Abstract
Dysregulation of lipid metabolism is common in cancer cells, but the underlying mechanisms are poorly understood. Sterol regulatory element–binding proteins (SREBPs) stimulate lipid biosynthesis through transcriptional activation of lipogenic enzymes. However, SREBPs' roles and potential interacting partners in cancer cells are not fully defined. Using a biochemical approach, we found here that pyruvate kinase M2 (PKM2) physically interacts with the nuclear form of SREBP-1a (nBP1a), by binding to amino acids 43–56 in nBP1a. We also found that PKM2 activates SREBP target gene expression and lipid biosynthesis by stabilizing nBP1a proteins. Using a competitive peptide inhibitor to block the formation of the SREBP-1a/PKM2 complex, we observed that this blockade inhibited lipogenic gene expression. Of note, nBP1a phosphorylation at Thr-59 enhanced the binding to PKM2 and promoted cancer cell growth. Moreover, we show that PKM2 phosphorylates Thr-59 in vitro. Lastly, in human patients with hepatocellular carcinoma, nBP1a phosphorylation at Thr-59 was negatively correlated with clinical outcomes. Together, our results reveal that nBP1a/PKM2 interaction activates lipid metabolism genes in cancer cells and that Thr-59 phosphorylation of SREBP-1a plays an important role in cancer cell proliferation.
Keywords: lipid metabolism, gene transcription, hepatocellular carcinoma, lipid synthesis, lipogenesis, cell proliferation, liver cancer, pyruvate kinase, PKM2, SREBP-1a, sterol regulatory element–binding protein
Introduction
Lipids are essential for cell proliferation. In fact, it has been estimated that more than 90% fatty acids in tumors cells are derived from de novo biosynthesis, whereas normal cells obtain lipids primarily from the circulation (1). In several types of cancer, including breast and prostate cancer, the fatty-acid synthase (Fasn) gene is up-regulated (2), suggesting that fatty acid biosynthesis may play a role in cancer pathogenesis. Sterol regulatory element–binding proteins (SREBPs)6 are conserved transcription factors that can activate the transcription of genes encoding the key lipogenic enzymes, including Fasn (3, 4). In mammalian cells, there are three SREBP isoforms (SREBP-1a, -1c, and -2) encoded by two different genes, Srebf1 and Srebf2 (5). Two distinct promoters generate SREBP-1a and SREBP-1c isoforms from the Srebf1 gene (5). Interestingly, Srebf1a is expressed at higher levels in cancer cells as compared with normal tissues, whereas Srebf1c is the predominant product of Srebf1 in normal tissues (6). In addition, SREBP-1a is a potent transcriptional activator for all known SREBP target genes (7). The SREBP-1 protein levels are often correlated with tumor size, histological grade, and metastasis, and SREBP-1 loss-of-function inhibits cell proliferation and induces apoptosis, cell migration, and invasion in liver, ovarian, and endometrial cancers (8–11). Furthermore, genetic depletion or pharmacological inhibition of SREBP-1 has been shown to suppress the epidermal growth factor receptor-induced glioblastoma (12). Blocking the SREBP pathway prevents hepatocellular carcinoma (HCC) in mouse models (13). Thus, SREBP-1 is required to support proliferation in some cancer cells.
Previous studies have shown that pyruvate kinase isoform M2 (PKM2) plays an important role in the Warburg effect (14). PKM2 and its isoform PKM1 are products of the Pkm gene through alternative splicing (15). It has been reported that in most tumors, the Pkm2 mRNA is elevated (16, 17), and in some cases, the Pkm gene expression is switched from Pkm1 to Pkm2 (17). Several studies have revealed that PKM2 can translocate into the nucleus and acts as a transcriptional cofactor to promote tumor development (18–20).
Here, we identified a novel SREBP-1a/PKM2 protein complex. We show that PKM2 stimulates SREBP-1-dependent cancer cell proliferation and activates the SREBP target gene expression by stabilizing nuclear SREBP-1a proteins.
Results
Nuclear SREBP-1a interacts with PKM2 in cancer cells
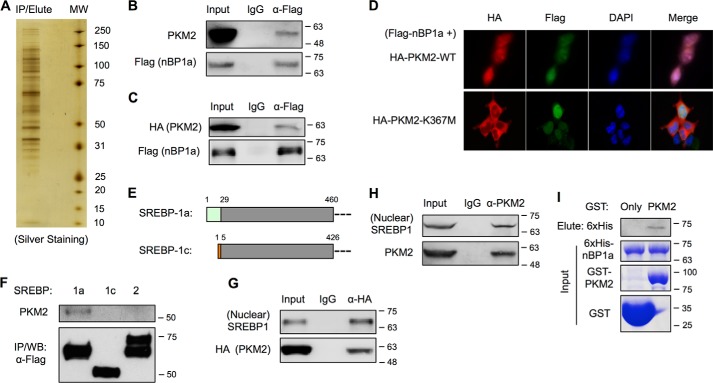
Protein/protein interaction is one of the key regulatory mechanisms in biology. To better understand the regulation of SREBP-1a in the nucleus, we overexpressed a FLAG-tagged nuclear form of SREBP-1a (FLAG–nBP1a) in HEK293T cells and screened for novel SREBP-1a–binding proteins by co-immunoprecipitation (co-IP) of nuclear extracts (Fig. 1A). Among the eluted proteins following IP with anti-FLAG antibody, we detected multiple peptides of PKM2 proteins by LC-tandem MS (data not shown), indicating that PKM2 binds to FLAG–nBP1a. The interaction between SREBP-1a and PKM2 was independently confirmed by multiple lines of evidence. By immunoblotting of IP samples, we found that overexpressed FLAG–nBP1a could pull down endogenous PKM2 (Fig. 1B) and overexpressed WT PKM2 (HA–PKM2–WT) (Fig. 1C). Immunostaining analyses revealed that FLAG–nBP1a proteins were co-localized with HA–PKM2–WT in the nuclei, whereas FLAG–nBP1a and cytoplasmically localized K367M mutant PKM2 proteins (HA–PKM–K367M) (19) were rarely co-localized (Fig. 1D). Moreover, although SREBP-1a and SREBP-1c differ only at the very N-terminal ends (Fig. 1E), PKM2 only interacted with SREBP-1a but not SREBP-1c or SREBP-2 (Fig. 1F). Conversely, both overexpressed HA–PKM2–WT (Fig. 1G) and endogenous PKM2 (Fig. 1H) could pull down the endogenous nuclear form of SREBP-1 proteins, which are presumably the SREBP-1a isoform because of the higher expression of its mRNA (6). To determine whether PKM2 directly interacts with SREBP-1a, we prepared GST fusion proteins of PKM2 and 6xHis-tagged nuclear form of SREBP-1a. As shown in Fig. 1I, recombinant PKM2 could also bind to recombinant SREBP-1a in GST pulldown assays. Thus, our data demonstrate that PKM2 directly interacts with the nuclear form of SREBP-1a in cancer cells.
Figure 1.
SREBP-1a interacts with PKM2. A, silver staining of nBP1a-binding proteins that were immunoprecipitated from nuclear extracts of HEK293T cells stably transfected with FLAG–nBP1a. B, co-IP analysis of endogenous PKM2 binding to overexpressed FLAG–nBP1a in HepG2 cells. The presence of PKM2 in IP eluates was analyzed by immunoblotting using anti-PKM2 antibody. C, co-IP analysis of overexpressed HA-PKM2 binding to overexpressed FLAG–nBP1a in HepG2 cells. D, immunostaining to analyze the localization of co-transfected HA-PKM2 and FLAG–nBP1a in HepG2 cells. E, diagram shows the sequence difference between SREBP-1a and SREBP-1c. F, co-IP analysis of endogenous PKM2 binding to overexpressed FLAG-tagged nuclear forms of SREBP-1a, SREBP-1c, and SREBP-2 in HEK293T cells. G, co-IP analysis of overexpressed HA-PKM2 binding to endogenous nuclear form of SREBP-1 in HepG2 cells. H, co-IP analysis of endogenous PKM2 binding to endogenous nuclear form of SREBP-1 in HepG2 cells. I, GST pulldown analysis of interaction between recombinant PKM2 and SREBP-1a. WB, Western blotting.
