Abstract
The strigolactone (SL) family of plant hormones regulates a broad range of physiological processes affecting plant growth and development and also plays essential roles in controlling interactions with parasitic weeds and symbiotic fungi. Recent progress elucidating details of SL biosynthesis, signaling, and transport offers many opportunities for discovering new plant-growth regulators via chemical interference. Here, using high-throughput screening and downstream biochemical assays, we identified N-phenylanthranilic acid derivatives as potent inhibitors of the SL receptors from petunia (DAD2), rice (OsD14), and Arabidopsis (AtD14). Crystal structures of DAD2 and OsD14 in complex with inhibitors further provided detailed insights into the inhibition mechanism, and in silico modeling of 19 other plant strigolactone receptors suggested that these compounds are active across a large range of plant species. Altogether, these results provide chemical tools for investigating SL signaling and further define a framework for structure-based approaches to design and validate optimized inhibitors of SL receptors for specific plant targets.
Keywords: plant hormone, hormone receptor, hydrolase, inhibition mechanism, crystal structure, chemical biology, strigolactone, strigolactone receptor inhibitor
Introduction
Numerous aspects of plant growth and development are tightly regulated by hormones, among which strigolactones (SLs)3 are critically involved in controlling shoot branching, root development, and leaf senescence (1–7). In particular, SLs inhibit the growth of axillary buds, resulting in reduced branching, and mutants with impaired SL production or that are unable to perceive SL consequently show increased levels of branching (3, 6–13). Through a combination of genetic, biochemical, and structural studies, the initial recognition events of the strigolactone signaling pathway in plants have now largely been resolved. The strigolactone receptor is an α/β-hydrolase (decreased apical dominance 2 in petunia (DAD2), DWARF14 in Arabidopsis (AtD14) and rice (OsD14), and RAMOSUS3 in pea (RMS3)) with remarkably slow, but absolutely essential, enzymatic activity toward its hormone substrate (11, 14–16). During catalysis, a bond between the ABC tricyclic lactone part of the SL molecule and the strictly conserved butenolide ring (D ring) is cleaved (11, 17) (Fig. 1C), with a covalent intermediate formed between the D ring and the catalytic histidine of the receptor that probably explains the very slow rate of catalysis (16, 18). Upon catalysis, the receptor undergoes a large conformational change in its lid domain that triggers its interaction with the F-box component (more axillary growth 2A in petunia (PhMAX2A), MAX2 in Arabidopsis, 3 in rice, and RMS4 in pea) of a Skp-Cullin-Fbox (SCF) complex (18, 19). In the presence of SL, the receptor also interacts with downstream repressor proteins of SL signaling belonging to the D53/suppressor of MAX2-like (SMXL) family, leading to their recruitment to the SCF complex and subsequent ubiquitination (14, 20, 21). Once polyubiquitinated, these repressors are targeted for degradation through the 26S proteasome, yielding a transcriptional response to the initial presence of the hormone signal.
Figure 1.
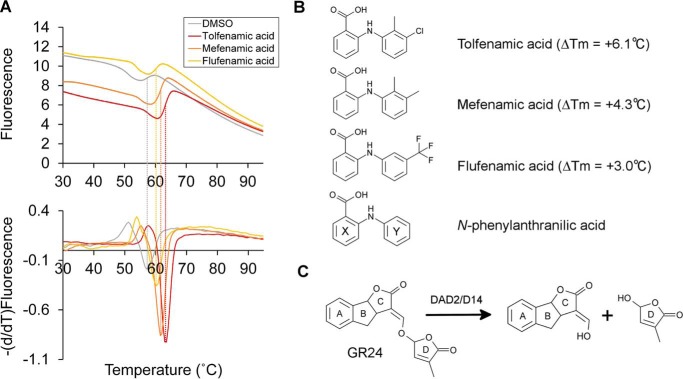
High-throughput screening of DAD2 inhibitors using the DSF assay. A, experimental melting curves (top) and derivatives of the melting curves (bottom) obtained for DAD2 in the presence of DMSO, tolfenamic acid, mefenamic acid, and flufenamic acid. Dashed lines, measured melting temperatures of DAD2 from which melting shifts (ΔTm = Tm(compound) − Tm(DMSO)) were calculated. B, chemical structures of the three identified inhibitors, tolfenamic acid, mefenamic acid, flufenamic acid, and of the parent compound, N-phenylanthranilic acid. The melting temperature shift of DAD2 (ΔTm) in the presence of inhibitors is indicated. Ring labels (X and Y) used throughout are labeled on the N-phenylanthranilic acid structure. C, hydrolysis reaction of the synthetic strigolactone GR24 by DAD2/D14 proteins.
In addition to their role as a plant hormone, SLs are exuded from the roots of plants to the rhizosphere, where they promote symbiotic interactions with arbuscular mycorrhizal fungi (22) and also stimulate the germination of parasitic weeds (8, 9, 15, 23). Hence, SLs are multifunctional compounds regulating a broad range of essential physiological processes that affect plant growth and development. Manipulation of the strigolactone biosynthetic and/or signaling pathways therefore offers tremendous opportunities for optimizing crop yields and for crop protection (24). Whereas numerous synthetic strigolactone analogues, including the widely used compound GR24 (25), have successfully been developed (26–31), only a few compounds inhibiting proteins involved in the biosynthesis of carotenoids (SL precursors), in the SL biosynthetic pathway and in SL signaling have recently been proposed or described (18, 24, 32–35). Among these, three compounds have been characterized as antagonists of SL receptors (33, 35, 36). Soporidine (ethyl 1-[4-(4-hydroxy-1-butyn-1-yl)benzyl]-4-[3-(trifluoromethyl)benzyl]-4-piperidinecarboxylate) was identified from a chemical screen based on the SL-dependent hypocotyl growth of Arabidopsis (33). This compound binds in vitro to Arabidopsis hyposensitive to light (AtHTL)/karrikin-insensitive (AtKAI2) (33), an α/β-hydrolase related to the plant SL receptors that responds to the smoke-derived karrikin compound instead of SLs (37). AtHTL orthologues from the parasitic weed Striga hermonthica (ShHTLs), however, do respond to SLs as signals for germination, and all three pathways (DAD2/D14, AtHTL, and ShHTL) probably converge at MAX2 for downstream signaling (13, 38, 39). Besides AtHTL, soporidin inhibits the hydrolytic activity of one of the Striga HTL orthologues (ShHTL7) in vitro and reduces the SL-induced germination of Striga seeds in a concentration-dependent manner (33). The second compound, 2-methoxy-1-naphthaldehyde (2-MN), was identified from an in silico virtual screening approach using the OsD14 structure as receptor (35). Although the direct effects of 2-MN on the binding and catalytic activities of OsD14 were not characterized, this compound was found to inhibit the SL-dependent interaction between OsD14 and D53 and between OsD14 and the rice-specific DELLA protein SLENDER RICE 1 (40) at concentrations above 25 μm in yeast two-hybrid assays (35). In an enhanced branching mutant of rice (d10), 2-MN was further able to restore the growth of rice tillering buds suppressed by exogenous application of strigolactone (35). Finally, and very recently, β-lactones were described as a class of compounds acting as irreversible antagonists for strigolactone receptors (36). Due to their specific mode of action involving acylation of the catalytic serine, these compounds successfully inhibit both plant (AtD14) and parasitic weed (ShHTL7) receptors with respective IC50 values in the 0.16–7.9 and 0.47–77 μm range, depending on side chain variations at positions 3 and 4 of the lactone ring (36).
To date, no crystal structure of any antagonist bound to SL receptor targets has been reported, and details of their corresponding inhibition mechanisms therefore remain largely unknown. Here we report the identification and detailed biochemical characterization of N-phenylanthranilic acid derivatives as novel inhibitors of plant SL receptors. High-resolution crystal structures of receptor-inhibitor complexes elucidate the binding mode of these compounds inside the internal cavities of the petunia and rice SL receptors to provide an understanding of the inhibition mechanism at the atomic level. Besides providing new chemical tools for investigating the various roles played by SLs, our results define a framework for structure-based approaches to design and validate optimized inhibitors of SL receptors for specific plant targets.
Results
High-throughput screening for compounds interacting with DAD2
Using differential scanning fluorimetry (DSF (41)), we previously showed that DAD2 undergoes strong thermal destabilization in the presence of the synthetic SL rac-GR24, characterized by a shift in DAD2's melting temperature (ΔTm) of ∼−9 °C (11). Conversely, we hypothesized that binding of candidate inhibitors inside the DAD2-binding pocket should stabilize the protein and trigger positive shifts in its melting temperature, measurable by DSF. We therefore screened the MicroSource Spectrum library of 2000 compounds (Discovery Systems, Inc.), consisting of drug components and natural products, for their ability to trigger a positive shift in DAD2's melting temperature in the DSF assay. Overall, 92% of experimental conditions yielded interpretable data where DAD2's melting temperature in the presence of compounds could be measured. Three of the compounds triggering the strongest stabilization of DAD2 (ΔTm ≥ +3.0 °C) were N-phenylanthranilic acid derivatives, namely tolfenamic acid (ΔTm = +6.1, best compound), mefenamic acid (ΔTm = +4.3, second best), and flufenamic acid (ΔTm = +3.0, fifth best) (Fig. 1, A and B). These were therefore selected for downstream assays.
N-Phenylanthranilic acid derivatives inhibit DAD2 catalytic activity and also inhibit the GR24-dependent interaction between DAD2 and SL signaling downstream targets, PhMAX2A and PhD53A
The ability of tolfenamic acid, mefenamic acid, and flufenamic acid to inhibit DAD2's catalytic activity was next investigated. DAD2 was incubated with a 4-fold molar excess of each compound for 30 min before the addition of a 20-fold molar excess of rac-GR24. Hydrolysis of rac-GR24 was subsequently analyzed after 3 and 16 h at 25 °C using TLC, as described previously (11). As seen in Fig. 2, partial and total hydrolysis of rac-GR24 by DAD2 was achieved after 3 and 16 h of incubation, respectively. In the presence of tolfenamic acid and mefenamic acid, however, the amount of product formed after 3 h of incubation was reduced, and a detectable amount of intact rac-GR24 remained in solution after 16 h of incubation, indicating that these two compounds effectively inhibit DAD2 catalytic activity. In comparison, flufenamic acid, which triggers a smaller increase in DAD2's melting temperature than the two previous compounds, was less efficacious (Fig. 2).
