown to a play a role in modulating interactions with DivIVA (homolog of Wag31) (59). We speculate that the phosphorylation of RodA in the carboxyl-terminal region, specifically at Thr-463, may be important for modulating its interactions with other cell division proteins, thus regulating the cell division process.
Bacterial shape and its evolution have been related to better survival in diverse milieus (72). Bacterial cell shape and dimensions are determined by the cell wall and its composition. The composition of the Mtb cell wall undergoes changes to adapt to and impart drug tolerance under stress conditions, and its remodeling events are critical for infection in macrophages (73). The composition of the bacterial cell wall is an important factor in modulating fitness by regulating adherence to biotic surfaces, efficient survival under nutrient-deprived and stress conditions, efficient nutrient uptake, and passive dispersal (74). Enzymes involved in PG biosynthesis have been shown to play an important role in maintaining cell shape and survival under stress (75). The deletion of enzyme decaprenol pyrophosphatase (UppP) involved in PG precursor Lipid II synthesis results in severe attenuation in a mouse model of infection (76). Deletion of ponA1, a PBP protein with transglycosylase and transpeptidase activities, displayed compromised survival in mice (70).
RodA and PbpA are involved at different steps of peptidoglycan synthesis. In Streptococcus thermophilus, RodA and PbpA play a role in combating oxidative stress (77–79). Mycobacterial PbpA has been found to be up-regulated under nutrient starvation conditions mediated via stringent response regulator RelA, which is required for the long-term survival of pathogen in mice and guinea pigs (80, 81). Moreover, expression of both Mtb rodA and pbpA is up-regulated under SDS and diamide stress, possibly due to their role in stress adaptation. The observed differences in the cell length phenotypes observed upon overexpression and deletion of RodA and PbpA are not very large but are observed consistently (Figs. 2 and 3). Thus, we speculated that either rodA or pbpA or both might play a role in survival of pathogen in the host.
Interestingly, we did not observe any significant difference between WT and mutant strains in the bacillary loads in mouse infection experiments at any time point post-infection (Fig. 9). Because hypoxic granulomas with necrotic lesions are not detected in the lungs of infected mice, they are viewed as inappropriate models for investigating the paucibacillary state observed in the human infections (53, 82). On the other hand, guinea pigs harbor the hallmark hypoxic granuloma structures that mimic the diseased state in humans (54). Importantly, in our guinea pig model infection studies, we observed a ∼0.5–1-log-fold (5–10-fold) decrease in the survival of pathogen in the lungs of guinea pigs infected with MtbΔr or MtbΔrp or MtbΔrp mutants (Fig. 9). Moreover, the corresponding granuloma scores for mutants were almost 50% lower compared with the WT (Fig 10). In the case of spleen, whereas the infection loads were similar, the granuloma scores mirrored the compromised scores for lungs. The granuloma architecture in Mtb-infected guinea pig is highly organized, hypoxic with necrotic lesions, and harbors recalcitrant nonreplicating bacilli, close to the diseased state in humans (54). Thus, we suggest that rodA and pbpA play a role in long-term mycobacterial survival in the host. Further experiments in a guinea pig model with protracted time points would be required to conclusively demonstrate their roles in long-term survival.
We also observed decreased survival of the pbpA mutant in an in vitro Wayne model of hypoxia (43) (Fig. 5e). Persisters analysis for deletion mutants of rodA and pbpA exhibited a 10-fold decline in cfu upon exposure to 10 μg/ml isoniazid (Fig. 5f). The data collectively underscore the importance of RodA and PbpA in mediating survival under hypoxic conditions in vivo. In line with this thinking, muramyl dipeptides that are a part of PG have previously been shown to activate macrophages, and when associated with branched fatty acids, they promote granuloma formation (83). Taken together, using the guinea pig infection model, we report here for the first time a role for the PG biosynthetic proteins RodA and PbpA in modulating survival of the human pathogen Mtb in the host.
Experimental procedures
Bacterial strains and reagents
Constructs and strains used in this study are listed in Table 3. Oligonucleotides and fine chemicals were purchased from Sigma, Amresco, Merck, and Bio Basic. Restriction and modification enzymes were purchased from New England Biolabs or MBI-Fermentas (Thermo Scientific). The pENTR/Directional TOPO cloning kit and BocillinTM FL penicillin, sodium salt were purchased from Invitrogen, and growth medium was from BD Difco. pMAL-c2x (New England Biolabs) kinase clone constructs already available in the laboratory were used for purification (49). [3H]mDAP was purchased from American Radiolabeled Chemicals, and [γ-32P]ATP was purchased from PerkinElmer Life Sciences. pNiT-I vector was a kind gift from Prof. Christopher M. Sassetti (84). pNit-ET was a kind gift from Prof. Eric Rubin (85). α-FLAG mAb was purchased from Sigma. α-PknB, α-PknA, and α-GroEL-I antibodies were generated in the laboratory, and α-PstP antibody was a kind gift of Dr. Yogendra Singh (CSIR-Institute of Genomics and Integrative Biology) (86). EM chemicals were purchased from Electron Microscopy Sciences.
Table 3.