PKM2 regulates nuclear SREBP-1 protein stability
To understand the significance of PKM2 binding to SREBP-1a in cancer cells, we analyzed PKM2 and SREBP-1 proteins (likely including both SREBP-1a and SREBP-1c) in tumor samples from patients with HCC by immunohistochemistry (Fig. 2A). Semi-quantitative analyses of samples from 75 patients revealed that the protein levels of SREBP-1 were significantly correlated with those of PKM2 in human HCC (Fig. 2B). These data suggest that PKM2 may increase SREBP-1 protein stability in HCC cells, because nuclear SREBP-1 proteins are normally unstable. To examine this possibility, we knocked down PKM2 in HepG2 cells by PKM2-specific siRNA. Nonspecific (NS) siRNA was used as control. Indeed, PKM2 knockdown significantly reduced the protein levels of nuclear SREBP-1, whereas the precursor form of SREBP-1 was essentially not affected (Fig. 2C). Moreover, PKM2 knockdown did not change the mRNA levels of the Srebf1 gene in HepG2 cells (data not shown). These data indicate that PKM2 positively regulates nuclear SREBP-1 protein levels.
Figure 2.
Nuclear SREBP-1 protein stability is enhanced by PKM2. A, sections of human HCC tissues were stained with anti-SREBP-1 or anti-PKM2 antibody. Representative tissue images are shown (scale bar, 50 μm). B, semi-quantitative analyses of immunohistochemistry data of human HCC tissues for PKM2 or SREBP-1. Correlation between PKM2 and SREBP-1a was analyzed by Spearman rank correlation analysis. C, effects of PKM2 knockdown by siRNA on endogenous SREBP-1 protein levels in HepG2 cells. D, effects of PKM2 knockdown on the degradation of overexpressed FLAG–nBP1a in the presence of CHX. E, semi-quantitative analyses by densitometry of the effects of PKM2 knockdown on the degradation of overexpressed FLAG–nBP1a (n = 3). F, effects of the proteasome inhibitor MG132 on PKM2 regulation of overexpressed FLAG–nBP1a.
To determine whether PKM2 increases nuclear SREBP-1 protein stability, we overexpressed FLAG–nBP1a in HepG2 cells and treated with cycloheximide (CHX), a protein synthesis inhibitor, for various periods of time with or without PKM2 knockdown. As shown in Fig. 2, D and E, PKM2 knockdown accelerated nBP1a degradation. It has been shown that nuclear SREBP-1 proteins are degraded by proteasomes (22). Therefore, we examined whether inhibition of proteasomes could abolish PKM2 regulation of nBP1a. For that purpose, we overexpressed FLAG–nBP1a in HepG2 cells and then treated with MG132, a proteasome inhibitor, with or without PKM2 knockdown. As shown in Fig. 2F, PKM2 knockdown-caused reduction of nBP1a was blocked in the presence of MG132, suggesting that PKM2 inhibits proteasome-mediated degradation of nuclear SREBP-1a.
Thr-59 phosphorylation of nuclear SREBP-1a enhances its protein stability and binding to PKM2
Because serine or threonine phosphorylation regulates nuclear SREBP-1 protein stability (22, 23), we wondered whether PKM2 regulates nBP1a through phosphorylation. To this end, we immunoprecipitated endogenous SREBP-1 from HepG2 cells that were treated with PKM2-specific or control (NS) siRNA, and we analyzed serine or threonine phosphorylation using anti-phosphoserine (pSer) or anti-phosphothreonine (pThr) antibody. As shown in Fig. 3A, when compared with a similar amount of SREBP-1 proteins, PKM2 knockdown in HepG2 cells did not affect serine phosphorylation of nuclear SREBP-1, although it reduced threonine phosphorylation. Together with the data in Fig. 2C, PKM2 positively controls the protein levels and threonine phosphorylation of nuclear SREBP-1a. Therefore, it is less likely that PKM2 regulates phosphorylation of the previously identified Thr-426 residue of nBP1a (22, 23), as reducing Thr-426 phosphorylation would increase nBP1a protein levels.
Figure 3.
Thr-59 phosphorylation of nuclear SREBP-1a enhances its protein stability and binding to PKM2. A, effects of PKM2 knockdown on the phosphorylation of nuclear SREBP-1 in HepG2 cells. B, GST pulldown analyses of FLAG–PKM2 binding to the indicated fragments of SREBP-1a. C, immunoblotting of the indicated mutants of FLAG–nBP1a in HepG2 cells. D, effects of PKM2 knockdown on the protein levels of WT and T59A mutant FLAG–nBP1a. E, effects of PKM2 knockdown on the phosphorylation of FLAG–nBP1a at threonine residues. (The immunoprecipitated FLAG–nBP1a proteins were normalized to the same level before comparison for phosphorylation.) F, effects of PKM2 knockdown and T27A or T59A mutation on FLAG–nBP1a ubiquitination. G, effects of PKM2 knockdown on the degradation of overexpressed WT and T59A mutant FLAG–nBP1a in the presence of CHX (n = 3). H, effects of PKM2 on SREBP-1a phosphorylation by in vitro kinase assays. I, effects of WT and K367M mutant PKM2 on WT, T27A, and T59A mutant SREBP-1a phosphorylation in vitro.
To identify the novel threonine residue(s) in nBP1a whose phosphorylation is regulated by binding to PKM2, we decided to identify which domain(s) of nBP1a are responsible for interacting with PKM2. For that purpose, we generated three truncated forms of nBP1a as GST fusion proteins (Fig. 3B), and we examined their ability to interact with PKM2 by GST pulldown assays. As shown in Fig. 3B, when incubated with recombinant FLAG-tagged PKM2 proteins followed by an extensive wash, the full-length and amino acids (aa) 1–209, but not aa 61–273 or 323–490, fragment of nBP1a could pull down PKM2. These data suggests that the 1–60-aa region of SREBP-1a is required for interaction with PKM2.
There are two threonine residues (Thr-27 and Thr-59) within the 1–60-aa region of SREBP-1a. To determine whether Thr-27 and/or Thr-59 phosphorylation affects nBP1a protein stability and its binding to PKM2, we generated phospho-defective mutant (T27A or T59A) of nBP1a. As shown in Fig. 3C, the T59A, but not T27A, mutant of FLAG–nBP1a was expressed at a level significantly lower than wildtype (WT) in HepG2 cells, indicating that Thr-59 phosphorylation is responsible for enhancing the nBP1a protein stability. Moreover, the T59A mutation not only decreased nBP1a protein levels but also abolished PKM2 knockdown-induced reduction of nBP1a proteins (Fig. 3D). To compare the phosphorylation levels, we normalized the immunoprecipitated FLAG–nBP1a proteins to the same level among different samples before immunoblotting. As shown in Fig. 3E, the T59A mutation reduced threonine phosphorylation in nBP1a and abolished the PKM2 knockdown-caused reduction of threonine phosphorylation (Fig. 3E).
Because ubiquitination controls nuclear SREBP-1 protein degradation (22), we examined the effects of PKM2 knockdown on ubiquitination of FLAG–nBP1a. To examine the short-lived polyubiquitinated FLAG–nBP1a, we co-expressed HA-tagged ubiquitin (HA-Ub) with FLAG–nBP1a in HepG2 cells followed by MG132 treatment. The ubiquitination levels of immunoprecipitated FLAG–nBP1a proteins were analyzed by immunoblotting using anti-HA antibody. As shown in Fig. 3F, PKM2 knockdown significantly increased nBP1a ubiquitination, correlated with the reduced levels of nBP1a proteins (Fig. 3C). Consistent with the data in Fig. 3C, T59A, but not T27A, mutation also increased nBP1a ubiquitination, whereas PKM2 knockdown did not result in any further increase of nBP1a ubiquitination (Fig. 3F), suggesting that PKM2-regulated Thr-59 phosphorylation blocks ubiquitination and thus increases the stability of nBP1a proteins. To further examine this possibility, we transfected HepG2 cells with FLAG–nBP1a (WT or T59A) after PKM2 knockdown, and we then treated cells with CHX for various periods of time, similar to Fig. 2D. Semi-quantitative analyses of immunoblotting data indicate that PKM2 knockdown or T59A mutation similarly accelerated the degradation of nBP1a (Fig. 3G). Together, these results indicate that Thr-59 phosphorylation is a novel mechanism for enhancing the nBP1a protein stability.
