Abstract
Chromatin consists of DNA and histones, and specific histone modifications that determine chromatin structure and activity are regulated by three types of proteins, called writer, reader, and eraser. Histone reader proteins from vertebrates, vertebrate-infecting parasites, and higher plants possess a CW domain, which has been reported to read histone H3 lysine 4 (H3K4). The CW domain of Arabidopsis SDG8 (also called ASHH2), a histone H3 lysine 36 methyltransferase, preferentially binds monomethylated H3K4 (H3K4me1), unlike the mammalian CW domain protein, which binds trimethylated H3K4 (H3K4me3). However, the molecular basis of the selective binding by the CW domain of SDG8 (SDG8-CW) remains unclear. Here, we solved the 1.6-Å-resolution structure of SDG8-CW in complex with H3K4me1, which revealed that residues in the C-terminal α-helix of SDG8-CW determine binding specificity for low methylation levels at H3K4. Moreover, substitutions of key residues, specifically Ile-915 and Asn-916, converted SDG8-CW binding preference from H3K4me1 to H3K4me3. Sequence alignment and mutagenesis studies revealed that the CW domain of SDG725, the homolog of SDG8 in rice, shares the same binding preference with SDG8-CW, indicating that preference for low methylated H3K4 by the CW domain of ASHH2 homologs is conserved among higher-order plants. Our findings provide first structural insights into the molecular basis for specific recognition of monomethylated H3K4 by the H3K4me1 reader protein SDG8 from Arabidopsis.
Keywords: histone methylation, histone modification, epigenetics, Arabidopsis thaliana, chromatin modification, ASHH2, CW domain, H3K4me1 reader, recognition of low methylation level, SDG8
Introduction
The basic unit of chromatin is the nucleosome, which contains eight histone proteins and 147 bp of DNA (1). The histone proteins have tails that protrude from the nucleosome, and many residues in these tails can be covalently modified (2). A number of specific modifications of histones have been identified, including methylation, acetylation, ubiquitination, phosphorylation, SUMOylation,2 deamination, and ADP-ribosylation (2–4). Combinations of these modifications play important roles in many biological processes, such as regulation of gene activity and cell fate determination (5–8). Most of these modifications have been found to be dynamic (9, 10). Therefore, establishment, recognition, and removal of histone modifications are carefully regulated by three types of proteins, called “writer,” “reader,” and “eraser,” respectively (11, 12). Investigations of these proteins show that more than one functional domain may occur in one protein, and they may play multiple roles in chromatin-associated processes (13). The N termini of histones are rich in lysine residues. Methylation markers can be deposited on particular lysine residues with different degrees of methylation (mono-, di-, and trimethylation) and can be recognized by various functional domains of histone readers (14). The “royal family” of proteins is well known for its function in the recognition of methylated histones, including chromodomain, Tudor domain, malignant brain tumor domain, and PWWP domain (15, 16). Plant homeodomain and WD40 domain are also capable of binding methylated or unmethylated histones (17–19). Recently, a domain family, called CW domain, was found to function as a H3K4 reader (20–23).
The CW domain is a zinc-binding domain with conserved cysteines and tryptophans (hence the name CW) and has been found in vertebrates, vertebrate-infecting parasites, and higher-order plants (24–27). The CW domains are usually found in chromatin-related proteins associated with other domains, such as PWWP domain, SET domain, and amine oxidase domain, suggesting a gene regulation role for this domain. There are seven CW domain–containing proteins in humans and 11 in Arabidopsis. Previous studies indicate that the CW domains in various proteins show different preference for the degree of methylation of H3K4 (20, 21, 23, 28). The CW domains in mammalian MORC1, MORC2, and LSD2 have been reported to have no ability to bind any histone H3K4 peptides, whereas the CW domains of mammalian ZCWPW1, ZCWPW2, MORC3, and MORC4 bind to H3K4me3 (20, 23, 29). Interestingly, the CW domain of Arabidopsis SDG8 (also called ASHH2/CCR1/EFS) was reported to preferentially bind H3K4me1 (21). Moreover, SDG8 bears sequence homology to SET2 (the sole yeast H3K36 methyltransferase), catalyzing the di- and trimethylation of H3K36 from the monomethylated state (30, 31). The sdg8 mutant plants exhibit early flowering with a global reduction of H3K36me2/me3 level and an increase of H3K36me1 level. SDG8 is not the sole H3K36 methyltransferase in Arabidopsis. SDG26, which lacks the N-terminal CW domain, is also homologous to SET2. In addition, SDG8 is involved in many biological processes, including shoot branching, ovule and anther development, carotenoid biosynthesis, defense response, seed development, brassinosteroid-regulated gene expression, and light- and/or carbon-responsive gene expression, indicating the nonredundant role of SDG8 in chromatin modification and gene regulation (32–39). Therefore, SDG8 may serve as a platform for downstream H3K36 methylation via the recognition of H3K4me1 by the CW domain.
So far, several structures of mammalian CW domains complexed with histone H3K4 peptides have been reported (20, 21, 23, 40). However, mammalian CW domains prefer to bind unmethylated or trimethylated H3K4 (20, 23, 40). The molecular mechanism by which the Arabidopsis SDG8 CW domain specifically recognizes low-level methylation of H3K4 remains unclear. Here, we determined the crystal structure of SDG8-CW in complex with H3K4me1 peptide at 1.6-Å resolution. The structural and biochemical data provide the molecular basis for the selective recognition of H3K4me1/2. Key residues that determine the specificity were identified. Furthermore, we also tested the binding specificity of SDG725, the homolog of SDG8 in rice, for various histone peptides. Sequence alignment and biochemical data indicate that the preference for low-level methylation of H3K4 by the CW domain of SDG8 is conserved in green plants. Our findings may provide new insights into the molecular mechanism of the recruitment of SDG8 to its target genes and shed light on the conserved role played by an incomplete aromatic cage in plants in recognizing low-level methylation of H3K4.
Results
The CW domain of SDG8 preferentially binds monomethylated H3K4
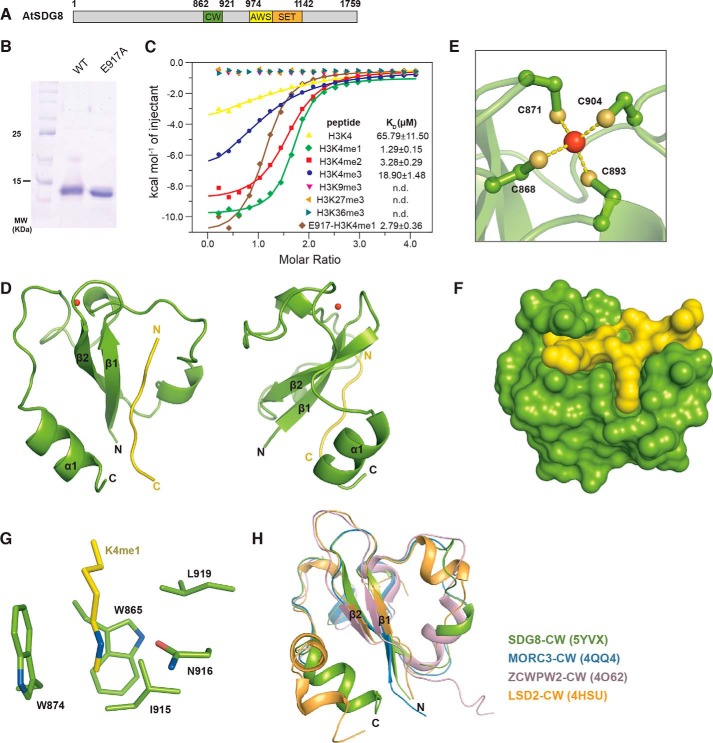
SDG8 harbors a CW domain in the middle of its amino acid sequence and a SET domain that is C-terminal to the CW domain (Fig. 1A). To systematically explore the binding affinity of the SDG8 CW domain to different histone markers, we performed isothermal titration calorimetry (ITC) using label-free histone peptides. SDG8-CW (residues 862–921) was purified with its N-terminal His-SUMO fusion protein removed to rule out any impact introduced by this fusion protein (Fig. 1B). As shown in Fig. 1C, SDG8-CW showed an ability to bind to H3K4me peptides but not H3K9me3, H3K27me3, and H3K36me3, consistent with a previous report (21). The equilibrium disassociation constants (KD) determined for SDG8 to various H3K4me peptides are 1.3 ± 0.2 μm for H3K4me1, 3.3 ± 0.3 μm for H3K4me2, 18.9 ± 1.5 μm for H3K4me3, and 65.8 ± 11.5 μm for H3K4me0 (Fig. 1B and Table S1). Compared with monomethylated H3K4, di- and trimethylation decreased the binding affinity 2.5- and 14.5-fold, respectively, indicating the SDG8 CW domain's strong preference for low levels of methylation of H3K4, especially H3K4me1.