Constructs and strains used in the study
| Description | |
|---|---|
| Constructs | |
| pENTRCmr | pENTR chloramphenicol cloned in KpnI and SnaBI amplified from pVR1 |
| pNit1 (84) | Nitrile-inducible vector kanr |
| pNitchl | pNit1 vector modified by inserting cmr gene |
| pST-KT (38) | Anhydrotetracyclin-inducible vector (ref) kanr |
| pJV53 (41) | Plasmid encoding phage phage Che9c 60 and 61 genes under acetamide-inducible promoter; kanr |
| pNit-ET (85) | Plasmid encoding phage Che9c 60 and 61 genes cloned in pNit1; kanr |
| Strains | |
| DH5α | E. coli strain used for cloning experiments |
| Msm | WT M. smegmatis mc2155 strain |
| Mtb | WT H37Rv M. tuberculosis strain |
| Mtb::Vc | Mtb electroporated with pST-KT; kanr |
| Mtb::rodA | Mtb electroporated with pST-KT-RodAMtb; kanr |
| Mtb::pbpA | Mtb electroporated with pST-KT-PbpAMtb; kanr |
| Mtb::pNitET | Mtb electroporated with pNit-ET; kanr |
| MtbΔr | rodAMtb gene replacement mutant; hygr |
| MtbΔp | pbpAMtb gene replacement mutant; hygr |
| MtbΔrp | rodAMtb-pbpAMtb gene replacement mutant; hygr |
| MtbΔr::r | MtbΔr complemented with pNitchl-RodAMtb; hygr, cmr |
| MtbΔp::p | MtbΔp complemented with pNitchl-PbpAMtb; hygr, cmr |
| MtbΔr::r | MtbΔr complemented with pST-KT-RodAMtb; hygr, kanr |
| MtbΔr::rT463A | MtbΔr complemented with pST-KT-RodAMtb-T463A; hygr, kanr |
| MtbΔr::rT463E | MtbΔr complemented with pST-KT-RodAMtb-T463E; hygr, kanr |
| Msm::pJV53 | Msm electroporated with pJV53 plasmid; kanr |
| MsmΔr | Msm rodA gene replacement mutant; hygr |
| MsmΔp | Msm pbpA gene replacement mutant; hygr |
| MsmΔrp | Msm rodA-pbpA gene replacement mutant; hygr |
| Msm::pN | Msm electroporated with pNitchl; cmr |
| Msm::pN-r | Msm electroporated with pNitchl-RodAMtb; cmr |
| Msm::pN-p | Msm electroporated with pNitchl-PbpA; cmr |
| MsmΔp::pMtb | MsmΔp complemented with pST-KT-PbpAMtb; hygr, kanr |
| MsmΔp::p | MsmΔp electroporated with pNit-PbpA; hygr, kanr |
| MsmΔp::pS281A | MsmΔp electroporated with pNit-PbpAS281A; hygr, kanr |
| MsmΔp::pK424G | MsmΔp electroporated with pNit-PbpAK424G; hygr, kanr |
| MsmΔr::rMtb | MsmΔr complemented with pST-KT-RodAMtb; hygr, kanr |
| MsmΔr::rMsm | MsmΔr complemented with pST-KT-RodAMtb; hygr, kanr |
| MsmΔr::ftsW | MsmΔr complemented with pST-KT-FtsWMtb; hygr, kanr |
| MsmΔr::mviN | MsmΔr complemented with pST-KT-MviNMtb; hygr, kanr |
| MsmΔp::pMtb | MsmΔp complemented with pST-KT-PbpAMtb; hygr, kanr |
| MsmΔr::rMtb-W175R | MsmΔr complemented with pST-KT-RodAMtb-W175R; hygr, kanr |
| MsmΔr::rMtb-D343R | MsmΔr complemented with pST-KT-RodAMtb-D343R; hygr, kanr |
| MsmΔr::rMtb-DM | MtbΔr complemented with pST-KT-RodAMtb-W175R -D343R; hygr, kanr |
| MsmΔr::rMsm-W176R | MtbΔr complemented with pST-KT-RodAMsm-W176R; hygr, kanr |
| MsmΔr::rMsm-D344R | MtbΔr complemented with pST-KT-RodAMsm-D344R; hygr, kanr |
| MsmΔr::rMsm-DM | MtbΔr complemented with pST-KT-RodAMsm-W176R -D344R; hygr, kanr |
Generation of plasmid constructs
pNit-Cm was generated by inserting a blunt-ended chloramphenicol resistance gene (cmr) from pENTR-Cmr into PflMI-linearized and filled-in pNit vector. rodA, pbpA, ftsW, and mviN genes were amplified using H37Rv genomic DNA as the template, and the amplicons obtained were cloned into pENTR vector. The pENTR clones of rodA and pbpA were digested with NdeI and HindIII enzymes to release the rodA and pbpA genes, which were then subcloned into the corresponding sites in pST-KT (38) and/or pNit-Cm vectors. rodA(411–469) was amplified using specific forward primer containing an NdeI site and 3× FLAG tag and gene-specific reverse primer. The 3X-rodA(411–469) amplicon was digested with NdeI–HindIII and was cloned into pET28b vector. Noncanonical transglycosylase active site residue mutants of Mtb and Msm RodA were generated with the help of overlapping PCR. Point mutants of PbpA, pbpAS281A and pbpAK424G, were generated using Mtb genomic DNA with the help of overlapping PCR
Generation of gene replacement mutant construct
Gene replacement mutants of rodA, pbpA, and rodA-pbpA together were generated in both M. tuberculosis H37Rv and M. smegmatis mc2155 with the help of recombineering (41). Toward the generation of allelic exchange substrates, the upstream flank (flank 1) and downstream flank (flank 2) were amplified using genomic DNA as the template. The apramycin resistance gene (amr) was amplified from pMV261-apra (87), and hygromycin resistance gene (hygr) and oriE+λ cos fragments were amplified from pYUB1471 (88). Amplicons obtained were digested with PflMI/BstAPI and ligated in a five-piece ligation reaction. M. smegmatis and M. tuberculosis were electroporated with pJV53 and pNit-ET constructs to generate Msm::JV53 and Mtb::ET strains. Allelic exchange substrates were digested with restriction enzyme SnaBI or EcoRV to generate linear blunt-ended recombineering-proficient (containing flank1-hygr-flank2) DNA fragments that were electroporated into recombineering-proficient Msm::JV53 and Mtb::ET strains. The recombinant deletion strains were confirmed by performing PCRs across the deletion junctions, and the positive strains were cured of recombineering plasmids pJV53 and pNit-ET.