PKM2 has been reported as a threonine/serine kinase of proteins, including histone H3 (24) and MLC2 (25). Thus, we examined whether PKM2 could directly phosphorylate SREBP-1a. For that purpose, we performed in vitro kinase assays using FLAG-tagged PKM2 proteins that were overexpressed and immunopurified from HEK293T cell lysates as kinase and recombinant GST–nBP1a proteins as substrate (24). We detected phosphothreonine signals in recombinant SREBP-1a only when the PKM2 substrate phosphoenolpyruvate was added to the reactions (Fig. 3H). Importantly, WT PKM2 becomes unable to phosphorylate SREBP-1a when Thr-59 is mutated to alanine (Fig. 3I), suggesting that Thr-59 contributes to the phosphorylation signals. Moreover, PKM2 with K367M mutation, which is kinase-dead according to the previous studies (19), is unable to increase threonine phosphorylation in SREBP-1a. Together, our data suggest that PKM2 is able to directly phosphorylate SREBP-1a at Thr-59.
Thr-59 phosphorylation of nuclear SREBP-1a is correlated with poor prognosis in HCC patients
To determine the clinical significance of nBP1a phosphorylation at the Thr-59 residue, we generated an antibody against Thr-59-phosphorylated SREBP-1 (p-Thr-59–BP1). The specificity of this antibody for immunohistochemistry was first validated in HepG2 cells with overexpression of the FLAG–nBP1a–WT or –T59A mutant, which serves as a negative control (Fig. 4A). Then we performed phosphopeptide competition analyses in immunohistochemistry assays of HepG2 cells (data not shown) and liver samples from patients with HCC (Fig. 4B) to further confirm its specificity. Using this antibody, we examined the p-Thr-59–BP1 levels in both tumors and the adjacent normal tissues from a total of 90 patients with HCC by immunohistochemistry. Overall, the p-Thr-59–BP1 signals were detected mainly in the nuclei, and tumor samples displayed significantly stronger p-Thr-59–BP1 signals than adjacent normal liver tissues (Fig. 4C), indicating that Thr-59 phosphorylation occurs mainly in nuclear SREBP-1, and Thr-59 phosphorylation of SREBP-1 is increased in human HCC. Using a scale of 0 to 7 (in which 0 to 3 stands for low and 4 to 7 for high p-Thr-59–BP1 signals) to quantify SREBP-1 phosphorylation, we found that the median survival duration was 41.9 and 28.1 months for patients with low and high scores of p-Thr-59–BP1, respectively (Fig. 4D). The Kaplan-Meier survival curve showed that the p-Thr-59–BP1 levels were negatively and significantly correlated with the survival duration of HCC patients (Fig. 4D). Moreover, the p-Thr-59–BP1 levels were significantly higher in tumor tissues than in adjacent normal tissues from this group of patients with HCC (Fig. 4E). By multivariate statistical analyses, we found that the p-Thr-59–BP1 levels were significantly correlated with histologic grade (p = 0.008) and TNM stage (p = 0.030) (Table 1 and Fig. 4, F and G). sing a Cox multivariate model, we found that the p-Thr-59–BP1 level was an independent predictor of HCC patient survival (hazard ratio = 2.166) (95% confidence interval: 1.262U-3.567). Together, our results revealed a good correlation between Thr-59 phosphorylation of nuclear SREBP-1 proteins and the clinical outcomes of patients with HCC.
Figure 4.
Phosphorylation of SREBP-1a at the Thr-59 residue predicts the poor prognosis of HCC. A, validation of the anti-p-Thr-59–SREBP-1 antibody by immunohistochemistry analyses of HepG2 cells with transfections of vector, wildtype (WT), or T59A mutant of nBP1a. B, effects of pretreatment with nonphosphopeptides and phosphopeptides on the immunohistochemistry signals, which were generated using the anti-p-Thr-59–SREBP-1 antibody, in liver samples from HCC patients. C, representative images of histopathologic sections of human HCC and adjacent normal tissues that were analyzed by immunohistochemistry using anti-p-Thr-59–SREBP-1 antibody or H&E staining. Low and high staining groups were categorized according to the immunohistochemistry scores of SREBP-1 Thr-59 phosphorylation. (Scale bar, 50 μm.) D, Kaplan-Meier analysis of overall survival in correlation with the levels of SREBP-1 Thr-59 phosphorylation. E, semi-quantitative analyses of immunohistochemistry data of human HCC and adjacent normal tissues for SREBP-1 Thr-59 phosphorylation. F, correlation analysis of staining score of SREBP-1 Thr-59 phosphorylation and tumor grade of human HCC. G, correlation analysis of staining score of SREBP-1 Thr-59 phosphorylation and tumor stage of human HCC.
Table 1.
SREBP-1 phosphorylation at Thr-59 in HCC
| Variable | N | Thr-59 phosphorylation |
p value | |
|---|---|---|---|---|
| Low | High | |||
| Age (years) | ||||
| <50 | 31 | 16 | 15 | |
| ≥50 | 59 | 20 | 39 | 0.896 |
| Gender | ||||
| Female | 9 | 4 | 5 | |
| Male | 81 | 32 | 49 | 0.142 |
| Tumor size (cm) | ||||
| ≤5 | 36 | 15 | 21 | |
| >5 | 54 | 21 | 33 | 0.085 |
| Histologic grade | ||||
| I and II | 64 | 32 | 32 | |
| III | 26 | 4 | 22 | 0.008 |
| TNM stage | ||||
| I and II | 38 | 21 | 17 | |
| III and IV | 52 | 15 | 37 | 0.030 |
| Liver cirrhosis | ||||
| + | 57 | 23 | 34 | |
| − | 33 | 13 | 20 | 0.629 |
Thr-59 phosphorylation of nuclear SREBP-1a is critical for cancer cell proliferation
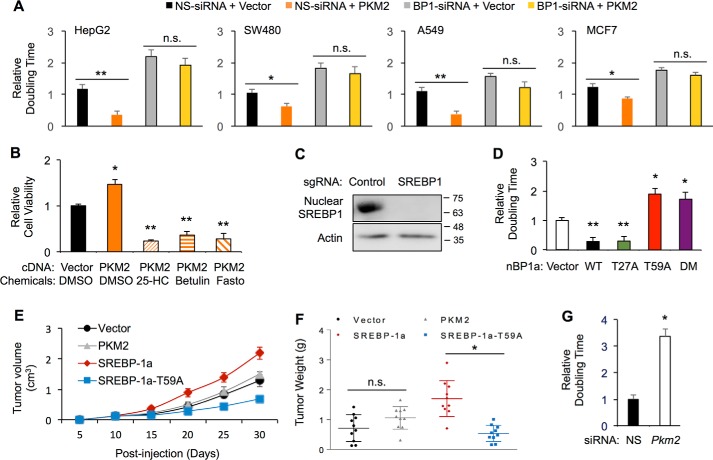
Consistent with the oncogenic property of PKM2, overexpression of PKM2 in HepG2 cells decreased the cell doubling time, indicating an increase of cell proliferation (Fig. 5A). However, when SREBP-1 was depleted by specific siRNA, PKM2 overexpression was no longer able to enhance HepG2 cell proliferation (Fig. 5A), suggesting that SREBP-1 is required for PKM2-induced cell proliferation. The SREBP-1 dependence for PKM2-enhanced cell proliferation may not be limited to HCC, as we obtained similar results in SW480, A549, and MCF7 cells (Fig. 5A). To further determine the requirement of SREBP-1 for PKM2-induced cell proliferation, we treated HepG2 cells with chemical SREBP inhibitors, 25-hydroxyl cholesterol, betulin, or fatostatin. As shown in Fig. 5B, in the presence of these SREBP inhibitors, PKM2 overexpression was unable to promote HepG2 cell growth.
