Abstract
Under various conditions of liver injury, the intrahepatic biliary epithelium undergoes dynamic tissue expansion and remodeling, a process known as ductular reaction. Mouse models defective in inducing such a tissue-remodeling process are more susceptible to liver injury, suggesting a crucial role of this process in liver regeneration. However, the molecular mechanisms regulating the biliary epithelial cell (BEC) dynamics in the ductular reaction remain largely unclear. Here, we demonstrate that the transcription factor Krüppel-like factor 5 (Klf5) is highly enriched in mouse liver BECs and plays a key role in regulating the ductular reaction, specifically under cholestatic injury conditions. Although mice lacking Klf5 in the entire liver epithelium, including both hepatocytes and BECs (Klf5-LKO (liver epithelial-specific knockout) mice), did not exhibit any apparent phenotype in the hepatobiliary system under normal conditions, they exhibited significant defects in biliary epithelial tissue remodeling upon 3,5-diethoxycarbonyl-1,4-dihydrocollidine-induced cholangitis, concomitantly with exacerbated cholestasis and reduced survival rate. In contrast, mice lacking Klf5 solely in hepatocytes did not exhibit any such phenotypes, confirming Klf5's specific role in BECs. RNA-sequencing analyses of BECs isolated from the Klf5-LKO mouse livers revealed that the Klf5 deficiency primarily affected expression of cell cycle-related genes. Moreover, immunostaining analysis with the proliferation marker Ki67 disclosed that the Klf5-LKO mice had significantly reduced BEC proliferation levels upon injury. These results indicate that Klf5 plays a critical role in the ductular reaction and biliary epithelial tissue expansion and remodeling by inducing BEC proliferation and thereby contributing to liver regeneration.
Keywords: epithelial cell, regeneration, liver injury, homeostasis, cell proliferation, bioinformatics, bile duct, ductular reaction (DR), kruppel-like factor 5 (KLF5)
Introduction
The liver is a vital organ for life and plays an array of critical biological functions, including metabolism, detoxification, serum protein production, and bile secretion. Being inherently susceptible to a wide range of chemicals, toxins, and xenobiotics entering from the intestinal tract through the portal venous flow, the liver has tremendous capability to regenerate itself in response to various types of injury. Although hepatocytes, parenchymal cells of the liver, elicit vigorous regenerative activity through proliferation and self-duplication (1, 2), other nonparenchymal cells and tissues in the liver also contribute significantly to the regenerative process, such as by inducing fibrogenesis for temporal repair of tissue architecture, evoking and modulating the immune and inflammatory responses, and supporting hepatocyte renewal.
Among liver nonparenchymal cells, biliary epithelial cells (BECs)6 compose the bile duct, a conduit system that collects bile produced by hepatocytes and excretes it to the intestine. Although the bile duct is a quiescent tissue under normal conditions, it undergoes dynamic tissue remodeling once the liver is injured; the biliary tree structure arborizes and transforms adaptively so that the branches extend toward the sites of parenchymal injury (3). This remodeling process is called the ductular reaction (DR) and is associated with various liver disorders in human patients, such as viral hepatitis, acute and chronic cholestasis, alcoholic liver disease, and nonalcoholic fatty liver disease (4). Although the DR has been classically assumed to correspond to the emergence and expansion of liver stem/progenitor cells (LPC) that differentiate into hepatocytes, thereby contributing to renewal of the parenchymal tissue (5, 6), this notion has been challenged in recent years based on the results obtained from in vivo genetic lineage-tracing studies in mice (6, 7). Thus, in most, if not all, cases of liver regeneration upon chronic injury in mice, newly formed hepatocytes are derived almost exclusively from pre-existing hepatocytes rather than LPCs or BECs. Nevertheless, mouse models with attenuated or diminished DR generally suffer from more aggravated liver injury, suggesting that DR is a fundamental physiological reaction for the liver to counter toxic attacks. DR is induced by coordinated actions of BECs and other liver cell types, and accordingly, several kinds of humoral factors and extracellular signals have been identified that act on BECs and regulate their proliferation and differentiation (8–10). In contrast, BEC intrinsic genetic programs and gene regulatory networks that underlie DR regulation still remain largely unknown.
To reveal the BEC intrinsic mechanisms regulating DR, we sought to identify and reveal the role of BEC-enriched transcription factors, and hence, we focused on Krüppel-like factor 5 (Klf5). Klf5 is a member of Krüppel-like factors, which are versatile transcription factors that play diverse roles in processes such as cell proliferation, differentiation, development, and regeneration in a wide range of tissues and cell types (11). Notably, Klf5 has been shown to be involved in the development and maintenance of several kinds of epithelial tissues and organs, including the intestine, lung, and renal collecting duct (12–14). In the small intestine, for example, Klf5 is locally expressed in the crypt and maintains tissue morphology by contributing to the maintenance of intestinal stem cells (15). With regard to the liver, however, there are few reports addressing the role of Klf5 in organ homeostasis and regeneration, although its involvement in hepatocarcinogenesis has been well documented (16). In this study, we revealed that in the mouse liver Klf5 is a transcription factor whose expression was highly enriched in BECs. In vivo studies employing liver cell type-specific knockout mouse models, in combination with multiple liver injury protocols with different etiologies, delineated a previously unidentified role of Klf5 in the biliary epithelium under cholestatic injury conditions.
Results
Klf5 is expressed predominantly in biliary epithelial cells in the liver
To identify candidate transcription factors that are expressed in BECs and are potentially involved in DR regulation, we utilized publicly available BEC transcriptome datasets. A previous study by Dorrell et al. (17) examined mRNA profiles of the BEC-enriched nonparenchymal cell fractions (“ductal NPC” fractions) sorted from the liver of both normal and 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC)–treated mice based on the expression of surface markers. DDC administration is a well-established model for chronic and cholestatic liver injury in mice that accompanies typical DR induction. Upon examining the gene expression profile data, with a particular focus on transcription factors, we noticed that expression of Klf5 was highly enriched in MIC1C3+/CD133+/CD26− BEC fractions, particularly under DDC-induced injury conditions (data not shown).
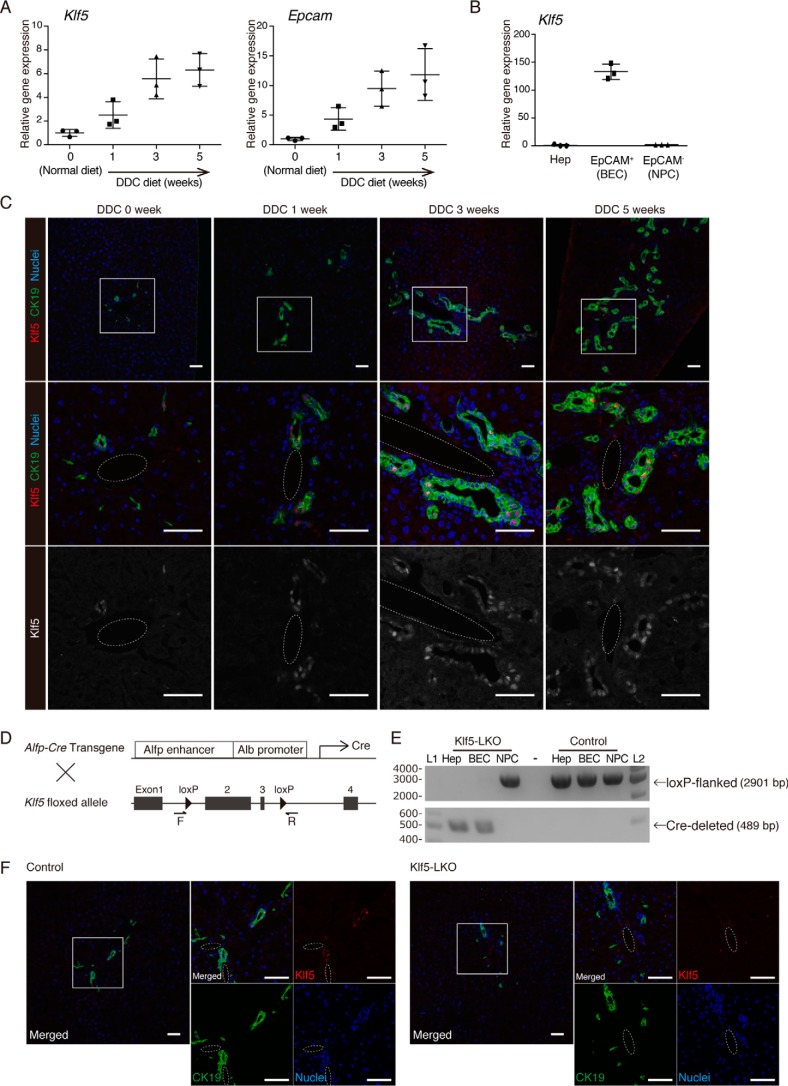
To reveal a potential role of Klf5 in regulating DR in injured livers, we first confirmed its expression profile in the DDC-induced mouse liver injury model. Quantitative reverse transcription-PCR (RT-PCR) analysis using whole-liver samples revealed that Klf5 was expressed in the liver and that its expression level increased significantly in the time course of injury, along with that of the BEC marker Epcam (Fig. 1A). To determine whether Klf5 is expressed in BECs, we isolated BECs using a cell sorter based on the expression of EpCAM as the cell-surface antigen. RT-PCR analysis revealed that Klf5 expression was highly enriched in the EpCAM+ BEC fraction, whereas it was barely detected in other nonparenchymal cells or hepatocytes, both under normal conditions and upon DDC injury (Fig. 1B and Fig. S1A). Of note, the levels of Klf5 expression in BECs were comparable under the normal and injured conditions. Immunostaining analysis of liver sections also showed that Klf5 was predominantly expressed in BECs in both normal and injured livers (Fig. 1C).
Figure 1.
Expression pattern of Klf5 in the mouse liver. A, expression levels of Klf5 and Epcam in whole-liver mRNA samples prepared from WT mice were determined by quantitative RT-PCR. B, expression of Klf5 in liver cell fractions. Hepatocytes (Hep), EpCAM+ BEC fraction, and EpCAM− NPC fractions were collected from livers of WT mice upon DDC administration for 3 weeks and analyzed by quantitative RT-PCR. C, immunostaining for Klf5 (red in the upper and center panels; gray scale in the lower panels) and CK19 (green) in the WT mouse liver. Counterstaining for nuclei is shown in blue (upper and center panels). Regions indicated by white boxes in the upper panels are magnified in the middle and lower panels. Dashed lines show portal veins. Scale bar, 50 μm. D, scheme for production of Klf5-LKO mice. F and R denote the positions of primers used for genomic PCR analyses shown in E and Fig. 4B. E, genomic PCR analysis for Cre-mediated recombination in the Klf5 locus. The upper and lower panels show amplicons corresponding to nonrecombined (floxed) and recombined (Cre-deleted) alleles, respectively. Lanes L1 and L2 were loaded with 100-bp ladder and 1-kb ladder DNA size markers, respectively. The sizes of markers (bp) are indicated to the left. F, loss of Klf5 expression in the Klf5-LKO mouse liver was confirmed at the protein level. Immunostaining results for Klf5 (red) and CK19 (green) are shown with counterstaining for nuclei (blue). Regions indicated by white boxes in the left panels are magnified in the right panels together with single color channel images. Dashed lines show portal veins. Scale bar, 50 μm.