Figure 1.
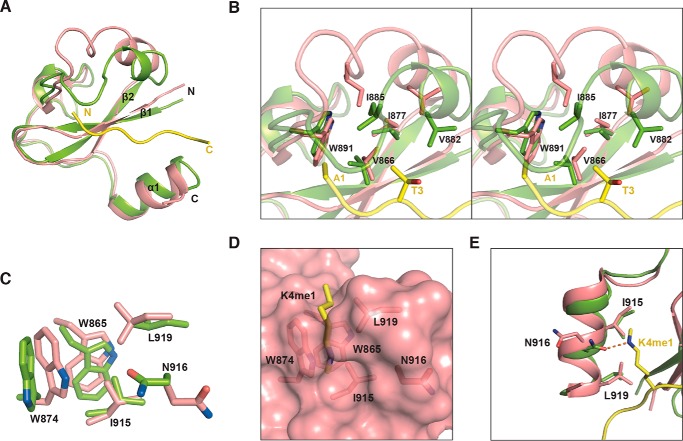
Overall structure of SDG8-CW in complex with H3K4me1. A, domain architecture of A. thaliana (At) SDG8. The amino acid positions at domain junctions are indicated. The CW domain is colored green. The AWS and SET domains are colored yellow and orange, respectively. B, purified WT and E917A mutant SDG8-CW (residues 862–921) stained by Coomassie Blue. C, ITC measurements of the interaction of WT and E917A mutant SDG8-CW with the indicated histone peptides. D, schematic representation of overall structure of SDG8-CW (green) in complex with H3K4me1 peptide (yellow). The view of 90° rotation around the vertical axis is shown at right. E, ball and stick representation of the conserved zinc finger of SDG8-CW. The zinc atom is colored red, and the sulfur atoms are orange. The chelate bonds are indicated by yellow dashes. F, surface representation of SDG8-CW and H3K4me1 peptide. G, residues consisting of aromatic cages are shown as sticks. H, superimposition of CW domains of SDG8 (green), MORC3 (blue), ZCWPW2 (pink), and LSD2 (wheat).
Overall structure of SDG8-CW in complex with H3K4me1
To elucidate the molecular mechanism for the specific recognition of H3K4me1, we tried to cocrystallize SDG8-CW and an H3K4me1 peptide (residues 1–9). However, our initial attempts at crystallizing the complex failed. By analyzing the sequence of SDG8-CW using the SERp server (41), we mutated Glu-917 to alanine to reduce the potential surface entropy for crystallization. The binding affinity of the E917A mutant to H3K4me1 was also measured (Fig. 1C). The KD is 2.79 ± 0.36 μm, indicating that the alanine mutation at Glu-917 has little impact on the binding affinity. E917A was successfully cocrystallized with H3K4me1. Thus, for convenience, we treated the E917A mutant as SDG8-CW.
The CW domain is a zinc-binding domain. We determined the structure of SDG8-CW in complex with H3K4me1 (residues 1–9) by the single-wavelength anomalous dispersion method using the anomalous zinc ion signal. The structure was refined to 1.6-Å resolution with all the statistics within a reasonable range (Table 1). There are three SDG8-CW molecules in one asymmetric unit with each molecule binding one H3K4me1 peptide. The overall structure of SDG-CW comprises a β-hairpin core (β1 and β2), a 310-helical turn (η1), and a C-terminal α-helix (α1) (Fig. 1D). The whole structure is stabilized by four highly conserved cysteine residues, Cys-868, Cys-871, Cys-893, and Cys-904, chelating a zinc ion (Fig. 1E). The H3K4me1 peptide is bound on the concave surface of SDG8-CW formed by β1 and α1 with a buried surface area of 557 Å2, calculated by PISA (Fig. 1F) (42). The monomethylated side chain of Lys-4 inserts into a pocket formed by five residues, Trp-865 on β1, Trp-874 on β2, Ile-915, Asn-916, and Leu-919 on α1 (Fig. 1G). The overall structure of SDG8-CW–H3K4me1 complex is similar to those of previously reported CW domain complex structures. The root mean square deviation (r.m.s.d.) of SDG8 CW domain to ZCWPW2-CW (Protein Data Bank code 4O62), MORC3-CW (Protein Data Bank code 4QQ4), and LSD2-CW (Protein Data Bank code 4HSU) is 1.70, 0.28, and 2.25 Å, respectively (Fig. 1H) (23, 43).
Table 1.
Crystallographic statistics of SDG8-CW and H3K4me1 complex
| Data collection | |
| Space group | P63 |
| Cell dimension | |
| a, b, c (Å) | 66.4, 66.4, 29.2 |
| α, β, γ (°) | 90, 90, 120 |
| Wavelength (Å) | 1.2824 |
| Resolution range (Å)a | 30.00–1.59 (1.65–1.59) |
| Completeness (%)a | 98.6 (89.0) |
| Rmerge (%)a | 12.2 (58.1) |
| I/σ(I) | 26.6 (2.3) |
| Redundancy | 6.4 (2.9) |
| Refinement | |
| Resolution range (Å) | 28.76–1.59 |
| No. of reflections | 19,132 |
| Rwork (%)/Rfree (%) | 16.75/18.45 |
| No. atoms | |
| Protein | 475 |
| Peptide | 54 |
| Water | 84 |
| B-factors | |
| Protein | 25.7 |
| Peptide | 33.0 |
| Water | 35.2 |
| r.m.s.b deviations | |
| Bond length (Å) | 0.007 |
| Bond angles (°) | 0.991 |
| Ramachandran plot | |
| Most favored region (%) | 100 |
| Allowed region (%) | 0 |
| Outliers (%) | 0 |
| MolProbity analysis | |
| Clashscore | 0.97 |
| Overall score | 0.97 |
| Rotamer outliers (%) | 1.8 |
| C-β outliers (%) | 0.0 |
a Values in parentheses are for the highest-resolution shell.
b Root mean square.