Figure 5.
PKM2 regulates SREBP-1-dependent cancer cell growth. A, effects of SREBP-1 depletion and PKM2 overexpression on cell growth of HepG2, SW480, A549, or MCF7 cells. B, effects of the indicated compounds on PKM2-induced increase of HepG2 cell growth. C, generation of SREBP-1-knockout HepG2 cells. D, effects of overexpressed WT or the indicated mutant nBP1a on HepG2 cell growth. E, growth curve of xenograft tumors from SREBP-1 knockout HepG2 cells that were stably transfected with vector control, PKM2, WT, or T59A mutant nBP1a in nude mice. F, weight of tumors from stably transfected SREBP-1 knockout HepG2 cells when mice were sacrificed. G, effects of PKM2 knockdown on HepG2 cell growth. (*, p < 0.05; **, p < 0.01 versus control; n.s., not significant.)
To further study the function of SREBP-1, we generated a HepG2 cell line of SREBP-1-knockout (BP1-KO) using the CRISPR/Cas9 technology (Fig. 5C). To determine the role of PKM2 regulating nBP1a in cell proliferation, we transfected HepG2 cells with WT, T27A, and T59A or T27A/T59A double mutant of FLAG–nBP1a (vector as transfection control). As shown in Fig. 5D, overexpression of either WT or T27A mutant nBP1a significantly increased cell growth as evidenced by the reduced cell doubling time. In contrast, overexpression of T59A single mutant or T59A/T27A double mutant nBP1a inhibited the growth of HeG2 cells (Fig. 5D).
To determine the role of PKM2 binding to nBP1a in tumor development in vivo, we established mouse xenograft models by subcutaneously injecting nude mice with stable BP1-KO HepG2 cells that express FLAG–PKM2 (WT) or FLAG–nBP1a (WT or T59A). Stable transfection with the empty vector served as a control. As shown in Fig. 5E, when compared with vector alone, PKM2 overexpression on the SREBP-1-knockout background did not alter the rate of tumor growth and thus resulted in tumors with weights similar to those from vector control (Fig. 5F). These data are consistent with that from cell culture (Fig. 5A) and indicate that PKM2 regulation of cell proliferation is SREBP-1-dependent in vivo. Moreover, overexpression of nBP1a–WT on the SREBP-1-knockout background significantly accelerated tumor growth (Fig. 5E) and gave rise to larger tumors than vector control (Fig. 5F), suggesting a positive role of SREBP-1a on HepG2 cell proliferation in vivo. In contrast, tumors originated from nBP1a–T59A expressing cells grew with slower rates (Fig. 5E) and gave rise to smaller tumors (Fig. 5F) than those from nBP1a–WT-expressing cells, consistent with the cell culture data (Fig. 5D). Moreover, PKM2 knockdown in HepG2 cells by specific siRNA increased the cell doubling time, indicating reduced cell proliferation (Fig. 5G). Together, our in vitro and in vivo results suggest that Thr-59 phosphorylation of SREBP-1a is required for HCC cancer cell proliferation.
PKM2 activates lipogenic gene expression and lipid biosynthesis
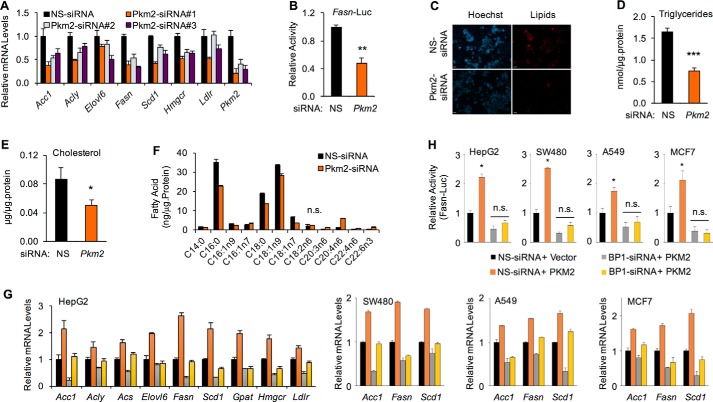
The well-defined SREBP-1a target genes include those encoding enzymes in both fatty acids and cholesterol biosynthesis pathways (3, 4). Because PKM2 regulates the abundance of nuclear SREBP-1a (Fig. 2C), we examined whether PKM2 regulates SREBP-1a–mediated transcription. First, we analyzed the mRNA levels of SREBP-1a target genes after PKM2 knockdown. We used three different PKM2–siRNAs to exclude potential off-target effects, and the PKM2 knockdown was efficient (Figs. 2C and 6A). As shown in Fig. 6A, PKM2 knockdown significantly reduced the mRNA levels of previously defined SREBP-1a target genes, including Acc1, Acly, Elovl6, Fasn, Scd1, Hmgcr, and Ldlr, in HepG2 cells, and the change of gene expression was correlated with the PKM2 knockdown efficiency. By luciferase reporter assays, we found that PKM2 knockdown significantly reduced the activity of human Fasn promoter (Fig. 6B), which has a size of 175 bp, but includes all SREBP-1-binding sequences. Consistent with the decrease of lipogenic gene expression, PKM2 knockdown caused a significant reduction of lipid levels in HepG2 cells as assayed by both lipid staining (Fig. 6C) and quantitative measurements of cellular triglycerides (Fig. 6D) as well as cholesterol (Fig. 6E). To further determine the effect of PKM2 knockdown on lipid levels, we quantitatively analyzed fatty acid profiles. Consistent with the gene expression data, the amount of some fatty acids, including C16:0, C18:0, and C18:1, was significantly reduced after PKM2 knockdown (Fig. 6F).
Figure 6.
PKM2 stimulates lipid biosynthesis in cancer cells. A, real-time RT-PCR analysis of the effects of PKM2 knockdown in HepG2 cells by three independent siRNA (NS-siRNA as control) on the mRNA of indicated genes. (All significantly different except as indicated.) B, effects of PKM2 knockdown in HepG2 cells by siRNA on the activity of human Fasn promoter as detected by luciferase reporter assay. C, effects of PKM2 depletion by siRNA on cellular lipid levels in HepG2 cells, which were stained with LipidTOX (red). The nuclei were stained with Hoechst 3342 (blue). (Scale bar, 50 μm.) D, effects of PKM2 depletion on cellular triglyceride levels in HepG2 cells. E, effects of PKM2 depletion on cellular cholesterol levels in HepG2 cells. F, LC-MS analysis of the effects of PKM2 depletion on fatty acid profiles in HepG2 cells. (All significantly different except as indicated.). G, real-time RT-PCR analysis of the effects of SREBP-1 depletion and PKM2 overexpression in HepG2, SW480, A549, or MCF7 cells on the mRNA of indicated genes. H, effects of SREBP-1 depletion and PKM2 overexpression in HepG2, SW480, A549, or MCF7 cells on the activity of human Fasn promoter as detected by luciferase reporter assay. (*, p < 0.05; **, p < 0.01; ***, p < 0.01 versus control; n.s., not significant).
To further understand the PKM2 regulation of lipogenic gene expression, we overexpressed PKM2 in HepG2 cells and observed a significant up-regulation of SREBP-1a target genes (Fig. 6G). However, when SREBP-1 was depleted by specific siRNA, PKM2-enhanced lipogenic gene expression was compromised for some of those genes, including Acly, Elvol6, Gpat, and Hmgcr (Fig. 6G). Interestingly, PKM2 could still stimulate the expression of some genes, including Fasn, from lower backgrounds that were caused by SREBP-1 depletion (Fig. 6G). To determine whether PKM2 regulation of SREBP-1a target genes is common in cancer cells, we also examined SW480, A549, and MCF-7 cells by qRT-PCR. As shown in Fig. 6G, we obtained similar results from different cell lines, indicating that PKM2 is a novel activator for lipogenic gene expression in cancer cells. Furthermore, PKM2 overexpression also stimulated the promoter activity of Fasn in those cancer cells (Fig. 6H). However, different from the regulation of endogenous Fasn (Fig. 6G), PKM2 overexpression was unable to activate the short Fasn promoter when SREBP-1 was depleted (Fig. 6H), suggesting that PKM2 regulates endogenous Fasn gene likely through both SREBP-1-dependent and -independent mechanisms. Nevertheless, our results reveal a novel function of PKM2 in regulating lipogenic gene expression and lipid biosynthesis in cancer cells.