Growth analysis
Mtb (WT), rodA deletion mutant (MtbΔr), pbpA deletion mutant (MtbΔp), and rodA-pbpA double deletion mutant (MtbΔrp) were grown in Middlebrook 7H9 medium containing 10% ADC (albumin, dextrose, and catalase) in the presence or absence of 100 μg/ml hygromycin. Cultures were grown until absorbance (A600) reached ∼0.8, and the cells were harvested and washed with PBS containing 0.05% Tween 80 (PBST80). For growth analyses, cultures were initiated at A600 ∼0.1 in 7H9/Sauton's medium without antibiotics and incubated at 37 °C with shaking (100 rpm). A600 was measured every 24 h for 6–10 days, and serial dilutions of cultures were plated on 7H11 agar. cfu were enumerated. Experiments were performed in triplicates, and obtained average cfu were plotted as a function of time. S.E. values were calculated using GraphPad Prism version 6 for each time point.
SEM and TEM
Cells grown in either 7H9 or Sauton's medium were diluted to A600 ∼0.5. 10 ml of the cultures were harvested, and SEM and TEM were performed as described previously (69). For the quantification of cell length, ∼200 individual cells were measured from the acquired SEM images using Smart Tiff software. The mean cell length and statistical significance (ANOVA test) were determined using GraphPad Prism version 6 software.
Hypoxia experiments
Mtb, MtbΔr, and MtbΔp cells were subjected to hypoxic conditions as described earlier (69). Bacterial survival was assessed at the indicated time points by cfu enumeration. Persisters analyses of Msm, MsmΔr, and MsmΔp strains were performed as described previously (89).
MIC determination
MIC determination was performed as described earlier with the help of a resazurin microtiter assay (REMA) (90). Briefly, different dilutions of antibiotics in Dubos medium with appropriate controls were added to a flat-bottomed 96-well plate. Cells grown in Dubos medium until early or late exponential phase were diluted to A600 of 0.006 in the same medium, and 100 μl of each was added to each well of the 96-well plate containing different dilutions of antibiotics. The plates were incubated at 37 °C for 24 h, and 20 μl of resazurin solution (0.02%) was added to each well, followed by overnight incubation at 37 °C. Color transition from blue to pink was an indicator of bacterial growth. MIC was defined as lowest concentration of antibiotic that prevented this change in color.
Bocillin-FL labeling assay
Cultures of Msm, MsmΔp, MsmΔp::p MsmΔp::pS281A, and MsmΔp::pK424G strains were induced with 5 μm IVN, and the membrane fractions were prepared as described previously (91). Protein concentrations were estimated using a Bio-Rad Bradford assay. For each strain, ∼20 μg of transmembrane preparations in a 24-μl reaction volume was incubated with Bocillin-FL (Invitrogen) in PBS for 20 min at 37 °C in the dark. Reactions were stopped by the addition of SDS-sample buffer, followed by boiling for 5 min and incubation on ice for 5 min (92). Samples were resolved on 8% SDS-PAGE, gel was washed with MQ water for 1 h, and fluorescence of the Bocillin-labeled proteins was detected using a Typhoon scanner (GE Healthcare) under 488-nm excitation and 532-nm emission conditions. A similar amount of membrane extracts were processed for immunoblot analysis.
RodA Lipid II accumulation assay
Msm and MsmΔr strains were grown in Sauton's medium until A600 ∼0.2–0.3 and pulsed with 1.5 μCi/ml [3H]mDAP (procured from American Radiolabeled Chemicals) for 4–5 h. Equal amounts of cultures were processed for small-scale Lipid II accumulation analysis as described (47). Obtained lipid fractions were dried using a SpeedVac, followed by the addition of scintillation fluid (Betaplate Scint, PerkinElmer Life Sciences) and overnight incubation. Radiolabeled [3H]Lipid II in the samples was quantified by scintillation counting using a β-counter. The reading obtained for Msm in each independent experiment was normalized to 100%, and the counts obtained for MsmΔr were represented as percentage change with respect to counts obtained for Msm.
Identification of phosphorylation site
Mtb H37Rv strain was grown in 7H9-ADC medium until A600 ∼0.8–1.0. Whole-cell lysates (WCLs) were prepared in lysis buffer containing 8 m urea in the presence of protease and phosphatase inhibitors. Trypsinization and strong cation exchange (SCX) chromatography of 10 mg of WCLs was performed as described previously (93). SCX fractions were phosphoenriched using TiO2 beads and eluted in 0.3 m ammonium hydroxide, followed by desalting. Fractions were resuspended in 20 μl of solution containing 5% acetonitrile and 0.1% formic acid. LC-MS analysis using LTQ Orbitrap Velos was performed as described earlier (94), and the data were analyzed using the Proteome Discoverer Software Suite (version 1.3). For the search using SEQUEST, carbamidomethylation on cysteine residues was used as a fixed modification, and oxidation of methionine and phosphorylation of serine, threonine, and tyrosine were used as variable modifications. Spectra were queried against the Mtb UniProt database.
In vitro kinase assay
pMAL-c2X constructs expressing Mtb STPKs generated by us previously (49) and the pET-RodA(441–469) construct were transformed into E. coli BL21 (DE3) Codon Plus cells followed by expression and purification as described previously (49). An in vitro kinase assay was performed as described earlier using ∼10 pmol of purified STPKs and 2 μg of purified RodA(441–469) as substrate (95).
