Abstract
Background
Ambulances may represent a potential source of infection to patients, patients’ relatives, and paramedical staffs. In this study, we analyzed the extent of bacterial contamination in ambulance vehicles and measured the degree of antimicrobial resistance among isolated pathogens.
Materials and methods
Twenty-five vehicles were included and 16 sampling points were swabbed in each vehicle. Then the swabs were immediately transferred to the laboratory to identify bacterial contaminants utilizing standard microbiological procedures and API® systems. Antibiotic susceptibility testing and screening for methicillin-resistant staphylococci and extended spectrum β-lactamases (ESBLs)-producing Gram-negative rods were carried out.
Results
A total of 400 samples were collected, 589 bacteria were isolated and 286 (48.6%) of the isolates were potentially pathogenic. The highest contamination rate with pathogenic bacteria was detected in suction devices (75.8%) and stethoscopes (67.7%). Staphylococci were the most frequently detected microorganisms (n=184) followed by Klebsiella spp. (49), Escherichia coli (40), Citrobacter spp. (7), and Proteus spp. (6). Staphylococci were mostly sensitive to vancomycin, whereas Gram-negative bacteria were sensitive to imipenem. Overall, 46.1% of Staphylococcus aureus were methicillin resistant, whereas 20.4% of the coagulase-negative staphylococci were methicillin resistant. Moreover, 36.7% of Klebsiella spp. and 27.5% of E. coli were ESBL producers.
Conclusion
Our study provides evidence that ambulances represent a source of prehospital multidrug-resistant infections.
Keywords: ambulance, contamination, resistance, methicillin-resistant Staphylococcus aureus, extended spectrum β-lactamases
Introduction
Ambulance vehicles are an integral part of emergency medical services. Emergency ambulances aim to help critically ill patients and to prevent the development of life-threatening complications in seriously injured cases.1 Ambulances can respond to thousands of cases per year and after each mission, each ambulance vehicle must be cleaned and decontaminated to be ready for use for the next mission. Effective cleaning protocols for this emergency medical environment are essential in order to avoid the presence of pathogenic microorganisms that represent a potential risk of infection for patients or even the accompanying emergency medical professionals. Although different infection control procedures are employed, ambulances remain a potential source of infection to patients, relatives of the patients, and paramedical staff during transport to a health care facility, or during an interfacility transfer.1,2 Recent research has demonstrated that ambulance surfaces that are directly surrounding patients are frequently contaminated with microorganisms.1 Moreover, Eibicht and Vogel reported that ambulance cars were contaminated with methicillin-resistant Staphylococcus aureus (MRSA) after even a short transport of MRSA-colonized patients.3 Multidrug-resistant bacteria as MRSA are common pathogens isolated from hospitalized patients as well as outpatient settings. Nowadays, the spread of some multiresistant clones in the health care settings and the emergence of community-acquired MRSA are causes of major concern for public health specialists and clinicians.4,5
Since patients transported on rescue vehicles are generally critically ill, great care should be taken to avoid the presence of contaminating bacteria that are resistant to antibiotics and therefore difficult to treat such as MRSA or the extended spectrum β-lactamase (ESBL)-producing Gram-negative rods. Of great concern is the fact that previous studies have reported that MRSA colonization was detected among emergency services personnel.6
Microorganisms were detected not only in the interior of ambulances but also on the emergency services equipment. Some reports showed that medical devices such as stethoscopes showed a high rate of MRSA contamination.7 Other studies by Brown et al8 and Roline et al9 showed that around half of samples from emergency ambulances tested positive for MRSA contamination. Therefore, microbiological evaluation of ambulance vehicles is an essential infection control step that must be considered in order to develop valuable risk reduction interventions.
To date, there has been no data about contamination of ambulance vehicles in Egypt. Therefore, the goal of this observational, cross-sectional study was to determine the extent of bacterial contamination in different ambulance vehicles in Assiut city, Egypt and to describe the degree of antimicrobial resistance for the isolated pathogens.
Materials and methods
In our study, we aimed to determine the extent of bacterial contamination in different ambulance vehicles in Assiut city and Assiut University hospitals in Egypt and to describe the degree of antimicrobial resistance for the isolated pathogens. This cross-sectional study was carried out in the period from January to June 2016. The investigations were carried out with the full cooperation of the Ministry of Health and University hospital officers.
Sampling process
A total of 25 rescue stations throughout Assiut city were included in the study. Unannounced visits were performed. The ambulance crews had no advance knowledge about the visits. Sixteen sampling points were selected based on their well-known high frequency of contact by emergency personnel and patients. These areas were as follows: carrying handle of cart; sidebar of cart; DC shock apparatus; stethoscope; cervical collar; inside wall of the vehicle; emergency personnel seats; portable ventilator; blood pressure cuff; monitor; door grip; oxygen humidifier glass; suction device; headboard of patient stretcher; and steering wheel.