Figure 2.
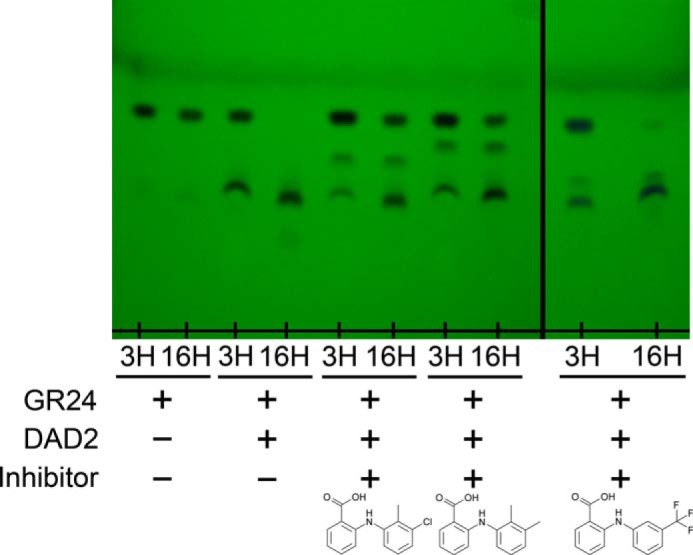
TLC analysis of the rac-GR24 hydrolysis by DAD2 in the presence of tolfenamic, mefenamic, and flufenamic acid. DAD2 was first incubated with a 4-fold molar excess of inhibitors for 30 min at 20 °C. GR24 was then added to a final 20-fold molar excess, and reactions were incubated at 25 °C. A positive control consisted of DAD2 (25 μm), GR24 (500 μm), 5% DMSO in PBS, and a negative control was GR24 (500 μm), 5% DMSO in PBS. Compounds were extracted after 3 and 16 h of incubation and analyzed by TLC.
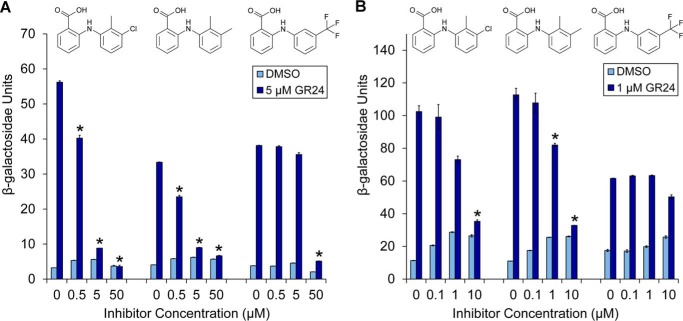
In the current model of strigolactone signaling, it is proposed that SL perception by DAD2/D14 recruits downstream proteins from the D53/SMXL family and targets them for degradation by the SCFMAX2 complex to promote shoot branching (14, 21, 42, 43). Indeed, direct SL-dependent interaction between DAD2/D14 and both D53/SMXL and MAX2 was observed using yeast two-hybrid assays (11, 14, 21, 42, 43). In agreement with this model and with the DSF and TLC assays described above, the addition of tolfenamic acid, mefenamic acid, and, to a lesser extent, flufenamic acid in the yeast two-hybrid assay also inhibited the rac-GR24-induced interaction between DAD2 and PhMAX2A and between DAD2 and a petunia ortholog of D53 (PhD53A; see “Experimental procedures”) in a concentration-dependent manner (Fig. 3).
Figure 3.
Yeast two-hybrid analysis of DAD2 interactions with downstream partners in the presence of inhibitors. Shown is inhibition of rac-GR24–induced DAD2/PhMAX2A (A) and DAD2/PhD53A (B) interactions by tolfenamic acid, mefenamic acid, and flufenamic acid. Protein-protein interactions are quantified by assaying β-gal activity in a yeast two-hybrid liquid culture system. Data points are the mean ± S.E. (error bars) of three technical replicates. *, GR24 treatments where the means are significantly different from the relevant 0 control (p < 0.01, Student's t test). Western blotting controls for expression of proteins in yeast are shown in Fig. S12.
Tolfenamic acid binds inside DAD2's cavity
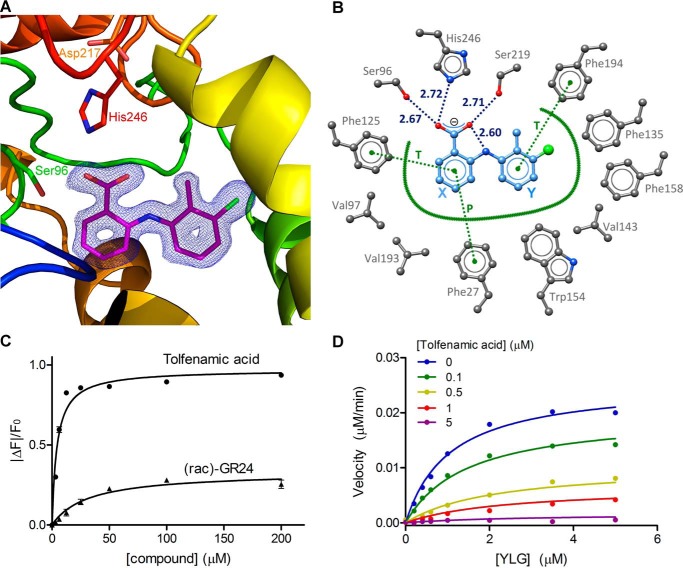
To obtain a detailed understanding of the inhibition mechanism, DAD2 was co-crystallized with tolfenamic acid, and the structure of the complex was solved to 1.68 Å resolution (Table 1, Figs. 4 and 5, and Figs. S1 and S2). To facilitate crystallization, a surface cysteine (Cys89) located on the other side of the protein compared with the entrance of the internal binding cavity was mutated to a glutamine. This mutation was confirmed to have no detectable influence on DAD2 catalytic activity and allowed a new triclinic crystal form diffracting to high resolution to be obtained (see “Experimental procedures”). Electron density maps of excellent quality were observed for tolfenamic acid–bound molecules in both DAD2 molecules of the asymmetric unit (Fig. 4A and Fig. S2). Tolfenamic acid fully occupies the DAD2-binding cavity with excellent shape complementarities (Fig. 5) resulting from small positional shifts (≤1 Å) of Val143 and Val193, and reorientations of a few side chains lining the internal cavity (Phe125, Ile140, Phe194, His218, and Ser219) compared with the apo-structure (Fig. S1). Among these residues, the largest movement is observed for the side chain of His218 that is displaced by tolfenamic acid from pointing toward the center of the cavity through a ∼90° rotation along the Cα-Cβ axis (Fig. S1). Overall, tolfenamic acid binds through a combination of electrostatic and hydrophobic interactions (Fig. 4B). The carboxylic group of tolfenamic acid directly interacts with the side chains of two residues of DAD2's catalytic triad, Ser96 and His246, and with Ser219 to anchor the compound deep inside the cavity (Figs. 4B and 5C). Notably, the relative position of the negatively charged carboxylic group of tolfenamic acid and of His246 suggests His246 to be protonated and positively charged, allowing the formation of an ionic interaction between tolfenamic acid and His246. In addition, tolfenamic acid is sandwiched between the side chains of four conserved phenylalanine residues of the binding pocket that form π–π hydrophobic interactions with both rings of tolfenamic acid; Phe27 and Phe125 clamp the X ring through parallel and T-stack interactions, respectively, whereas Phe194 and Phe158 form respective T-stack and distorted T-stack interactions from each side of the Y ring (Fig. 4B). The crucial role of Phe27 in binding tolfenamic acid was confirmed by site-directed mutagenesis. DAD2F27V was expressed and purified as for WT and assessed in the DSF assay. In agreement with the structural results, DAD2F27V completely lost its ability to bind tolfenamic acid (Fig. S3), yet the mutated protein retained its ability to be destabilized, similarly to WT, in the presence of a range of strigolactone compounds, including racemic mixtures of strigol, orobanchol, 5-deoxystrigol, and GR24, as well as with pure enantiomers of 5-deoxystrigol or GR24 (Fig. S3 and Table S1).
Table 1.
Data collection and refinement statistics
Values in parentheses are for the highest-resolution shell. RMSD, root mean square deviation.
| DAD2C89Q-tolfenamic acid | DAD2C89Q-MNAB | OsD14-MNAB | |
|---|---|---|---|
| PDB code | 6AP6 | 6AP7 | 6AP8 |
| Space group | P1 | P1 | P212121 |
| Cell parameters | |||
| a, b, c (Å) | 36.86, 55.83, 69.23 | 36.68, 48.31, 71.94 | 48.01, 88.43, 119.03 |
| α, β, γ (°) | 95.76, 95.13, 108.46 | 82.67, 86.76, 69.95 | β = ɣ = 90.0 |
| Data set | |||
| λ (Å) | 0.9537 | 0.9537 | 0.9537 |
| Unique reflections | 56,704 | 69,577 | 134,179 |
| Resolution range (Å) | 34.67–1.65 (1.68–1.65) | 45.08–1.51 (1.53–1.51) | 48.01–1.27 (1.29–1.27) |
| Rmerge | 0.09 (0.87) | 0.08 (0.83) | 0.15 (1.99) |
| CC½ | 0.998 (0.648) | 0.998 (0.593) | 0.999 (0.549) |
| I/σ(I) | 10.9 (1.6) | 12.1 (1.6) | 12.0 (1.6) |
| Completeness | 91.2 (89.8) | 95.9 (74.9) | 100.0 (100.0) |
| Multiplicity | 3.9 (3.8) | 4.0 (3.7) | 14.5 (14.3) |
| Bwilson (Å2) | 16.3 | 10.3 | 10.3 |
| Refinement | |||
| Resolution (Å) | 30.42–1.65 (1.69–1.65) | 36.27–1.51 (1.55–1.51) | 44.52–1.27 (1.30–1.27) |
| Reflections | 53,834 | 66,090 | 127,214 |
| Total number of atoms | 4737 | 4738 | 5026 |
| TLS groups | 2 (one for each monomer) | 2 (one for each monomer) | |
| Rwork/Rfree | 16.8/20.0 (28.6/29.8) | 16.4/19.7 (28.0/30.4) | 12.1/15.4 (25.5/27.0) |
| RMSD, bonds (Å)/angles (degrees) | 0.0107/1.45 | 0.0168/1.76 | 0.0205/1.96 |
| Average B-factors (Å2) (protein/water/inhibitor) | 21.6/28.6/12.0 | 18.7/26.3/13.3 | 12.0/28.5/18.6 |
| Ramachandran statistics of φ/ψ angles (%) | |||
| Most favored | 90.5 | 89.6 | 92.7 |
| Additional favored | 9.1 | 10.0 | 6.8 |
| Generously favored | 0.4 | 0.4 | 0.4 |
| Disallowed | 0 | 0 | 0 |
Figure 4.