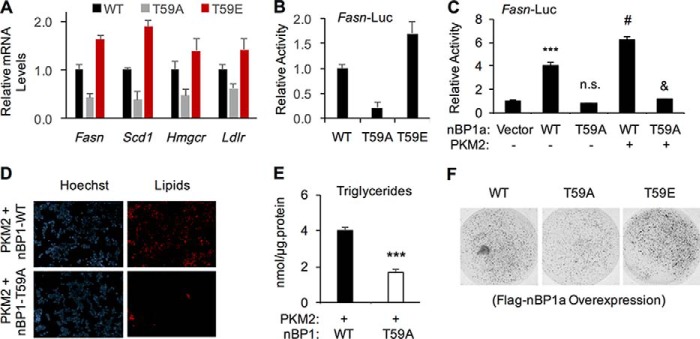
To determine the role of Thr-59 phosphorylation in regulating SREBP-1a target gene expression, we overexpressed FLAG–nBP1a (WT, T59A, or T59E) in SREBP-1 knockout HepG2 cells (Fig. 5C) and examined the mRNA levels by qRT-PCR. When compared with WT, phospho-defective T59A mutant was less potent, whereas the phosphomimic T59E mutant was more potent, to activate SREBP-1a target genes (Fig. 7A). Similarly, T59A mutant was less potent, whereas T59E mutant was more potent, than WT-nBP1a to induce the Fasn promoter in luciferase reporter assays (Fig. 7B). The increased function of the T59E mutant is likely due to the increased stability/levels of this mutant. In addition, overexpression of PKM2 could further increase the transcriptional output of nBP1a–WT but not the T59A mutant (Fig. 7C). Consistent with the gene expression data, PKM2 overexpression in BP1-KO HepG2 cells expressing nBP1a–WT resulted in significantly more lipid accumulation than in those expressing nBP1a–T59A (Fig. 7, D and E). We also established stable cell lines that overexpress FLAG–nBP1a (WT, T59A, or T59E) or FLAG–PKM2 (WT) in BP1-KO HepG2 cells. When compared with HepG2 cells expressing nBP1a–WT, those expressing the T59A mutant were less proliferative in three-dimensional (3D) culture, whereas those expressing T59E mutant were more proliferative (Fig. 7F), which correlated with the pattern of nBP1 protein stability/levels (Fig. 3C).
Figure 7.
Thr-59 phosphorylation of nuclear SREBP-1a promotes lipid biosynthesis and cancer cell proliferation. A, real-time RT-PCR analysis of the effects of overexpressing wildtype (WT), T59A, or T59E mutant nBP1a in SREBP-1-knockout cells on the mRNA levels of indicated genes. B, effects of overexpressing wildtype (WT), T59A, or T59E mutant nBP1a in SREBP-1-knockout cells on the activity of human Fasn promoter as detected by luciferase reporter assay. C, effects of PKM2 overexpression on WT or T59A mutant FLAG–nBP1a -induced activation of human Fasn promoter in HepG2 cells as examined by luciferase reporter assays. D, effects of PKM2 overexpression and/or T59A mutation in nBP1a on lipid accumulation HepG2 cells. Cellular lipids were stained with LipidTOX, and nuclei were stained with Hoechst 3342. (Scale bar, 50 μm.) E, effects of PKM2 overexpression and/or T59A mutation in nBP1a on triglyceride levels in HepG2 cells. F, effects of overexpressed WT, T59A, or T59E mutant nBP1a in SREBP-1 knockout HepG2 cell growth in 3D culture. (***, p < 0.001; n.s., p > 0.05 versus vector alone; #, p < 0.01; &, p < 0.001 versus T59A-nBP1a alone, n = 3.)
Together, these results suggest a model in which PKM2 interaction with nuclear SREBP-1a increases SREBP target gene expression and subsequent lipid biosynthesis in cancer cells, which is at least in part correlated with cancer cell proliferation.
Blocking PKM2 binding to SREBP-1a by a small peptide inhibits lipogenic gene expression
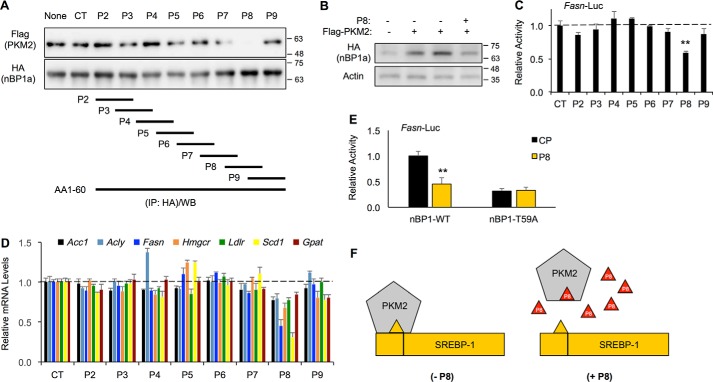
To further determine the role of PKM2 binding to nBP1a, we synthesized eight small peptides covering the 1–60-aa sequence of SREBP-1a and a control peptide (termed CP) (Fig. 8A). The C-terminal ends of those peptides were fused to a cell-penetrating sequence (GRKKRRQRRR) from the Tat proteins to enhance the efficiency of entering into cells. We first examined their effects on the interaction between PKM2 and nBP1a by co-IP. As shown in Fig. 8A, peptide 8 (termed P8), which corresponds to the 43–56-aa sequence of SREBP-1a, could efficiently block endogenous PKM2 binding to overexpressed HA-tagged nBP1a in HepG2 cells. Consistent with the data that PKM2 interaction with nBP1a increases the nBP1a protein stability, P8 could inhibit PKM2 overexpression-induced accumulation nBP1a proteins (Fig. 8B). To determine whether those peptides affect SREBP target gene expression, we examined gene expression in HepG2 cells after the treatment with those peptides. Consistent with the co-IP data, only P8 significantly reduced the Fasn promoter activity in luciferase reporter assays (Fig. 8C). Similarly, only P8 significantly decreased the mRNA levels of endogenous SREBP-1a target genes, including Acc1, Acly, Fasn, Hmgcr, Ldlr, Scd1, and Gpat, as detected by qRT-PCR (Fig. 8D). Moreover, P8 efficiently inhibited WT, but not T59A mutant, nuclear SREBP-1a–induced activation of the Fasn promoter in luciferase reporter assays (Fig. 8E). Thus, we generated a small peptide inhibitor that was able to functionally block the interaction between PKM2 and nBP1a, likely through a mechanism of competition, and our data also suggest that the 43–56-aa sequence of SREBP-1a constitutes a binding surface for PKM2 (Fig. 8F).
Figure 8.
Blocking nBP1a binding to PKM2 by small peptides inhibits gene expression. A, co-IP analysis of overexpressed FLAG–PKM2 binding to HA-nBP1a in HepG2 cells in the presence of peptides P2–P8 (8 μm) or without peptide (CT, peptide solvent control; None, no-treatment control). The lower panel shows the peptide sequences corresponding to aa 1–60 of SREBP-1a. B, effects of 8 μm peptide P8 on overexpressed HA-nBP1a protein levels in HepG2 cells. C, effects of peptides P2–P9 (8 μm) on the Fasn promoter activity in HepG2 cells as detected by luciferase reporter assay. D, effects of treatment with peptides P2–P9 (8 μm) for 24 h on the mRNA levels of indicated genes in HepG2 cells. E, effects of P8 or control peptide CP (8 μm) on the Fasn promoter activity upon overexpressing WT or T59A mutant SREBP-1a in HepG2 cells as detected by luciferase reporter assay. F, suggested model shows how the small peptide P8 may work.