Ambulance car surfaces were swabbed with cotton wool swabs moistened with sterile 0.9% NaCl solution that has been shown to be superior in recovering bacteria from ambulance surfaces to sponges and dry cotton wool swabs.3 During the sampling process, swabs were passed over the entire sampling point and rotated to collect as much material as possible. Swabs were inserted into sterile tubes containing normal saline to avoid desiccation during transport.
Microbiological analysis and identification of contaminating microorganisms
Upon arrival at the microbiology laboratory, swabs were immediately transferred into brain heart infusion broth (Becton Dickinson GmbH, Heidelberg, Germany) and incubated for 20–24 h at 37°C (5% CO2). Thereafter, the enriched cultures were streaked onto microbial culture plates of blood agar, mannitol salt agar, MacConkey’s agar, and eosin methylene blue agar to be identified utilizing standard microbiological procedures. All positive cultures were stained by Gram stain for preliminary identification of Gram-positive and Gram-negative microorganisms. Catalase and coagulase production were used to classify Gram-positive cocci. Staphylococci were identified by standard protocols including the colony morphology, Gram staining, catalase test, and tube coagulase test.10 The API® system (API® 20E and API® 20NE, bioMérieux, Marcy l’Etoile, France) were used to further identify Gram-negative organisms.
After isolation of the contaminating bacteria, antibiotic sensitivity for 15 antibiotics (ampicillin, amoxicillin/clavulanic acid, ceftriaxone, cefotaxime, ceftazidime, cefpodoxime, imipenem, amikacin, gentamicin, tetracycline, ciprofloxacin, norfloxacin, chloramphenicol, trimethoprim/sulfamethoxazole, and vancomycin) was tested by the disc diffusion method on Thermo Scientific™ Oxoid™ Mueller-Hinton agar (Thermo Fisher Scientific, Waltham, MA, USA) using the modified Kirby–Bauer disk-susceptibility method.11 Only Staphylococci were tested for vancomycin sensitivity. Inhibition zone diameters were measured and results were interpreted according to the susceptibility breakpoints recommended by the Clinical and Laboratory Standards Institute (CLSI) guidelines.12
In addition, standard microbiological techniques were employed to rapidly screen for methicillin resistance in staphylococci and ESBL-producing Gram-negative rods. These organisms were selected because these pathogens have well-known robust antibiotic resistance profiles and they can cause serious life-threatening nosocomial infections.
Screening and confirmatory tests for ESBL production in Gram-negative rods
Testing for ESBL production is considered for all Gram-negative rods according to the recommendations of the CLSI guidelines.12 Initially, isolates were screened for ESBL production by detecting microorganisms that exhibited reduced susceptibility to one or more of cefpodoxoime, ceftazidime, ceftriaxone, and cefotaxime. These bacteria were considered as potential producers of ESBL. Confirmation of ESBL production was performed phenotypically by the combined disc synergy testing between ceftazidime versus ceftazidime-clavulanate and cefotaxime versus cefotaxime-clavulanate where a ≥5 mm increase in the zone diameter for the antimicrobial agent tested in combination with β-lactamase inhibitor versus its zone when tested alone indicates ESBL production. In addition, minimal inhibitory concentration (MIC) values for ceftazidime and cefotaxime in the presence of clavulanic acid are reduced by ≥3 twofold dilutions as tested by the Etest® (bioMérieux, Marcy l’Etoile, France).13
Screening for methicillin resistance among staphylococcal isolates
For the detection of methicillin resistance among staphylococci, suspected colonies on mannitol salt agar were subcultured on oxacillin resistance screening agar base (Oxoid™ Thermo Fisher Scientific, Waltham, MA, USA) that contains 6 µg/mL oxacillin.14,15 For confirmation of methicillin resistance, MRSA-screen latex agglutination test was carried out. This test was used for the detection of penicillin-binding protein 2a (PBP2a), and based on agglutination of latex particles sensitized with monocloclonal antibodies against PBP2a in accordance with the manufacturer’s protocol (Denka Seiken Co, Niigata, Japan).16 In addition, MIC values for cefoxitine were tested using the commercially available cefoxitine Etest. MIC values >4 µg/mL were considered positive for mecA-mediated resistance.12
Results
Analysis of microbial samples
In our study, a total number of 25 ambulance cars were screened for bacterial contamination without the crews’ advance knowledge. Samples were taken from 16 sites within each vehicle. In total, 400 samples were collected and examined. Remarkably, all sites within ambulance vehicles showed bacterial growth. A total of 589 bacteria were isolated. About half of the isolates 286 (48.6%) were potentially pathogenic organisms as shown in Table 1. When comparing different areas within each vehicle, it was clear that suction devices (75.8%) and stethoscopes (67.7%) showed the highest pathogenic ratios, whereas the lowest contamination rates with pathogenic bacteria were detected in steering wheels and DC shock apparatus.