DAD2 inhibition by tolfenamic acid. A, structure of DAD2 bound to tolfenamic acid. DAD2 is drawn in ribbon mode and rainbow-colored from blue (N terminus) to red (C terminus). The catalytic triad residues and tolfenamic acid (pink) are shown in stick mode. The final σA-weighted map contoured at 1.3σ around tolfenamic acid is shown in dark blue (the corresponding omit map is shown in Fig. S2). B, tolfenamic acid binding mode within DAD2's internal cavity. Oxygen, nitrogen, and chlorine atoms are represented in red, blue, and green spheres, respectively. Carbon atoms are shown as gray spheres for protein atoms, and light blue spheres for tolfenamic acid. Hydrogen bonds are shown as dotted blue lines, with distances (in Å) between polar atoms indicated. Hydrophobic interactions are represented by a thick green line. Specific π–π stacking interactions are shown as dotted green lines and labeled T and P for perpendicular T-stack and parallel stack, respectively. Residue numbers are indicated in gray. Ring labels (X and Y) used throughout are indicated in blue. C, intrinsic fluorescence of DAD2 in the presence of tolfenamic acid and rac-GR24. Each data point is the mean ± S.E. (error bars) of three technical replicates. D, competition assay of YLG hydrolysis by DAD2 using tolfenamic acid. Each data point is the average of three technical replicates. All of the individual replicates for each compound concentration were included during the nonlinear global fit analysis using a mixed-inhibition model. See also Table S2.
Figure 5.
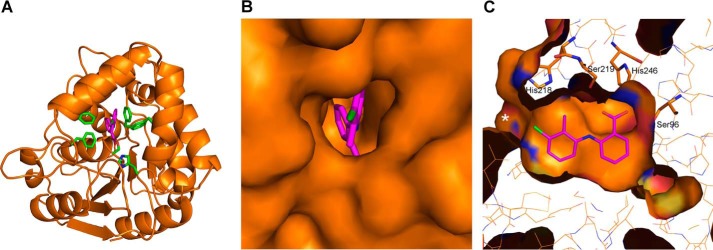
Structure of tolfenamic acid bound inside DAD2's internal cavity. A, overview of the crystal structure of DAD2 bound to tolfenamic acid. Tolfenamic acid is drawn in pink, whereas key DAD2-binding residues (Phe27, Ser96, Phe125, Phe158, Phe194, and His248 are drawn in green). B, close-up of DAD2 surface looking at the cavity entrance, in the same orientation as A. C, side view of tolfenamic acid bound inside DAD2's cavity. The view is rotated ∼90° along the y axis and 180° along the x axis compared with A and B. His218, Ser219, His246, and Ser96 are drawn in stick mode. The cavity entrance is indicated by a white asterisk.
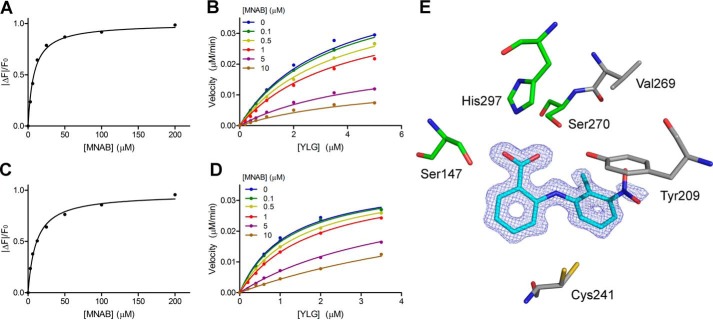
Intrinsic fluorescence experiments indicated that tolfenamic acid, mefenamic acid, and flufenamic acid bind to DAD2 with low micromolar affinities (Kd = 4.3, 4.7, and 10.8 μm, respectively; Fig. 4C, Fig. S4 (A and B), and Table S2). For tolfenamic acid, this is about 7 times lower than the Kd of 31.6 and 28.1 μm measured for rac-GR24 and (+)-GR24, respectively (Fig. 4C, Fig. S4C, and Table S2). The potencies of tolfenamic acid and mefenamic acid were further quantified with kinetic experiments using the Yoshimulactone Green (YLG) probe, which releases a fluorescein compound upon hydrolysis by AtD14 (15, 33). The progress curve of YLG hydrolysis by DAD2 was similar to the one obtained with AtD14 (15), characterized by a sharp increase in fluorescein formation over the first 50–60 min, followed by a slowdown in hydrolysis rate (Fig. S4D). Similar nonclassical Michaelis–Menten kinetics have been described, albeit at a faster rate, for the pea ortholog of DAD2, RMS3, with other profluorescent probes (16). Because these progress curves do not reflect classical steady-state kinetics, all subsequent analysis was performed in the pre-steady-state phase, using a 16-min time point. As described previously for RMS3 (16), we will therefore also refer hereafter to kcat as the rate constant of the pre-steady-state phase and to K½ as the probe concentration that gives half-maximal velocity. With these limits, the K½ for YLG hydrolysis by DAD2 was 1.12 μm, and kcat was 0.076 min−1. For competition assays, tolfenamic acid and mefenamic acid were added at increasing concentration to the reaction mixture, yielding apparent Ki values of 0.12 and 0.39 μm, respectively (Fig. 4D, Fig. S4E, and Table S2).
Extended structure-activity relationship (SAR) study
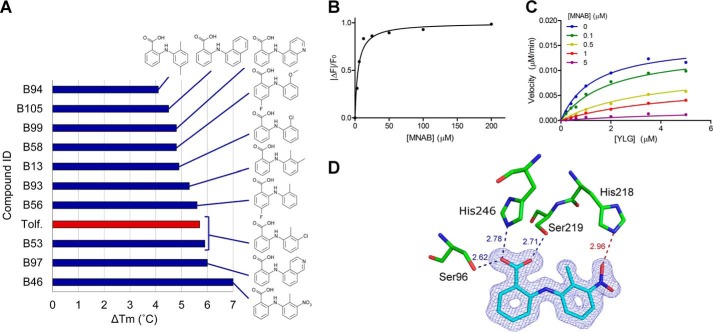
A set of 119 N-phenylanthranilic acid derivatives plus 19 heteroanalog 2-phenoxybenzoic acid derivatives (Table S3) were screened and compared with tolfenamic acid for their ability to bind to DAD2 in the DSF assay. As seen in Fig. 6A and Fig. S5, only one compound, 2-(2′-methyl-3′-nitroanilino)benzoic acid (MNAB), triggered a stronger stabilization of DAD2 than tolfenamic acid. This compound differs from tolfenamic acid by having a nitro group replacing the chlorine (Fig. 6A and Table S3). The crystal structure of DAD2 bound to MNAB confirmed that this compound binds in the same pose as tolfenamic acid inside DAD2's cavity, with the nitro group forming an additional hydrogen bond with the side chain of the displaced His218, possibly reinforcing the interaction with DAD2 (Fig. 6D). However, both binding and kinetic parameters obtained with MNAB are very similar to the ones obtained for tolfenamic acid (Fig. 6 (B and C) and Table S2).
Figure 6.
SAR study of DAD2 inhibitors. A, structures of the top 10 compounds of the SAR study, as assessed by decreasing values of DAD2's melting temperature shifts in the DSF assay. The red bar corresponds to known tolfenamic acid (Tolf.) used as reference. The experimental melting curves and derivatives of the melting curves for DAD2 in the presence of the top 10 compounds are shown in Fig. S5. Compound IDs for the SAR study were B1–B136, as detailed in Table S3. B, binding of MNAB to DAD2 using intrinsic fluorescence experiments. Each data point is the mean ± S.E. (error bars) of three technical replicates. C, competition of YLG hydrolysis by DAD2 using MNAB. Each data point is the average of three technical replicates. All of the individual replicates for each compound concentration were included during the nonlinear global fit analysis using a mixed-inhibition model. D, MNAB bound to DAD2. The final σA-weighted map contoured at 1.0σ around MNAB is shown in dark blue (the corresponding omit map is shown in Fig. S2). DAD2 residues involved in polar interactions with MNAB are shown. Hydrogen bonds are shown as dotted lines, with distances (in Å) between polar atoms indicated. The additional hydrogen bond between the nitro group of MNAB and His246 is shown as a dotted red line. See also Table S2.
Inhibition of DAD2 orthologues by N-phenylanthranilic acid derivatives
To assess the potential of N-phenylanthranilic acid derivatives to inhibit strigolactone receptors from other plant species, the structures of 19 close DAD2 orthologues (the “D14 clade”; Fig. S6) were modeled based on the structure of DAD2 bound to tolfenamic acid. For each model, the orientations of individual side chains pointing toward the internal binding cavity were checked and manually corrected to match those from the DAD2/tolfenamic acid structure as closely as possible. Docking scores of tolfenamic acid in a DAD2 “empty” crystal structure (i.e. the DAD2/tolfenamic acid crystal structure in which tolfenamic acid had been removed) and in the same empty structure where the F27V mutation was introduced were −10.6 and −8.0 kcal/mol, respectively, with poses for the docked tolfenamic acid moieties perfectly matching the crystal structure in both cases. These docking scores were subsequently used as benchmarks for binding versus nonbinding conditions, against which docking of the same compound in the 19 D14-clade models could be evaluated. As seen in Table S4, docking scores of tolfenamic acid in 16 of 19 models ranged between −10 and −9.1 kcal/mol, suggesting that these proteins may also have the ability to bind tolfenamic acid. By contrast, the three remaining models had docking scores ranging from −8.6 to −8.0 kcal/mol. Among these models, two (Morchella esculenta D14 and Populus trichocarpa D14_2) have the F27V mutation previously identified as preventing binding of tolfenamic acid in DAD2, whereas the remaining one (M. guttatus D14) harbors a V143F mutation in which the large phenylalanine side chain would protrude toward the center of the internal cavity, therefore probably preventing binding of tolfenamic acid inside the cavity.