Mycobacterial infection of mice and guinea pigs
Mtb, MtbΔr, MtbΔp, and MtbΔrp strains were grown in 7H9-ADC broth at 37 °C, 100 rpm until A600 ∼0.6. Cells were harvested at 3000 × g at room temperature, and cell pellets were washed twice with sterile PBST80 before resuspension in neutral buffered saline. BALB/c mice of either sex (6–8 weeks old) were obtained from the animal breeding facility at the National Institute of Immunology. Mice (n = 6) were infected with 2 × 108 cfu by the aerosol route. Bacillary loads in lungs were enumerated after day 1 and after 1, 2, 4, and 8 weeks postinfection (33). For infecting guinea pigs, outbred female guinea pigs of the Duncan–Hartley strain (weight 200–300 g) were infected with 108 cfu through the aerosol route to implant 100 cfu/lung. Bacillary loads in lung and spleen were enumerated 4 weeks postinfection (69). Infected lungs and spleens were fixed in neutral buffered saline followed by hematoxylin and eosin staining. Histopathological evaluation and granuloma grading was performed as described in an earlier study (69). The total granuloma score was obtained by adding the individual score for each type of granuloma.
Ethics statement
The experimental protocol for the animal experiments was approved by the Animal Ethics Committee of the National Institute of Immunology, New Delhi, India (approval IAEC 389/15). The approval is as per the guidelines issued by Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Government of India.
Author contributions
D. A. and V. K. N. conceived and designed the experiments. D. A. performed most of the experiments, Y. C. performed the Guinea pig infection experiment, B. M. performed mass spectrometry studies, and A. S. performed TEM studies. D. A. and V. K. N. analyzed the data and wrote the paper. All authors read and approved the manuscript.
Supplementary Material
Acknowledgments
We thank the Tuberculosis Aerosol Challenge Facility (DBT-TACF) staff Dr. Lakshyaveer Singh, Sudesh Rathour and Mahendra Singh at International Centre for Genetic Engineering and Biotechnology (New Delhi, India) for kind help. We thank Dr. Dhiraj Kumar for extending the use of the TACF facility. We thank Suresh Kumar for help in purification of MBP-tagged STPKs. We thank the TEM facility at CSIR-Institute of Genomics and Integrative Biology, scanning EM facility, and the biocontainment facility (BSL3) at the National Institute of Immunology. We thank Rekha Rani and Bhanu Mantri for support in SEM/TEM. We thank Dr. Christopher Sassetti, Dr. Graham Hatfull, Dr. Eric Rubin, and Dr. William Jacobs for providing the constructs used in this study. We thank Dr. Swati Saha for critical reading of the manuscript.
This work was supported by the funding provided by SERB, Department of Science and Technology, Government of India Project EMR/2014/000877 (to V. K. N.). The authors declare that they have no conflicts of interest with the contents of this article. Supported by an INSPIRE faculty grant IFA12-LSBM35 from the Department of Science and Technology, Government of India.
This article contains Fig. S1.
- Mtb
- M. tuberculosis
- Msm
- M. smegmatis; peptidoglycan
- STPK
- serine/threonine protein kinase
- SEM
- scanning EM
- TEM
- transmission EM
- PBP
- penicillin-binding protein
- SEDS
- shape, elongation, division, and sporulation
- mDAP
- meso-diaminopimelic acid
- ANOVA
- analysis of variance
- MIC
- minimum inhibitory concentration
- REMA
- resazurin microtiter assay
- WCL
- whole-cell lysate
- SCX
- strong cation exchange
- IVN
- isovaleronitrile
- H&E
- hematoxylin and eosin.
References
- 1. Barry C. E., Crick D. C., and McNeil M. R. (2007) Targeting the formation of the cell wall core of M. tuberculosis. Infect. Disord. Drug Targets 7, 182–202 10.2174/187152607781001808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaur D., Guerin M. E., Skovierová H., Brennan P. J., and Jackson M. (2009) Chapter 2: Biogenesis of the cell wall and other glycoconjugates of Mycobacterium tuberculosis. Adv. Appl. Microbiol. 69, 23–78 10.1016/S0065-2164(09)69002-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bansal-Mutalik R., and Nikaido H. (2014) Mycobacterial outer membrane is a lipid bilayer and the inner membrane is unusually rich in diacyl phosphatidylinositol dimannosides. Proc. Natl. Acad. Sci. U.S.A. 111, 4958–4963 10.1073/pnas.1403078111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barry C. E. 3rd, Boshoff H. I., Dartois V., Dick T., Ehrt S., Flynn J., Schnappinger D., Wilkinson R. J., and Young D. (2009) The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7, 845–855 10.1038/nrmicro2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boshoff H. I., and Barry C. E. 3rd (2005) Tuberculosis: metabolism and respiration in the absence of growth. Nat. Rev. Microbiol. 3, 70–80 10.1038/nrmicro1065 [DOI] [PubMed] [Google Scholar]
- 6. Nathan C., Gold B., Lin G., Stegman M., de Carvalho L. P., Vandal O., Venugopal A., and Bryk R. (2008) A philosophy of anti-infectives as a guide in the search for new drugs for tuberculosis. Tuberculosis 88, S25–S33 10.1016/S1472-9792(08)70034-9 [DOI] [PubMed] [Google Scholar]
- 7. Rustad T. R., Harrell M. I., Liao R., and Sherman D. R. (2008) The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One 3, e1502 10.1371/journal.pone.0001502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rustad T. R., Sherrid A. M., Minch K. J., and Sherman D. R. (2009) Hypoxia: a window into Mycobacterium tuberculosis latency. Cell Microbiol. 11, 1151–1159 10.1111/j.1462-5822.2009.01325.x [DOI] [PubMed] [Google Scholar]