Table 1.
Bacterial contamination of different sites in ambulance vehicles
| Site | Potential pathogenic bacteria n (%) | Non pathogenic bacteria n (%) | Total number of isolated bacteria per site |
|---|---|---|---|
| Carrying handle of cart | 21 (63.6) | 12 (36.4) | 33 |
| Sidebar of cart | 14 (48.3) | 15 (51.7) | 29 |
| Bed | 32 (54.2) | 27 (45.8) | 59 |
| DC shock apparatus | 2 (8.3) | 22 (91.7) | 24 |
| Stethoscope | 21 (67.7) | 10 (32.3) | 31 |
| Cervical collar | 22 (59.5) | 15 (40.5) | 37 |
| Inside wall of the vehicle | 14 (43.8) | 18 (56.3) | 32 |
| Chair | 17 (42.5) | 23 (57.5) | 40 |
| Portable ventilator | 29 (53.7) | 25 (46.3) | 54 |
| Blood pressure cuff | 8 (24.2) | 25 (75.8) | 33 |
| Monitor | 13 (34.2) | 25 (65.8) | 38 |
| Door grip | 15 (40.5) | 22 (59.5) | 37 |
| Oxygen humidifier glass | 27 (61.4) | 17 (38.6) | 44 |
| Suction device | 25 (75.8) | 8 (24.2) | 33 |
| Headboard of patient stretcher | 25 (62.5) | 15 (37.5) | 40 |
| Steering wheel | 1 (4.0) | 24 (96.0) | 25 |
| Sum | 286 (48.6) | 303 (51.4) | 589 |
Isolated microorganisms according to site of collection
With respect to the isolated bacteria, Staphylococci (184) were the most frequently detected microorganisms from the collection sites followed by Klebsiella pneumoniae (49) and Escherichia coli (40). Only a few sites were contaminated with Citrobacter spp. (7) and Proteus spp. (6). In addition, all specimens showed contamination with non pathogenic contaminants such as Bacillus spp., Corynebacterium spp., and Micrococcus spp. Of great concern is the fact that the headboard of the patient stretcher showed the highest yield of staphylococcal contamination (20). On the other hand, most of the K. pneumoniae and E. coli isolates were detected in suction devices (14) and beds (11), respectively (Table 2). Of the 184 isolated staphylococci, 76 (41.3%) were identified as S. aureus and 108 (58.7%) were coagulase-negative staphylococci (CNS).
Table 2.
Frequency of different isolated microorganisms in each site of the vehicles
| Site | Potentially pathogenic bacteria
|
Non pathogenic bacteria
|
||||||
|---|---|---|---|---|---|---|---|---|
| Staphylococcus spp. | Klebsiella pneumoniae | Escherichia coli | Proteus spp. | Citrobacter spp. | Corynebacterium spp. | Bacillus spp. | Micrococcus spp. | |
| Carrying handle of cart | 14 | 2 | 5 | 0 | 0 | 5 | 4 | 3 |
| Sidebar of cart | 14 | 0 | 0 | 0 | 0 | 0 | 15 | 0 |
| Bed | 12 | 7 | 11 | 2 | 0 | 2 | 25 | 0 |
| DC shock apparatus | 2 | 0 | 0 | 0 | 0 | 7 | 9 | 6 |
| Stethoscope | 18 | 0 | 0 | 0 | 3 | 0 | 10 | 0 |
| Cervical collar | 17 | 0 | 3 | 0 | 2 | 3 | 8 | 4 |
| Inside wall of the vehicle | 10 | 0 | 2 | 2 | 0 | 0 | 13 | 5 |
| Chair | 14 | 1 | 0 | 0 | 2 | 0 | 15 | 8 |
| Portable ventilator | 9 | 9 | 9 | 2 | 0 | 0 | 25 | 0 |
| Blood pressure cuff | 8 | 0 | 0 | 0 | 0 | 12 | 13 | 0 |
| Monitor | 13 | 0 | 0 | 0 | 0 | 12 | 13 | 0 |
| Door grip | 15 | 0 | 0 | 0 | 0 | 0 | 17 | 5 |
| Oxygen humidifier glass | 12 | 11 | 4 | 0 | 0 | 5 | 12 | 0 |
| Suction device | 5 | 14 | 6 | 0 | 0 | 0 | 8 | 0 |
| Headboard of patient stretcher | 20 | 5 | 0 | 0 | 0 | 5 | 10 | 0 |
| Steering wheel | 1 | 0 | 0 | 0 | 0 | 0 | 24 | 0 |
| Sum | 184/286 (64.3%) | 49/286 (17.1%) | 40/286 (13.9%) | 6/286 (2.1%) | 7/286 (2.5%) | 51/303 (16.8%) | 221/303 (73%) | 31/303 (10.2%) |
Rate of microbial resistance
Antibiotic sensitivities of isolated organisms showed that the isolated staphylococci were mostly sensitive to vancomycin (100%), Klebsiella spp. and E. coli were sensitive to imipenem (95.9% and 92.5%, respectively); however, Proteus spp. and Citrobacter spp. were the least resistant organisms as all strains were sensitive to the third-generation cephalosporins, cefotaxime, ceftriaxone, ceftazidime, and cefpodoxime in addition to imipenem and amikacin antibiotics (Table 3).