To validate these results, the rice and Arabidopsis D14 orthologues (OsD14 and AtD14, with docking scores for tolfenamic acid of −9.7 and −9.5 kcal/mol, respectively) were expressed, purified, and assayed against the 138 N-phenylanthranilic acid derivatives using DSF. As seen in Fig. S5 (panels B and E and panels C and F), the compound triggering the largest thermal shift for OsD14 and AtD14 was again MNAB (ΔTm = +3.5 and +4.4 °C, respectively). Furthermore, seven compounds are shared within the top 10 hits of all three proteins (Fig. S5, D–F). Despite melting temperature shifts observed for OsD14 lower than those observed for DAD2, OsD14 could readily be co-crystallized with MNAB. The structure was solved at 1.27 Å resolution, and excellent electron density maps were obtained for MNAB bound inside the OsD14 cavity in a position almost identical to the one observed in DAD2 (Fig. 7E). Only a minor rotation of the Y ring of MNAB is seen in OsD14 compared with DAD2, most likely resulting from additional steric constraints created by the presence of Cys241 at the bottom of the cavity in OsD14 instead of Ser190 in DAD2 (Fig. 7E and Table S4). In the absence of His218, replaced by Val269 in OsD14, no additional hydrogen bond is directly formed between the protein and the nitro group of the bound compound. This may allow for some rotational flexibility of the nitro group, in agreement with the weaker density observed for the nitro group in molecule A of the asymmetric unit (Fig. 7E). However, this flexibility may still be partly restrained by a water molecule–mediated interaction between the nitro group of MNAB and Tyr209 of OsD14 (replacing Phe158 in DAD2), which provides additional hydrogen bonding options within the protein's cavity (Fig. 7E and Table S4). In line with these results, the Kd values of MNAB and tolfenamic acid to OsD14 were 8.0 and 9.3 μm, respectively (approximately double those observed for DAD2), whereas the apparent Ki values obtained for these two compounds were 2.4 and 3.23 μm, respectively, an order of magnitude higher than those obtained for DAD2 (Fig. 7 (A and B), Fig. S7, and Table S2). Likewise, Kd values of MNAB and tolfenamic acid to AtD14 were 11.0 and 11.2 μm, respectively, whereas the corresponding Ki values for these compounds were 1.9 and 2.52 μm, respectively (Fig. 7 (C and D), Fig. S8, and Table S2).
Figure 7.
OsD14 and AtD14 inhibition by MNAB. A, binding of MNAB to OsD14 using intrinsic fluorescence experiments. Each data point is the mean ± S.E. (error bars) of three technical replicates. B, competition of YLG hydrolysis by OsD14 using MNAB. Each data point is the average of three technical replicates. All of the individual replicates for each compound concentration were included during the nonlinear global fit analysis using a mixed-inhibition model. C and D, same as A and B, respectively, for AtD14. E, MNAB bound to OsD14 in the same orientation as in Fig. 6. The final σA-weighted map contoured at 1.0σ around MNAB is shown in dark blue (the corresponding omit map is shown in Fig. S2). Nonconserved residues between OsD14 and DAD2 are shown in gray: Val269 (His218 in DAD2), Tyr209 (Phe158), and Cys241 (Ser190). All other residues lining the internal cavity are conserved between the two proteins. See also Table S2.
In vivo effects of tolfenamic acid on bud growth
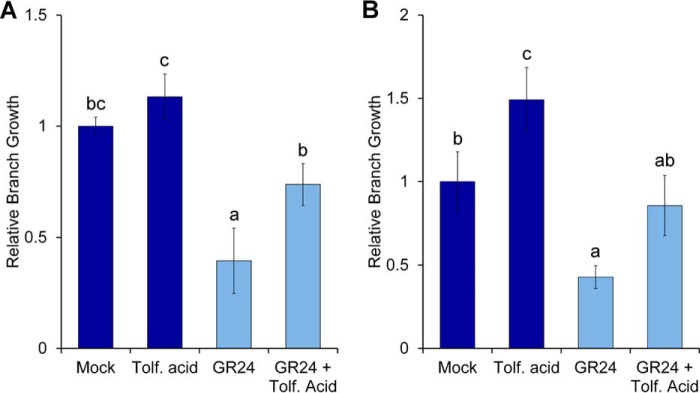
In vivo activity of tolfenamic acid was tested in bud growth assays in petunia and Arabidopsis as well as in a seed germination assay on the parasitic weed Orobanche minor. Tolfenamic acid showed activity on bud growth in petunia and Arabidopsis (Fig. 8), but not on germination of O. minor (Fig. S9). The bud assay used in petunia involves decapitation of whole plants after the second leaf, whereas in Arabidopsis, it uses excised stem fragments with two nodes (see “Experimental procedures”). In normal conditions, these treatments stimulate strong growth of branches, which can be inhibited by GR24 (Fig. 8). When tolfenamic acid is combined with the application of GR24, we observed that the GR24-induced inhibition of branch growth is reduced in both species (Fig. 8). Tolfenamic acid treatment on its own was also found to stimulate growth over untreated controls in Arabidopsis. However, this effect was not significantly different in petunia (Fig. 8).
Figure 8.
In vivo activity of tolfenamic acid. A, bud growth assay of petunia plants decapitated after leaf 2, treated with 0.5 μm GR24 and/or 50 μm tolfenamic acid. The numbers of leaves produced at the two leaf axils for each treatment were normalized relative to the mock-treated control (n = 10–11; data shown are mean ± S.E. (error bars)). B, bud growth assay of Arabidopsis two-node stem segments, treated with 0.5 μm GR24 and/or 5 μm tolfenamic acid. Total bud lengths for the two nodes were normalized relative to the mock-treated control (n = 16; data shown are mean ± S.E.). Statistical tests of differences between treatments were calculated by analysis of variance and Fisher's protected LSD multiple comparisons test (p = 0.05).
Discussion
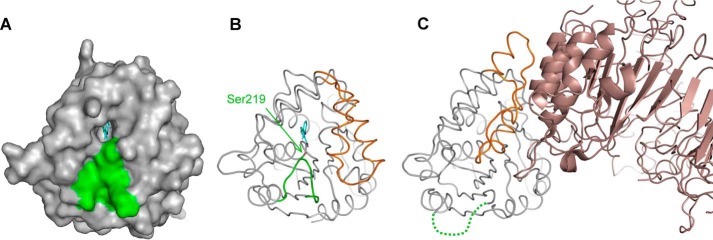
The development of novel plant-growth regulators targeting the SL pathway is anticipated to result in significant improvements in crop management and yield (24). Taking advantage of previous biochemical and structural knowledge of the petunia SL receptor, DAD2 (11), we designed high throughput assays that identified tolfenamic acid and other N-phenylanthranilic acid derivatives as lead DAD2 inhibitors with low micromolar binding affinities and submicromolar apparent inhibitory constants. Structural analysis showed that tolfenamic acid binds inside DAD2's internal cavity and interacts with highly conserved residues among SL receptors, including Ser96 and His246 from the catalytic triad, Ser219, and conserved Phe residues lining the internal cavity. Among these, Ser219 sits at a critical position at the tip of the loop (Gln213–Ala222 in DAD2) that shapes the entrance of the internal cavity (Fig. 9). In the crystal structure of the D14-D3 complex where D14 has undergone large conformational change in its lid domain to interact with D3, this loop is disordered (18), suggesting that movements of this loop upon SL binding and catalysis facilitate the conformational change of the lid. The direct electrostatic interaction observed between Ser219 and the carboxylic group of tolfenamic acid is consistent with such a hypothesis and provides a mechanistic understanding of the inhibition process (Fig. 9); by locking the activation loop in its “ground” conformation, the inhibitor prevents the conformational change of the lid domain that triggers downstream interactions with MAX2 and, in agreement with the positive melting temperature shift observed in the DSF assay, stabilizes the whole protein.
Figure 9.
Importance of the activation loop. A, surface representation of DAD2 in complex with tolfenamic acid. Tolfenamic acid bound inside the cavity is drawn in cyan, and residues from the activation loop (Gln213–Ala222) are shown in green. B, same as A with DAD2 drawn in ribbon mode. Ser219 is drawn in stick mode (green) and indicated. The part of the lid that undergoes conformation change when interacting with MAX2 is colored in orange. C, structure of the D14-D3 complex (PDB entry 5HZG). D14 is colored with the same color scheme as DAD2 in B. The disordered activation loop is represented by a green dashed line. D3 is shown in brown.
Based on the structural data, an in silico study of a set of 19 SL receptors from other plant species indicated that most of these receptors are likely to be inhibited by N-phenylanthranilic acid derivatives. Only two mutations were identified in the binding cavity of DAD2 orthologues that would prevent binding of the inhibitor: F27V, already identified through site-directed mutagenesis (Fig. S3), and V143F, most likely due to steric hindrance. Apart from these, mutations of H218V/C, F158Y, V143L, S190C, V193I, and V97I (Table S4) were all predicted to allow inhibitor binding. In vitro experiments using the Arabidopsis and rice orthologues of DAD2, carrying one and three amino acid differences in their respective binding cavities compared with DAD2, confirmed these results. Altogether, our studies therefore indicate that derivatives of N-phenylanthranilic acid inhibit SL receptors from a broad range of species and further provide a framework for structure-based drug design approaches to improve the potencies of modified compounds for SL receptors.