Liver-specific Klf5-knockout mice develop normally with no obvious defect in the liver under normal conditions
To assess the functional involvement of Klf5 in DR regulation in vivo, we crossed Klf5 flox mice (18) with the Alfp-Cre transgenic mice (19) to produce liver epithelial cell-specific Klf5 conditional knockout mice (Alfp-CreTg/+;Klf5flox/flox) (Fig. 1D). In the Alfp-Cre transgenic line, the Cre recombinase under the control of an α-fetoprotein (Alfp) gene enhancer and an albumin (Alb) gene promoter starts to be expressed in fetal liver hepatoblasts, which are bi-potential stem/progenitor cells giving rise to hepatocytes and BECs in the postnatal liver; thus, Cre-mediated recombination at a target locus is initially induced in hepatoblasts and hence is inherited and present in the entire epithelial cell lineages in the adult liver, including hepatocytes and BECs, as well as LPCs upon liver injury (10). It should be noted that expression of Alfp-Cre in the adult liver, which might possibly be induced more strongly in cells with stem/progenitor-like characters, does not affect this recombination pattern in principle. In addition, the recombination does not occur in other nonepithelial lineages, such as sinusoidal endothelial cells or hepatic stellate cells, as they originate from distinct types of lineage-specific progenitor cells other than hepatoblasts and do not express Alfp or Alb.
Nevertheless, we empirically determined the recombination profile in our compound mutant mice, and as expected, genomic PCR analysis revealed that deletion of Klf5 was achieved efficiently and specifically in both hepatocytes and BECs but not in other NPCs, in the Alfp-CreTg/+;Klf5flox/flox mouse liver (Fig. 1E). Immunostaining analysis also confirmed the complete absence of Klf5 protein expression in the liver of these mice (Fig. 1F), which are referred to as Klf5-LKO mice hereafter.
Although the systemic deletion of the Klf5 gene has been reported to result in embryonic lethality (20), Klf5-LKO mice were viable and developed normally. No significant difference was observed between the Klf5-LKO and control mice in terms of body weight (Fig. S1B), liver weight (Fig. S1C), liver-to-body weight ratio (Fig. S1D), and serum tests for liver injury markers (Fig. S1E). Hematoxylin and eosin staining (Fig. S1F) and immunostaining for BEC markers (Fig. 1F and Fig. S1G) also revealed no histological abnormality in the liver and bile ducts in the Klf5-LKO mice. These results indicate that although expression of Klf5 is clearly detected in BECs, it is likely dispensable for development and function under physiological conditions of the hepatobiliary system.
Ductular reaction upon cholestatic liver injury is suppressed in Klf5-LKO mice
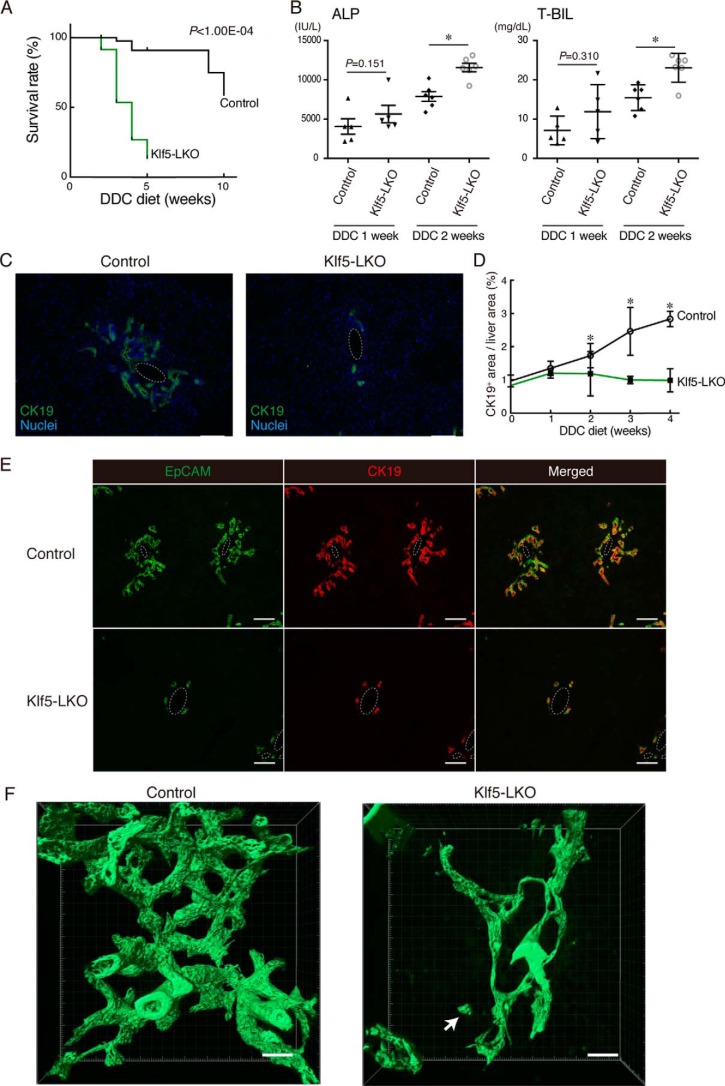
To test whether any functional requirement of Klf5 could be manifested upon liver injury, we next applied the hepatotoxin DDC–induced injury protocol to Klf5-LKO mice. Upon DDC administration, the Klf5-LKO mice exhibited significantly increased mortality compared with the control mice (Fig. 2A). In accordance with this observation, the level of DDC-induced cholestasis was aggravated in Klf5-LKO mice as represented by increased serum cholestatic marker levels, alkaline phosphatase (ALP), and total bilirubin (T-BIL) (Fig. 2B), whereas the serum levels of hepatocyte injury markers (alanine aminotransferase and aspartate aminotransferase) were not different between the cohorts (Fig. S2A).
Figure 2.
DR induction upon DDC-induced liver injury is suppressed in Klf5-LKO mice. A, Kaplan-Meier survival curves of control (n = 49) and Klf5-LKO (n = 70) mice treated with DDC. B, serum ALP and T-BIL levels were measured in control and Klf5-LKO mice treated with DDC for 1 week (n = 5 mice) or 2 weeks (n = 6 mice). p values were calculated by Mann-Whitney U test. p values comparing the control and Klf5-LKO mice (DDC 2 weeks) are 0.00430 (ALP) and 0.0130 (T-BIL). C, immunostaining for CK19 (green) in the Klf5-LKO and control livers treated with DDC for 4 weeks shown with counterstaining for nuclei (blue). Scale bar, 100 μm. D, quantification of CK19+ areas in whole-liver sections. Data represent the mean ± S.D. n ≥ 4 mice for each time point. p values calculated by Mann-Whitney U test for each time point compare the control and Klf5-LKO mice and are as follows: 0.343 (DDC 0 week); 0.700 (DDC 1 week); 0.0207 (DDC 2 weeks); 0.0238 (DDC 3 weeks); and 0.0286 (DDC 4 weeks). E, immunostaining for EpCAM (green) and CK19 (red) in the Klf5-LKO and control livers treated with DDC for 4 weeks. Scale bar, 100 μm. F, 3D immunostaining for CK19 (green) in the Klf5-LKO and control livers treated with DDC for 4 weeks. Stacked images were obtained with confocal microscopy and used to reconstruct a 3D image using the IMARIS software. The image is shown in surface mode. Note that a CK19+ cell cluster separated from the biliary tree structure is observed in the Klf5-LKO liver (white arrow). Scale bar, 50 μm. Asterisks indicate that the p values are <0.05 (*, p < 0.06).
In control mice, DDC-induced liver injury caused a massive DR, which is remodeling of bile ducts that can be observed in liver sections as expansion and parenchymal invasion of cells expressing BEC markers such as CK19 (Fig. 2C and Fig. S2B). In Klf5-LKO mouse livers, however, DR was completely suppressed at 4 weeks after administration of DDC. Quantitative analyses of the level of DR induction along the time course of DDC administration showed that there was a significant difference in DR induction between the Klf5-LKO and control mice at 2 weeks and thereafter (Fig. 2D). Immunostaining analyses employing other BEC markers, EpCAM and promonin-1 (Prom1), also showed essentially the same expression pattern as that of CK19 (Fig. 2E and Fig. S2C), confirming that DR was severely suppressed in Klf5-LKO mice upon DDC-induced liver injury.
As DR reflects structural transformation of the biliary tree, we also examined the biliary epithelial tissue morphology at the three-dimensional (3D) level. Immunostaining for CK19 using 200-μm-thick tissue sections and subsequent analysis with confocal microscopy (3) revealed that bile ducts in the Klf5-LKO mouse liver contain fewer numbers of branches than those in the control liver, whereas that the length or thickness of the branches was not significantly different between the cohorts (Fig. 2F and Fig. S2, D–G). Of note, CK19+ cell clusters that were spatially separated from the biliary tree were observed in the Klf5-LKO liver, implicating disorganization of the biliary structure in the absence of Klf5 function.
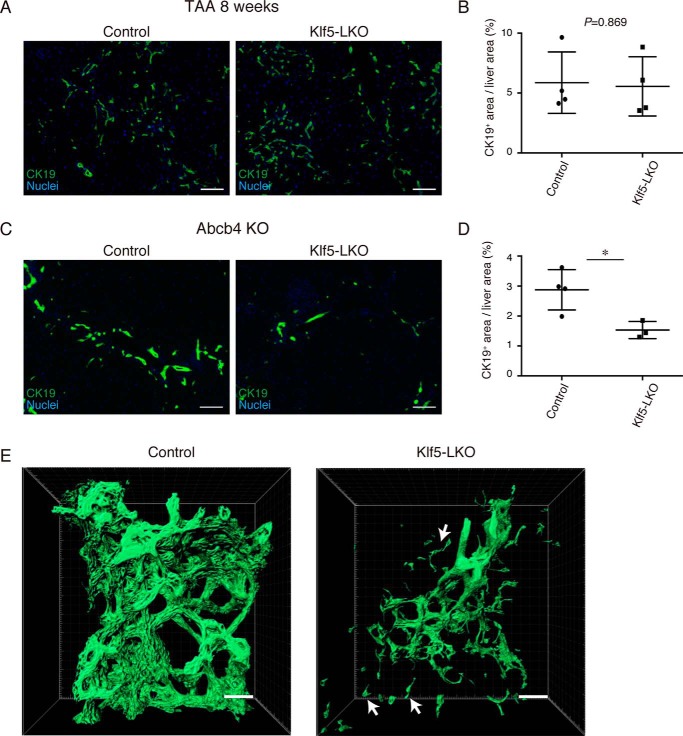
To determine whether the role of Klf5 in DR regulation can be more generalized, we next assessed the DR phenotype in the Klf5-LKO liver by applying other types of liver injury protocols. Thioacetamide (TAA) is known to induce local hepatocyte injury around the central vein caused by reactive oxygen species, leading to DR induction in the absence of apparent cholestasis (21). Upon induction of the TAA-induced liver injury for 8 weeks, the Klf5 expression in BECs was confirmed to be present both at the mRNA and protein levels in wildtype (WT) mice (Fig. S3, A and B). We then administered TAA to Klf5-LKO and control mice and evaluated the level of DR induction by CK19 immunostaining. Expansion of the CK19+ cells was not affected in the Klf5-LKO liver, indicating that Klf5 is dispensable for DR upon TAA injury, and hence is not likely a universal regulator for DR (Fig. 3, A and B). As a model for the cholestatic liver injury alternative to the DDC protocol, we utilized the Abcb4 KO mouse model and crossed it with the Klf5-LKO mouse strain. Abcb4 (also known as Mdr2 in mice) is homologous to human MDR3, the genetic mutations of which lead to a chronic and cholestatic disorder called progressive familial intrahepatic cholestasis type 3. Accordingly, Abcb4 KO mice exhibit the cholestatic injury phenotype with DR induction as they develop (22, 23), where the expression of Klf5 in the remodeling biliary epithelium was again confirmed (Fig. S3C). In the livers of the Abcb4 KO;Klf5-LKO double KO mice, DR was significantly suppressed when compared with the control liver (Fig. 3, C–E), and CK19+ cell clusters that were spatially separated from the biliary tree were also observed (Fig. 3E). Taken together, these results suggest that Klf5 plays an essential role in DR induction specifically upon cholestatic liver injury, thereby contributing to amelioration of cholestasis.