Table 3.
Antibiogram for the microorganisms isolated from different sites in ambulances
| Antibiotic |
Staphylococcus spp.
|
Klebsiella pneumoniae
|
Escherichia coli
|
Proteus spp.
|
Citrobacter spp.
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n=184 | % | n=49 | % | n=40 | % | n=6 | % | n=7 | % | ||
| Ampicillin | s | 40 | 21.7 | 4 | 8.2 | 5 | 12.5 | 2 | 33.3 | 3 | 42.9 |
| r | 144 | 78.3 | 45 | 91.8 | 35 | 87.5 | 4 | 66.7 | 4 | 57.1 | |
| Amoxicillin/clavulanic acid | s | 66 | 35.9 | 7 | 14.3 | 7 | 17.5 | 4 | 66.7 | 5 | 71.4 |
| r | 118 | 64.1 | 42 | 85.7 | 33 | 82.5 | 2 | 33.3 | 2 | 28.6 | |
| Ceftriaxone | s | 155 | 84.2 | 25 | 51.0 | 19 | 47.5 | 6 | 100 | 7 | 100 |
| r | 29 | 15.8 | 24 | 49.0 | 21 | 52.5 | 0 | 0.0 | 0 | 0.0 | |
| cefotaxime | s | 160 | 87.0 | 23 | 46.9 | 25 | 62.5 | 6 | 100 | 7 | 100 |
| r | 79 | 42.9 | 26 | 53.1 | 15 | 37.5 | 0 | 0.0 | 0 | 0.0 | |
| Ceftazidime | s | 113 | 61.4 | 23 | 46.9 | 23 | 57.5 | 6 | 100 | 7 | 100 |
| r | 71 | 38.6 | 26 | 53.1 | 17 | 42.5 | 0 | 0.0 | 0 | 0.0 | |
| Cefpodoxime | s | 99 | 53.8 | 18 | 36.7 | 23 | 57.5 | 6 | 100 | 7 | 100 |
| r | 85 | 46.2 | 31 | 63.3 | 17 | 42.5 | 0 | 0.0 | 0 | 0.0 | |
| Imipenem | s | 150 | 81.5 | 47 | 95.9 | 37 | 92.5 | 6 | 100 | 7 | 100 |
| r | 34 | 18.5 | 2 | 4.1 | 3 | 7.5 | 0 | 0.0 | 0 | 0.0 | |
| Amikacin | s | 110 | 59.8 | 33 | 67.3 | 35 | 87.5 | 6 | 100 | 7 | 100 |
| r | 74 | 40.2 | 16 | 32.7 | 5 | 12.5 | 0 | 0.0 | 0 | 0.0 | |
| Gentamicin | s | 100 | 54.3 | 20 | 40.8 | 13 | 32.5 | 5 | 83.3 | 3 | 42.9 |
| r | 84 | 45.7 | 29 | 59.2 | 27 | 67.5 | 1 | 16.7 | 4 | 57.1 | |
| Tetracycline | s | 40 | 21.7 | 5 | 10.2 | 4 | 10.0 | 1 | 16.7 | 1 | 14.3 |
| r | 144 | 78.3 | 44 | 89.8 | 36 | 90.0 | 5 | 83.3 | 6 | 85.7 | |
| Ciprofloxacin | s | 50 | 27.2 | 30 | 61.2 | 10 | 25.0 | 4 | 66.7 | 5 | 71.4 |
| r | 134 | 72.8 | 19 | 38.8 | 30 | 75.0 | 2 | 33.3 | 2 | 28.6 | |
| Norfloxacin | s | 55 | 29.9 | 33 | 67.3 | 15 | 37.5 | 5 | 83.3 | 4 | 57.1 |
| r | 129 | 70.1 | 16 | 32.7 | 25 | 62.5 | 1 | 16.7 | 3 | 42.9 | |
| Chloramphenicol | s | 14 | 7.6 | 5 | 10.2 | 4 | 10.0 | 5 | 83.3 | 5 | 71.4 |
| r | 170 | 92.4 | 44 | 89.8 | 36 | 90.0 | 1 | 16.7 | 2 | 28.6 | |
| Trimethoprim/sulfamethoxazole | s | 23 | 12.5 | 9 | 18.4 | 6 | 15.0 | 4 | 66.7 | 6 | 85.7 |
| r | 161 | 87.5 | 40 | 81.6 | 34 | 85.0 | 2 | 33.3 | 1 | 14.3 | |
| Vancomycin | s | 184 | 100 | na | na | na | na | na | na | na | na |
| r | 0 | 0.0 | na | na | na | na | na | na | na | na | |
Abbreviations: s, sensitive; r, resistant; na, not applicable.