- 9. Shi L., Sohaskey C. D., Pfeiffer C., Datta P., Parks M., McFadden J., North R. J., and Gennaro M. L. (2010) Carbon flux rerouting during Mycobacterium tuberculosis growth arrest. Mol. Microbiol. 78, 1199–1215 10.1111/j.1365-2958.2010.07399.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Typas A., Banzhaf M., Gross C. A., and Vollmer W. (2011) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 10, 123–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kramer N. E., Smid E. J., Kok J., de Kruijff B., Kuipers O. P., and Breukink E. (2004) Resistance of Gram-positive bacteria to nisin is not determined by lipid II levels. FEMS Microbiol. Lett. 239, 157–161 10.1016/j.femsle.2004.08.033 [DOI] [PubMed] [Google Scholar]
- 12. Storm D. R., and Strominger J. L. (1974) Binding of bacitracin to cells and protoplasts of Micrococcus lysodeikticus. J. Biol. Chem. 249, 1823–1827 [PubMed] [Google Scholar]
- 13. de Kruijff B., van Dam V., and Breukink E. (2008) Lipid II: a central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot. Essent. Fatty Acids 79, 117–121 10.1016/j.plefa.2008.09.020 [DOI] [PubMed] [Google Scholar]
- 14. Breukink E., and de Kruijff B. (2006) Lipid II as a target for antibiotics. Nat. Rev. Drug Discov. 5, 321–332 10.1038/nrd2004 [DOI] [PubMed] [Google Scholar]
- 15. Liu Y., and Breukink E. (2016) The membrane steps of bacterial cell wall synthesis as antibiotic targets. Antibiotics 5, E28 10.3390/antibiotics5030028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ling L. L., Schneider T., Peoples A. J., Spoering A. L., Engels I., Conlon B. P., Mueller A., Schäberle T. F., Hughes D. E., Epstein S., Jones M., Lazarides L., Steadman V. A., Cohen D. R., Felix C. R., et al. (2015) A new antibiotic kills pathogens without detectable resistance. Nature 517, 455–459 10.1038/nature14098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elhenawy W., Davis R. M., Fero J., Salama N. R., Felman M. F., and Ruiz N. (2016) The O-antigen flippase Wzk can substitute for MurJ in peptidoglycan synthesis in Helicobacter pylori and Escherichia coli. PLoS One 11, e0161587 10.1371/journal.pone.0161587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meeske A. J., Sham L. T., Kimsey H., Koo B. M., Gross C. A., Bernhardt T. G., and Rudner D. Z. (2015) MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 112, 6437–6442 10.1073/pnas.1504967112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boyle D. S., Khattar M. M., Addinall S. G., Lutkenhaus J., and Donachie W. D. (1997) ftsW is an essential cell-division gene in Escherichia coli. Mol. Microbiol. 24, 1263–1273 10.1046/j.1365-2958.1997.4091773.x [DOI] [PubMed] [Google Scholar]
- 20. Sieger B., Schubert K., Donovan C., and Bramkamp M. (2013) The lipid II flippase RodA determines morphology and growth in Corynebacterium glutamicum. Mol. Microbiol. 90, 966–982 10.1111/mmi.12411 [DOI] [PubMed] [Google Scholar]
- 21. Sham L. T., Butler E. K., Lebar M. D., Kahne D., Bernhardt T. G., and Ruiz N. (2014) Bacterial cell wall: MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345, 220–222 10.1126/science.1254522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fay A., and Dworkin J. (2009) Bacillus subtilis homologs of MviN (MurJ), the putative Escherichia coli lipid II flippase, are not essential for growth. J. Bacteriol. 191, 6020–6028 10.1128/JB.00605-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gee C. L., Papavinasasundaram K. G., Blair S. R., Baer C. E., Falick A. M., King D. S., Griffin J. E., Venghatakrishnan H., Zukauskas A., Wei J. R., Dhiman R. K., Crick D. C., Rubin E. J., Sassetti C. M., and Alber T. (2012) A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci. Signal. 5, ra7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ikeda M., Sato T., Wachi M., Jung H. K., Ishino F., Kobayashi Y., and Matsuhashi M. (1989) Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol. 171, 6375–6378 10.1128/jb.171.11.6375-6378.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Meeske A. J., Riley E. P., Robins W. P., Uehara T., Mekalanos J. J., Kahne D., Walker S., Kruse A. C., Bernhardt T. G., and Rudner D. Z. (2016) SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 10.1038/nature19331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leclercq S., Derouaux A., Olatunji S., Fraipont C., Egan A. J., Vollmer W., Breukink E., and Terrak M. (2017) Interplay between penicillin-binding proteins and SEDS proteins promotes bacterial cell wall synthesis. Sci. Rep. 7, 43306 10.1038/srep43306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emami K., Guyet A., Kawai Y., Devi J., Wu L. J., Allenby N., Daniel R. A., and Errington J. (2017) RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat. Microbiol. 2, 16253 10.1038/nmicrobiol.2016.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Born P., Breukink E., and Vollmer W. (2006) In vitro synthesis of cross-linked murein and its attachment to sacculi by PBP1A from Escherichia coli. J. Biol. Chem. 281, 26985–26993 10.1074/jbc.M604083200 [DOI] [PubMed] [Google Scholar]
- 29. Datta P., Dasgupta A., Singh A. K., Mukherjee P., Kundu M., and Basu J. (2006) Interaction between FtsW and penicillin-binding protein 3 (PBP3) directs PBP3 to mid-cell, controls cell septation and mediates the formation of a trimeric complex involving FtsZ, FtsW and PBP3 in mycobacteria. Mol. Microbiol. 62, 1655–1673 10.1111/j.1365-2958.2006.05491.x [DOI] [PubMed] [Google Scholar]
- 30. Fraipont C., Alexeeva S., Wolf B., van der Ploeg R., Schloesser M., den Blaauwen T., and Nguyen-Distèche M. (2011) The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology 157, 251–259 10.1099/mic.0.040071-0 [DOI] [PubMed] [Google Scholar]