Figure 3.
Loss of Klf5 affects DR induction specifically under cholestatic liver injury conditions. A, representative images of immunostaining for CK19 (green) in the Klf5-LKO and control livers treated with TAA for 8 weeks. Counterstaining for nuclei is also included (blue). Scale bar, 100 μm. B, quantification of CK19+ areas in whole-liver sections prepared from the TAA-treated mice as in A. n = 4 mice for each group. p values were calculated by Student's t test. C, representative images of immunostaining for CK19 (green) in the Abcb4 KO and Klf5-LKO double knockout and control (Abcb4 single knockout) livers at 8 weeks after birth. Counterstaining for nuclei is also shown (blue). Scale bar, 100 μm. D, quantification of CK19+ areas in whole-liver sections prepared from the Abcb4 KO cohorts as in C. n = 4 and 3 mice for the control (Abcb4 single knockout) and the Klf5-LKO (Abcb4 KO; Klf5-LKO double knockout) groups, respectively. p values were calculated by Student's t test. p values = 0.0209. E, 3D immunostaining for CK19 (green) in the Abcb4 KO and Klf5-LKO double knockout and control (Abcb4 single knockout) livers at 8 weeks after birth. Stacked images were obtained with confocal microscopy and used to reconstruct a 3D image using the IMARIS software. The image is shown in surface mode. White arrows indicate CK19+ cells separated from the biliary tree structure. Scale bar, 50 μm. Asterisks indicate that the p values are <0.05 (*, p < 0.06).
BEC-intrinsic expression of Klf5 is responsible for DR regulation
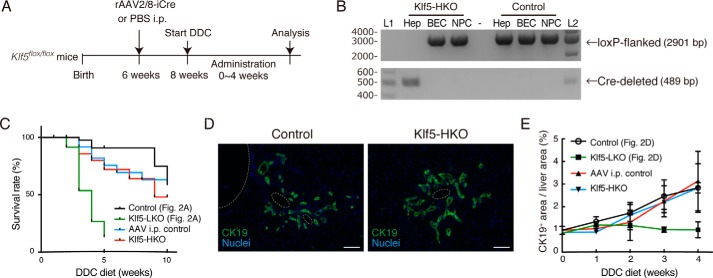
Our results from RT-PCR and immunostaining analyses showed that Klf5 was predominantly expressed in BECs and was barely detected in hepatocytes (Fig. 1, B and C), which makes it likely that the aforementioned phenotypes in Klf5-LKO mice are attributable to the loss of Klf5 in BECs. A previous study, however, reported expression and a metabolic function of Klf5 in hepatocytes using primary cultured mouse hepatocytes (24). To eliminate the possibility that Klf5 in hepatocytes may have caused the DR phenotype in Klf5-LKO mice, we assessed the function of Klf5 particularly in hepatocytes. It has been well-established that recombinant human adeno-associated virus 2 vector pseudo-serotyped with type 8 capsid (rAAV2/8) can specifically and efficiently transduce in vivo in mouse hepatocytes but not BECs. Therefore, we applied rAAV2/8 expressing an improved version of the Cre recombinase (iCre) under the control of a hepatocyte-specific promoter (7) to Klf5flox/flox mice to achieve hepatocyte-specific deletion of Klf5 (Fig. 4A). Genomic PCR analysis revealed that the deletion was achieved efficiently in hepatocytes, but not in BECs (Fig. 4B), and these mice (hereafter referred to as Klf5-HKO mice) were then subjected to the DDC liver injury protocol. Induction of DR upon DDC administration was not at all affected in Klf5-HKO mice (Fig. 4, D and E), and the levels of serum markers for liver injury as well as the survival rate were indistinguishable between Klf5-HKO mice and the control (Fig. 4C and data not shown). These results strongly suggest that Klf5 expressed in BECs per se plays a role in DR regulation in a cell-intrinsic manner.
Figure 4.
Deletion of Klf5 in hepatocytes does not affect DR induction. A, experimental scheme for analyses on the effect of hepatocyte-specific loss of Klf5. B, genomic PCR analysis for Cre-mediated recombination in the Klf5 locus using primers shown in Fig. 1D. The upper and lower panels show amplicons corresponding to nonrecombined (floxed) and recombined (Cre-deleted) alleles, respectively. Lanes L1 and L2 were loaded with 100-bp ladder and 1-kb ladder DNA size markers, respectively. The sizes of markers (bp) are indicated to the left. C, Kaplan-Meier survival curves of Klf5-HKO (n = 30) and AAV i.p. control (n = 27) mice treated with DDC. For comparison, the survival curves of Klf5-LKO and the control mice shown in Fig. 2A are also overlaid. D, immunostaining for CK19 (green) in the Klf5-HKO and control livers treated with DDC for 4 weeks shown with counterstaining for nuclei (blue). Scale bar, 100 μm. E, quantification of CK19+ areas in whole-liver sections prepared from Klf5-HKO and AAV i.p. control mice. For comparison, the data of Klf5-LKO and the control mice shown in Fig. 2D are also overlaid. Data represent mean ± S.D. n ≥ 3 mice for each time point. p values were calculated by Mann-Whitney U test for each time point comparing the Klf5-HKO and control mice and are as follows: 0.100 (DDC 0 week); 0.100 (DDC 1 week); 0.400 (DDC 2 weeks); 0.900 (DDC 3 weeks); and 0.857 (DDC 4 weeks).
It has been well-documented that induction and expansion of the DR is controlled by several kinds of humoral factors, such as fibroblast growth factor 7 (Fgf7), tumor necrosis factor-like weak inducer of apoptosis (Tweak), and hepatocyte growth factor (8–10). These growth factors and cytokines are produced by liver nonparenchymal cells such as mesenchymal cells, immune cells, and endothelial cells in the wake of injury and inflammatory responses and can act directly on BECs. In the livers of DDC-treated Klf5-LKO mice, the expression level of neither Fgf7, Tweak, nor Hgf was significantly affected (Fig. S5A). This further supports the notion that loss of Klf5 affected DR primarily in a biliary epithelial tissue-intrinsic manner, rather than through tissue microenvironment or the niche surrounding BECs.
Klf5 regulates proliferation of BECs upon DDC-induced liver injury
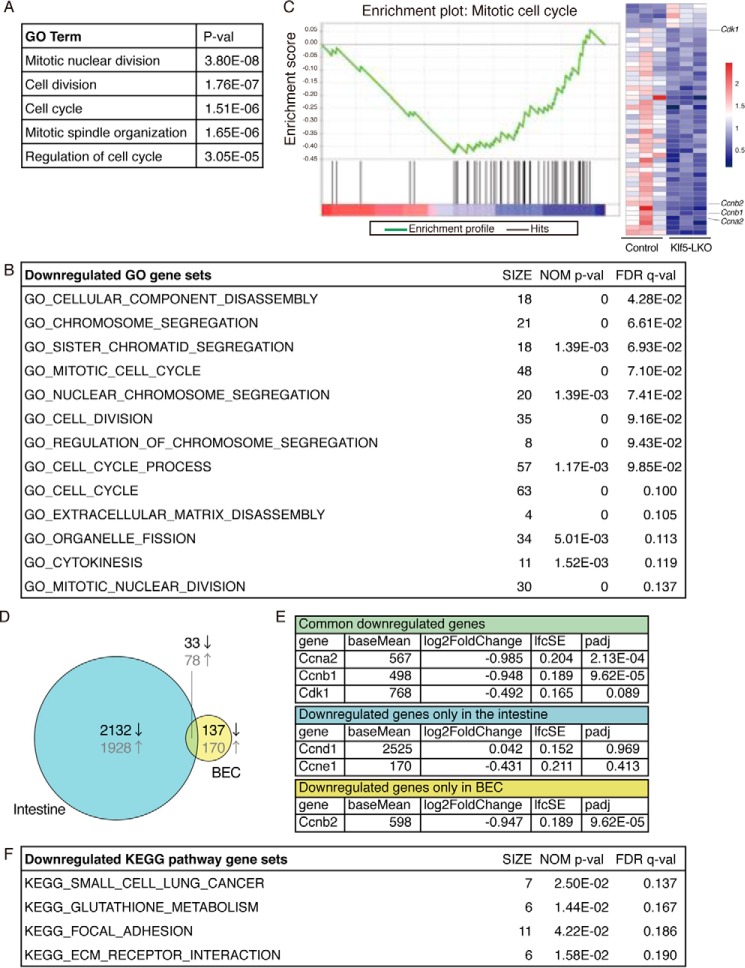
To address the mechanism whereby Klf5 induces and regulates DR, we compared gene expression profiles between WT and Klf5-deficient BECs under DDC-induced cholestatic conditions. As the DR suppression phenotype and exacerbated cholestasis in Klf5-LKO mice were already evident at 2 weeks of DDC administration (Fig. 2B), we focused on analyzing samples from 1-week injured animals to detect the earliest changes at the initial stage of DR. EpCAM+ cells were sorted from livers of Klf5-LKO mice and control mice after 1 week of DDC treatment and subjected to RNA sequencing (RNA-seq) analysis (n = 3 library samples for each genotype). According to the procedures described under “Experimental procedures,” we were able to identify 440 differential expression genes (DEG) (25), and DAVID gene ontology (GO) analysis (26) showed that they belonged primarily to categories involved in cell cycle regulation (e.g. cell division and mitotic nuclear division) in the Biological Process category (Fig. 5A). To further estimate the biological processes that could contribute to DR suppression in Klf5-LKO mice, we performed Gene Set Enrichment Analysis (GSEA) using DEG, which allowed us to compare the DEG to particular gene sets that are pre-made according to previously reported information about genes (27). The results also indicated that Klf5 regulated cellular processes involved in cell cycle progression (Fig. 5, B and C, and Fig. S4A).