Further testing of staphylococci for methicillin resistance showed that about half of S. aureus isolates (35/76, 46.1%) were MRSA, whereas 22/108 (20.4%) of the CNS were methicillin-resistant coagulase-negative staphylococci (MRCNS). All sites were contaminated with either MRSA or MRCNS except for the DC shock apparatus and steering wheels.
This high level of methicillin resistance among staphylococci diverted our attention to further test the isolated Gram-negative rods for production of the ESBL enzyme. A total of 18/49 (36.7%) of Klebsiella spp. and 11/40 (27.5%) of E. coli were ESBL producers. However, no ESBL-producing organisms were detected among Proteus spp. and Citrobacter spp.
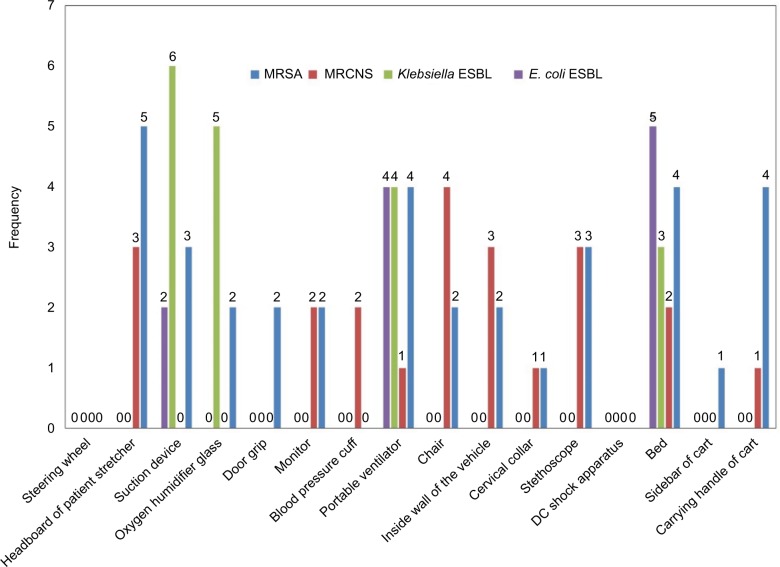
When comparing the frequency of MRSA, MRCNS, and ESBLs among the different sites of vehicles, there were no pronounced trends in their distribution (Figure 1). However when taken together, portable ventilators and beds had higher counts, whereas DC shock apparatuses and steering wheels had neither of them.
Figure 1.
Frequency of methicillin-resistant staphylococci and ESBL-producing Klebsiella spp. and Escherichia coli in each site of ambulance vehicles.
Note: When comparing the frequency of MRSA, MRCNS, and ESBLs among the different sites of vehicles, there were no pronounced trends in their distribution. However when taken together, portable ventilators and beds had higher counts, whereas DC shock apparatus and steering wheels had neither of them.
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; MRCNS, methicillin-resistant coagulase-negative staphylococci; ESBL, extended spectrum β-lactamase.
Discussion
This study is the first work to describe the level of bacterial contamination in ambulance vehicles in Assiut city in Egypt. There seems to be a failure in effective cleaning and disinfection of the vehicles, demonstrating a possible lack of consistency in frequency and/or method of cleaning.
Bacterial contamination of inanimate surfaces and equipment in the patient zone and health care area is a major health problem worldwide as it may play a role in cross-transmission of pathogens that may lead to infection or colonization. This colonization may generate a reservoir of potential multidrug-resistant pathogens.2 Biofilm formation is one of the principle factors that enable bacteria to survive on inanimate surfaces and equipment. Bacterial biofilm may reduce the efficacy of terminal cleaning procedures as it enables bacteria to resist disinfectants and survive in the environment for a long time.17
A total of 25 ambulance vehicles were examined, 400 swabs were cultured, and 286 potential pathogenic organisms were isolated. Sites where the highest frequency of pathogenic bacteria were detected included beds, portable ventilators, oxygen humidifier glass, suction devices, headboards of patient stretchers, cervical collars, stethoscopes, and the carrying handles of carts. The presence of pathogenic organisms in the equipment and sites was expected because they are usually touched with the hands and come frequently in contact with patients or their body secretions such as pus, blood, and sweat. The relatively lower frequency of pathogenic bacteria detected on DC shock apparatus and steering wheels is probably due to the infrequent use of them compared with other equipment and the fact that steering wheels does not come in direct contact with patients. Consistent with our results, the SEKURE study that analyzed the bacterial contamination of German ambulances has reported that contact surfaces directly surrounding patients or staff were most frequently contaminated with microorganisms.1
In total, 184 isolates of staphylococci, 49 of K. pneumoniae, 40 E. coli, 6 Proteus spp., and 7 Citrobacter spp. were detected. From this abundant distribution of staphylococci and coliforms, it may be inferred that there was no time-tabled organized cleaning schedules. Furthermore, such a high level of pathogenic microorganisms is worrying since these organisms have been implicated in different serious community and nosocomial infections.18–21 Similar to our findings, other studies have pointed out the widespread microbial populations in ambulances and their role as a potential source of prehospital acquired infections.1,8,22–26
The presence of pathogenic organisms in rescue vehicles is also very dangerous, especially when transporting immunocompromised and severely ill patients who are more liable to infections. Moreover, such a high frequency of microorganisms is a potential risk of transmission not only to patients but also to emergency personnel. In addition, patients transported by ambulance may act as a vector and disseminate those disease-causing microorganisms to hospital environments and therefore contribute to the development of health care associated infections.