- 31. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E. 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 32. Sharma A. K., Arora D., Singh L. K., Gangwal A., Sajid A., Molle V., Singh Y., and Nandicoori V. K. (2016) Serine/threonine protein phosphatase PstP of Mycobacterium tuberculosis is necessary for accurate cell division and survival of pathogen. J. Biol. Chem. 291, 24215–24230 10.1074/jbc.M116.754531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chawla Y., Upadhyay S., Khan S., Nagarajan S. N., Forti F., and Nandicoori V. K. (2014) Protein kinase B (PknB) of Mycobacterium tuberculosis is essential for growth of the pathogen in vitro as well as for survival within the host. J. Biol. Chem. 289, 13858–13875 10.1074/jbc.M114.563536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nagarajan S. N., Upadhyay S., Chawla Y., Khan S., Naz S., Subramanian J., Gandotra S., and Nandicoori V. K. (2015) Protein kinase A (PknA) of Mycobacterium tuberculosis is independently activated and is critical for growth in vitro and survival of the pathogen in the host. J. Biol. Chem. 290, 9626–9645 10.1074/jbc.M114.611822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kang C. M., Abbott D. W., Park S. T., Dascher C. C., Cantley L. C., and Husson R. N. (2005) The Mycobacterium tuberculosis serine/threonine kinases PknA and PknB: substrate identification and regulation of cell shape. Genes Dev. 19, 1692–1704 10.1101/gad.1311105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Molle V., and Kremer L. (2010) Division and cell envelope regulation by Ser/Thr phosphorylation: Mycobacterium shows the way. Mol. Microbiol. 75, 1064–1077 10.1111/j.1365-2958.2009.07041.x [DOI] [PubMed] [Google Scholar]
- 37. Richard-Greenblatt M., and Av-Gay Y. (2017) Epigenetic phosphorylation control of Mycobacterium tuberculosis infection and persistence. Microbiol. Spectrum 5 10.1128/microbiolspec.TBTB2-0005-2015 [DOI] [PubMed] [Google Scholar]
- 38. Parikh A., Kumar D., Chawla Y., Kurthkoti K., Khan S., Varshney U., and Nandicoori V. K. (2013) Development of a new generation of vectors for gene expression, gene replacement, and protein-protein interaction studies in mycobacteria. Appl. Environ. Microbiol. 79, 1718–1729 10.1128/AEM.03695-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Matsuzawa H., Asoh S., Kunai K., Muraiso K., Takasuga A., and Ohta T. (1989) Nucleotide sequence of the rodA gene, responsible for the rod shape of Escherichia coli: rodA and the pbpA gene, encoding penicillin-binding protein 2, constitute the rodA operon. J. Bacteriol. 171, 558–560 10.1128/jb.171.1.558-560.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. DeJesus M. A., Gerrick E. R., Xu W., Park S. W., Long J. E., Boutte C. C., Rubin E. J., Schnappinger D., Ehrt S., Fortune S. M., Sassetti C. M., and Ioerger T. R. (2017) Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio 8, e02133–16 10.1128/mBio.02133-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Kessel J. C., and Hatfull G. F. (2007) Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152 10.1038/nmeth996 [DOI] [PubMed] [Google Scholar]
- 42. Begg K. J., and Donachie W. D. (1985) Cell shape and division in Escherichia coli: experiments with shape and division mutants. J. Bacteriol. 163, 615–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wayne L. G., and Hayes L. G. (1996) An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64, 2062–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fedarovich A., Nicholas R. A., and Davies C. (2010) Unusual conformation of the SxN motif in the crystal structure of penicillin-binding protein A from Mycobacterium tuberculosis. J. Mol. Biol. 398, 54–65 10.1016/j.jmb.2010.02.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao G., Meier T. I., Kahl S. D., Gee K. R., and Blaszczak L. C. (1999) BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 43, 1124–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Breukink E., Wiedemann I., van Kraaij C., Kuipers O. P., Sahl H. G., and de Kruijff B. (1999) Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286, 2361–2364 10.1126/science.286.5448.2361 [DOI] [PubMed] [Google Scholar]
- 47. Qiao Y., Srisuknimit V., Rubino F., Schaefer K., Ruiz N., Walker S., and Kahne D. (2017) Lipid II overproduction allows direct assay of transpeptidase inhibition by β-lactams. Nat. Chem. Biol. 13, 793–798 10.1038/nchembio.2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ostash B., and Walker S. (2010) Moenomycin family antibiotics: chemical synthesis, biosynthesis, and biological activity. Nat. Prod Rep. 27, 1594–1617 10.1039/c001461n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Khan S., Nagarajan S. N., Parikh A., Samantaray S., Singh A., Kumar D., Roy R. P., Bhatt A., and Nandicoori V. K. (2010) Phosphorylation of enoyl-acyl carrier protein reductase InhA impacts mycobacterial growth and survival. J. Biol. Chem. 285, 37860–37871 10.1074/jbc.M110.143131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jani C., Eoh H., Lee J. J., Hamasha K., Sahana M. B., Han J. S., Nyayapathy S., Lee J. Y., Suh J. W., Lee S. H., Rehse S. J., Crick D. C., and Kang C. M. (2010) Regulation of polar peptidoglycan biosynthesis by Wag31 phosphorylation in mycobacteria. BMC Microbiol. 10, 327 10.1186/1471-2180-10-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krogh A., Larsson B., von Heijne G., and Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 52. Stoker N. G., Pratt J. M., and Spratt B. G. (1983) Identification of the rodA gene product of Escherichia coli. J. Bacteriol. 155, 854–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aly S., Wagner K., Keller C., Malm S., Malzan A., Brandau S., Bange F. C., and Ehlers S. (2006) Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J. Pathol 210, 298–305 10.1002/path.2055 [DOI] [PubMed] [Google Scholar]