A SAR study of 138 N-phenylanthranilic acid closely related compounds using the DSF assay on DAD2, OsD14, and AtD14 suggested that one compound, MNAB, is able to bind all three receptors with higher affinity than tolfenamic acid. However, further characterization using intrinsic fluorescence and inhibition kinetics yielded similar binding and inhibition parameters for this compound and for tolfenamic acid (Table S2). Hence, whereas the DSF assay is highly amenable to a high throughput format and therefore well adapted for the screening of large libraries of compounds, it may lack sensitivity to robustly differentiate the binding abilities of closely related compounds. The DSF assay also presents the drawback of being protein-sensitive. This is particularly evident with the OsD14 data set, where the observed melting temperature shifts in the presence of compounds are smaller than those observed for AtD14 despite comparable Kd values. Nevertheless, detailed analysis of binding versus nonbinding compounds within the SAR data sets provided the following insights. (i) Substituents other than H or F on the X ring are not favored; this is not surprising, given the tight steric constraints around the X ring seen in the crystal structures. (ii) All of the best compounds have a substituent in position 2 of the Y ring (2-Me or 2-CH(ring) in most cases), most likely to prevent rotation of the Y ring within the cavity. Altogether, this defines the core pharmacophore as N-(2-methylphenyl)anthranilic acid (Fig. S5G), with possible substituents to be located in positions 2, 3, and 5 of the Y ring.
In vivo activity showed promising initial results in petunia and Arabidopsis, with tolfenamic acid able to interfere with the GR24-induced inhibition of branch growth in bud assays. Tolfenamic acid on its own did not stimulate much growth over untreated controls in petunia, in contrast to Arabidopsis. This result may not be unexpected, however, given the plant treatments (decapitation versus excision of stem segments) both stimulate branch growth through complex mechanisms that probably involve multiple hormone signaling pathways (10, 12). As seen in Fig. 8, relatively high concentrations of tolfenamic acid were needed to observe a physiological response in plants. In both cases, this is in contrast to the submicromolar inhibitory constants measured on the receptors in vitro, suggesting that there may be issues with compound uptake and/or transport in planta. One area for future work would therefore be to assess bioavailability and potentially improve uptake of these compounds in various plant species and in various experimental conditions. The low solubility of tolfenamic acid derivatives in aqueous solutions could, for example, be mitigated by formulating these compounds as salts (44) or by taking advantage of the fact that position 3 of the Y ring is pointing toward the entrance of the cavity in the crystal structure. The attachment of large polar groups extending from that position could therefore alter the biophysical properties of the compounds without affecting activity. Alternatively, a recently described innovative approach to increase the uptake of charged compounds into plants using photocaged precursors (45) could also be envisaged. In vitro, however, our data show that N-phenylanthranilic derivatives are readily compatible with a broad range of biochemical assays and therefore present excellent potential as novel chemical tools to dissect the mechanisms underlying the wide spectrum of SL function.
Parasitic weeds from the Striga and Orobanche genera have an expansion of the HTL/KAI2 clade, related to, but distinct from, the D14 clade of SL receptors. In S. hermonthica, a combination of 11 receptors, structurally related to the plant SL receptors from the D14 clade, was recently found to be involved in SL perception (15, 46). Structural superposition of the S. hermonthica HTL5 (ShHTL5) receptor with DAD2 highlights significant differences in the size and shape of their respective binding cavities, with key DAD2 residues involved in tolfenamic acid binding being altered in ShHTL5 (notably F125V and F194S, but also S219L, V193I, and F158Y). These result in a much larger cavity on one side of the compound, which is likely to strongly reduce or abolish tolfenamic acid binding to ShHTL5 (Fig. S10). These residues, however, show a relatively high degree of variability among ShHTL receptors (46). For example, Phe125 is conserved in ShHTL2 and -3 and replaced by Tyr in ShHTL1, whereas Phe194 is conserved in ShHTL1, -2, -3, -8, and -11 and replaced by Tyr in ShHTL10 (46). Hence, it is possible that N-phenylanthranilic acid derivatives may inhibit some but not all ShHTL receptors. Consistent with these results, we observed that tolfenamic acid was unsuccessful in inhibiting the GR24-induced germination of O. minor seeds (Fig. S9). Ultimately, the most beneficial situation would be to have compounds specific for either plant or weed receptors, but not for both. The structural knowledge of the different class of SL receptors, together with detailed understanding of their inhibition mechanisms, will help to achieve this goal in the longer term.
Experimental procedures
Protein expression and purification
DAD2 was codon-optimized for bacterial expression and expressed as a cleavable His-MBP fusion protein in Escherichia coli Rosetta-Gami 2. Cells were resuspended in 20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 2 mm imidazole and homogenized twice at 10,000 p.s.i. using an Emusiflex C3 (Avestin). The soluble fraction was applied to a 15-ml TALON resin column (Clontech) and eluted with 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 300 mm imidazole. After dialysis at 4 °C for 16 h against 20 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm DTT, His-MBP-DAD2 was further purified by anion exchange using a HiTrapQ HP 5-ml column (GE Healthcare). Elution was achieved using a 50–500 mm NaCl linear gradient in 20 mm Tris-HCl, pH 8.0, 1 mm DTT. Cleavage of the His-MBP tag was done using tobacco etch virus protease at a 1:50 (w/w) protease/protein ratio, with the reaction mixture being dialyzed against 20 mm Tris-HCl, 100 mm NaCl, 50 mm l-glutamic acid, 50 mm l-arginine, 1 mm DTT, 1 mm EDTA (pH 8.0) at 4 °C for 16 h. The dialysis bag was then transferred to the same buffer lacking DTT and EDTA for 2 h before being applied to two His-Trap HP 5-ml columns mounted in series. The flow-through was collected, concentrated to 5.9 mg/ml, aliquoted, and stored at −80 °C.
Amino acid mutations (C89Q and F27V) were generated using the QuikChange® Lightning kit (Agilent Technologies). Mutants were purified in the same manner as WT protein (although no DTT was used for purification of DAD2C89Q).
OsD14 was codon-optimized for bacterial expression and produced as a cleavable His-MBP fusion protein in E. coli Rosetta–Gami 2. Cells were resuspended in 20 mm Tris-HCl, pH 8.0, 300 mm NaCl, 2 mm imidazole and homogenized twice at 10,000 p.s.i. using an Emusiflex C3 (Avestin). The soluble fraction was applied to a 7-ml TALON resin column (Clontech) and eluted with 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 300 mm imidazole. The imidazole was removed by a 2-h dialysis against 20 mm Tris, pH 8.0, 150 mm NaCl at 4 °C, and cleavage of the His-MBP tag was performed using tobacco etch virus protease at a 1:50 (w/w) protease/protein ratio in 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm DTT, 1 mm EDTA at 4 °C for 16 h. The digestion reaction was then dialyzed against 20 mm Tris, pH 8.0, 150 mm NaCl, 10 mm imidazole for 2 h before being applied to a His-Trap HP 5-ml column. The flow-through was collected and further purified by gel filtration using a Superdex 75 HighLoad 10/60 column (GE Healthcare) using 20 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm DTT as elution buffer. OsD14 was collected, concentrated to 6.0 mg/ml, aliquoted, and stored at −80 °C.
AtD14 was codon-optimized for bacterial expression, produced as a cleavable His-MBP fusion protein in E. coli Rosetta-Gami 2, and purified following the same protocol as for OsD14. Gel filtration fractions were pooled, concentrated to 2.8 mg/ml, and stored at −80 °C.
Protein concentrations were determined by UV absorption at 280 nm, using extinction coefficients calculated from the protein sequences using the ProtParam tool from the ExPASy server (https://web.expasy.org/protparam/).
Crystallization and structure determination
Although DAD2 crystals could easily be obtained in previously determined conditions (11), most if not all crystals showed no or very poor diffraction. Close examination of the two previously determined crystal forms in I23 (PDB entry 4DNP) and P31 (PDB entry 4DNQ) space groups showed peculiar features at their respective packing interface. In the I23 crystal form, two Cys89 residues from neighboring molecules were found to be covalently linked via a DTT molecule. In the P31 crystal form, however, two Cys89 residues of neighboring molecule were also covalently linked, but this time through a direct intermolecular disulfide bridge, resulting in shorter protein–protein distances in the crystal and alternative space group. It was then reasoned that poorly diffracting crystals could result from a “mixture” between the two crystal forms, creating long-range static disorder within newly formed crystals. To test this hypothesis, it was decided to mutate the cysteine 89 to glutamine. With Cys89 sitting at the surface of the α/β-hydrolase domain of DAD2, opposite from the lid, cavity entrance, or active site, it was likely that the C89Q mutation would not interfere with the activity of the protein. Indeed, in the DSF assay, DAD2C89Q had its melting temperature and melting shifts in presence of rac-GR24 and tolfenamic acid almost identical to the ones observed for DAD2 (57.0 versus 56.5, −8.8 versus −8.0, and +6.4 versus +6.1 °C, respectively).
Before crystallization (and as done previously with WT DAD2), DAD2C89Q was buffer-exchanged into 20 mm Tris, pH 8.0, 150 mm NaCl using a Superdex 200 10/300 GL column (GE Healthcare) and concentrated to ∼8 mg/ml. A new triclinic crystal form was obtained in new crystallization conditions by hanging-drop vapor diffusion at 18 °C. Drops consisted of 1 μl of protein solution and 1 μl of reservoir solution consisting of 0.1 m Tris acetate, pH 8.5, 30% PEG 4000, 0.2 m MgCl2 (Structure Screen 1–33, Molecular Dimensions) or 0.1 m Tris acetate, pH 8.5, 20% PEG 8000, 0.2 m MgCl2 (JCSG+ 1.42, Molecular Dimensions). For co-crystallization, DAD2C89Q was incubated with a ∼6-fold molar excess of inhibitor (tolfenamic acid or MNAB) in 20 mm Tris, pH 8.0, 150 mm NaCl, 2% DMSO for 30 min at 18 °C before crystallization. Crystals used for data collection were obtained in the following conditions: 1 μl of DAD2C89Q (4.2 mg/ml) in the presence of 0.83 mm tolfenamic acid and 1 μl of reservoir solution consisting of 0.1 m Tris acetate, pH 8.5, 24% PEG 8000, 0.2 m MgCl2; 1 μl of DAD2C89Q (6.2 mg/ml) in the presence of 1.25 mm MNAB and 1 μl of reservoir solution consisting of 0.1 m Tris acetate, pH 8.5, 27% PEG 3350, 0.2 m MgCl2. In both cases, crystals were cryoprotected by successive transfer to a reservoir solution containing 10, 15, and 20% glycerol. Full data sets were collected at the Australian Synchrotron MX1 beamline to 1.65 and 1.51 Å resolution for the tolfenamic acid and MNAB complexes, respectively.