Structural basis for the interaction of SDG-CW with H3K4me1
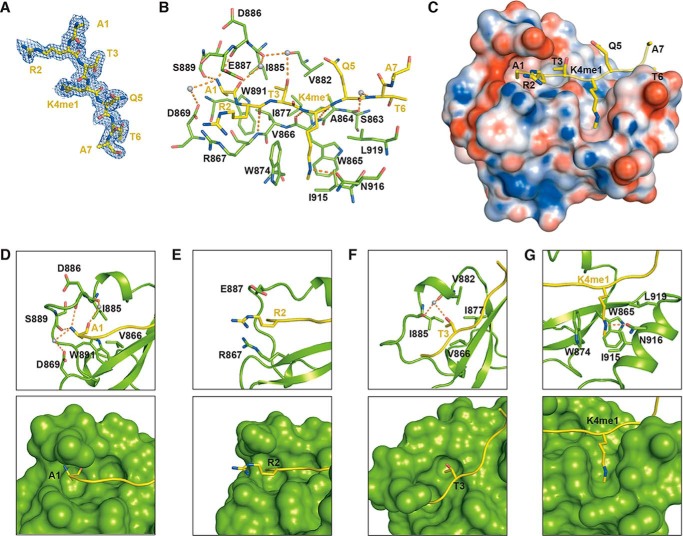
Residues 1–7 of the histone H3K4me1 peptide show a defined electron density and can be successfully modeled (Fig. 2A). Multiple interactions were observed between SDG8-CW and H3K4me1 (Fig. 2, B and C). The methyl group of Ala-1 inserts into a hydrophobic pocket formed by Val-866, Ile-885, and Trp-891 (Fig. 2D). The free amine group of Ala-1 interacts with the main chain carbonyl groups of Asp-886 and Ser-889 via hydrogen bonds. The carboxyl group of Asp-869 further stabilizes A1 via water-mediated hydrogen bonds. Moreover, Arg-2 and K4me1 form hydrogen bonds with the main chains of Arg-867 and Trp-865 on β1, respectively (Fig. 2B). Arg-2 stretched toward the solvent with its guanidinium moiety sandwiched by Arg-867 and Glu-887 (Fig. 2E). The methyl group of Thr-3 is anchored in a shallow hydrophobic pocket formed by Val-866, Ile-877, Val-882, and Ile-885 (Fig. 2F). In addition, the hydroxyl group of Thr-3 is involved in water-mediated hydrogen bond interactions with the main chains of Val-882 and Ile-885. Intriguingly, SDG8-CW adopts a unique cage to accommodate the monomethylated Lys-4 (Fig. 2G). Two conserved tryptophans, Trp-865 and Trp-874, occupy the back and left walls of the cage, respectively. Ile-915 and Leu-919 on α1 form the right wall of the cage, and Asn-916 constitutes the floor of the cage, leaving the front side of the cage unshielded. In addition, the amino group of Asn-916 contacts the guanidino group of K4me1 via a hydrogen bond. The K4me1 binds in a straight concave surface of SDG-CW, which is different from the canonical trimethyllysine-binding pocket formed by three or more aromatic residues to increase the hydrophobicity and space to facilitate the accommodation of a bulky trimethyllysine.
Figure 2.
Interactions of SDG8-CW and H3K4me1 peptide. A, 2Fo − Fc electron density map (contoured at 0.5 σ cutoff; blue mesh) of H3K4me1 peptide. B, details of intermolecular interactions between SDG8-CW and H3K4me1 peptide. Residues are shown as sticks, and water molecules are shown as gray spheres. Hydrogen bonds are indicated as orange dashes. C, electrostatic potential of the surface of SDG8-CW, calculated by the PDB2PQR server with H3K4me1 binding on the concave surface (60). D–G, detailed interactions between N-terminal-residues ARTK of H3K4me1 and SDG8-CW. Residues in sticks are shown in the top panel, and surface structures are shown in the bottom panel.
Validation of the key residues that determine the specific recognition of sequence and lysine methylation level
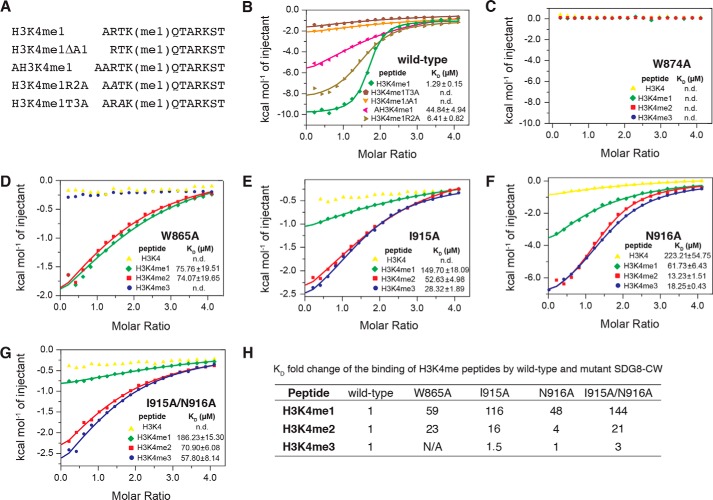
To validate the intermolecular interactions between SDG8-CW and H3K4me1, we performed ITC assays to detect changes in binding affinity that were introduced by site-specific mutagenesis. Structural analysis showed that the first three residues, ART, shared most of the interactions between SDG8-CW and the H3K4me1, indicating that residues ART may be important for the sequence-specific binding. We designed and synthesized four H3K4me1 mutant peptides, including H3K4me1ΔΑ1, which lacks the first Ala-1 residue; AH3K4me1, which adds an additional alanine to the N terminus of the peptide; and two alanine substitution mutants, namely H3K4me1R2A and H3K4me1T3A, which substitute Arg-2 and Thr-3 with alanine, respectively (Fig. 3A). H3K4me1ΔA1 peptide completely lost its ability to bind SDG8-CW, whereas AH3K4me1 exhibited severely diminished binding, underscoring the importance of residue Ala-1 in the recognition of H3K4me1 (Fig. 3B and Table S1). Mutation of Arg-2 to alanine only mildly reduced the binding about 5-fold, whereas mutation of Thr-3 abolished the binding, consistent with the structural analysis result. Thus, Ala-1 and Thr-3 are two key residues that determine the sequence-specific recognition of H3K4me1 by SDG8-CW.
Figure 3.
SDG8-CW interacts with histone H3 tail with specific recognition of sequence and preference of lysine methylation level. A, sequence of H3K4me1 mutant peptides. Residues substituted by alanine are in italics. B, ITC measurements of the interaction between SDG8-CW and H3K4me1 mutant peptides. C–G, ITC measurements of the interaction between SDG8-CW mutant and H3K4 peptides. H, KD -fold change of the binding of H3K4me peptides by WT and mutant SDG8-CW.