Discussion
In this study, we identified a functional link between oncogenic PKM2 and lipogenic transcription factor SREBP-1a. PKM2 increases the stability of nuclear SREBP-1a proteins by binding to SREBP-1a 43–56 aa. PKM2 binding to nuclear SREBP-1a also enhances the phosphorylation of SREBP-1a at the Thr-59 residue. As a result, PKM2 activates lipogenic gene expression and lipid synthesis and promotes SREBP-1-dependent cancer cell proliferation in cell culture and in xenograft mouse models. Clinically, Thr-59 phosphorylation of nuclear SREBP-1a in tumors is inversely correlated with the overall survival of patients with hepatocellular carcinoma, and the protein levels of PKM2 and SREBP-1 are correlated in HCC tumors, suggesting that the SREBP-1a/PKM2 complex may play a significant role in HCC. Using the biochemical information, we developed a small peptide to disrupt the SREBP-1a/PKM2 complex. Interestingly, this small peptide inhibitor could inhibit SREBP target gene expression in cancer cells. Thus, our data reveal a novel mechanism of PKM2 regulating cancer cell proliferation by stimulating SREBP-1-dependent lipid biosynthesis.
Among the transcriptional activators of lipogenic genes, SREBP-1a is highly expressed in many cancer cell lines and multiple types of cancer, including endometrial cancer, colorectal cancer, and pancreatic cancer (11, 26, 27). The SREBP-1a level is also correlated with cancer progression and metastasis (28, 29). Moreover, the SREBP-1-dependent lipogenesis is critically involved in p53 mutation-associated changes of mammary tissue architecture and tumorigenesis of breast cancer (8). Thus, the SREBP-1a isoform may be more involved in the developmental processes and tumor growth. Consistent with the oncogenic roles of PKM2 and SREBP-1a, our data show that PKM2 only interacts with SREBP-1a, but not SREBP-1c or SREBP-2 (Fig. 1F). The abundance of transcriptionally active SREBP-1a is controlled at the multiple steps, including transcription, proteolytic processing of the precursors, and protein stability of the nuclear forms. In addition to the transcriptional up-regulation of Srebf1a in cancer cells, we have recently shown that increasing the nuclear SREBP-1a abundance by p54nrb/NONO promotes tumorigenesis of breast cancer (21). Multiple lines of evidence in this study indicate that PKM2 also increases the nuclear SREBP-1a protein stability. Thus, regulation of nuclear SREBP-1a protein stability may be an important mechanism in controlling cancer cell proliferation.
Previous studies have demonstrated that nuclear SREBP proteins are degraded through a ubiquitination–proteasome mechanism (22), in which phosphorylation of nuclear SREBP-1a at Thr-426 by GSK3β (22) or CDK8 (23) provides the docking surface for the E3 ligase Fbw7. Although the mechanism(s) underlying PKM2 regulation of nuclear SREBP-1a protein stability are currently unknown, Thr-59 phosphorylation is apparently correlated with the increased stability of nuclear SREBP-1a. Thus, PKM2 regulation of nuclear SREBP-1a abundance is less likely through these mechanisms. Currently, it is unclear how Thr-59 phosphorylation controls the nuclear SREBP-1a protein stability, but phosphorylation at Thr-59 may change SREBP-1a conformation to make it less accessible to Fbw7, as Thr-59 phosphorylation of nuclear SREBP-1a dramatically reduces its ubiquitination. Although PKM2 can phosphorylate SREBP-1a at Thr-59 in vitro, future studies are necessary to further study the kinase(s) that are responsible for Thr-59 phosphorylation. Nevertheless, the accumulated nuclear SREBP-1a proteins seem functional, as we observed an increase of lipogenic gene expression and lipid accumulation in cancer cells. Importantly, our data show that the levels of Thr-59–phosphorylated SREBP-1a are closely correlated with the overall outcome of HCC in human patients, suggesting the clinical significance of Thr-59 phosphorylation of SREBP-1a.
In summary, this study uncovered a novel SREBP-1a/PKM2 protein complex, which promotes cancer cell proliferation by increasing the abundance of nuclear SREBP-1a and subsequent lipid biosynthesis. Our data suggest that the SREBP-1a/PKM2 complex may represent a novel target for fighting some types of cancer.
Experimental procedures
Tissue culture and antibodies
HEK293T, HepG2, SW480, A549, and MCF7 cell lines were purchased from the ATCC and maintained in Dulbecco's modified Eagle's medium or RPMI 1640 medium with 25 mm glucose supplemented with 10% fetal bovine serum, 2 mm glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin sulfate (Gibco). All cells were cultured in humidified incubators at 37 °C and 5% CO2. For gene expression experiments, cells were cultured in lipid-depleted medium. The following antibodies were used: anti-FLAG M2 from Sigma; anti-HA from Covance; anti-SREBP-1 from Santa Cruz Biotechnology; anti-β-actin, anti-PKM2, and anti-phosphoserine (or threonine) from Cell Signaling. MG132 and cycloheximide were purchased from Sigma. Lipofectamine 2000 from Invitrogen was used in transfection according to the manufacturer's protocol.
Co-IP
For endogenous proteins, 5 μl of anti-SREBP-1 antibodies were bound to 20 μl of mixed protein A/G beads (Thermo Fisher Scientific) and incubated with 1 ml of cell extracts in IP buffer for 3 h at 4 °C. For overexpressed FLAG or HA-tagged proteins, 30 μl of anti-FLAG M2 affinity gel (Sigma) or anti-HA–agarose (Thermo Fisher Scientific) was used. The beads were washed for five times with IP buffer. Eluted proteins were subjected to SDS-PAGE and analyzed by immunoblotting using relevant antibodies.
GST pulldown assay
GST-fused PKM2 or SREBP-1a full-length or mutants were expressed in Escherichia coli BL21 and purified using GSH-Sepharose 4B beads (GE Healthcare) according to the manufacturer's protocol. A small amount of proteins was used to verify its expression by Coomassie Blue staining. 6xHis-tagged proteins were prepared and purified using nickel-affinity resins (GE Healthcare). Recombinant 6xHis–nSREBP-1 or cell lysates were mixed with GST or GST–PKM2 beads at 4 °C for 1 h. The beads were then washed with IP buffer five times. The resulting beads were analyzed by immunoblotting using relevant antibodies.
Immunoblotting
Cells were lysed into the RIPA buffer containing protease inhibitor mixtures and ALLN by incubating on ice for 30 min followed by centrifugation at 10,000 × g for 15 min. The extracted proteins were subjected to electrophoresis on SDS-PAGE and transferred to polyvinylidene difluoride membranes (GE Healthcare), which were blocked and probed with specific primary antibodies with appropriate dilution at 4 °C overnight. After three washes with 1× TBST buffer, the membranes were then incubated with the horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature, and following three washes with 1× TBST buffer, the immunoreactive bands were visualized using the ECL Plus System.
Transfection of siRNA
Cells were transfected with oligonucleotide siRNAs using Lipofectamine 2000. The sense sequences of siRNA oligonucleotides used are as follows: Pkm2 #1, AGGCAGAGGCUGCCAUCUATT; Pkm2 #2, CCAUAAUCGUCCUCACCAATT; Pkm2 #3, GCCCGAGGCUUCUUCAAGAAGTT; and Srebf1, GCUGAAUAAAUCUGCUGUCUUTT; NS control, UUCUCCGAACGUGUCACGUTT.
Peptide design and synthesis
The following small peptides were designed based on the 1–60-aa sequence of SREBP-1a: P2, MDEPPFSEAALEQA; P3, EAALEQALGEPCDL; P4, LGEPCDLDAALLTD; P5, DAALLTDIEDMLQL; P6, IEDMLQLINNQDSD; P7, INNQDSDFPGLFDP; P8, FPGLFDPPYAGSGA; and P9, PYAGSGAGGTD.
A transmembrane domain (YGRKKRRQRRR) was conjugated to the C terminus of each peptide to increase the cell penetration. All peptides were synthesized; reversed-phase HPLC was purified to >98% by China Peptides (Shanghai, China), and their molecular weights were verified by electrospray ionization MS.