Before crystallization, OsD14 was buffer-exchanged to 20 mm Tris, pH 8.5, 200 mm ammonium acetate, 2 mm DTT using Superdex 200 10/300 GL and concentrated to 3.9 mg/ml. The protein was incubated with 1 mm (7.7-fold molar excess) MNAB in 20 mm Tris, pH 8.5, 200 mm ammonium acetate, 2 mm DTT, 2% DMSO for 30 min at 18 °C. Drops consisted of 1 μl of protein solution and 1 μl of reservoir solution consisting of 0.1 m HEPES, pH 7.5, 5% 2-methyl-2,4-pentanediol, 8% PEG 6000. Crystals were cryoprotected by successive transfer to a reservoir solution containing 5, 10, 15, and 20% glycerol. A full data set was collected at the Australian Synchrotron MX2 beamline to 1.27 Å resolution.
Data were processed with iMOSFLM (47) or XDS (48) and Pointless/Aimless (49). Structure determinations were achieved using programs from the CCP4 package (50). Structures were solved by molecular replacement using the DAD2 structure (PDB entry 4DNP) or the OsD14 structure (PDB entry 3W04) as starting models in Phaser (51). Refinement cycles were carried out using Refmac5 (52) and Coot (53). Optimized refinement parameters obtained from the PDB_redo server (54) were used in the final stages of refinement. Data collection and refinement statistics are listed in Table 1.
Modeling and docking
Sequences of DAD2/D14 homologs were aligned using Clustal Omega (55), and a phylogenetic tree is presented in Fig. S6 (and nexus file in supporting data 3). Models were generated in Modeler (56) using the structure of the DAD2-tolfenamic acid complex as template. Side chains of residues pointing toward the internal binding cavity were checked and manually corrected in Coot (53) to match those from the DAD2-tolfenamic acid structure as closely as possible. Docking was performed using Autodock Vina (57) using a 25 × 25 × 25-Å3 box centered on the oxygen atom of Ser96.
Differential scanning fluorimetry
The DSF experiments were performed as described previously (11). The MicroSource spectrum library (Discovery Systems) was purchased from Compounds Australia (Griffith University, Queensland, Australia), with each of the 2000 compounds (5 mm in DMSO) supplied as a 0.4-μl aliquot in 384-well plates. DAD2 was buffer-exchanged into PBS using Superdex 75 HiLoad 16/60. 18.6 μl of a solution containing 6.65 μm DAD2, SyproTangerine 10.25× in PBS were added to each compound using a BIOMEK 3000 pipetting robot (Beckman Coulter) to yield a final 19-μl reaction consisting of 6.5 μm DAD2, SyproTangerine 10×, 105 μm compound, 2.1% DMSO in PBS. Reactions were incubated for 30 min at 18 °C in the absence of light before DSF analysis.
For any subsequent DSF analysis, proteins (DAD2, DAD2F27V, OsD14, and AtD14) were buffer-exchanged into 20 mm Tris, pH 8.0, 200 mm NaCl. 0.5 μl of compounds (strigolactones and inhibitors) at 10 mm in DMSO were dispensed in a 384-well plate, and final reactions (19 μl) consisted of 10 μm protein, SyproTangerine 10×, 260 μm compound, 2.6% DMSO in 20 mm Tris, pH 8.0, 200 mm NaCl.
Thin-layer chromatography
Final reactions (200 μl) consisted of 25 μm DAD2, 100 μm inhibitor, 500 μm GR24, 5% DMSO in PBS. DAD2 (buffer-exchanged into PBS) was first incubated with a 4-fold molar excess of inhibitors (tolfenamic acid, mefenamic acid, and flufenamic acid) for 30 min at 4 °C. GR24 was then added to a final 20-fold molar excess, compared with DAD2, and reactions were incubated at 25 °C. A positive control consisted of DAD2 (25 μm), GR24 (500 μm), 5% DMSO in PBS, and a negative control was GR24 (500 μm), 5% DMSO in PBS. After a 3-h incubation at 25 °C, 100 μl of each reaction was transferred to 4-ml glass vials and extracted with 1 ml of ethyl acetate by vortexing for 1 min. After a 5-min centrifugation at 3500 × g, the organic phase was collected and transferred into a 1-ml glass vial containing 5 μl of acetic acid and stored at −20 °C. The remaining 100-μl reactions were further incubated for 15 h at 25 °C and extracted in a similar manner. All extracts were evaporated under nitrogen, resuspended in 20 μl of acetone containing 5:1000 (v/v) acetic acid, and analyzed by TLC on a precoated silica gel 60 F254 plate (Merck) using chloroform/acetone (4:1, v/v), containing 5:1000 (v/v) acetic acid as developing solvent. Spots were visualized under UV light (254 nm).
Yeast two-hybrid assays
We searched the publically available Petunia axillaris (58) and rice genome (59) data using BLAST to identify genes with sequence similarity to the SMAX1 and SMXL genes of Arabidopsis (20) and to the D53 gene from rice (14, 21). We identified 11 genes from rice and 13 genes from petunia. These are listed in supporting data 1 with the genome DB locus numbers. Using the gene models given in the locus records, the genes were conceptually translated to give the presumptive amino acid sequences for nine Arabidopsis genes, 10 rice genes, and nine petunia genes. The protein sequences were aligned using Geneious align (Geneious R10), and a phylogenetic tree was calculated with RAxML (60). On the basis of the tree, we selected the D53A and D53B genes for further study. Both genomic and mRNA copies of these two genes were sequenced from Petunia hybrida. This led us to correct the gene models for these genes, and the cloned cDNA sequence data are available in GenBankTM under accession numbers KY549358 and KY549359. The alignment was updated with these sequences, and the trees were recalculated (supporting data 2 and Fig. S11). Both genes were cloned from cDNA using CloneAmp HiFi polymerase and cloned into pCR8 using In-Fusion (Clontech) following the manufacturer's instructions.
Yeast two-hybrid experiments were performed following methods from the Clontech Yeast Protocols Handbook (2009), using yeast PJ69-4 (61), with DAD2 cloned into pBD vector (62), and MAX2A and D53A cloned into pAD vector (62). The presence of the binding domain and activation domain fusion proteins in selected diploid yeast strains was confirmed by Western blot analysis (Fig. S12). Liquid culture assays using orthonitrophenyl-β-galactopyranoside (Sigma-Aldrich) as the substrate were used to quantify the strength of the interactions between DAD2 and target proteins. DAD2/MAX2A and DAD2/D53A interactions were detected in the presence of 5 and 1 μm GR24, respectively, and compared with DMSO controls. Each inhibitor was tested at 0.1-, 1-, and 10-fold molar ratios compared with GR24. To control that the addition of the tested inhibitors did not interfere with the assay, the compound-independent interaction between PhMAX2A and PSK3 (11) was verified in the absence and presence of 5 μm GR24 and, in both cases, in the presence of 0.5, 5, and 50 μm concentrations of each inhibitor (data not shown).
Intrinsic fluorescence assays
Experiments were performed on a FLUOStar Omega microplate reader (BMG LabTech) using a 280 ± 10-nm excitation filter and 340 ± 10-nm emission filters. The gain was set to 1800 and the number of flashes to 50. Compound stocks were prepared at 10× final concentration in 20 mm Tris, pH 8.0, 150 mm NaCl, 20% DMSO. DAD2 was buffer-exchanged in 20 mm Tris, pH 8.0, 150 mm NaCl using gel filtration (Superdex 75 10/300 GL, GE Healthcare), and its concentration was adjusted to 11.11 μm. For experiments, 10 μl of compounds were manually dispensed and then mixed with either 90 μl of buffer or 90 μl of DAD2 protein solution in flat-bottomed, black 96-well plates using a Biomek 3000 robot (Beckman Coulter). Final protein concentration was 10 μm, with compounds ranging from 0 to 200 μm in 20 mm Tris, pH 8.0, 150 mm NaCl, 2% DMSO. All experiments (protein and buffer alike) were performed in triplicate. The plate was incubated for 30 min at 25 °C before measurements. Binding curves were obtained by plotting the relative fluorescence (|ΔF|/F0) versus compound concentration, where F0 is the fluorescence of the DMSO control, and |ΔF| = |F − F0|. GraphPad Prism was used to perform nonlinear regressions and determine Kd values using the “one site specific binding” model.
Enzymatic inhibition assays
Experiments were performed on a FLUOStar Omega microplate reader (BMG LabTech) using a 485 ± 12-nm excitation filter and 520 ± 10-nm emission filters. The temperature was set to 25 °C, and the gain was set to 920. Measurements were performed in black 96-well plates at 2-min intervals, with 20 flashes per cycle, over 240 min for progress curves and 90 min for kinetic experiments. For progress curves, final reactions (100 μl) consisted of 0.34 μm protein (DAD2, OsD14, or AtD14) and 1 μm YLG (TCI Chemicals) in 20 mm Tris/HCl, pH 8.0, 150 mm NaCl, 1% DMSO. Controls consisted of 1 μm YLG in the same buffer. All experiments were performed in triplicate. Fluorescence units were converted to fluorescein concentrations using fluorescein standard curves. For enzyme inhibition assays, reactions were performed at eight YLG concentrations (0, 0.2, 0.4, 0.6, 1, 2, 3.5, and 5 μm) and five (or six) inhibitor concentrations (0, 0.1, 0.5, 1, and 5 (and 10) μm). In all cases, protein concentration was 0.34 μm, and reaction buffer was 20 mm Tris/HCl, pH 8.0, 150 mm NaCl, 1% DMSO. All experiments were performed in triplicate. YLG-only controls were measured in the same buffer, and fluorescent units were converted to fluorescein concentrations using fluorescein standard curves. Michaelis–Menten analyses were performed by nonlinear regression in GraphPad Prism using the YLG series in the absence of inhibitor at the 16-min time point. For inhibition kinetics, global nonlinear regression analyses were performed in GraphPad Prism using a mixed inhibition model.