Unlike other CW domains, such as MORC3-CW, which has additional negatively charged residues, or ZCWPW2-CW, which has a third aromatic residue, the monomethyllysine-binding cage of SDG8-CW comprises some hydrophobic residues (see Fig. 7, A and B). To further investigate how SDG8-CW prefers to bind monomethylated lysine, we used the same strategy as above to monitor the impact of point mutations on binding affinity (Table S1). Alanine substitution of Trp-865 and Trp-874 significantly affected binding ability. W874A mutant exhibited no ability to bind any of the four H3K4me peptides (Fig. 3C). W865A mutant lost its ability to bind H3K4me0 and H3K4me3 and showed dramatically decreased binding to H3K4me1 and H3K4me2, about 60- and 23-fold lower compared with WT, respectively (Fig. 3D). This is consistent with previous studies, which indicated that the corresponding residue of Trp-865 is substituted by isoleucine and threonine in ZCWPW2, resulting in the absence of binding to any histone H3K4me peptide (23). In the crystal structure, the monomethyl group of K4me1 faces toward Ile-915, and Asn-916 contacts the methylammonium ion via a hydrogen bond (Fig. 2G). Using this structural information as a guide, we generated two single mutants, I915A and N916A, and the double mutant I915A/N916A. Both I915A and N916A mutants reduced the ability to bind to H3K4me peptides (Fig. 3, E and F). However, for various degrees of methylation, I915A and N916A mutants both exhibit drastic differences in the reduction of ability to bind to histone H3K4me peptides. Compared with WT, I915A reduced the binding about 116-fold for H3K4me1, 16-fold for H3K4me2, and 1.5-fold for H3K4me3 (Fig. 3E). N916A and the double mutant I915A/N916A showed selectively diminished binding to H3K4me peptides in a manner similar to I915A (Fig. 3, F and G). Therefore, mutation of Ile-915 and Asn-916 leads to a conversion of ligand binding preference from K4me1 to K4me3, indicating that Ile-915 and Asn-916 are determinants of the selectivity for the recognition of monomethyllysine (Fig. 3H).
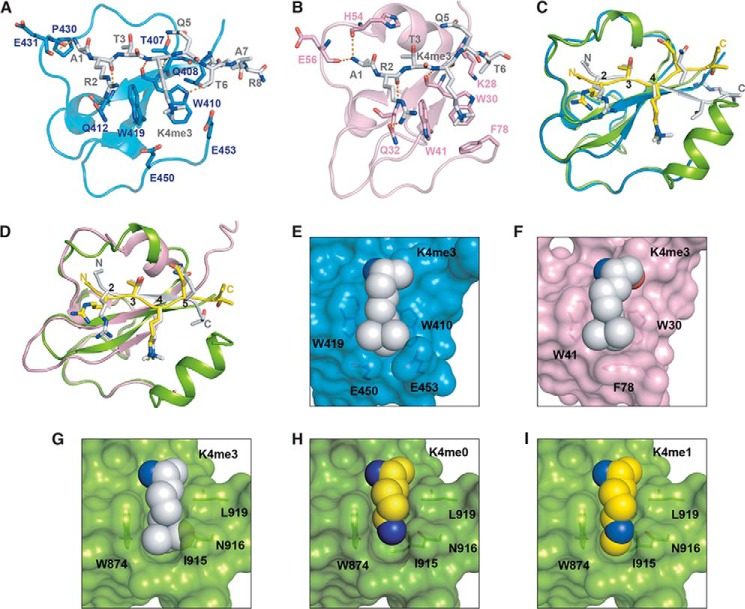
Figure 7.
Structure comparison of SDG8-CW–H3K4me1 complex with MORC3-CW–H3K4me3 complex and ZCWPW-CW–H3K4me3 complex. A, structure of MORC3-CW (blue) binding with H3K4mfe3 peptide (gray) (Protein Data Bank code 4QQ4). B, structure of ZCWPW2-CW (pink) binding with H3K4mfe3 peptide (gray) (Protein Data Bank code 4O62). C, superimposition of SDG8-CW–H3K4me1 complex with MORC3-CW–H3K4me3 complex. D, superimposition of SDG8-CW–H3K4me1 complex with ZCWPW2-CW–H3K4me3 complex. E, surface representation of MORC3-CW with trimethylated lysine binding on the concave surface shown in spheres. F, surface representation of ZCWPW2-CW with trimethylated lysine binding on the concave surface shown in spheres. G, surface representation of SDG8-CW with docked trimethylated lysine of ZCWPW2-CW–H3K4me3 complex shown in spheres. H, surface representation of SDG8-CW with unmethylated lysine shown in spheres. I, surface representation of SDG8-CW with monomethylated lysine binding on the concave surface shown in spheres.
SDG8-CW undergoes conformational change upon binding to H3K4me1
The solution structure of the apo form of the SDG8 CW domain was reported previously (21). However, superimposition of the apo form (Protein Data Bank code 2L7P) and the complex form reveals a significant conformational change at the η1 turn (Fig. 4A). The r.m.s.d. between the apo form and the complex is 1.18 Å. In the complex structure, the loop connecting β2 and α1, containing the η1 in between, is closer to the β-hairpin core, thus facilitating the binding to H3K4me1 peptide. In the apo form, residues Val-882, Ile-885, and Trp-891 that are involved in the recognition of Ala-1 and Thr-3 were far away from Val-866 on β1 and Ile-877 on β2, failing to form the hydrophobic pockets for the accommodation of the methyl groups of Ala-1 and Thr-3 (Fig. 4B). In addition, conformational changes were also observed in the methyllysine-binding cage. The cage is more open in the complex structure compared with that in the apo form due to the deviation of the indole ring of Trp-874 (Fig. 4, C and D). Moreover, the orientation of the side chain of Asn-916, which forms a hydrogen bond with the methylammonium of K4me1, turns away from the peptide (Fig. 4E). Together, we suggest that the methyllysine-binding pocket is closed in the absence of histone ligand. Upon binding, SDG-CW undergoes conformational changes, including forming the open state of the methyllysine-binding cage, the flip of Asn-916, and the closer distance of 310-turn η1, to facilitate the binding of histone H3K4me1 peptide.
Figure 4.
SDG-CW undergoes conformational change upon binding to H3K4me1. A, superimposition of SDG8-CW in apo form (salmon) and SDG8-CW (green) in complex with H3K4me1 peptide (yellow). B, a stereoview of the conformational change at SDG8-CW η1 turn due to H3K4me1 peptide binding. Key residues involved in binding are shown in sticks. C, superimposition of the aromatic cage of SDG8-CW in apo form and in complex with H3K4me1 peptide. D, surface representation of the aromatic cage of SDG8-CW in apo form with residues shown in sticks. E, conformational change at SDG8-CW C-terminal α-helix due to H3K4me1 peptide binding.
The C-terminal α-helix of SDG-CW domain that is important for the K4me1 recognition is conserved in green plants
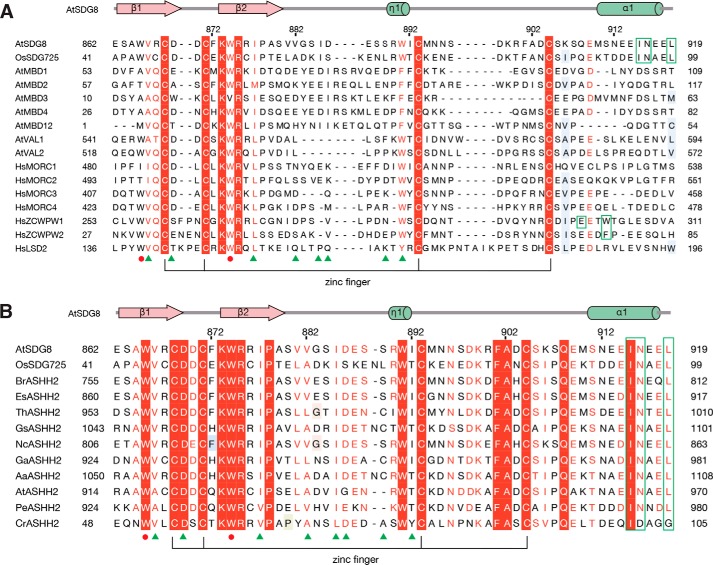
Ile-915, Asn-916, and Leu-919 are the three key residues that form the monomethyllysine-binding pocket. The mutagenesis study and ITC assays also indicated the critical role of Ile-915 and Asn-916 in the selection of the level of lysine methylation. These three residues are located on the C-terminal α-helix of SDG8-CW. We searched the structural homology of SDG8 CW domain on the Dali server (44), which showed that the most homologous structure is ZCWPW2 with an r.m.s.d. of 1.6 Å. However, we found that in all previous reported CW structures no C-terminal α-helix is observed, including the CW domains of ZCWPW1–3, MORC1–3, and LSD2 (20, 23, 43), indicating that the presence of the C-terminal α-helix is unique in SDG8-CW (Fig. 1G). To investigate whether the C-terminal α-helix may exist in a wider range of species, we performed sequence alignments (Fig. 5A). The alignment results indicated that the three key residues (Ile-915, Asn-916, and Leu-919) involved in the monomethyllysine-binding pocket were not conserved in human CW domains or in other CW-containing proteins in Arabidopsis. Therefore, we speculated that the specific recognition of monomethylated lysine is a unique feature to SDG8 protein in green plants.