Figure 5.
Transcriptome analysis by RNA-seq reveals that Klf5 regulates cell proliferation in BECs. A, DEG were categorized in biological GO terms using DAVID. B and C, GSEA using DEG to identify enriched biological GO terms. B, entire list of enriched gene sets categorized in GO biological process terms that were down-regulated by Klf5 deletion in BECs. The SIZE column indicates the number of genes hit in each gene set. Gene sets that meet the criteria for both NOM p value of <0.05 and FDR q value of <0.25 are considered to be significantly enriched and are listed. C, representative enrichment plot, corresponding to the “GO_MITOTIC_CELL_CYCLE” set in B. The heat map shows expression levels of genes included in the gene set. The left three and right three columns correspond to BEC samples from the control and Klf5-LKO livers, respectively. The expression levels are indicated according to the scale bar at right. D, Venn diagram showing DEG identified in the intestine (left circle) and BECs (right circle) upon Klf5 deletion. Numbers shown in black and gray characters in the diagram indicate the counts of down-regulated and up-regulated genes, respectively. E, RNA-seq data for expression levels for cell cycle-related genes. Three categories correspond to those in the Venn diagram shown in D. F, entire list of enriched KEGG pathway gene sets. The SIZE column indicates the number of genes hit in each gene set. Gene sets that meet the criteria for both NOM p value <0.05 and FDR q value <0.25 are considered to be significantly enriched and are listed.
These results from GO analyses are consistent with the notion that Klf5 is a pro-proliferation factor in many types of epithelial cells, including those in normal and cancer tissues, regulating components of both direct accelerators and breaks in the cell cycle (28, 29). Thus, many cyclins, cyclin-dependent kinases (Cdks), and Cdk inhibitors have been reported to be targets of Klf5 (28–30). Therefore, we compared the DEG identified herein with the one reported in the GEO database from a study on an intestine-specific deletion of Klf5 (Fig. 5D) (30). Our RNA-seq data showed that expression of some cell cycle–related genes was indeed down-regulated in BECs upon Klf5 deficiency, whereas the set of genes affected in BECs was not necessarily consistent with those in the intestine (Fig. 5, D and E). Quantitative RT-PCR analysis using isolated BEC fractions confirmed that expression of cyclin genes, Ccna2, Ccnb1, and Ccnb2, was significantly suppressed in Klf5-LKO mice under the DDC-induced injury condition (Fig. S5E).
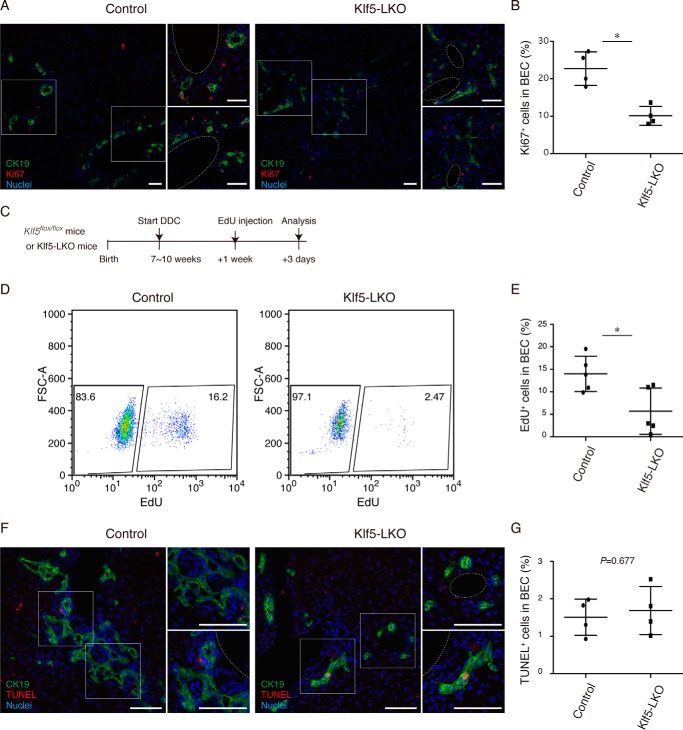
To empirically determine the role of Klf5 as a regulator of BEC proliferation in DR, we immunostained liver sections for the proliferation marker Ki67 and quantitated the level of BEC proliferation upon DDC injury (Fig. 6, A and B). Ki67+ cells among the CK19+ BEC population reduced significantly in Klf5-LKO mice, suggesting that suppression of DR upon the loss of Klf5 is, in part, due to the reduced proliferation rate of BECs. To further strengthen this notion, we also performed an in vivo 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay (Fig. 6C). Flow cytometric analysis of EdU+ cells in BEC fractions isolated from the livers of DDC-injured animals showed that those BECs that entered the cell cycle and had undergone the S phase were significantly reduced in Klf5-LKO mice (Fig. 6, D and E). As Klf5 has also been implicated in cell survival by suppressing apoptosis (29–31), we also investigated whether DR suppression in Klf5-LKO mice was associated with aberrant induction of apoptosis in BECs. In situ terminal deoxynucleotidyltransferase dUTP nick end labeling (TUNEL) staining assays using liver sections revealed that TUNEL+ apoptotic cells among the CK19+ BEC population did not increase significantly in Klf5-LKO mice compared with those in the control mice (Fig. 6, F and G). In addition, the result of GSEA did not show that Klf5 was involved in apoptosis or cell death (Fig. 5B and Fig. S4A).
Figure 6.
BEC proliferation upon DDC-induced liver injury is suppressed in Klf5-LKO mice. A, immunostaining for Ki67 (red) and CK19 (green) in the Klf5-LKO and control livers treated with DDC for 1 week shown with counterstaining for nuclei (blue). Two regions of interest indicated by white boxes in the left panels are magnified in the right panels. Scale bar, 50 μm. B, quantification of Ki67+ cells in the CK19+ BEC population. n = 4 mice. p value was calculated by Student's t test to be 0.00520. C, experimental scheme of in vivo EdU incorporation assays. D, flow cytometry analysis of EdU incorporation in BECs in Klf5-LKO and control mice upon DDC injury. Representative dot plots for the BEC population (EpCAM+ CD45− cell fraction) gated out from NPCs are shown. E, quantification of EdU+ cells in BECs as revealed by flow cytometry analyses. n = 5 mice. p value was calculated by Student's t test to be 0.0224. F, TUNEL staining (red) was performed to detect apoptotic cells with co-immunostaining for CK19 (green) in the Klf5-LKO and control livers treated with DDC for 2 weeks. Counterstaining for nuclei is also shown (blue). Scale bar, 50 μm. G, quantification of TUNEL+ cells in the CK19+ BEC population. n = 4 mice. p value calculated by Student's t test. Asterisks indicate that the p values are <0.05 (*, p < 0.06).
Among the known signaling molecules and pathways involved in DR regulation, the roles of Fgf7 and Tweak are quite remarkable in that simple overexpression of either of these factors alone in the healthy adult mouse liver can sufficiently induce DR through BEC proliferation (3, 8, 10). To address a possible role of Klf5 as a downstream effector of these pro-proliferative signals in BECs, we overexpressed each of these humoral factors by hydrodynamic delivery of the gene expression plasmids into the liver of Klf5-LKO mice and assessed the BEC proliferation rates by the EdU incorporation assay (Fig. S5B). The rate of EdU+ BECs did not significantly reduce in Klf5-LKO mice (Fig. S5, C and D), suggesting that these factors can affect BEC proliferation independent of Klf5.
The Notch- and Wnt-signaling pathways are also known to play important roles in promoting DR and BEC proliferation. However, our RNA-seq data and subsequent pathway analyses using KEGG pathway gene sets revealed that neither Notch nor Wnt signaling was significantly affected in the absence of Klf5 expression (Fig. 5F and Fig. S4C). More specifically, although Notch1, Notch2, and Jag1 have been reported to be involved in BEC proliferation (32–34), expression levels of these genes as well as their target genes, Hes1 and Hey1, remained unchanged in Klf5-deficient BECs (Fig. S4B). Indeed, none of the genes listed in the “KEGG_NOTCH_SIGNALING_PATHWAY” dataset showed differential expression (data not shown). DEG were not enriched in Wnt signaling pathways either (Fig. S4C). These results suggest that Klf5 functions as a more proximal factor controlling cell cycle progression in BECs, rather than by acting upstream of or modulating the activities of these signaling pathways in DR induction.
We also subjected the RNA-seq data to leading edge analysis (27), which enables us to extract the core genes from particular gene sets, and we found that the core genes contributing to the “KEGG_FOCAL_ADHESION” and “KEGG_ECM-RECEPTOR_INTERACTION” pathways were Lama3 andLamb3 (Fig. S5F). Quantitative RT-PCR analysis confirmed that expression of these genes in BECs was certainly reduced in Klf5-LKO mice (Fig. S5G). Both of the Lama3 and Lamb3 genes encode the components of laminin-332 (the α3 and β3 subunits, respectively), an extracellular matrix (ECM) protein that is well known to be critically involved in the maintenance of skin architecture by composing hemidesmosome and thereby bridging the epidermis and the underlying dermis (35). Intriguingly, expression of these genes in BECs tended to be augmented specifically under the DDC-induced cholestatic liver injury condition but not under the TAA-induced injury condition (Fig. S5H). This gene expression pattern correlates well with the DR phenotype in Klf5-LKO mice and hence suggests that laminin-332 may be a critical target molecule of Klf5 to maintain biliary architecture under the cholestatic liver injury conditions.
Discussion
The transcription factor Klf5 has been shown to play diverse roles in various types of tissues and cells, including embryonic stem cells, vascular endothelial cells, fibroblasts, and epithelial cells. In vivo studies using several kinds of tissue-specific conditional knockout mice have revealed that Klf5 is critically involved in developmental morphogenesis and/or in the maintenance of tissue morphology and functions against injury in multiple epithelial tissues, such as tissues in the intestine, lung, and kidney. Here, we have demonstrated for the first time that Klf5 also plays a physiologically important role in yet another epithelial tissue of the intrahepatic biliary epithelium, specifically under cholestatic liver injury conditions. Notably, based on a transcriptomic meta-analysis that compared a large number of microarray data, a very recent report by Passman et al. (36) suggested that Klf5 is a candidate marker molecule for LPCs. This notion is consistent with our present finding that expression of the gene was highly enriched in BECs in the injured liver, which corresponds to LPCs, among liver cell populations (Fig. 1B).