Plant methods
Petunia bud growth assays were performed using P. hybrida inbred line V26. Plants were sown on soil and grown in a glasshouse under natural light. One month after germination, plants were decapitated after the second true leaf, and any cotyledonary buds were removed. Treatment solutions were freshly prepared and contained 1% DMSO, 0.05% Silwet, 0.1% Tween 20, and (depending on treatment) 0.5 μm GR24 and/or 50 μm tolfenamic acid. Treatment solutions were applied directly to the axils (or axillary buds if present) of the two leaves at a rate of 4 μl of solution to each bud, once per day for 5 days. Leaf growth (number of leaves 3 mm in size or larger) of axillary buds was scored after a total of 7 days from the beginning of treatment.
Arabidopsis bud assays were performed as described previously (11). Briefly, two node sections of stems were excised from bolting plants. The base of the stem was placed in treatment solutions containing 2% DMSO in 0.5× HSS (11), including 0.5 μm GR24 and/or 5 μm tolfenamic acid. Total bud length from the two leaf axils was measured after 5 days.
Orobanche minor seed germination assays were based on the method of Matusova et al. (63). Seeds were surface-sterilized in 1.5% sodium hypochlorite containing 0.02% Tween 20 for 5 min, followed by four washes with H2O. Seeds were dried and then distributed onto 6-mm glass fiber filter disks, placed on damp filter paper in a Petri dish, sealed, and incubated at 24 °C in the dark for 6 days. Seeds on their glass fiber filter disks were then transferred to the wells of a 96-well plate to which 20 μl of treatment solution was added. Treatment solutions all included 2% DMSO and the indicated compounds, and all treatments were repeated in triplicate. The plates were sealed and then incubated at 24 °C in the dark for 5 days, after which the individual treatments were photographed. Seed germination and total seed counts were calculated from the photographs.
Author contributions
C.H., R. S. M. D., Z.L., P.S., H.W.L., B.J.J., and K.C.S. performed experiments. W.A.D. supplied the 138 N-phenylanthranilic acid derivatives and analyzed the SAR data. C.H., B.J.J., N.B.P., and K.C.S. were involved in discussions throughout the project and revised the manuscript. C.H. wrote the manuscript with input from all authors. C.H. and K.C.S. conceived and supervised the project.
Supplementary Material
Acknowledgments
We thank B. Hawkins (University of Otago) for fruitful discussions, M. Simpson (Compound Australia) for help with supplying the compound library, members of the Laboratory of Structural Biology (University of Auckland) for help with X-ray data collection, and Y. Zhang and H. Bouwmeester (University of Wageningen) for providing the 5DS enantiomers. This research was undertaken on the MX1 and MX2 beamlines at the Australian Synchrotron, part of ANSTO.
This work was supported by funding from AGMARDT (Agribusiness Innovation Grant 1323), Plant and Food Research, and a Marsden grant (contract PAF1301). C. H., K. C. S., and the New Zealand Institute for Plant and Food Research Limited have filed a New Zealand provisional patent application (734620) that relates to the specific topic “Strigolactone receptor antagonists.”
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) KY549358 and KY549359.
The atomic coordinates and structure factors (codes 6AP6, 6AP7, and 6AP8) have been deposited in the Protein Data Bank (http://wwpdb.org/).
This article contains Tables S1–S4 and Figs. S1–S11.
- SL
- strigolactone
- 2-MN
- 2-methoxy-1-naphthaldehyde
- DSF
- differential scanning fluorimetry
- MNAB
- 2-(2′-methyl-3′-nitroanilino)benzoic acid
- SAR
- structure-activity relationship
- SCF
- Skp-Cullin-Fbox
- YLG
- Yoshimulactone Green
- PDB
- Protein Data Bank
- MBP
- maltose-binding protein.
References
- 1. Woo H. R., Chung K. M., Park J.-H., Oh S. A., Ahn T., Hong S. H., Jang S. K., and Nam H. G. (2001) ORE9, an F-Box protein that regulates leaf senescence in Arabidopsis. Plant Cell 13, 1779–1790 10.2307/3871318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yamada Y., Furusawa S., Nagasaka S., Shimomura K., Yamaguchi S., and Umehara M. (2014) Strigolactone signaling regulates rice leaf senescence in response to a phosphate deficiency. Planta 240, 399–408 10.1007/s00425-014-2096-0 [DOI] [PubMed] [Google Scholar]
- 3. Gomez-Roldan V., Fermas S., Brewer P. B., Puech-Pagès V., Dun E. A., Pillot J.-P., Letisse F., Matusova R., Danoun S., Portais J.-C., Bouwmeester H., Becard G., Beveridge C. A., Rameau C., and Rochange S. F. (2008) Strigolactone inhibition of shoot branching. Nature 455, 189–194 10.1038/nature07271 [DOI] [PubMed] [Google Scholar]
- 4. Kapulnik Y., Delaux P. M., Resnick N., Mayzlish-Gati E., Wininger S., Bhattacharya C., Séjalon-Delmas N., Combier J. P., Bécard G., Belausov E., Beeckman T., Dor E., Hershenhorn J., and Koltai H. (2011) Strigolactones affect lateral root formation and root-hair elongation in Arabidopsis. Planta 233, 209–216 10.1007/s00425-010-1310-y [DOI] [PubMed] [Google Scholar]
- 5. Rasmussen A., Mason M. G., De Cuyper C., Brewer P. B., Herold S., Agusti J., Geelen D., Greb T., Goormachtig S., Beeckman T., and Beveridge C. A. (2012) Strigolactones suppress adventitious rooting in Arabidopsis and pea. Plant Physiol. 158, 1976–1987 10.1104/pp.111.187104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Snowden K. C., Simkin A. J., Janssen B. J., Templeton K. R., Loucas H. M., Simons J. L., Karunairetnam S., Gleave A. P., Clark D. G., and Klee H. J. (2005) The decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17, 746–759 10.1105/tpc.104.027714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Umehara M., Hanada A., Yoshida S., Akiyama K., Arite T., Takeda-Kamiya N., Magome H., Kamiya Y., Shirasu K., Yoneyama K., Kyozuka J., and Yamaguchi S. (2008) Inhibition of shoot branching by new terpenoid plant hormones. Nature 455, 195–200 10.1038/nature07272 [DOI] [PubMed] [Google Scholar]
- 8. Al-Babili S., and Bouwmeester H. J. (2015) Strigolactones, a novel carotenoid-derived plant hormone. Annu. Rev. Plant Biol. 66, 161–186 10.1146/annurev-arplant-043014-114759 [DOI] [PubMed] [Google Scholar]
- 9. Brewer P. B., Koltai H., and Beveridge C. A. (2013) Diverse roles of strigolactones in plant development. Mol. Plant 6, 18–28 10.1093/mp/sss130 [DOI] [PubMed] [Google Scholar]
- 10. Chatfield S. P., Stirnberg P., Forde B. G., and Leyser O. (2000) The hormonal regulation of axillary bud growth in Arabidopsis. Plant J. 24, 159–169 10.1046/j.1365-313x.2000.00862.x [DOI] [PubMed] [Google Scholar]
- 11. Hamiaux C., Drummond R. S., Janssen B. J., Ledger S. E., Cooney J. M., Newcomb R. D., and Snowden K. C. (2012) DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 22, 2032–2036 10.1016/j.cub.2012.08.007 [DOI] [PubMed] [Google Scholar]
- 12. Janssen B. J., Drummond R. S., and Snowden K. C. (2014) Regulation of axillary shoot development. Curr. Opin. Plant Biol. 17, 28–35 10.1016/j.pbi.2013.11.004 [DOI] [PubMed] [Google Scholar]
- 13. Stirnberg P., Furner I. J., and Ottoline Leyser H. M. (2007) MAX2 participates in an SCF complex which acts locally at the node to suppress shoot branching. Plant J. 50, 80–94 10.1111/j.1365-313X.2007.03032.x [DOI] [PubMed] [Google Scholar]
- 14. Jiang L., Liu X., Xiong G., Liu H., Chen F., Wang L., Meng X., Liu G., Yu H., Yuan Y., Yi W., Zhao L., Ma H., He Y., Wu Z., et al. (2013) DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature 504, 401–405 10.1038/nature12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsuchiya Y., Yoshimura M., Sato Y., Kuwata K., Toh S., Holbrook-Smith D., Zhang H., McCourt P., Itami K., Kinoshita T., and Hagihara S. (2015) Parasitic Plants: probing strigolactone receptors in Striga hermonthica with fluorescence. Science 349, 864–868 10.1126/science.aab3831 [DOI] [PubMed] [Google Scholar]
- 16. de Saint Germain A., Clavé G., Badet-Denisot M. A., Pillot J. P., Cornu D., Le Caer J. P., Burger M., Pelissier F., Retailleau P., Turnbull C., Bonhomme S., Chory J., Rameau C., and Boyer F. D. (2016) An histidine covalent receptor and butenolide complex mediates strigolactone perception. Nat. Chem. Biol. 12, 787–794 10.1038/nchembio.2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao L. H., Zhou X. E., Wu Z. S., Yi W., Xu Y., Li S., Xu T. H., Liu Y., Chen R. Z., Kovach A., Kang Y., Hou L., He Y., Xie C., Song W., et al. (2013) Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 23, 436–439 10.1038/cr.2013.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yao R., Ming Z., Yan L., Li S., Wang F., Ma S., Yu C., Yang M., Chen L., Chen L., Li Y., Yan C., Miao D., Sun Z., Yan J., et al. (2016) DWARF14 is a non-canonical hormone receptor for strigolactone. Nature 536, 469–473 10.1038/nature19073 [DOI] [PubMed] [Google Scholar]
- 19. Zhao L. H., Zhou X. E., Yi W., Wu Z., Liu Y., Kang Y., Hou L., de Waal P. W., Li S., Jiang Y., Scaffidi A., Flematti G. R., Smith S. M., Lam V. Q., Griffin P. R., et al. (2015) Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 25, 1219–1236 10.1038/cr.2015.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stanga J. P., Morffy N., and Nelson D. C. (2016) Functional redundancy in the control of seedling growth by the karrikin signaling pathway. Planta 243, 1397–1406 10.1007/s00425-015-2458-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhou F., Lin Q., Zhu L., Ren Y., Zhou K., Shabek N., Wu F., Mao H., Dong W., Gan L., Ma W., Gao H., Chen J., Yang C., Wang D., et al. (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504, 406–410 10.1038/nature12878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Akiyama K., Matsuzaki K., and Hayashi H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435, 824–827 10.1038/nature03608 [DOI] [PubMed] [Google Scholar]