Figure 5.
Sequence alignment of CW proteins in species. Sequence alignment was calculated using Jalview (53), and amino acids were shaded according to the ESPript server (54). Secondary structural elements of SDG8-CW are displayed above the sequence alignment. The conserved zinc-binding mode is shown by lines at the bottom of the alignment. The residues that form the conserved aromatic cage are marked by red circles, and the various residues of the aromatic cage are marked by green frames. The key residues involved in histone tail sequence-specific recognition are marked by green triangles. A, sequence alignment of CW domains in human (Hs) and plant (Os and At). B, sequence alignment of SDG8 CW domain with its homologs from other plants. Br, Brassica rapa, XP_018508679.1; Es, Eutrema salsugineum, XP_006390102.1; Th, Tarenaya hassleriana, XP_010534709.1; Gs, Glycine soja, KHN44299.1; Nc, Noccaea caerulescens, JAU85503.1; Ga, Gossypium arboreum, XP_017641601.1; Aa, Anthurium amnicola, JAT51201.1; At, Aegilops tauschii, EMT20452.1; Pe, Phalaenopsis equestris, XP_020573738.1; Cr, Chlamydomonas reinhardtii, XP_001694743.1.
The CW domain of OsSDG725 in rice shows a similar binding preference as SDG8-CW
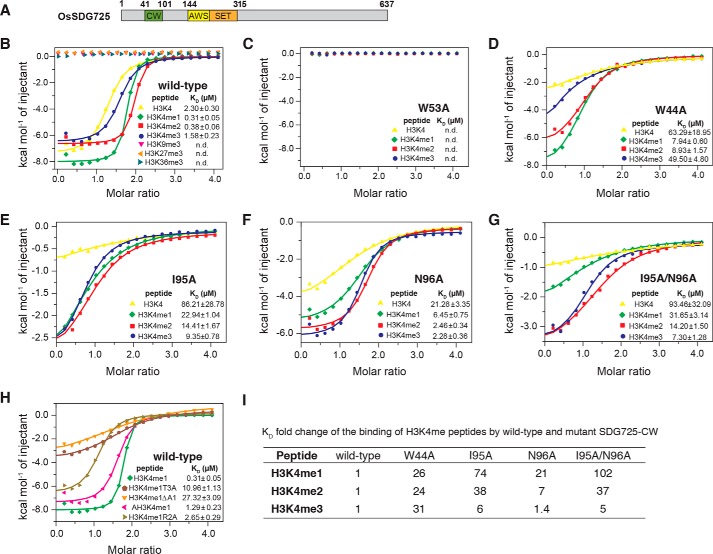
However, we found that the C-terminal α1 of SDG8-CW is highly conserved in ASHH2 proteins in most green plants, including dicots and monocots (Fig. 5B). To investigate whether the CW domain of SDG8 protein in other plant species has the same binding preference as Arabidopsis SDG8, we chose OsSDG725, the rice homolog of SDG8. OsSDG725 contains 637 amino acids with a CW domain and a SET domain (Fig. 6A). OsSDG725-CW domain (residues 41–101) exhibits high binding affinity for histone H3K4me peptides like SDG8-CW but no binding to H3K9me3, H3K27me3, and H3K36me3 (Fig. 6B and Table S2). The KD values for H3K4me peptides are 0.31 ± 0.05 μm for H3K4me1, 0.38 ± 0.06 μm for H3K4me2, 1.58 ± 0.23 μm for H3K4me3, and 2.30 ± 0.30 μm for H3K4me0. Therefore, the order of preference of OsSDG725 for H3K4me peptides is H3K4me1 ≥ H3K4me2 > H3K4me3 > H3K4me0. However, the binding affinity difference caused by the degree of methylation is narrower. Next, according to the mutagenesis studies in SDS8-CW, we generated five mutants in OsSDG725, that is W53A (corresponding to W874A in SDG8-CW), W44A (corresponding to W865A), I95A (corresponding to I915A), N96A (corresponding to N916A), and I95A/N96A (corresponding to the double mutant I915A/N916A). Similar to SDG8-CW mutants, the W53A mutant completely lost the ability to bind any of the four H3K4me peptides (Fig. 6C and Table S2). W44A reduced the binding ability about 26-fold for mono-, 24-fold for di-, 31-fold for trimethylated H3K4, and 28-fold for unmethylated peptide (Fig. 6D). Ile-95 and Asn-96 in OsSDG725 also play critical roles in the recognition of low-level methylation (Fig. 6, E, F, and G). Mutation at the C-terminal α-helix decreases the binding to H3K4me peptides at different levels. The binding affinities to H3K4me1 are most affected compared with H3K4me2 and H3K4me3. The -fold change of binding affinity caused by I95A is 74-fold for mono-, 38-fold for di-, and 6-fold for trimethylated H3K4 (Fig. 6, E and I). N96A reduced the binding about 21-, 6-, and 1.4-fold, respectively (Fig. 6, F and I), as did the double mutant I95A/N96A (Fig. 6, G and I). Mutation at the histone peptides was also performed (Fig. 6H). The results showed that ART (residues 1–3) of histone H3 is important for the recognition of H3K4me1 by OsSDG725-CW, the same as SDG8. Together, these results demonstrate that the sequence-specific recognition of H3K4me1 by the CW domain of ASHH2 is conserved in rice. According to the sequence alignment results, residues that specifically recognize Ala-1 and Thr-3 are highly conserved among green plants.
Figure 6.
The CW domain of OsSDG725 in rice showed a similar binding preference as SDG8-CW. A, domain architecture of O. sativa SDG725. The CW domain is colored blue. The AWS and SET domains are colored yellow and orange, respectively. B, ITC measurements of the interaction between SDG725-CW and the indicated histone peptides. C–G, ITC measurements of the interaction between the SDG725-CW mutant and H3K4 peptides. H, ITC measurements of the interaction of SDG725-CW and H3K4me1 mutant peptides. I, KD -fold change of the binding of H3K4me peptides by WT and mutant SDG725-CW.