DR is a histopathological phenomenon that can be typically recognized by microscopic observation of tissue sections as an ectopic emergence and expansion of biliary-like cells in the parenchymal region upon liver injury. Recent studies have established that it actually represents dynamic remodeling of the tree-like structure of the intrahepatic biliary epithelial tissue. A characteristic feature of this remodeling is that the structural transformations of the biliary tree are diverse and correlated with the parenchymal injury patterns (3). Thus, under the DDC-induced cholestatic injury condition, when hepatocyte damage occurs across the liver parenchyma, including the peri-portal venous region, biliary branches split intricately around the portal vein and expand randomly to the parenchymal area in all directions. In the TAA model, where zonal metabolizing activity restricts production of toxic metabolites and concomitant hepatocyte injury only to the peri-central venous region, the branches exhibited a different structure that extended in a relatively straight line toward the distant injured area. The present results showed that the involvement of Klf5 was essential for DR induction in the DDC model, but not in the TAA model, strongly suggesting that such a morphological diversity in the biliary remodeling is associated with distinct molecular mechanisms. Hence, our study has further substantiated the concept that DR actually involves phenotypically and mechanistically diverged heterogenic tissue remodeling processes not only at the cellular but also at the molecular level.
The results of quantitative gene expression analysis revealed that expression of the Klf5 mRNA in BECs was kept constant upon liver injury and under different types of liver injury conditions (Figs. S1A and S3A). It has been reported that Klf5 proteins undergo several types of post-translational modifications, including acetylation, sumoylation, and phosphorylation, that can modulate their stability, subcellular localization, and/or transactivation activities (37). It is thus possible that the Klf5 functions in BECs are regulated at the post-translational level in a cholestatic liver injury-specific manner, which needs to be addressed in future studies.
Our unbiased approach employing the whole transcriptome analysis based on RNA-seq has shown that Klf5 is primarily involved in regulation of cell proliferation in BECs under the DDC injury condition (Fig. 5, A–C). This was somewhat surprising, as BEC proliferation is induced not only under this particular injury condition but also upon TAA-induced liver injury (7). Our previous study has revealed that a select population of BECs proliferates continuously and makes a major contribution in DR induction under the TAA liver injury, although it is not yet defined whether this population is conserved among different types of mouse liver injury models (7). In rat liver injury models, there are different proliferative compartments that differentially respond to the types of liver injury (38–40). Klf5 might regulate cell type-specific proliferation signals in a subpopulation of BECs under cholestatic liver injury.
Studies using various KO mouse models have shown that the Notch-, Wnt-, Fgf7-, and Tweak-signaling pathways play important roles in DR induction, particularly under the DDC-induced liver injury condition (8, 10, 34, 41–43). In our RNA-seq analysis, neither of the Notch- nor Wnt-signaling pathways was significantly affected by the lack of Klf5 expression nor did the MAPK, Akt, or NF-κB pathways, which are the potential downstream signaling mechanisms for Fgf7 or Tweak (Fig. 5, Fig. S4, and data not shown). Hence, Klf5 does not likely function as an upstream regulator and may rather play a role as a downstream effector and/or a transcriptional co-activator in relation to these pathways. A very recent report suggested that the Wnt noncanonical signaling pathway, not the β-catenin-dependent canonical pathway, is involved in DDC-induced BEC proliferation (43). Notably, Klf5 is a biologically relevant target of Wnt1 signaling that is activated in a β-catenin–independent manner in a mammary epithelial cell line (44). This Wnt noncanonical signal, however, up-regulates Klf5 at the mRNA level. As the expression level of Klf5 mRNA in BECs did not increase in the DDC-injured liver compared with that in the normal liver (Fig. S1A), the relevance of the relationship between Wnt1 signaling and Klf5 in DR induction requires further investigation.
With regard to Fgf7 and Tweak, we examined their possible roles as upstream signals for Klf5 more directly by employing in vivo gene expression and DR induction experiments in the mouse liver. Contrary to our expectations, the results indicated that Fgf7 and Tweak were capable of inducing BEC proliferation even in the absence of Klf5 (Fig. S5, B–D), suggesting that Klf5 is not the primary target of these signals. It should be noteworthy that in those experimental settings DR and BEC proliferations were achieved without any induction of liver injury or concomitant inflammatory responses. A number of studies have pointed out a function for Klf5 as a mediator of external stress responses following tissue injuries, such as those provoked by the bacterial membrane component lipopolysaccharide or ionizing radiation (45, 46). Cholestatic injury conditions cause damages and stresses directly on BECs (47, 48) and hence could be the trigger for the Klf5 activation. Humoral signals such as Fgf7 and Tweak may act cooperatively with the stress-activated Klf5, and their modes of action on BECs could differ under liver injury conditions.
In organs and tissues containing epithelial cells, Klf5 regulates diverse biological functions. In many cases, the loss of Klf5 affects cell proliferation conspicuously, with several cell cycle–associated genes being identified as critical targets of this transcription factor. Ccnb1, Ccnd1, Cdk1, and Cdkn2b have been reported to be direct targets, substantiated by in vitro assay, and also in a study using intestine-specific Klf5 KO mice (28–30, 49–52), whereas our RNA-seq showed that only Ccnb1 and Cdk1 were significantly affected on loss of Klf5 in BECs. Moreover, Klf5 is involved in the maturation, and not the proliferation, of lung epithelial cells (14). These findings together indicate that Klf5 functions are not necessarily consistent among the different epithelial cell types and that even cell cycle-related genes critically regulated by Klf5 may vary depending on the context of the cell types. It has been reported that Klf5 regulates gene expression in concert with other cell type-specific transcription factors (53, 54), which may render Klf5 with such diverse transcriptional abilities and functions in a context-dependent manner.
In Klf5-LKO mice upon DDC administration, the level of DR induction as revealed by CK19+ area seemed to increase slightly during the very early phase and peaked around 1–2 weeks after the onset of injury, after which time point it tended to reduce gradually (Fig. 2D). As the proliferation of BECs was still weakly induced, albeit significantly suppressed, in the Klf5-LKO mice (Fig. 6, A–E), the observed DR phenotype suggests that Klf5 likely contributes in regulating cellular functions other than proliferation in BECs. Of note, 3D immunostaining analysis showed that some CK19+ cell clusters were certainly separated from the bile ducts upon DDC administration in Klf5-LKO mice (Fig. 2F), suggesting that Klf5 may be involved in maintaining tissue structural integrity of bile ducts against biliary pressure and/or cholangitis under the cholestatic conditions. Epithelial cells in general, including BECs, are structurally supported by ECMs and receive signals from ECM proteins. It is well established that in the course of bile duct development, ECMs, such as α1-containing laminin and α5-containing laminin, play fundamental roles by regulating initiation of tubulogenesis and maturation of the duct structure, respectively (55). At the adult stage, remodeling of ECMs such as collagens and laminins occurs along with DR induction when the liver is injured, and functional evidence has been accumulated stating that selective contribution of DR to biliary maintenance or hepatocyte differentiation depends on the types of ECMs, suggesting a close relationship between ECMs and DR regulation (56). Intriguingly, GSEA pathway analysis of our RNA-seq data revealed four pathways that were significantly affected by Klf5 deletion, among which “ECM–receptor interaction” and “focal adhesion” were included (Fig. 5F). Moreover, we identified the laminin-332 components Lama3 and Lamb3 as candidate Klf5 target genes in BECs (Fig. S5, F and G). This notion is consistent with a previous study by Shinoda et al. (57) showing by microarray analyses that expression of genes related to ECMs and adhesion molecules, including Lama3 and Lamb3, was up-regulated by overexpression of Klf5 in a chondrogenic cell line. Importantly, the expressions of these laminin component genes in BECs were up-regulated specifically under the DDC-induced cholestatic injury condition (Fig. S5H). Thus, it is tempting to speculate that Klf5 regulates certain aspects of interactions between BECs and the ECM microenvironment, plausibly via laminin-332 deposition, which is critical for maintaining the bile duct structure under conditions of biliary stress. Future studies aimed at elucidating the functional relevance and the modes of action of laminin-332 in the BEC–ECM interaction and the remodeling and maintenance of the biliary epithelial structure should lead to further understanding of the molecular mechanisms underlying DR induction under cholestatic liver injury conditions.
Experimental procedures
Animals and liver injury models
Wildtype (WT) C57BL/6J mice were purchased from CLEA Japan, Inc. To generate liver epithelial-specific Klf5 KO (Klf5-LKO) mice, mice harboring floxed alleles of the Klf5 gene (Klf5flox/flox) (18) were crossed with Alfp-Cre Tg mice (kindly provided by Dr. Klaus Kaestner, University of Pennsylvania) (19). For the hepatocyte-specific Klf5 KO (Klf5-HKO) experiments, the recombinant adeno-associated virus expressing a codon-optimized Cre variant (iCre) under the control of a hepatocyte-specific promoter (rAAV2/8-iCre) (7) was packaged in HEK293 cells according to the protocol described previously (58). iCre was acquired from the pDIRE plasmid, which was a gift from Rolf Zeller (Addgene plasmid no. 26745 (59)). The titered virus was delivered by intraperitoneal injection at a dose of 1 × 1011 vector genomes/mouse.
For injury models, mice were fed a 0.1% DDC-containing diet (F-4643; Bio-Serv) for the DDC model or administered TAA (204-00881; Wako; 300 mg/liter) in drinking water for the TAA model. The duration of each injury protocol is indicated in the figure legends. For in vivo EdU-labeling assays to monitor BEC proliferation, EdU was injected intraperitoneally at a dose of 2 mg/mouse. Overexpression of Fgf7 and Tweak in the mouse liver was achieved by hydrodynamic tail vein injection essentially as described previously (3), with 10 μg of plasmids per 20 g of body weight being injected into mice at the age of 6–8 weeks old.
All animals were maintained under standard specific pathogen-free conditions. All animal experiments were conducted in accordance with the Guideline for the Care and Use of Laboratory Animals of the University of Tokyo, under the approval of the Institutional Animal Care and Use Committee of the Institute of Molecular and Cellular Biosciences, University of Tokyo (approval numbers 2501, 2501-1, 2609, 2706, 2804, and 2904).
Immunostaining analyses with tissue sections
Dissected livers were directly embedded in Tissue-Tek O.C.T. Compound (4583; Sakura Finetek USA, Inc.), and snap-frozen. Frozen sections (8 μm) of the liver were prepared using an HM525 cryostat (Microm International) and placed on aminopropyltriethoxysilane-coated glass slides (Matsunami Glass). Fixation was performed with acetone and/or 4% paraformaldehyde after sectioning. After blocking in 3% BSA in phosphate-buffered saline (PBS) containing 0.1% Triton X-100 or 3% fetal bovine serum in PBS containing 0.1% Triton X-100, the samples were incubated with primary antibodies described in Table 1 and then with fluorescence-conjugated secondary antibodies. Nuclei were counterstained with Hoechst 33342 (Sigma). Liver sections were imaged with a fluorescence microscope (Axio Observer.Z1, Zeiss; IX83, Olympus) and a confocal laser-scanning microscope (Fluoview FV1200 and FV3000, Olympus). Immunostaining of 200-μm tissue sections was performed according to the protocol described previously (3). For 3D presentation, surfaces were virtually constructed using the “Surface” function in IMARIS software (Bitplane).