- 23. Khosla A., and Nelson D. C. (2016) Strigolactones, super hormones in the fight against Striga. Curr. Opin. Plant Biol. 33, 57–63 10.1016/j.pbi.2016.06.001 [DOI] [PubMed] [Google Scholar]
- 24. Nakamura H., and Asami T. (2014) Target sites for chemical regulation of strigolactone signaling. Front. Plant Sci. 5, 623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mangnus E. M., and Zwanenburg B. (1992) Tentative molecular mechanism for germination stimulation of Striga and Orobanche seeds by strigol and its synthetic analogs. J. Agric. Food Chem. 40, 1066–1070 10.1021/jf00018a032 [DOI] [Google Scholar]
- 26. Boyer F. D., de Saint Germain A., Pouvreau J. B., Clavé G., Pillot J. P., Roux A., Rasmussen A., Depuydt S., Lauressergues D., Frei Dit Frey N., Heugebaert T. S., Stevens C. V., Geelen D., Goormachtig S., and Rameau C. (2014) New strigolactone analogs as plant hormones with low activities in the rhizosphere. Mol. Plant 7, 675–690 10.1093/mp/sst163 [DOI] [PubMed] [Google Scholar]
- 27. Cohen M., Prandi C., Occhiato E. G., Tabasso S., Wininger S., Resnick N., Steinberger Y., Koltai H., and Kapulnik Y. (2013) Structure-function relations of strigolactone analogs: activity as plant hormones and plant interactions. Mol. Plant 6, 141–152 10.1093/mp/sss134 [DOI] [PubMed] [Google Scholar]
- 28. Fukui K., Yamagami D., Ito S., and Asami T. (2017) A taylor-made design of phenoxyfuranone-type strigolactone mimic. Front. Plant Sci. 8, 936 10.3389/fpls.2017.00936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lumba S., Bunsick M., and McCourt P. (2017) Chemical genetics and strigolactone perception. F1000Res 6, 975 10.12688/f1000research.11379.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zwanenburg B., Nayak S. K., Charnikhova T. V., and Bouwmeester H. J. (2013) New strigolactone mimics: structure-activity relationship and mode of action as germinating stimulants for parasitic weeds. Bioorg. Med. Chem. Lett. 23, 5182–5186 10.1016/j.bmcl.2013.07.004 [DOI] [PubMed] [Google Scholar]
- 31. Jamil M., Kountche B. A., Haider I., Guo X., Ntui V. O., Jia K. P., Ali S., Hameed U. S., Nakamura H., Lyu Y., Jiang K., Hirabayashi K., Tanokura M., Arold S. T., Asami T., and Al-Babili S. (2017) Methyl phenlactonoates are efficient strigolactone analogs with simple structure. J. Exp. Bot., in press 10.1093/jxb/erx438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harrison P. J., Newgas S. A., Descombes F., Shepherd S. A., Thompson A. J., and Bugg T. D. (2015) Biochemical characterization and selective inhibition of β-carotene cis-trans isomerase D27 and carotenoid cleavage dioxygenase CCD8 on the strigolactone biosynthetic pathway. FEBS J. 282, 3986–4000 10.1111/febs.13400 [DOI] [PubMed] [Google Scholar]
- 33. Holbrook-Smith D., Toh S., Tsuchiya Y., and McCourt P. (2016) Small-molecule antagonists of germination of the parasitic plant Striga hermonthica. Nat. Chem. Biol. 12, 724–729 10.1038/nchembio.2129 [DOI] [PubMed] [Google Scholar]
- 34. Jamil M., Charnikhova T., Verstappen F., and Bouwmeester H. (2010) Carotenoid inhibitors reduce strigolactone production and Striga hermonthica infection in rice. Arch. Biochem. Biophys. 504, 123–131 10.1016/j.abb.2010.08.005 [DOI] [PubMed] [Google Scholar]
- 35. Mashita O., Koishihara H., Fukui K., Nakamura H., and Asami T. (2016) Discovery and identification of 2-methoxy-1-naphthaldehyde as a novel strigolactone-signaling inhibitor. J. Pestic. Sci. 41, 71–78 10.1584/jpestics.D16-028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Xiang H., Yao R., Quan T., Wang F., Chen L., Du X., Zhang W., Deng H., Xie D., and Luo T. (2017) Simple beta-lactones are potent irreversible antagonists for strigolactone receptors. Cell Res. 27, 1525–1528 10.1038/cr.2017.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Waters M. T., Nelson D. C., Scaffidi A., Flematti G. R., Sun Y. K., Dixon K. W., and Smith S. M. (2012) Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development 139, 1285–1295 10.1242/dev.074567 [DOI] [PubMed] [Google Scholar]
- 38. Nelson D. C., Scaffidi A., Dun E. A., Waters M. T., Flematti G. R., Dixon K. W., Beveridge C. A., Ghisalberti E. L., and Smith S. M. (2011) F-box protein MAX2 has dual roles in karrikin and strigolactone signaling in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 108, 8897–8902 10.1073/pnas.1100987108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu Q., Zhang Y., Matusova R., Charnikhova T., Amini M., Jamil M., Fernandez-Aparicio M., Huang K., Timko M. P., Westwood J. H., Ruyter-Spira C., van der Krol S., and Bouwmeester H. J. (2014) Striga hermonthica MAX2 restores branching but not the very low fluence response in the Arabidopsis thaliana max2 mutant. New Phytol. 202, 531–541 10.1111/nph.12692 [DOI] [PubMed] [Google Scholar]
- 40. Nakamura H., Xue Y. L., Miyakawa T., Hou F., Qin H. M., Fukui K., Shi X., Ito E., Ito S., Park S. H., Miyauchi Y., Asano A., Totsuka N., Ueda T., Tanokura M., and Asami T. (2013) Molecular mechanism of strigolactone perception by DWARF14. Nat. Commun. 4, 2613 [DOI] [PubMed] [Google Scholar]
- 41. Niesen F. H., Berglund H., and Vedadi M. (2007) The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2, 2212–2221 10.1038/nprot.2007.321 [DOI] [PubMed] [Google Scholar]
- 42. Soundappan I., Bennett T., Morffy N., Liang Y., Stanga J. P., Abbas A., Leyser O., and Nelson D. C. (2015) SMAX1-LIKE/D53 family members enable distinct MAX2-dependent responses to strigolactones and karrikins in Arabidopsis. Plant Cell 27, 3143–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang L., Wang B., Jiang L., Liu X., Li X., Lu Z., Meng X., Wang Y., Smith S. M., and Li J. (2015) Strigolactone signaling in Arabidopsis regulates shoot development by targeting D53-like SMXL repressor proteins for ubiquitination and degradation. Plant Cell 27, 3128–3142 10.1105/tpc.15.00605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gaglioti K., Chierotti M. R., Grifasi F., Gobetto R., Griesser U. J., Hasa D., and Voinovich D. (2014) Improvement of the water solubility of tolfenamic acid by new multiple-component crystals produced by mechanochemical methods. Crystengcomm 16, 8252–8262 10.1039/C4CE00549J [DOI] [Google Scholar]
- 45. Griffiths C. A., Sagar R., Geng Y., Primavesi L. F., Patel M. K., Passarelli M. K., Gilmore I. S., Steven R. T., Bunch J., Paul M. J., and Davis B. G. (2016) Chemical intervention in plant sugar signalling increases yield and resilience. Nature 540, 574–578 10.1038/nature20591 [DOI] [PubMed] [Google Scholar]
- 46. Toh S., Holbrook-Smith D., Stogios P. J., Onopriyenko O., Lumba S., Tsuchiya Y., Savchenko A., and McCourt P. (2015) Structure-function analysis identifies highly sensitive strigolactone receptors in Striga. Science 350, 203–207 10.1126/science.aac9476 [DOI] [PubMed] [Google Scholar]
- 47. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., and Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 10.1107/S0907444910048675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Evans P. R., and Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., and Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 10.1107/S0021889807021206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Joosten R. P., Joosten K., Cohen S. X., Vriend G., and Perrakis A. (2011) Automatic rebuilding and optimization of crystallographic structures in the Protein Data Bank. Bioinformatics 27, 3392–3398 10.1093/bioinformatics/btr590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Webb B., and Sali A. (2016) Comparative protein structure modeling using MODELLER. Curr. Protoc. Protein Sci. 86, 2.9.1–2.9.37 10.1002/cpps.20 [DOI] [PubMed] [Google Scholar]
- 57. Trott O., and Olson A. J. (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bombarely A., Moser M., Amrad A., Bao M., Bapaume L., Barry C. S., Bliek M., Boersma M. R., Borghi L., Bruggmann R., Bucher M., D'Agostino N., Davies K., Druege U., Dudareva N., et al. (2016) Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida. Nat. Plants 2, 16074 10.1038/nplants.2016.74 [DOI] [PubMed] [Google Scholar]
- 59. International Rice Genome Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436, 793–800 10.1038/nature03895 [DOI] [PubMed] [Google Scholar]
- 60. Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. James P., Halladay J., and Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maier R., Brandner C., Hintner H., Bauer J., and Onder K. (2008) Construction of a reading frame-independent yeast two-hybrid vector system for site-specific recombinational cloning and protein interaction screening. BioTechniques 45, 235–244 10.2144/000112897 [DOI] [PubMed] [Google Scholar]
- 63. Matusova R., Rani K., Verstappen F. W. A., Franssen M. C. R., Beale M. H., and Bouwmeester H. J. (2005) The strigolactone germination stimulants of the plant-parasitic Striga and Orobanche spp. are derived from the carotenoid pathway. Plant Physiol. 139, 920–934 10.1104/pp.105.061382 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.