Discussion
In this work, we investigated the binding ability and preference of the CW domains of Arabidopsis SDG8 and rice SDG725 to the monomethylated histone H3K4 peptide and determined the crystal structure of SDG8-CW in complex with H3K4me1 peptide. We found that the N terminus of H3 is critical for the sequence-specific binding, and residues on the C-terminal α-helix α1 are the determinants of monomethyllysine recognition. Mutation on α1 leads to a conversion of the binding preference from monomethylated lysine to trimethylated lysine. By analyzing the sequences and structures of other CW domains, we found that the existence of α1 is a unique phenomenon in the CW domains of ASHH2 homologs among green plants. Our findings may provide structural insights into the molecular mechanism of the specific recognition of a histone H3K4me1 marker by the SDG8 CW domain.
So far, many crystal structures of histone reader and histone H3K4 peptides have been reported (45, 46). However, most of these structures are in complex with H3K4me3 or unmethylated H3K4. Two structures of histone readers in complex with H3K4me1 have been published previously in the Protein Data Bank code, including MORC3-CW–H3K4me1 (Protein Data Bank code 5SVY) (40) and WDR5–H3K4me1 (Protein Data Bank code 2H9N) (47). However, these two proteins are not H3K4me1 readers. They preferentially bind trimethylated H3K4 (MORC3-CW) and unmethylated H3K4 (WDR5). Therefore, our structure is the first H3K4me1-specific reader protein in complex with H3K4me1.
We compared our crystal structure with other reported human CW domain structures, such as MORC3 and ZCWPW2, revealing some differences between H3K4me1-reader CW domain and H3K4me3-reader CW domain (23). In our structure, Ala-1 and Thr-3 are key residues in the sequence-specific recognition. Arg-2 plays a less important role than Ala-1 and Thr-3. Arg-2 is sandwiched by Arg-867 and Glu-887 in SDG8 (Fig. 2, B and D). However, the guanidinium moiety is closer to Arg-867, which causes electrostatic repulsion. Mutation of Arg-2 to alanine only decreased the binding about 5-fold. In MORC3-CW–H3K4me3 complex and ZCWPW2-CW–H3K4me3 complex structures, the corresponding residues of Arg-867 are Gln-412 and Gln-32, respectively (23). Thus, Arg-2 forms hydrogen bonds with Gln-412 or Gln-32 (Fig. 7, A and B). Mutation of Arg-2 to alanine nearly abolishes the binding by ZCWPW2-CW (23). No binding could be detected in an NMR experiment of MORC3-CW titrated with the histone H3K4me3 peptides (residues 3–10) in the absence of Ala-1 and Arg-2 (40). We noticed that the orientation of histone H3K4me peptides starting from residue Gln-5 are quite different in the SDG8-CW–H3K4me1 complex structure compared with similar structures (Fig. 7, C and D). In the MORC3-CW or ZCWPW2-CW complex structure, the C-terminal histone peptides form antiparallel β-strands with β1. However, in our structure, the C-terminal α1 helix blocks the position so that the direction of the peptide flips away and cannot form antiparallel β-strands. Moreover, the existence of α1 results in narrower and tighter binding to the methylated lysine. By contrast, in the MORC3-CW–H3K4me3 and ZCWPW2-CW–H3K4me3 complex structures, the trimethylated lysine is surrounded by three aromatic residues in a much more open and large pocket (Fig. 7, E and F). If we model the trimethylated lysine in the structure of SDG8-CW, a steric hindrance between the trimethyl group and the binding pocket is observed (Fig. 7G). Similarly, unmethylated lysine does not fit well into the pocket, resulting in the formation of a cavity in the pocket (Fig. 7H). Our crystal structure reveals that the hydrophobic, narrow pocket of SDG8-CW excludes binding of a more highly methylated state of lysine due to steric hindrance (Fig. 7I).
In Arabidopsis, SDG8 is an H3K36 methyltransferase, which catalyzes the dimethylation and trimethylation of H3K36 and functions in nutrient and energy metabolism, cell differentiation, timing of flowering, and other processes (30, 48, 49). The function of the CW domain in SDG8 remains unclear. In animals, the H3K4me1 marker is associated with enhancers (50). However, the H3K4me1 in Arabidopsis is predominantly located on gene bodies, especially the transcribed regions correlated with CG DNA methylation (51–53). How the combination of a histone H3K4me1 reader and an H3K36me2/3 writer interprets the effect on gene regulation, plant development, and stress response remains to be elucidated. Based on our study, we propose that SDG8 is recruited via the CW domain to the H3K4me1-labeled transcribed region to deposit the H3K36me2/3 mark and is subject to gene regulation.
Experimental procedures
Protein expression and purification
The cDNAs encoding the CW domain of Arabidopsis thaliana SDG8 (residues 862–921) and Oryza sativa SDG725 (residues 41–101) were amplified by PCR and cloned into the modified pET28-SMT3 vector with an N-terminal His-SUMO tag. Site-specific mutants were generated using a site-directed mutagenesis kit (New England Biolabs) according to the manufacturer's instructions. The plasmid was transformed into Escherichia coli strain BL21 (DE3). The cells were cultured in LB medium with 50 μg/ml kanamycin and 0.1 mm ZnSO4. The cells were induced by isopropyl β-d-thiogalactopyranoside (IPTG) at a final concentration of 0.2 mm and continued growing at 18 °C for 18 h. Cells were harvested by centrifugation and resuspended in 20 mm Tris, pH 8.0, 500 mm NaCl, 25 mm imidazole. Cells were lysed by French press (JNBIO). WT and mutant proteins were purified by a HisTrap column (GE Healthcare) followed by removal of the His-SUMO tag by Ulp1 digestion. The target protein was further purified by ion exchange chromatography using a HiTrap Q column (GE Healthcare) and size exclusive chromatography using a Superdex G75 HiLoad 16/60 column (GE Healthcare). Fractions with target proteins were pooled and concentrated to 50 mg/ml in buffer containing 10 mm Tris, pH 8.0, 100 mm NaCl, 1 mm DTT for structural and biochemical studies. Sequence alignment was calculated using Jalview (54), and amino acid residues were shaded according to ESPript server (55).
Crystallization, data collection, and structure determination
To crystallize the SDG8-CW E917A and H3K4me1 complex, purified E917A protein (40 mg/ml) and H3K4me1 (residues 1–9) were mixed at a 1:2 ratio and incubated on ice for 1 h. The SDG8-CW E917A–H3K4me1 complex was crystallized by the hanging drop vapor diffusion method. The well buffer contained 0.1 m Tris, pH 8.5, 30% PEG 3350, 30% isopropanol. X-ray diffraction data were collected at BL19U1 of the Shanghai Synchrotron Radiation Facility. Crystals were flash frozen under a cold nitrogen stream (100 K) during data collection. The data were processed using the HKL3000 program suite (56). Initial phases were determined by the single-wavelength anomalous dispersion method using zinc anomalous scattering. The PHENIX program suite was used for location of zinc positions, phasing, and density modification (57). The graphics program Coot was used for model building (58), and refinement was performed using PHENIX. The structure was analyzed using the MolProbity server (59). Phasing and refinement statistics are listed in Table 1. Buried surface area was calculated using PISA (42). Figures were generated using PyMOL (The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC). Electrostatic surface potential was calculated by the PDB2PQR server (60).
Isothermal titration calorimetry assays
ITC experiments were performed at 20 °C on a MicroCal iTC200 (Malven). Proteins and peptides were kept in an identical buffer of 20 mm Tris, pH 8.0, 100 mm NaCl. The sample cell was filled with a 0.05 mm solution of protein, and the injection syringe was filled with 1 mm titrating ligand. Each titration consisted of 20 2-μl injections with 2-min intervals. Binding isotherms were analyzed by fitting data into the one-site model using the ITC data analysis module in Origin 7.0 software.