- 54. Via L. E., Lin P. L., Ray S. M., Carrillo J., Allen S. S., Eum S. Y., Taylor K., Klein E., Manjunatha U., Gonzales J., Lee E. G., Park S. K., Raleigh J. A., Cho S. N., McMurray D. N., Flynn J. L., and Barry C. E. 3rd. (2008) Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 76, 2333–2340 10.1128/IAI.01515-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Seiler P., Ulrichs T., Bandermann S., Pradl L., Jörg S., Krenn V., Morawietz L., Kaufmann S. H., and Aichele P. (2003) Cell-wall alterations as an attribute of Mycobacterium tuberculosis in latent infection. J. Infect. Dis. 188, 1326–1331 10.1086/378563 [DOI] [PubMed] [Google Scholar]
- 56. Kieser K. J., and Rubin E. J. (2014) How sisters grow apart: mycobacterial growth and division. Nat. Rev. Microbiol 12, 550–562 10.1038/nrmicro3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Straume D., Stamsås G. A., Berg K. H., Salehian Z., and Håvarstein L. S. (2017) Identification of pneumococcal proteins that are functionally linked to penicillin-binding protein 2b (PBP2b). Mol. Microbiol. 103, 99–116 10.1111/mmi.13543 [DOI] [PubMed] [Google Scholar]
- 58. van der Ploeg R., Goudelis S. T., and den Blaauwen T. (2015) Validation of FRET assay for the screening of growth inhibitors of Escherichia coli reveals elongasome assembly dynamics. Int. J. Mol. Sci. 16, 17637–17654 10.3390/ijms160817637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sieger B., and Bramkamp M. (2014) Interaction sites of DivIVA and RodA from Corynebacterium glutamicum. Front. Microbiol. 5, 738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Viswanathan G., Yadav S., Joshi S. V., Raghunand T. R., and Jass J. (2017) Insights into the function of FhaA, a cell division-associated protein in mycobacteria. FEMS Microbiol. Lett. 364 10.1093/femsle/fnw294 [DOI] [PubMed] [Google Scholar]
- 61. Plocinski P., Ziolkiewicz M., Kiran M., Vadrevu S. I., Nguyen H. B., Hugonnet J., Veckerle C., Arthur M., Dziadek J., Cross T. A., Madiraju M., and Rajagopalan M. (2011) Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. J. Bacteriol. 193, 3246–3256 10.1128/JB.00188-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cunningham A. F., and Spreadbury C. L. (1998) Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton α-crystallin homolog. J. Bacteriol. 180, 801–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Henriques A. O., Glaser P., Piggot P. J., and Moran C. P. Jr. (1998) Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol. 28, 235–247 10.1046/j.1365-2958.1998.00766.x [DOI] [PubMed] [Google Scholar]
- 64. Costa C. S., and Antón D. N. (1999) Conditional lethality of cell shape mutations of Salmonella typhimurium: rodA and mre mutants are lethal on solid but not in liquid medium. Curr. Microbiol. 38, 137–142 10.1007/PL00006777 [DOI] [PubMed] [Google Scholar]
- 65. Mohammadi T., Sijbrandi R., Lutters M., Verheul J., Martin N. I., den Blaauwen T., de Kruijff B., and Breukink E. (2014) Specificity of the transport of lipid II by FtsW in Escherichia coli. J. Biol. Chem. 289, 14707–14718 10.1074/jbc.M114.557371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. McPherson D. C., and Popham D. L. (2003) Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J. Bacteriol. 185, 1423–1431 10.1128/JB.185.4.1423-1431.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sureka K., Hossain T., Mukherjee P., Chatterjee P., Datta P., Kundu M., and Basu J. (2010) Novel role of phosphorylation-dependent interaction between FtsZ and FipA in mycobacterial cell division. PLoS One 5, e8590 10.1371/journal.pone.0008590 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68. Thakur M., and Chakraborti P. K. (2008) Ability of PknA, a mycobacterial eukaryotic-type serine/threonine kinase, to transphosphorylate MurD, a ligase involved in the process of peptidoglycan biosynthesis. Biochem. J. 415, 27–33 10.1042/BJ20080234 [DOI] [PubMed] [Google Scholar]
- 69. Soni V., Upadhayay S., Suryadevara P., Samla G., Singh A., Yogeeswari P., Sriram D., and Nandicoori V. K. (2015) Depletion of M. tuberculosis GlmU from infected murine lungs effects the clearance of the pathogen. PLoS Pathog. 11, e1005235 10.1371/journal.ppat.1005235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kieser K. J., Boutte C. C., Kester J. C., Baer C. E., Barczak A. K., Meniche X., Chao M. C., Rego E. H., Sassetti C. M., Fortune S. M., and Rubin E. J. (2015) Phosphorylation of the peptidoglycan synthase PonA1 governs the rate of polar elongation in mycobacteria. PLoS Pathog. 11, e1005010 10.1371/journal.ppat.1005010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nakedi K. C., Nel A. J., Garnett S., Blackburn J. M., and Soares N. C. (2015) Comparative Ser/Thr/Tyr phosphoproteomics between two mycobacterial species: the fast growing Mycobacterium smegmatis and the slow growing Mycobacterium bovis BCG. Front. Microbiol. 6, 237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yang D. C., Blair K. M., and Salama N. R. (2016) Staying in shape: the impact of cell shape on bacterial survival in diverse environments. Microbiol. Mol. Biol. Rev. 80, 187–203 10.1128/MMBR.00031-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Doerks T., van Noort V., Minguez P., and Bork P. (2012) Annotation of the M. tuberculosis hypothetical orfeome: adding functional information to more than half of the uncharacterized proteins. PLoS One 7, e34302 10.1371/journal.pone.0034302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. van Teeseling M. C. F., de Pedro M. A., and Cava F. (2017) Determinants of bacterial morphology: from fundamentals to possibilities for antimicrobial targeting. Front. Microbiol. 8, 1264 10.3389/fmicb.2017.01264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Boutte C. C., Baer C. E., Papavinasasundaram K., Liu W., Chase M. R., Meniche X., Fortune S. M., Sassetti C. M., Ioerger T. R., and Rubin E. J. (2016) A cytoplasmic peptidoglycan amidase homologue controls mycobacterial cell wall synthesis. Elife 5, e14590 10.7554/eLife.14590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Vandal O. H., Nathan C. F., and Ehrt S. (2009) Acid resistance in Mycobacterium tuberculosis. J. Bacteriol 191, 4714–4721 10.1128/JB.00305-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pang X., Vu P., Byrd T. F., Ghanny S., Soteropoulos P., Mukamolova G. V., Wu S., Samten B., and Howard S. T. (2007) Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 153, 1229–1242 10.1099/mic.0.29281-0 [DOI] [PubMed] [Google Scholar]