Luciferase reporter assay
A plasmid containing the human Fasn promoter (175 bp) fused to the firefly luciferase gene in the pGL3 vector (Promega) was used. HepG2 cells with a density of 1 × 104 per well were cultured in 24-well tissue culture plates and co-transfected with the firefly luciferase plasmid and a Renilla luciferase plasmid (as the control) at a ratio of 10:1 in addition to overexpression plasmids or siRNA. About 24 h post-transfection, cell lysates were analyzed by the Dual-LuciferaseTM assay system (Promega) according to the manufacturer's instructions. The ratio of firefly luciferase to Renilla activity was calculated for each of the triplicates.
Real-time RT-PCR (qRT-PCR)
Total RNA was isolated using a TRIzol kit (Omega) and converted to cDNA with a cDNA synthesis kit (Takara). Quantitative real-time PCR was performed using a SYBR Green PCR Master Mix (Takara), and the transcript levels were detected by using the StepOnePlus Real-Time PCR System (Applied Biosystems). The PCR primer sequences are available upon request.
Proliferation and colony formation assay
Cells were plated in 24-well plates with a density of 2 × 104 cells per well. One day after transfection, cells were harvested every 24 h, and live cells were counted using a hemocytometer after trypan blue exclusion. For colony formation assays, cells with various treatments were suspended in culture medium containing 0.3% agarose (Sigma) and placed on top of solidified 0.6% agarose in 24-well plates. After culture for 14 days, colonies were stained with 0.25% crystal violet, and three random regions for each treatment were analyzed under a microscope for the number of colonies.
Three-dimensional (3D) culture
For 3D cell culture, type I collagen (Cell Matrix, Japan), F-12 medium, and reconstitution buffer were mixed in ice-cold water with a ratio of 8:1:1. Cell suspension was added to the collagen solution with a final density of 1.5 × 105 cells/ml. Three drops of the collagen-cell mixture (30 μl per drop) were dropped in each well of a 6-well plate on ice, which was placed at 37 °C in the incubator containing 5% CO2 for 1 h to gelate the collagen-cell mixture. The final concentration was about 3 × 103 cells per collagen-gel droplet. For each well, 3 ml of DF medium (Nissui Pharmaceutical) containing 10% FBS was added, and the plates were incubated in a CO2 incubator at 37 °C overnight. The culture medium was changed every other day. At the end of the experiment, 30 μl of Neutral Red (Kurabo, Japan) solution was added to each well. After incubation at 37 °C for 2 h, cells were washed twice with 1× PBS, fixed 45 min with 10% neutral formalin buffer, washed 20 min in water, air-dried, and analyzed by a high-resolution microscope (VH-5910, Keyence, Japan).
SREBP-1 knockout
The CRISPR/Cas9 target sequence for Srebf1 was TCCAAAGCCGCCTCGCTGAA. Self-complementary gRNA oligonucleotides corresponding to the target site were synthesized by Sangon Biotech (Shanghai, China), annealed, and ligated into pCas9/gRNA using T4 DNA ligase (New England Biolabs). HepG2 cells were transfected with pCas9/gRNA-Srebf1 plasmids using Lipofectamine 2000 before selection with 1 μg/ml puromycin. Single colonies were isolated by limited dilution and expansion. Clones were then genotyped by sequencing and validated by immunoblotting.
Lipid staining
Two days after transfection, HepG2 cells were washed three times with 1× PBS and then fixed with 4% formalin for 20 min at room temperature. After gently rinsing with 1× PBS three times, cells were incubated with 1× LipidTOX (Invitrogen) for 30 min at room temperature. Images were examined and recorded by an Axio Observer Z1 microscope.
Xenograft tumor studies
All experimental procedures using animals were in accordance with the protocol approved by the Animal Ethics Committee of Renji Hospital of Shanghai Jiao Tong University School of Medicine. Tumor cells (5 × 106 per mouse) were harvested by trypsinization, resuspended in culture medium and Matrigel (BD Biosciences), and inoculated subcutaneously in the flank of 4-week-old male BALB/c SCID mice (Shanghai Laboratory Animal Center, Shanghai, China). Eight to 10 mice were used in each group. Tumor growth was assessed twice a week with caliper measurement. Tumor volume was calculated according to the following formula: V = (length × width2)/2. At the end of the experiments, mice were euthanized, and tumor tissues were harvested and weighed. For peptide treatments, mice were randomly divided into two groups when tumor mass was established (∼50 mm in diameter).
Immunohistochemistry
All experiments involving human tissues were approved by the Human Assurance Committee of Renji Hospital of Shanghai Jiao Tong University School of Medicine and were performed in compliance with the Declaration of Helsinki. All tissue samples were obtained from surgical resections agreed upon by patients of Renji Hospital. For immunohistochemical analyses, sections were de-waxed, hydrated, and washed. After microwave antigen retrieval, the slides were treated with 3% H2O2 to block endogenous peroxidase activity and then incubated overnight with anti-PKM2 antibody (1:3000), anti-p-Thr-59 of SREBP-1 (1:1000), or anti-SREBP-1 antibody (1:500). The sections were then incubated with the horseradish peroxidase-conjugated secondary antibody, and the signals were visualized with diaminobenzidine as the chromogen and counter-stained by hematoxylin. The signal intensity of immunohistochemistry was blandly scored by two independent researchers without prior knowledge about the patient and sample. The signal intensity was scored on a scale of 0–3: 0 = negative; 1 = weak; 2 = moderate; and 3 = strong. The percentage of stained cells was also scored on a scale of 0–3: 0 = no staining; 1 = <25%; 2 = 25–50%; and 3 = >50% staining. Thus, the final scores for indicated protein in those liver tissues were on a scale of 0–9, in which a score of 3 or more was defined as high expression and a score less than 3 as low expression.
Statistical analysis
All data were statistically analyzed using Graphpad Prism 6 software or SPSS 20, and presented as mean ± S.E. Unpaired two-tailed t test or analysis of variance test was used to analyze the difference between the two groups. Multivariate logistic regression was used to identify independent risk factors for the expression of p-Thr-59 phosphorylation of SREBP-1 proteins in human liver tissues. Spearman's rank correlation was applied to determine the correlation between PKM2 and SREBP-1 proteins. p values < 0.05 were considered as statistically significant.
Author contributions
X. Z., F. Y., and J. Liu conceptualization; X. Z., L. Z., H. Y., J. Li, and X. M. data curation; X. Z., L. Z., F. Y., J. Liu, and G. H. formal analysis; X. Z., F. Y., J. Liu, and G. H. funding acquisition; X. Z., H. Y., J. Li, and X. M. investigation; X. Z., H. Y., J. Li, and X. M. methodology; X. Z., F. Y., J. Liu, and G. H. writing-original draft; J. Li and X. M. validation; X. M. visualization; F. Y. and J. Liu resources; F. Y., J. Liu, and G. H. project administration; F. Y., J. Liu, and G. H. writing-review and editing; J. Liu supervision.
This work was supported by National Natural Science Foundation of China Grants 81372195, 81471685, 81471687, 81572719, and 81530053; Science Program for Professor of Special Appointment 1410000157; and National Institutes of Health Grants DK110063 and DK020541. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
- SREBP
- sterol regulatory element–binding protein
- aa
- amino acid
- co-IP
- co-immunoprecipitation
- CHX
- cycloheximide
- HCC
- hepatocellular carcinoma
- IP
- immunoprecipitation
- GST
- glutathione S-transferase
- NS
- nonspecific
- qRT
- quantitative PCR.