Author contributions
Y. L. data curation; Y. L. formal analysis; Y. H. supervision; Y. H. funding acquisition; Y. H. writing-original draft; Y. H. project administration.
Supplementary Material
Acknowledgments
We thank Prof. Wenhui Shen at Institut de Biologie Moléculaire des Plantes du CNRS, Université de Strasbourg for providing the cDNA of SDG8 and Prof. Aiwu Dong at Fudan University for providing the cDNA of OsSDG725. We thank the National Center for Protein Science Shanghai (NCPSS) for instrument support and technical assistance. We thank the staff from BL19U1 beamline of the National Facility for Protein Science Shanghai at the Shanghai Synchrotron Radiation Facility for assistance during data collection.
This work was supported by grants from the National Natural Science Foundation of China (91640102 to Y. H.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB08010202 to Y. H.), the Ministry of Science and Technology of the People's Republic of China (2012CB910500 to Y. H.), the Chinese Academy of Sciences Facility-based Open Research Program, and the State Key Laboratory of Molecular Biology. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Tables S1 and S2.
The atomic coordinates and structure factors (code 5YVX) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SUMO
- small ubiquitin-like modifier
- H3K4
- histone H3 lysine 4
- H3K4me1
- monomethylated H3K4
- me2
- dimethylated
- me3
- trimethylated
- me0
- unmethylated
- SDG8-CW
- CW domain of SDG8
- ITC
- isothermal titration calorimetry
- KD
- equilibrium disassociation constants
- r.m.s.d.
- root mean square deviation
- Os
- O. sativa.
References
- 1. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., and Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- 2. Allfrey V. G., Faulkner R., and Mirsky A. E. (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 51, 786–794 10.1073/pnas.51.5.786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cosgrove M. S. (2007) Histone proteomics and the epigenetic regulation of nucleosome mobility. Expert Rev. Proteomics 4, 465–478 10.1586/14789450.4.4.465 [DOI] [PubMed] [Google Scholar]
- 4. Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., and Bonner W. M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- 5. Jenuwein T., and Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- 6. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- 7. Shilatifard A. (2006) Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 75, 243–269 10.1146/annurev.biochem.75.103004.142422 [DOI] [PubMed] [Google Scholar]
- 8. Strahl B. D., and Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 9. Bannister A. J., Schneider R., and Kouzarides T. (2002) Histone methylation: dynamic or static? Cell 109, 801–806 10.1016/S0092-8674(02)00798-5 [DOI] [PubMed] [Google Scholar]
- 10. Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature 389, 349–352 10.1038/38664 [DOI] [PubMed] [Google Scholar]
- 11. Goldberg A. D., Allis C. D., and Bernstein E. (2007) Epigenetics: a landscape takes shape. Cell 128, 635–638 10.1016/j.cell.2007.02.006 [DOI] [PubMed] [Google Scholar]
- 12. Yun M., Wu J., Workman J. L., and Li B. (2011) Readers of histone modifications. Cell Res. 21, 564–578 10.1038/cr.2011.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ruthenburg A. J., Li H., Patel D. J., and Allis C. D. (2007) Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 10.1038/nrm2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Y., and Reinberg D. (2001) Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15, 2343–2360 10.1101/gad.927301 [DOI] [PubMed] [Google Scholar]
- 15. Maurer-Stroh S., Dickens N. J., Hughes-Davies L., Kouzarides T., Eisenhaber F., and Ponting C. P. (2003) The Tudor domain 'Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem. Sci. 28, 69–74 10.1016/S0968-0004(03)00004-5 [DOI] [PubMed] [Google Scholar]
- 16. Musselman C. A., Lalonde M. E., Côté J., and Kutateladze T. G. (2012) Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 10.1038/nsmb.2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peña P. V., Davrazou F., Shi X., Walter K. L., Verkhusha V. V., Gozani O., Zhao R., and Kutateladze T. G. (2006) Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature 442, 100–103 10.1038/nature04814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rajakumara E., Wang Z., Ma H., Hu L., Chen H., Lin Y., Guo R., Wu F., Li H., Lan F., Shi Y. G., Xu Y., Patel D. J., and Shi Y. (2011) PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol. Cell 43, 275–284 10.1016/j.molcel.2011.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmitges F. W., Prusty A. B., Faty M., Stützer A., Lingaraju G. M., Aiwazian J., Sack R., Hess D., Li L., Zhou S., Bunker R. D., Wirth U., Bouwmeester T., Bauer A., Ly-Hartig N., et al. (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 42, 330–341 10.1016/j.molcel.2011.03.025 [DOI] [PubMed] [Google Scholar]
- 20. He F., Umehara T., Saito K., Harada T., Watanabe S., Yabuki T., Kigawa T., Takahashi M., Kuwasako K., Tsuda K., Matsuda T., Aoki M., Seki E., Kobayashi N., Guntert P., et al. (2010) Structural insight into the zinc finger CW domain as a histone modification reader. Structure 18, 1127–1139 10.1016/j.str.2010.06.012 [DOI] [PubMed] [Google Scholar]
- 21. Hoppmann V., Thorstensen T., Kristiansen P. E., Veiseth S. V., Rahman M. A., Finne K., Aalen R. B., and Aasland R. (2011) The CW domain, a new histone recognition module in chromatin proteins. EMBO J. 30, 1939–1952 10.1038/emboj.2011.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu Y., Liu S., Zhang X., Liang X., Zahid K. R., Liu K., Liu J., Deng L., Yang J., and Qi C. (2016) Structure and function of CW domain containing proteins. Curr. Protein Pept. Sci. 17, 497–506 10.2174/1389203717666160125115130 [DOI] [PubMed] [Google Scholar]
- 23. Liu Y., Tempel W., Zhang Q., Liang X., Loppnau P., Qin S., and Min J. (2016) Family-wide characterization of histone binding abilities of human CW domain-containing proteins. J. Biol. Chem. 291, 9000–9013 10.1074/jbc.M116.718973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perry J., and Zhao Y. (2003) The CW domain, a structural module shared amongst vertebrates, vertebrate-infecting parasites and higher plants. Trends Biochem. Sci. 28, 576–580 10.1016/j.tibs.2003.09.007 [DOI] [PubMed] [Google Scholar]
- 25. Liggins A. P., Cooper C. D., Lawrie C. H., Brown P. J., Collins G. P., Hatton C. S., Pulford K., and Banham A. H. (2007) MORC4, a novel member of the MORC family, is highly expressed in a subset of diffuse large B-cell lymphomas. Br. J. Haematol. 138, 479–486 10.1111/j.1365-2141.2007.06680.x [DOI] [PubMed] [Google Scholar]
- 26. Suzuki M., Wang H. H., and McCarty D. R. (2007) Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiol. 143, 902–911 10.1104/pp.106.092320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang G. L., Wang C. Y., Cai X. Z., Chen W., Wang X. H., and Li F. (2010) Identification and expression analysis of a novel CW-type zinc finger protein MORC2 in cancer cells. Anat. Rec. 293, 1002–1009 10.1002/ar.21119 [DOI] [PubMed] [Google Scholar]
- 28. Zhang Z., Zhang F., Cheng Z. J., Liu L. L., Lin Q. B., Wu F. Q., Zhang H., Wang J. L., Wang J., Guo X. P., Zhang X., Lei C. L., Zhao Z. C., Zhu S. S., and Wan J. M. (2017) Functional characterization of rice CW-domain containing zinc finger proteins involved in histone recognition. Plant Sci. 263, 168–176 10.1016/j.plantsci.2017.06.013 [DOI] [PubMed] [Google Scholar]