In the present study, antibiogram for the isolated environmental pathogens was determined. Alarmingly, isolated staphylococci showed high levels of resistance to different antibiotics, such as chloramphenicol (92.4%), trimethoprim/sulfamethoxazole (87.5%), ampicillin (78.3%), and tetracycline (78.3%), while no resistance to vancomycin was observed. A similar high level of resistance was also observed among Klebsiella spp. and E. coli but not among Proteus spp. and Citrobacter spp. About 90% of Klebsiella spp. and E. coli were resistant to ampicillin, tetracycline, and chloramphenicol, and about half of them were resistant to the third-generation cephalosporins while Proteus spp. and Citrobacter spp. showed only remarkable resistance to ampicillin antibiotic (66.7% and 47.1%, respectively) probably due to the limited frequency of Proteus spp. and Citrobacter spp. detected. Detection of such high counts of multidrug-resistant staphylococci, Klebsiella spp., and E. coli may be explained by the absence of infection control programs that monitor the hygienic measures in our ambulances leading to the consequent existence and spread of these organisms. If these organisms initiate an infection, only limited therapeutic choices will be available.
Since 64.3% of the isolates were identified as Staphylococcus spp., the potential for the existence of methicillin resistance was high. Further testing of these colonies detected methicillin resistance not only among S. aureus but also among S. epidermidis strains. About half of the S. aureus were MRSA and 20% of S. epidermidis were MRCNS. In addition, our study was the first study that described the prevalence of ESBL among environmental samples from emergency ambulances. ESBL-producing K. pneumoniae and E. coli were isolated from suction devices and portable ventilators. These multidrug-resistant bacteria may contribute to life-threatening respiratory tract infections if these types of equipment are reused for critically ill patients without being disinfected. No ESBL bacteria were detected among Proteus spp. and Citrobacter spp., most likely due to the limited number of isolates.
Both methicillin-resistant staphylococci and ESBL colonies were recovered from different sites within the ambulances, with the exception of DC shock apparatus and steering wheels that contained methicillin-sensitive S. epidermidis isolates that were probably skin contaminants. Our results showed no relationship between sites inside ambulances and methicillin resistance or ESBLs distribution indicating a possible cross-contamination within the vehicle.
Detection of MRSA, MRCNS, and ESBL raises concerns since these pathogens may cause severe life-threatening complications. It was shown that MRSA was detected in ambulance vehicles immediately after transport of MRSA-colonized or -infected patients.3 Other studies showed that ambulances have a significant degree of MRSA contamination and may therefore represent an important factor for transmission of serious infections to patients.9,27 Generally, ESBL and MRSA testing in the ambulance environment is useful for infection control purposes. Documenting these potential hazards may promote the development of best practices to reduce risks of transmission to transported patients.
Interestingly, studies have shown that a high density of bacterial contamination was not only detected in ground emergency ambulances1,8,9,28–30 but also detected in Emergency Service Helicopters.31,32
Based on our observations, it can be concluded that the level of microbial contamination in ambulances within Assiut city in Egypt is not acceptable. It is highly recommended that standard operating procedures of cleaning, decontamination, and disinfection of emergency vehicles and their equipment be implemented. This includes decreasing the contamination rate to the environment by colonized patients and contaminated health care workers by employing regular time-tabled cleaning schedules that are carried out by specialized infection control members, the use of appropriate disinfectants, appropriate cleaning of medical equipment in vehicles, management of water quality introduced into vehicles, avoiding non essential contact of patients’ relatives in the ambulance environment to limit concentrations of shed and airborne bacteria.33,34
Additional studies are also planned to evaluate and validate the disinfection protocols. Further research should be carried out to analyze the effect of seasonal climate changes on the prevalence of pathogens in ambulances. The reason for that is the well-known effect of temperature and environmental conditions on microbial growth. Our planned future studies will highlight the difference in microbial populations in the summer versus the winter.