Table 1.
List of antibodies used in this study
| Antigen | Source/supplier | Host | Fixation | Dilution | Refs. |
|---|---|---|---|---|---|
| CD45a | Pharmingen, catalog no. 553081 | Rat | No fixation | 1:100 | |
| CK19 | In-house | Rabbit | PFA only or acetone and PFA | 1:2000 | 62 |
| EpCAM | Pharmingen, catalog no. 552370 | Rat | Acetone and PFA | 1:200 | |
| EpCAMa | In-house | Rat | No fixation | 1:100 | 60 |
| Ki67 | Invitrogen, catalog no. 14-5698-82 | Rabbit | PFA | 1:200 | |
| Klf5 | Generous gift from Drs. Ryozo Nagai (Jichi Medical University, Tochigi, Japan) and Ichiro Manabe (Chiba University, Chiba, Japan) (Km1784) | Rat | PFA | 1:300 | 63 |
| Prominin1 | Biolegend (San Diego), catalog no. 141207 | Rat | Acetone | 1:100 |
a These antibody were used for FACS. The others were used for immunostaining. PFA is paraformaldehyde.
TUNEL assay was performed using the In Situ Apoptosis Detection kit (MK500; TaKaRa) according to the manufacturer's instructions, and the fluorescence signal was enhanced by treating the samples with AlexaFluor488-conjugated anti-fluorescein antibodies (200-542-037, Jackson ImmunoResearch, Inc.).
Quantitative analyses in tissue sections
For the quantification of positive areas with CK19 expression (Figs. 2D, 3, B and D, and 4E), immunostained whole-liver sections were imaged and quantified using an IN Cell Analyzer 2000 (GE Healthcare). The ratio of CK19+ area per total liver area was calculated. More than three mice were used for each of the control and Klf5-LKO groups in all experiments. For BEC proliferation and apoptosis assays in Fig. 6, numbers of Ki67+/CK19+ and TUNEL+/CK19+ cells were manually counted, and the ratios to the total numbers of CK19+ cells were calculated. More than 10 fields in each section were randomly selected for analysis.
For the quantification of biliary branches in the 3D images (Fig. S2, E–G), confocal image stacks were recorded with a confocal microscope (FV3000, Olympus) using a 20×/0.75 NA objective lens (UPLSAPO 20X; Olympus). Settings used were as follows: 800 × 800-nm pixel frame size; 795-nm pixel size; 1.02-μm z-distances between sections; and a 12.5-ms/pixel scan speed. Each visual field was acquired in a uniform size, 636.396 × 636.396 μm in the x-y plane, throughout the full length of the z axis to represent the entire structure in 200-μm-thick tissue sections. For 3D images, Gaussian smoothing (0.795 voxel radius) was performed. Biliary branches were skeletonized as filaments (as exemplified in Fig. S2E), and the length and volume size of each branch segment were quantified using the “Filament Tracer” application attached to IMARIS. When comparing the thickness of biliary branches (Fig. S2G), “branch thickness index” was calculated as the square root of the quotient obtained by dividing the branch volume size by the branch length.
Cell preparation and flow cytometry
Preparation of cell fractions from adult mouse livers was performed as described previously (60). Briefly, a single cell suspension from the mouse liver was obtained by a two-step collagenase perfusion method and the parenchymal (hepatocyte), and NPC fractions were prepared by centrifugal separation. To prepare the BEC fraction, NPCs were treated with anti-EpCAM mAb (Table 1), and the samples were sorted by Moflo XDP (Beckman-Coulter). Nonviable cells were excluded by propidium iodide staining. Typically, we were able to obtain around 1–2 × 104 EpCAM+ BECs per mouse.
In the EdU incorporation experiments, NPCs were co-stained with anti-EpCAM and anti-CD45 antibodies, and BEC fractions were identified as an EpCAM+ CD45− cell population. EdU detection was performed by using the Click-iT Plus EdU AlexaFluor488 cytometry assay kit (Life Technologies, Inc.), following the manufacturer's instructions. The samples were analyzed by FACSCanto II (BD Biosciences). Nonviable cells were excluded by Fixable Viability Stain 450 (BD Biosciences).
Gene expression analysis by quantitative RT-PCR
Total RNA was isolated from whole-liver samples or sorted cell populations using TRIzol reagent (Invitrogen), treated with DNase I (Invitrogen), and then used for cDNA synthesis with PrimeScript RT Master Mix (Takara). Quantitative RT-PCR analyses were performed using LightCycler (Roche Applied Science) with SYBR Premix Ex Taq (Takara). Gapdh was used as the internal control. Primer sequences are listed in Table 2.
Table 2.
List of primers used for quantitative RT-PCR in this study
| Gene | Direction | Sequence (5′ to 3′) |
|---|---|---|
| Ccna2 | Forward | CTTGGCTGCACCAACAGTAA |
| Reverse | CAAACTCAGTTCTCCCAAAAACA | |
| Ccnb1 | Forward | ACCAGAGGTGGAACTTGCTG |
| Reverse | GGCTTGGAGAGGGATTATCA | |
| Ccnb2 | Forward | TGAAACCAGTGCAGATGGAG |
| Reverse | CAGAGAAAGCTTGGCAGAGG | |
| Ccne1 | Forward | TTGCAAGACCCAGATGAAGA |
| Reverse | TCCACGCATGCTGAATTATC | |
| Cdk1 | Forward | TCCGTCGTAACCTGTTGAGT |
| Reverse | TGGCCAGTGACTCTGTGTCT | |
| Epcam | Forward | AGGGGCGATCCAGAACAACG |
| Reverse | ATGGTCGTAGGGGCTTTCTC | |
| Fgf7 | Forward | TTTGGAAAGAGCGACGACTT |
| Reverse | GGCAGGATCCGTGTCAGTAT | |
| Gapdh | Forward | TGTGTCCGTCGTGGATCTGA |
| Reverse | TTGCTGTTGAAGTCGCAGGAG | |
| Hgf | Forward | CCCGAGAACTTCAAATGCAA |
| Reverse | TATGACGGTGTAAATCCTCCA | |
| Klf5 | Forward | TGCGATTATAATGGTTGCACA |
| Reverse | GGTGCACTTGTAGGGCTTCT | |
| Lama3 | Forward | TGTACCTTGGGAATAAGGATGC |
| Reverse | CGTCAGGACCTGGTCTATCTG | |
| Lamb3 | Forward | AGCCAGCAGGCAATGAAT |
| Reverse | GCCGGTCCTTCAACTCTGTAT | |
| Tat | Forward | CATCTGGAGCCATGTACCTT |
| Reverse | TCCAGCATCATCACCTCG | |
| Tweak (Tnfsf12) | Forward | GCCCATTATGAGGTTCATCC |
| Reverse | TCACTGTCCCATCCACACC |
Genomic PCR analysis for Klf5 deletion
Isolated hepatocytes, BECs, and NPCs (1.0 × 102 cells for each) were lysed in a buffer containing 65 mm Tris-HCl, pH 8.8, 16.6 mm ammonium sulfate, 1 mm 2-mercaptoethanol, 6.7 μm EDTA, and 0.5% Triton X-100. The lysates were subjected to genomic PCR amplification of the Klf5 gene locus with Tks Gflex DNA polymerase (Takara) using thermal cycler (Bio-Rad). Primer sequences are listed in Table 3. Cycling conditions were as follows: 98 °C for 1 min; followed by 40 cycles of 98 °C for 10 s, 62 °C for 15 s, and 68 °C for 3 min; and a final 10-min elongation step at 68 °C. The amplified samples were analyzed by agarose gel electrophoresis along with DNA size markers (100-bp ladder and 1-kb ladder: New England Biolabs).
Table 3.
List of primers used for genomic PCR in this study
| Gene | Direction | Sequence (5′ to 3′) |
|---|---|---|
| Klf5 | Forward | GTAATGGATGTGAACAGATTTGAGG |
| Reverse | GTAAACACTGCCGTTTACGTTTTGA |
RNA-seq
BEC populations were prepared by cell sorting from the control and Klf5-LKO mice treated with DDC for 1 week (n = 3 mice for each genotype; 1.0–1.7 × 104 cells per sample). The cells obtained from each mouse was individually used to prepare a RNA-seq library (resulting in a total of six libraries). Total RNA was isolated using the TRIzol reagent (Invitrogen), treated with DNase I (Invitrogen), and purified using TRIzol reagent again. Total RNA samples that met the quality control thresholds (RNA integrity RIN >8.5) were used to prepare barcoded libraries with the SMARTer ultra low input RNA kit for sequencing, version 3 (Takara), and subsequently with the Nextera XT DNA library preparation kit (Illumina) according to the manufacturers' instructions. Library samples were sequenced on HiSeq 2000 (Illumina).
RNA-seq data analysis
The first 14 bases from each read were trimmed, and the subsequent 52 bases were aligned to the Mus musculus genome (UCSC mm10) using Tophat. Only the reads that uniquely aligned to the transcripts were counted. Transcript counts were normalized, and differential gene expression was calculated using DESeq2 package in R (25). Significant DEG were selected based on a false-discovery q-value cutoff of 0.1. GO analysis was performed using the DAVID database (26). GSEA was employed to identify significantly affected biological pathways using the GO biological process modules, the KEGG gene sets modules, and the Reactome gene sets modules. After Kolmogorov-Smirnoff testing, the gene sets showing NOM p value of <0.05 and FDR q value of <0.25 were considered enriched.
RNA-seq fastq files on intestinal crypt samples from the intestine-specific Klf5 KO and control mice (30, 61) were obtained from GEO database under accession number GSE79758 (“Differential Gene Expression in Intestinal Mouse Crypts after Loss of KLF5”). These data were also analyzed as described above.
Statistical analysis
Data are expressed as the mean ± S.D. The Shapiro-Wilk test was used to assess the normality of distribution of investigated parameters, and significant differences were tested using the unpaired two-tailed Mann-Whitney U test or Student's t test accordingly. Statistical analyses were performed using the R software and the Prism software (GraphPad, San Diego). Differences were considered statistically significant at p < 0.05.
Author contributions
H. O. and T. I. conceptualization; H. O. and M. Y. data curation; H. O. formal analysis; H. O., A. M., and T. I. funding acquisition; H. O. and M. Y. validation; H. O. and M. Y. investigation; H. O., K. Kamimoto, and K. Kaneko visualization; H. O., A. M., and T. I. writing-original draft; H. O. and T. I. project administration; H. O., C. Y.-Y. K., M. E., A. M., and T. I. writing-review and editing; K. Kamimoto, C. Y.-Y. K., and K. Kaneko methodology; C. Y.-Y. K., M. E., and T. I. resources; M. E., A. M., and T. I. supervision.