- 78. Manganelli R., Voskuil M. I., Schoolnik G. K., and Smith I. (2001) The Mycobacterium tuberculosis ECF σ factor σE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41, 423–437 10.1046/j.1365-2958.2001.02525.x [DOI] [PubMed] [Google Scholar]
- 79. Manganelli R., Voskuil M. I., Schoolnik G. K., Dubnau E., Gomez M., and Smith I. (2002) Role of the extracytoplasmic-function σ factor σ(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45, 365–374 10.1046/j.1365-2958.2002.03005.x [DOI] [PubMed] [Google Scholar]
- 80. Dahl J. L., Kraus C. N., Boshoff H. I., Doan B., Foley K., Avarbock D., Kaplan G., Mizrahi V., Rubin H., and Barry C. E. 3rd (2003) The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl. Acad. Sci. U.S.A. 100, 10026–10031 10.1073/pnas.1631248100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Klinkenberg L. G., Lee J. H., Bishai W. R., and Karakousis P. C. (2010) The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J. Infect. Dis. 202, 1397–1404 10.1086/656524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Tsai M. C., Chakravarty S., Zhu G., Xu J., Tanaka K., Koch C., Tufariello J., Flynn J., and Chan J. (2006) Characterization of the tuberculous granuloma in murine and human lungs: cellular composition and relative tissue oxygen tension. Cell Microbiol. 8, 218–232 10.1111/j.1462-5822.2005.00612.x [DOI] [PubMed] [Google Scholar]
- 83. Emori K., and Tanaka A. (1978) Granuloma formation by synthetic bacterial cell wall fragment: muramyl dipeptide. Infect. Immun. 19, 613–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pandey A. K., Raman S., Proff R., Joshi S., Kang C. M., Rubin E. J., Husson R. N., and Sassetti C. M. (2009) Nitrile-inducible gene expression in mycobacteria. Tuberculosis 89, 12–16 10.1016/j.tube.2008.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wei J. R., Krishnamoorthy V., Murphy K., Kim J. H., Schnappinger D., Alber T., Sassetti C. M., Rhee K. Y., and Rubin E. J. (2011) Depletion of antibiotic targets has widely varying effects on growth. Proc. Natl. Acad. Sci. U.S.A. 108, 4176–4181 10.1073/pnas.1018301108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sajid A., Arora G., Gupta M., Upadhyay S., Nandicoori V. K., and Singh Y. (2011) Phosphorylation of Mycobacterium tuberculosis Ser/Thr phosphatase by PknA and PknB. PLoS One 6, e17871 10.1371/journal.pone.0017871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Parker A. E., and Bermudez L. E. (1997) Expression of the green fluorescent protein (GFP) in Mycobacterium avium as a tool to study the interaction between mycobacteria and host cells. Microb. Pathog. 22, 193–198 10.1006/mpat.1996.0106 [DOI] [PubMed] [Google Scholar]
- 88. Jain P., Hsu T., Arai M., Biermann K., Thaler D. S., Nguyen A., González P. A., Tufariello J. M., Kriakov J., Chen B., Larsen M. H., and Jacobs W. R. Jr. (2014) Specialized transduction designed for precise high-throughput unmarked deletions in Mycobacterium tuberculosis. MBio 5, e01245–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Khan M. Z., Bhaskar A., Upadhyay S., Kumari P., Rajmani R. S., Jain P., Singh A., Kumar D., Bhavesh N. S., and Nandicoori V. K. (2017) Protein kinase G confers survival advantage to Mycobacterium tuberculosis during latency-like conditions. J. Biol. Chem. 292, 16093–16108 10.1074/jbc.M117.797563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Martin A., Camacho M., Portaels F., and Palomino J. C. (2003) Resazurin microtiter assay plate testing of Mycobacterium tuberculosis susceptibilities to second-line drugs: rapid, simple, and inexpensive method. Antimicrob. Agents Chemother. 47, 3616–3619 10.1128/AAC.47.11.3616-3619.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Miesel L., Weisbrod T. R., Marcinkeviciene J. A., Bittman R., and Jacobs W. R. Jr. (1998) NADH dehydrogenase defects confer isoniazid resistance and conditional lethality in Mycobacterium smegmatis. J. Bacteriol. 180, 2459–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Patru M. M., and Pavelka M. S. Jr. (2010) A role for the class A penicillin-binding protein PonA2 in the survival of Mycobacterium smegmatis under conditions of nonreplication. J. Bacteriol. 192, 3043–3054 10.1128/JB.00025-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dephoure N., and Gygi S. P. (2011) A solid phase extraction-based platform for rapid phosphoproteomic analysis. Methods 54, 379–386 10.1016/j.ymeth.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Jhingan G. D., Kumari S., Jamwal S. V., Kalam H., Arora D., Jain N., KrishnaKumaar L., Samal A., Rao K. V., Kumar D., and Nandicoori V. K. (2016) Comparative proteomic analyses of avirulent, virulent and clinical strains of Mycobacterium tuberculosis identifies strain-specific patterns. J. Biol. Chem. 291, 14257–14273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Parikh A., Verma S. K., Khan S., Prakash B., and Nandicoori V. K. (2009) PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J. Mol. Biol. 386, 451–464 10.1016/j.jmb.2008.12.031 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.