Acknowledgments
We would like to thank all the participants in this study, particularly the ambulance workers.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Wepler M, Stahl W, von Baum H, et al. Prevalence of nosocomial pathogens in German ambulances: the SEKURE study. Emerg Med J. 2015;32(5):409–411. doi: 10.1136/emermed-2013-202551. [DOI] [PubMed] [Google Scholar]
- 2.Russotto V, Cortegiani A, Fasciana T, et al. What healthcare workers should know about environmental bacterial contamination in the intensive care unit. BioMed Res Int. 2017;2017:6905450. doi: 10.1155/2017/6905450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eibicht SJ, Vogel U. Methicillin-resistant Staphylococcus aureus (MRSA) contamination of ambulance cars after short term transport of MRSA-colonised patients is restricted to the stretcher. J Hosp Infect. 2011;78(3):221–225. doi: 10.1016/j.jhin.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 4.Mammina C, Cala C, Bonura C, et al. Polyclonal nonmultiresistant methicillin resistant Staphylococcus aureus isolates from clinical cases of infection occurring in Palermo, Italy, during a one-year surveillance period. Ann Clin Microbiol Antimicrob. 2012;11:17. doi: 10.1186/1476-0711-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geraci DM, Bonura C, Giuffre M, et al. Is the monoclonal spread of the ST258, KPC-3-producing clone being replaced in southern Italy by the dissemination of multiple clones of carbapenem-nonsusceptible, KPC-3-producing Klebsiella pneumoniae? Clin Microbiol Infect. 2015;21(3):e15–e17. doi: 10.1016/j.cmi.2014.08.022. [DOI] [PubMed] [Google Scholar]
- 6.Amiry Al A, Bissell RA, Maguire BJ, Alves DW. Methicillin-resistant Staphylococcus aureus nasal colonization prevalence among emergency medical services personnel. Prehosp Disaster Med. 2013;28(4):348–352. doi: 10.1017/S1049023X13003476. [DOI] [PubMed] [Google Scholar]
- 7.Merlin MA, Wong ML, Pryor PW, et al. Prevalence of methicillin-resistant Staphylococcus aureus on the stethoscopes of emergency medical services providers. Prehosp Emerg Care. 2009;13(1):71–74. doi: 10.1080/10903120802471972. [DOI] [PubMed] [Google Scholar]
- 8.Brown R, Minnon J, Schneider S, Vaughn J. Prevalence of methicillin-resistant Staphylococcus aureus in ambulances in southern Maine. Prehosp Emerg Care. 2010;14(2):176–181. doi: 10.3109/10903120903564480. [DOI] [PubMed] [Google Scholar]
- 9.Roline CE, Crumpecker C, Dunn TM. Can methicillin-resistant Staphylococcus aureus be found in an ambulance fleet? Prehosp Emerg Care. 2007;11(2):241–244. doi: 10.1080/10903120701205125. [DOI] [PubMed] [Google Scholar]
- 10.Winn WC, Allen SD, Janda W, et al. Koneman’s Color Atlas and Textbook of Diagnostic Microbiology. 6th ed. London: Lippincott, Williams & Wilkins; 2005. [Google Scholar]
- 11.Boyle VJ, Fancher ME, Ross RW., Jr Rapid, modified Kirby-Bauer susceptibility test with single, high-concentration antimicrobial disks. Antimicrob Agents Chemother. 1973;3(3):418–424. doi: 10.1128/aac.3.3.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing. Wayne: CLSI; 2014. CLSI document M100-S24. [Google Scholar]
- 13.Jarlier V, Nicolas MH, Fournier G, Philippon A. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev Infect Dis. 1988;10(4):867–878. doi: 10.1093/clinids/10.4.867. [DOI] [PubMed] [Google Scholar]
- 14.Simor AE, Goodfellow J, Louie L, Louie M. Evaluation of new medium, oxacillin resistance screening agar base, for the detection of methicillin-resistant Staphylococcus aureus from clinical specimens. J Clin Microbiol. 2001;39(9):3422. doi: 10.1128/JCM.39.9.3422.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed SH, Ahmed SA, Mohamed WA, et al. Nosocomial vancomycin and methicillin resistant staphylococcal infections in intensive care units in Assiut University Hospitals. Egyptian J Med Microbiol. 2011;20(2):127–140. [Google Scholar]
- 16.Felten A, Grandry B, Lagrange PH, Casin I. Evaluation of three techniques for detection of low-level methicillin-resistant Staphylococcus aureus (MRSA): a disk diffusion method with cefoxitin and moxalac-tam, the Vitek 2 system, and the MRSA-screen latex agglutination test. J Clin Microbiol. 2002;40(8):2766–2771. doi: 10.1128/JCM.40.8.2766-2771.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cala C, Amodio E, Di Carlo E, Virruso R, Fasciana T, Giammanco A. Biofilm production in Staphylococcus epidermidis strains, isolated from the skin of hospitalized patients: genetic and phenotypic characteristics. New Microbiol. 2015;38(4):521–529. [PubMed] [Google Scholar]