Supplementary Material
Acknowledgments
We thank Dr. L. W. Katsumata for technical instructions; N. Miyata and C. Koga for cell sorting; N. Imaizumi for animal care; and the members of the Miyajima lab for helpful discussions and advice. We thank Prof. Ryozo Nagai (Jichi Medical University, Tochigi, Japan) and Prof. Ichiro Manabe (Chiba University, Chiba, Japan) for the anti-Klf5 antibody; Prof. Klaus Kaestner (University of Pennsylvania) for the Alfp-Cre Tg mouse; Prof. Ian Alexander (Children's Medical Research Institute, Australia) for the pAM-LSP1-EGFP plasmid; Prof. R. Jude Samulski and the NGVB Biorepository (University of North Carolina at Chapel Hill, NC) for the XX6-80 plasmid; Prof. Rolf Zeller for the pDIRE plasmid; Penn Vector Core (University of Pennsylvania) for the p5E18-VD2/8 plasmid; and Dr. Yuki Kato and Prof. Katsuhiko Shirahige (Institute of Molecular and Cellular Biosciences, University of Tokyo, Japan) for RNA-seq. We also thank the University of Tokyo IMCB Olympus Bioimaging Center (TOBIC) for helping with microscopy and image acquisition.
This work was supported by the Japan Society for the Promotion of Science KAKENHI Grants 22118006 (to A. M.), 24112507 (to T. I.), 24590342 (to T. I.), 26112704 (to T. I.), 26253023 (to A. M.), 15H01369 (to T. I.), 15J08420 (to H. O.), and 17H05497 (to T. I.); CREST from the Japan Science and Technology Agency Grant 15652259 (to A. M.); a research grant from the Takeda Science Foundation (to T. I.); and a research grant from the Daiichi Sankyo Foundation of Life Science (to T. I.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
RNA-seq fastq files and processed data files were deposited into the NCBI database under accession no. GSE97167.
- BEC
- biliary epithelial cell
- DR
- ductular reaction
- LPC
- liver stem/progenitor cell
- NPC
- nonparenchymal cell
- DDC
- 3,5-diethoxycarbonyl-1,4-dihydrocollidine
- LKO
- liver epithelial-specific knockout
- ALP
- alkaline phosphatase
- T-BIL
- total bilirubin
- TAA
- thioacetamide
- HKO
- hepatocyte-specific knockout
- Tweak
- tumor necrosis factor-like weak inducer of apoptosis
- DEG
- differential expression genes
- GO
- Gene ontology
- GSEA
- Gene set enrichment analysis
- TUNEL
- terminal deoxynucleotidyltransferase dUTP nick end labeling
- ECM
- extracellular matrix
- RNA-seq
- RNA-sequencing
- EdU
- 5-ethynyl-2′-deoxyuridine
- FDR
- false discovery rate
- Cdk
- cyclin-dependent kinase.
References
- 1. Michalopoulos G. K., and DeFrances M. C. (1997) Liver regeneration. Science 276, 60–66 10.1126/science.276.5309.60 [DOI] [PubMed] [Google Scholar]
- 2. Michalopoulos G. K., and Khan Z. (2015) Liver stem cells: experimental findings and implications for human liver disease. Gastroenterology 149, 876–882 10.1053/j.gastro.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kaneko K., Kamimoto K., Miyajima A., and Itoh T. (2015) Adaptive remodeling of the biliary architecture underlies liver homeostasis. Hepatology 61, 2056–2066 10.1002/hep.27685 [DOI] [PubMed] [Google Scholar]
- 4. Gouw A. S., Clouston A. D., and Theise N. D. (2011) Ductular reactions in human liver: diversity at the interface. Hepatology 54, 1853–1863 10.1002/hep.24613 [DOI] [PubMed] [Google Scholar]
- 5. Duncan A. W., Dorrell C., and Grompe M. (2009) Stem cells and liver regeneration. Gastroenterology 137, 466–481 10.1053/j.gastro.2009.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miyajima A., Tanaka M., and Itoh T. (2014) Stem/progenitor cells in liver development, homeostasis, regeneration, and reprogramming. Cell Stem Cell 14, 561–574 10.1016/j.stem.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 7. Kamimoto K., Kaneko K., Kok C. Y., Okada H., Miyajima A., and Itoh T. (2016) Heterogeneity and stochastic growth regulation of biliary epithelial cells dictate dynamic epithelial tissue remodeling. Elife 5, e15034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jakubowski A., Ambrose C., Parr M., Lincecum J. M., Wang M. Z., Zheng T. S., Browning B., Michaelson J. S., Baetscher M., Wang B., Bissell D. M., and Burkly L. C. (2005) TWEAK induces liver progenitor cell proliferation. J. Clin. Invest. 115, 2330–2340 10.1172/JCI23486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ishikawa T., Factor V. M., Marquardt J. U., Raggi C., Seo D., Kitade M., Conner E. A., and Thorgeirsson S. S. (2012) Hepatocyte growth factor/c-met signaling is required for stem-cell-mediated liver regeneration in mice. Hepatology 55, 1215–1226 10.1002/hep.24796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takase H. M., Itoh T., Ino S., Wang T., Koji T., Akira S., Takikawa Y., and Miyajima A. (2013) FGF7 is a functional niche signal required for stimulation of adult liver progenitor cells that support liver regeneration. Genes Dev. 27, 169–181 10.1101/gad.204776.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bialkowska A. B., Yang V. W., and Mallipattu S. K. (2017) Krüppel-like factors in mammalian stem cells and development. Development 144, 737–754 10.1242/dev.145441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McConnell B. B., Kim S. S., Yu K., Ghaleb A. M., Takeda N., Manabe I., Nusrat A., Nagai R., and Yang V. W. (2011) Krüppel-like factor 5 is important for maintenance of crypt architecture and barrier function in mouse intestine. Gastroenterology 141, 1302–1313 10.1053/j.gastro.2011.06.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fujiu K., Manabe I., and Nagai R. (2011) Renal collecting duct epithelial cells regulate inflammation in tubulointerstitial damage in mice. J. Clin. Investig. 121, 3425–3441 10.1172/JCI57582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wan H., Luo F., Wert S. E., Zhang L., Xu Y., Ikegami M., Maeda Y., Bell S. M., and Whitsett J. (2008) Kruppel-like factor 5 is required for perinatal lung morphogenesis and function. Development 135, 2563–2572 10.1242/dev.021964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nakaya T., Ogawa S., Manabe I., Tanaka M., Sanada M., Sato T., Taketo M. M., Nakao K., Clevers H., Fukayama M., Kuroda M., and Nagai R. (2014) KLF5 regulates the integrity and oncogenicity of intestinal stem cells. Cancer Res. 74, 2882–2891 10.1158/0008-5472.CAN-13-2574 [DOI] [PubMed] [Google Scholar]
- 16. Maehara O., Sato F., Natsuizaka M., Asano A., Kubota Y., Itoh J., Tsunematsu S., Terashita K., Tsukuda Y., Nakai M., Sho T., Suda G., Morikawa K., Ogawa K., Chuma M., et al. (2015) A pivotal role of Kruppel-like factor 5 in regulation of cancer stem-like cells in hepatocellular carcinoma. Cancer Biol. Ther. 16, 1453–1461 10.1080/15384047.2015.1070992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dorrell C., Erker L., Schug J., Kopp J. L., Canaday P. S., Fox A. J., Smirnova O., Duncan A. W., Finegold M. J., Sander M., Kaestner K. H., and Grompe M. (2011) Prospective isolation of a bipotential clonogenic liver progenitor cell in adult mice. Genes Dev. 25, 1193–1203 10.1101/gad.2029411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Azami T., Waku T., Matsumoto K., Jeon H., Muratani M., Kawashima A., Yanagisawa J., Manabe I., Nagai R., Kunath T., Nakamura T., Kurimoto K., Saitou M., Takahashi S., and Ema M. (2017) Klf5 maintains the balance of primitive endoderm versus epiblast specification during mouse embryonic development by suppression of Fgf4. Development 144, 3706–3718 10.1242/dev.150755 [DOI] [PubMed] [Google Scholar]
- 19. Zhang L., Rubins N. E., Ahima R. S., Greenbaum L. E., and Kaestner K. H. (2005) Foxa2 integrates the transcriptional response of the hepatocyte to fasting. Cell Metab. 2, 141–148 10.1016/j.cmet.2005.07.002 [DOI] [PubMed] [Google Scholar]
- 20. Ema M., Mori D., Niwa H., Hasegawa Y., Yamanaka Y., Hitoshi S., Mimura J., Kawabe Y., Hosoya T., Morita M., Shimosato D., Uchida K., Suzuki N., Yanagisawa J., Sogawa K., Rossant J., Yamamoto M., Takahashi S., and Fujii-Kuriyama Y. (2008) Krüppel-like factor 5 is essential for blastocyst development and the normal self-renewal of mouse ESCs. Cell Stem Cell 3, 555–567 10.1016/j.stem.2008.09.003 [DOI] [PubMed] [Google Scholar]
- 21. Yeh C. N., Maitra A., Lee K. F., Jan Y. Y., and Chen M. F. (2004) Thioacetamide-induced intestinal-type cholangiocarcinoma in rat: an animal model recapitulating the multi-stage progression of human cholangiocarcinoma. Carcinogenesis 25, 631–636 [DOI] [PubMed] [Google Scholar]
- 22. Smit J. J., Schinkel A. H., Oude Elferink R. P., Groen A. K., Wagenaar E., van Deemter L., Mol C. A., Ottenhoff R., van der Lugt N. M., van Roon M. A., van der Valk M. A., Offerhaus G. J., Berns A. J., and Borst P. (1993) Homozygous disruption of the murine MDR2 P-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell 75, 451–462 10.1016/0092-8674(93)90380-9 [DOI] [PubMed] [Google Scholar]
- 23. Mauad T. H., van Nieuwkerk C. M., Dingemans K. P., Smit J. J., Schinkel A. H., Notenboom R. G., van den Bergh Weerman M. A., Verkruisen R. P., Groen A. K., and Oude Elferink R. P. (1994) Mice with homozygous disruption of the mdr2 P-glycoprotein gene. A novel animal model for studies of nonsuppurative inflammatory cholangitis and hepatocarcinogenesis. Am. J. Pathol. 145, 1237–1245 [PMC free article] [PubMed] [Google Scholar]
- 24. Kumadaki S., Karasawa T., Matsuzaka T., Ema M., Nakagawa Y., Nakakuki M., Saito R., Yahagi N., Iwasaki H., Sone H., Takekoshi K., Yatoh S., Kobayashi K., Takahashi A., Suzuki H., et al. (2011) Inhibition of ubiquitin ligase F-box and WD repeat domain-containing 7α (Fbw7α) causes hepatosteatosis through Krüppel-like factor 5 (KLF5)/peroxisome proliferator-activated receptor γ2 (PPARγ2) pathway but not SREBP-1c protein in mice. J. Biol. Chem. 286, 40835–40846 10.1074/jbc.M111.235283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 27. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., and Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tetreault M.-P., Yang Y., and Katz J. P. (2013) Krüppel-like factors in cancer. Nat. Rev. Cancer 13, 701–713 10.1038/nrc3582 [DOI] [PubMed] [Google Scholar]