- 29. Fang R., Chen F., Dong Z., Hu D., Barbera A. J., Clark E. A., Fang J., Yang Y., Mei P., Rutenberg M., Li Z., Zhang Y., Xu Y., Yang H., Wang P., et al. (2013) LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation. Mol. Cell 49, 558–570 10.1016/j.molcel.2012.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xu L., Zhao Z., Dong A., Soubigou-Taconnat L., Renou J. P., Steinmetz A., and Shen W. H. (2008) Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 28, 1348–1360 10.1128/MCB.01607-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao Z., Yu Y., Meyer D., Wu C., and Shen W. H. (2005) Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 7, 1256–1260 10.1038/ncb1329 [DOI] [PubMed] [Google Scholar]
- 32. Berr A., McCallum E. J., Alioua A., Heintz D., Heitz T., and Shen W. H. (2010) Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154, 1403–1414 10.1104/pp.110.161497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cazzonelli C. I., Cuttriss A. J., Cossetto S. B., Pye W., Crisp P., Whelan J., Finnegan E. J., Turnbull C., and Pogson B. J. (2009) Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21, 39–53 10.1105/tpc.108.063131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cazzonelli C. I., Roberts A. C., Carmody M. E., and Pogson B. J. (2010) Transcriptional control of SET DOMAIN GROUP 8 and CAROTENOID ISOMERASE during Arabidopsis development. Mol. Plant 3, 174–191 10.1093/mp/ssp092 [DOI] [PubMed] [Google Scholar]
- 35. Dong G., Ma D. P., and Li J. (2008) The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem. Biophys. Res. Commun. 373, 659–664 10.1016/j.bbrc.2008.06.096 [DOI] [PubMed] [Google Scholar]
- 36. Grini P. E., Thorstensen T., Alm V., Vizcay-Barrena G., Windju S. S., Jørstad T. S., Wilson Z. A., and Aalen R. B. (2009) The ASH1 HOMOLOG 2 (ASHH2) histone H3 methyltransferase is required for ovule and anther development in Arabidopsis. PLoS One 4, e7817 10.1371/journal.pone.0007817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y., Mukherjee I., Thum K. E., Tanurdzic M., Katari M. S., Obertello M., Edwards M. B., McCombie W. R., Martienssen R. A., and Coruzzi G. M. (2015) The histone methyltransferase SDG8 mediates the epigenetic modification of light and carbon responsive genes in plants. Genome Biol. 16, 79 10.1186/s13059-015-0640-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tang X., Lim M. H., Pelletier J., Tang M., Nguyen V., Keller W. A., Tsang E. W., Wang A., Rothstein S. J., Harada J. J., and Cui Y. (2012) Synergistic repression of the embryonic programme by SET DOMAIN GROUP 8 and EMBRYONIC FLOWER 2 in Arabidopsis seedlings. J. Exp. Bot. 63, 1391–1404 10.1093/jxb/err383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang X., Chen J., Xie Z., Liu S., Nolan T., Ye H., Zhang M., Guo H., Schnable P. S., Li Z., and Yin Y. (2014) Histone lysine methyltransferase SDG8 is involved in brassinosteroid-regulated gene expression in Arabidopsis thaliana. Mol. Plant 7, 1303–1315 10.1093/mp/ssu056 [DOI] [PubMed] [Google Scholar]
- 40. Andrews F. H., Tong Q., Sullivan K. D., Cornett E. M., Zhang Y., Ali M., Ahn J., Pandey A., Guo A. H., Strahl B. D., Costello J. C., Espinosa J. M., Rothbart S. B., and Kutateladze T. G. (2016) Multivalent chromatin engagement and inter-domain crosstalk regulate MORC3 ATPase. Cell Rep. 16, 3195–3207 10.1016/j.celrep.2016.08.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goldschmidt L., Cooper D. R., Derewenda Z. S., and Eisenberg D. (2007) Toward rational protein crystallization: a web server for the design of crystallizable protein variants. Protein Sci. 16, 1569–1576 10.1110/ps.072914007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 43. Chen F., Yang H., Dong Z., Fang J., Wang P., Zhu T., Gong W., Fang R., Shi Y. G., Li Z., and Xu Y. (2013) Structural insight into substrate recognition by histone demethylase LSD2/KDM1b. Cell Res. 23, 306–309 10.1038/cr.2013.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Holm L., and Laakso L. M. (2016) Dali server update. Nucleic Acids Res. 44, W351–W355 10.1093/nar/gkw357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Patel D. J. (2016) A structural perspective on readout of epigenetic histone and DNA methylation marks. Cold Spring Harb. Perspect. Biol. 8, a018754 10.1101/cshperspect.a018754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Patel D. J., and Wang Z. (2013) Readout of epigenetic modifications. Annu. Rev. Biochem. 82, 81–118 10.1146/annurev-biochem-072711-165700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schuetz A., Allali-Hassani A., Martín F., Loppnau P., Vedadi M., Bochkarev A., Plotnikov A. N., Arrowsmith C. H., and Min J. (2006) Structural basis for molecular recognition and presentation of histone H3 by WDR5. EMBO J. 25, 4245–4252 10.1038/sj.emboj.7601316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shafiq S., Berr A., and Shen W. H. (2014) Combinatorial functions of diverse histone methylations in Arabidopsis thaliana flowering time regulation. New Phytol. 201, 312–322 10.1111/nph.12493 [DOI] [PubMed] [Google Scholar]
- 49. Cazzonelli C. I., Nisar N., Roberts A. C., Murray K. D., Borevitz J. O., and Pogson B. J. (2014) A chromatin modifying enzyme, SDG8, is involved in morphological, gene expression, and epigenetic responses to mechanical stimulation. Front. Plant Sci. 5, 533 10.3389/fpls.2014.00533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheng J., Blum R., Bowman C., Hu D., Shilatifard A., Shen S., and Dynlacht B. D. (2014) A role for H3K4 monomethylation in gene repression and partitioning of chromatin readers. Mol. Cell 53, 979–992 10.1016/j.molcel.2014.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roudier F., Ahmed I., Bérard C., Sarazin A., Mary-Huard T., Cortijo S., Bouyer D., Caillieux E., Duvernois-Berthet E., Al-Shikhley L., Giraut L., Després B., Drevensek S., Barneche F., Dèrozier S., et al. (2011) Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30, 1928–1938 10.1038/emboj.2011.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sequeira-Mendes J., Aragüez I., Peiró R., Mendez-Giraldez R., Zhang X., Jacobsen S. E., Bastolla U., and Gutierrez C. (2014) The functional topography of the Arabidopsis genome is organized in a reduced number of linear motifs of chromatin states. Plant Cell 26, 2351–2366 10.1105/tpc.114.124578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang X., Bernatavichute Y. V., Cokus S., Pellegrini M., and Jacobsen S. E. (2009) Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10, R62 10.1186/gb-2009-10-6-r62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Waterhouse A. M., Procter J. B., Martin D. M., Clamp M., and Barton G. J. (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Robert X., and Gouet P. (2014) Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 57. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Emsley P., Lohkamp B., Scott W. G., and Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dolinsky T. J., Nielsen J. E., McCammon J. A., and Baker N. A. (2004) PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32, W665–W667 10.1093/nar/gkh381 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.