References
- 1. Medes G., Thomas A., and Weinhouse S. (1953) Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 13, 27–29 [PubMed] [Google Scholar]
- 2. Swinnen J. V., Brusselmans K., and Verhoeven G. (2006) Increased lipogenesis in cancer cells: new players, novel targets. Curr. Opin. Clin. Nutr. Metab. Care 9, 358–365 10.1097/01.mco.0000232894.28674.30 [DOI] [PubMed] [Google Scholar]
- 3. Osborne T. F., and Espenshade P. J. (2009) Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: what a long, strange tRIP it's been. Genes Dev. 23, 2578–2591 10.1101/gad.1854309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jeon T. I., and Osborne T. F. (2012) SREBPs: metabolic integrators in physiology and metabolism. Trends Endocrinol. Metab. 23, 65–72 10.1016/j.tem.2011.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xiaoli, and Yang F. (2013) Mediating lipid biosynthesis: Implications for cardiovascular disease. Trends Cardiovasc. Med. 23, 269–273 10.1016/j.tcm.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shimomura I., Shimano H., Horton J. D., Goldstein J. L., and Brown M. S. (1997) Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 99, 838–845 10.1172/JCI119247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amemiya-Kudo M., Shimano H., Hasty A. H., Yahagi N., Yoshikawa T., Matsuzaka T., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., Osuga J., Harada K., Gotoda T., Sato R., Kimura S., et al. (2002) Transcriptional activities of nuclear SREBP-1a, -1c, and -2 to different target promoters of lipogenic and cholesterogenic genes. J. Lipid Res. 43, 1220–1235 [PubMed] [Google Scholar]
- 8. Freed-Pastor W. A., Mizuno H., Zhao X., Langerød A., Moon S. H., Rodriguez-Barrueco R., Barsotti A., Chicas A., Li W., Polotskaia A., Bissell M. J., Osborne T. F., Tian B., Lowe S. W., Silva J. M., et al. (2012) Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148, 244–258 10.1016/j.cell.2011.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yecies J. L., Zhang H. H., Menon S., Liu S., Yecies D., Lipovsky A. I., Gorgun C., Kwiatkowski D. J., Hotamisligil G. S., Lee C. H., and Manning B. D. (2011) Akt stimulates hepatic SREBP-1c and lipogenesis through parallel mTORC1-dependent and -independent pathways. Cell Metab. 14, 21–32 10.1016/j.cmet.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Williams K. J., Argus J. P., Zhu Y., Wilks M. Q., Marbois B. N., York A. G., Kidani Y., Pourzia A. L., Akhavan D., Lisiero D. N., Komisopoulou E., Henkin A. H., Soto H., Chamberlain B. T., Vergnes L., et al. (2013) An essential requirement for the SCAP/SREBP signaling axis to protect cancer cells from lipotoxicity. Cancer Res. 73, 2850–2862 10.1158/0008-5472.CAN-13-0382-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li W., Tai Y., Zhou J., Gu W., Bai Z., Zhou T., Zhong Z., McCue P. A., Sang N., Ji J. Y., Kong B., Jiang J., and Wang C. (2012) Repression of endometrial tumor growth by targeting SREBP-1 and lipogenesis. Cell Cycle 11, 2348–2358 10.4161/cc.20811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guo D., Prins R. M., Dang J., Kuga D., Iwanami A., Soto H., Lin K. Y., Huang T. T., Akhavan D., Hock M. B., Zhu S., Kofman A. A., Bensinger S. J., Yong W. H., Vinters H. V., et al. (2009) EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci. Signal. 2, ra82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li N., Zhou Z. S., Shen Y., Xu J., Miao H. H., Xiong Y., Xu F., Li B. L., Luo J., and Song B. L. (2017) Inhibition of the SREBP pathway suppresses hepatocellular carcinoma through repressing inflammation. Hepatology 65, 1936–1947 [DOI] [PubMed] [Google Scholar]
- 14. Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., and Cantley L. C. (2008) The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature 452, 230–233 10.1038/nature06734 [DOI] [PubMed] [Google Scholar]
- 15. David C. J., Chen M., Assanah M., Canoll P., and Manley J. L. (2010) hnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 463, 364–368 10.1038/nature08697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bluemlein K., Grüning N. M., Feichtinger R. G., Lehrach H., Kofler B., and Ralser M. (2011) No evidence for a shift in pyruvate kinase PKM1 to PKM2 expression during tumorigenesis. Oncotarget 2, 393–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Desai S., Ding M., Wang B., Lu Z., Zhao Q., Shaw K., Yung W. K., Weinstein J. N., Tan M., and Yao J. (2014) Tissue-specific isoform switch and DNA hypomethylation of the pyruvate kinase PKM gene in human cancers. Oncotarget 5, 8202–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Luo W., Hu H., Chang R., Zhong J., Knabel M., O'Meally R., Cole R. N., Pandey A., and Semenza G. L. (2011) Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 145, 732–744 10.1016/j.cell.2011.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang W., Xia Y., Ji H., Zheng Y., Liang J., Huang W., Gao X., Aldape K., and Lu Z. (2011) Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature 480, 118–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao X., Wang H., Yang J. J., Liu X., and Liu Z. R. (2012) Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol. Cell 45, 598–609 10.1016/j.molcel.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu Z., Zhao X., Zhao L., Yang H., Liu L., Li J., Wu J., Yang F., Huang G., and Liu J. (2016) p54(nrb)/NONO regulates lipid metabolism and breast cancer growth through SREBP-1A. Oncogene 35, 1399–1410 10.1038/onc.2015.197 [DOI] [PubMed] [Google Scholar]
- 22. Sundqvist A., Bengoechea-Alonso M. T., Ye X., Lukiyanchuk V., Jin J., Harper J. W., and Ericsson J. (2005) Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab. 1, 379–391 10.1016/j.cmet.2005.04.010 [DOI] [PubMed] [Google Scholar]
- 23. Zhao X., Feng D., Wang Q., Abdulla A., Xie X. J., Zhou J., Sun Y., Yang E. S., Liu L. P., Vaitheesvaran B., Bridges L., Kurland I. J., Strich R., Ni J. Q., Wang C., et al. (2012) Regulation of lipogenesis by cyclin-dependent kinase 8-mediated control of SREBP-1. J. Clin. Invest. 122, 2417–2427 10.1172/JCI61462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang W., Xia Y., Hawke D., Li X., Liang J., Xing D., Aldape K., Hunter T., Alfred Yung W. K., and Lu Z. (2012) PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150, 685–696 10.1016/j.cell.2012.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiang Y., Wang Y., Wang T., Hawke D. H., Zheng Y., Li X., Zhou Q., Majumder S., Bi E., Liu D. X., Huang S., and Lu Z. (2014) PKM2 phosphorylates MLC2 and regulates cytokinesis of tumour cells. Nat. Commun. 5, 5566 10.1038/ncomms6566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ettinger S. L., Sobel R., Whitmore T. G., Akbari M., Bradley D. R., Gleave M. E., and Nelson C. C. (2004) Dysregulation of sterol response element–binding proteins and downstream effectors in prostate cancer during progression to androgen independence. Cancer Res. 64, 2212–2221 10.1158/0008-5472.CAN-2148-2 [DOI] [PubMed] [Google Scholar]
- 27. Sun Y., He W., Luo M., Zhou Y., Chang G., Ren W., Wu K., Li X., Shen J., Zhao X., and Hu Y. (2015) SREBP-1 regulates tumorigenesis and prognosis of pancreatic cancer through targeting lipid metabolism. Tumour Biol. 36, 4133–4141 10.1007/s13277-015-3047-5 [DOI] [PubMed] [Google Scholar]
- 28. Calvisi D. F., Wang C., Ho C., Ladu S., Lee S. A., Mattu S., Destefanis G., Delogu S., Zimmermann A., Ericsson J., Brozzetti S., Staniscia T., Chen X., Dombrowski F., and Evert M. (2011) Increased lipogenesis, induced by AKT-mTORC1-RPS6 signaling, promotes development of human hepatocellular carcinoma. Gastroenterology 140, 1071–1083 10.1053/j.gastro.2010.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yamashita T., Honda M., Takatori H., Nishino R., Minato H., Takamura H., Ohta T., and Kaneko S. (2009) Activation of lipogenic pathway correlates with cell proliferation and poor prognosis in hepatocellular carcinoma. J. Hepatol. 50, 100–110 10.1016/j.jhep.2008.07.036 [DOI] [PubMed] [Google Scholar]