- 29. McConnell B. B., Ghaleb A. M., Nandan M. O., and Yang V. W. (2007) The diverse functions of Krüppel-like factors 4 and 5 in epithelial biology and pathobiology. BioEssays 29, 549–557 10.1002/bies.20581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bell K. N. (2016) The Role of Krüppel-like Factor 5 in Normal Intestinal Homeostasis. Ph.D. thesis, University of Cincinnati, Cincinnati, OH [Google Scholar]
- 31. Liu R., Zheng H.-Q., Zhou Z., Dong J.-T., and Chen C. (2009) KLF5 promotes breast cell survival partially through fibroblast growth factor-binding protein 1-pERK-mediated dual specificity MKP-1 protein phosphorylation and stabilization. J. Biol. Chem. 284, 16791–16798 10.1074/jbc.M808919200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Geisler F., Nagl F., Mazur P. K., Lee M., Zimber-Strobl U., Strobl L. J., Radtke F., Schmid R. M., and Siveke J. T. (2008) Liver-specific inactivation of Notch2, but not Notch1, compromises intrahepatic bile duct development in mice. Hepatology 48, 607–616 10.1002/hep.22381 [DOI] [PubMed] [Google Scholar]
- 33. Zong Y., Panikkar A., Xu J., Antoniou A., Raynaud P., Lemaigre F., and Stanger B. Z. (2009) Notch signaling controls liver development by regulating biliary differentiation. Development 136, 1727–1739 10.1242/dev.029140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fiorotto R., Raizner A., Morell C. M., Torsello B., Scirpo R., Fabris L., Spirli C., and Strazzabosco M. (2013) Notch signaling regulates tubular morphogenesis during repair from biliary damage in mice. J. Hepatol. 59, 124–130 10.1016/j.jhep.2013.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kiritsi D., Has C., and Bruckner-Tuderman L. (2013) Laminin 332 in junctional epidermolysis bullosa. Cell Adh. Migr. 7, 135–141 10.4161/cam.22418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Passman A. M., Low J., London R., Tirnitz-Parker J. E., Miyajima A., Tanaka M., Strick-Marchand H., Darlington G. J., Finch-Edmondson M., Ochsner S., Zhu C., Whelan J., Callus B. A., and Yeoh G. C. (2016) A transcriptomic signature of mouse liver progenitor cells. Stem Cells Int. 2016, 5702873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dong J.-T., and Chen C. (2009) Essential role of KLF5 transcription factor in cell proliferation and differentiation and its implications for human diseases. Cell. Mol. Life Sci. 66, 2691–2706 10.1007/s00018-009-0045-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Alpini G., Glaser S. S., Ueno Y., Pham L., Podila P. V., Caligiuri A., LeSage G., and LaRusso N. F. (1998) Heterogeneity of the proliferative capacity of rat cholangiocytes after bile duct ligation. Am. J. Physiol. 274, G767–G75 [DOI] [PubMed] [Google Scholar]
- 39. LeSage G. D., Benedetti A., Glaser S., Marucci L., Tretjak Z., Caligiuri A., Rodgers R., Phinizy J. L., Baiocchi L., Francis H., Lasater J., Ugili L., and Alpini G. (1999) Acute carbon tetrachloride feeding selectively damages large, but not small, cholangiocytes from normal rat liver. Hepatology 29, 307–319 10.1002/hep.510290242 [DOI] [PubMed] [Google Scholar]
- 40. Lesage G., Glaser S., Ueno Y., Alvaro D., Baiocchi L., Kanno N., Phinizy J. L., Francis H., and Alpini G. (2001) Regression of cholangiocyte proliferation after cessation of ANIT feeding is coupled with increased apoptosis. Am. J. Physiol. Gastrointest. Liver Physiol. 281, G182–G190 10.1152/ajpgi.2001.281.1.G182 [DOI] [PubMed] [Google Scholar]
- 41. Lozier J., McCright B., and Gridley T. (2008) Notch signaling regulates bile duct morphogenesis in mice. PLoS ONE 3, e1851 10.1371/journal.pone.0001851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Apte U., Thompson M. D., Cui S., Liu B., Cieply B., and Monga S. P. (2008) Wnt/β-catenin signaling mediates oval cell response in rodents. Hepatology 47, 288–295 10.1002/hep.21973 [DOI] [PubMed] [Google Scholar]
- 43. Okabe H., Yang J., Sylakowski K., Yovchev M., Miyagawa Y., Nagarajan S., Chikina M., Thompson M., Oertel M., Baba H., Monga S. P., and Nejak-Bowen K. N. (2016) Wnt signaling regulates hepatobiliary repair following cholestatic liver injury in mice. Hepatology 64, 1652–1666 10.1002/hep.28774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Taneyhill L., and Pennica D. (2004) Identification of Wnt responsive genes using a murine mammary epithelial cell line model system. BMC Dev. Biol. 4, 6 10.1186/1471-213X-4-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chanchevalap S., Nandan M. O., McConnell B. B., Charrier L., Merlin D., Katz J. P., and Yang V. W. (2006) Krüppel-like factor 5 is an important mediator for lipopolysaccharide-induced proinflammatory response in intestinal epithelial cells. Nucleic Acids Res. 34, 1216–1223 10.1093/nar/gkl014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li M., Gu Y., Ma Y.-C., Shang Z.-F., Wang C., Liu F.-J., Cao J.-P., Wan H.-J., and Zhang X.-G. (2015) Krüppel-like factor 5 promotes epithelial proliferation and DNA damage repair in the intestine of irradiated mice. Int. J. Biol. Sci. 11, 1458–1468 10.7150/ijbs.13444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fickert P., Stöger U., Fuchsbichler A., Moustafa T., Marschall H.-U., Weiglein A. H., Tsybrovskyy O., Jaeschke H., Zatloukal K., Denk H., and Trauner M. (2007) A new xenobiotic-induced mouse model of sclerosing cholangitis and biliary fibrosis. Am. J. Pathol. 171, 525–536 10.2353/ajpath.2007.061133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Trauner M., Fickert P., and Wagner M. (2007) MDR3 (ABCB4) defects: a paradigm for the genetics of adult cholestatic syndromes. Semin. Liver Dis. 27, 77–98 10.1055/s-2006-960172 [DOI] [PubMed] [Google Scholar]
- 49. Nandan M. O., Chanchevalap S., Dalton W. B., and Yang V. W. (2005) Krüppel-like factor 5 promotes mitosis by activating the cyclin B1/Cdc2 complex during oncogenic Ras-mediated transformation. FEBS Lett. 579, 4757–4762 10.1016/j.febslet.2005.07.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Guo P., Zhao K.-W., Dong X.-Y., Sun X., and Dong J.-T. (2009) Acetylation of KLF5 alters the assembly of p15 transcription factors in transforming growth factor-β-mediated induction in epithelial cells. J. Biol. Chem. 284, 18184–18193 10.1074/jbc.M109.007096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nandan M. O., Yoon H. S., Zhao W., Ouko L. A., Chanchevalap S., and Yang V. W. (2004) Krüppel-like factor 5 mediates the transforming activity of oncogenic H-Ras. Oncogene 23, 3404–3413 10.1038/sj.onc.1207397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nandan M. O., Ghaleb A. M., Liu Y., Bialkowska A. B., McConnell B. B., Shroyer K. R., Robine S., and Yang V. W. (2014) Inducible intestine-specific deletion of Krüppel-like factor 5 is characterized by a regenerative response in adult mouse colon. Dev. Biol. 387, 191–202 10.1016/j.ydbio.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Aizawa K., Suzuki T., Kada N., Ishihara A., Kawai-Kowase K., Matsumura T., Sasaki K., Munemasa Y., Manabe I., Kurabayashi M., Collins T., and Nagai R. (2004) Regulation of platelet-derived growth factor-A chain by Krüppel-like factor 5. J. Biol. Chem. 279, 70–76 10.1074/jbc.M306621200 [DOI] [PubMed] [Google Scholar]
- 54. Hayashi S., Manabe I., Suzuki Y., Relaix F., and Oishi Y. (2016) Klf5 regulates muscle differentiation by directly targeting muscle-specific genes in cooperation with MyoD in mice. Elife 5, e17462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tanimizu N., Kikkawa Y., Mitaka T., and Miyajima A. (2012) α1- and α5-containing laminins regulate the development of bile ducts via β1 integrin signals. J. Biol. Chem. 287, 28586–28597 10.1074/jbc.M112.350488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Williams M. J., Clouston A. D., and Forbes S. J. (2014) Links between hepatic fibrosis, ductular reaction, and progenitor cell expansion. Gastroenterology 146, 349–356 10.1053/j.gastro.2013.11.034 [DOI] [PubMed] [Google Scholar]
- 57. Shinoda Y., Ogata N., Higashikawa A., Manabe I., Shindo T., Yamada T., Kugimiya F., Ikeda T., Kawamura N., Kawasaki Y., Tsushima K., Takeda N., Nagai R., Hoshi K., Nakamura K., et al. (2008) Krüppel-like factor 5 causes cartilage degradation through transactivation of matrix metalloproteinase 9. J. Biol. Chem. 283, 24682–24689 10.1074/jbc.M709857200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kok C. Y., Cunningham S. C., Carpenter K. H., Dane A. P., Siew S. M., Logan G. J., Kuchel P. W., and Alexander I. E. (2013) Adeno-associated virus-mediated rescue of neonatal lethality in argininosuccinate synthetase-deficient mice. Mol. Ther. 21, 1823–1831 10.1038/mt.2013.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Osterwalder M., Galli A., Rosen B., Skarnes W. C., Zeller R., and Lopez-Rios J. (2010) Dual RMCE for efficient re-engineering of mouse mutant alleles. Nat. Methods 7, 893–895 10.1038/nmeth.1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Okabe M., Tsukahara Y., Tanaka M., Suzuki K., Saito S., Kamiya Y., Tsujimura T., Nakamura K., and Miyajima A. (2009) Potential hepatic stem cells reside in EpCAM+ cells of normal and injured mouse liver. Development 136, 1951–1960 10.1242/dev.031369 [DOI] [PubMed] [Google Scholar]
- 61. el Marjou F., Janssen K.-P., Chang B. H., Li M., Hindie V., Chan L., Louvard D., Chambon P., Metzger D., and Robine S. (2004) Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 10.1002/gene.20042 [DOI] [PubMed] [Google Scholar]
- 62. Tanimizu N., Nishikawa M., Saito H., Tsujimura T., and Miyajima A. (2003) Isolation of hepatoblasts based on the expression of Dlk/Pref-1. J. Cell Sci. 116, 1775–1786 10.1242/jcs.00388 [DOI] [PubMed] [Google Scholar]
- 63. Oishi Y., Manabe I., Tobe K., Ohsugi M., Kubota T., Fujiu K., Maemura K., Kubota N., Kadowaki T., and Nagai R. (2008) SUMOylation of Krüppel-like transcription factor 5 acts as a molecular switch in transcriptional programs of lipid metabolism involving PPAR-δ. Nat. Med. 14, 656–666 10.1038/nm1756 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.