- 18.Mills JP, Talati NJ, Alby K, Han JH. The epidemiology of carbapenem-resistant Klebsiella pneumoniae colonization and infection among long-term acute care hospital residents. Infect Control Hosp Epidemiol. 2016;37(1):55–60. doi: 10.1017/ice.2015.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Latifpour M, Gholipour A, Damavandi MS. Prevalence of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates in nosocomial and community-acquired urinary tract infections. Jundishapur J Microbiol. 2016;9(3):e31179. doi: 10.5812/jjm.31179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martelius T, Jalava J, Karki T, et al. Nosocomial bloodstream infections caused by Escherichia coli and Klebsiella pneumoniae resistant to third-generation cephalosporins, Finland, 1999-2013 trends, patient characteristics and mortality. Infect Dis (Lond) 2016;48(3):229–234. doi: 10.3109/23744235.2015.1109135. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed SH, Daef EA, Badary MS, Mahmoud MA, Abd-Elsayed AA. Nosocomial blood stream infection in intensive care units at Assiut University Hospitals (Upper Egypt) with special reference to extended spectrum beta-lactamase producing organisms. BMC Res Notes. 2009;2:76. doi: 10.1186/1756-0500-2-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nigam Y, Cutter J. A preliminary investigation into bacterial contamination of Welsh emergency ambulances. Emerg Med J. 2003;20(5):479–482. doi: 10.1136/emj.20.5.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vikke HS, Giebner M. UniStatus – a cross-sectional study on the contamination of uniforms in the Danish ambulance service. BMC Res Notes. 2015;8:95. doi: 10.1186/s13104-015-1057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alves DW, Bissell RA. Bacterial pathogens in ambulances: results of unannounced sample collection. Prehosp Emerg Care. 2008;12(2):218–224. doi: 10.1080/10903120801906721. [DOI] [PubMed] [Google Scholar]
- 25.Alrazeeni D, Al Sufi MS. Nosocomial infections in ambulances and effectiveness of ambulance fumigation techniques in Saudi Arabia. Phase I study. Saudi Med J. 2014;35(11):1354–1360. [PMC free article] [PubMed] [Google Scholar]
- 26.Yoo IS, You Y, Kwon KC, Jeong TO. Bacterial contamination conditions in ambulances and their equipment in South Korea. J Korean Soc Emerg Med. 2012;23(1):1–7. [Google Scholar]
- 27.Ro YS, Shin SD, Noh H, Cho SI. Prevalence of positive carriage of tuberculosis, methicillin-resistant Staphylococcus aureus, and vancomycin-resistant Enterococci in patients transported by ambulance: a single center observational study. J Prev Med Public Health. 2012;45(3):174–180. doi: 10.3961/jpmph.2012.45.3.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mogensen CB, Kjaeldgaard P, Jensen C, Chen M. MRSA screening in emergency department detects a minority of MRSA carriers. Dan Med J. 2015;62(11):A5151. [PubMed] [Google Scholar]
- 29.Ribeiro J, Boyce JM, Zancanaro PQ. Prevalence of methicillin-resistant Staphylococcus aureus (MRSA) among patients visiting the emergency room at a tertiary hospital in Brazil. Braz J Infect Dis. 2005;9(1):52–55. doi: 10.1590/s1413-86702005000100009. [DOI] [PubMed] [Google Scholar]
- 30.Maguire BJ, Hunting KL, Guidotti TL, Smith GS. Occupational injuries among emergency medical services personnel. Prehosp Emerg Care. 2005;9(4):405–411. doi: 10.1080/10903120500255065. [DOI] [PubMed] [Google Scholar]
- 31.Makiela S, Taylor-Robinson AW, Weber A, Maguire BJ. A preliminary assessment of contamination of emergency service helicopters with MRSA and multi-resistant Staphylococcus aureus. Emerg Med Open Access. 2016;6:304. [Google Scholar]
- 32.Galtelli M, Deschamp C, Rogers J. An assessment of the prevalence of pathogenic microorganisms in the rotor wing air ambulance: one program’s findings. Air Med J. 2006;25(2):81–84. doi: 10.1016/j.amj.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Ruan S. Modeling nosocomial infections of methicillin-resistant Staphylococcus aureus with environment contamination. Sci Rep. 2017;7(1):580. doi: 10.1038/s41598-017-00261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Centers for Disease Control and Prevention Guidelines for environmental infection control in health-care facilities. Recommendations of CDC and the Healthcare Infection Control Practices Advisory Committee (HICPAC) MMWR. 2003;52(RR-10):1–48. [PubMed] [Google Scholar]