Abstract
Introduction
Coagulase-negative staphylococci (CNS) are normal commensals of human skin and mucous membranes. The objective of the study was to determine the prevalence of CNS among clinical isolates, characterize them up to species level, compare the three conventional methods for detection of biofilm formation, and study their antimicrobial susceptibility pattern.
Materials and methods
CNS were obtained from various clinical samples including blood, urine, central venous catheter tips, endotracheal tube aspirate, and pus during a 1-year period (July 1, 2014, to June 30, 2015). Characterization up to species level was done using biochemical tests, and biofilm formation was detected by tube adherence, Congo red agar, and tissue culture plate method. Antimicrobial susceptibility testing was performed following Clinical and Laboratory Standards Institute guidelines.
Results
A total of 71 CNS isolates, comprising of seven species were obtained. Staphylococcus epidermidis was the most common species followed by S. saprophyticus and S. haemolyticus. We detected biofilm formation in 71.8% of isolates. Considering the fact that tissue culture plate method is the gold standard, sensitivity of tube adherence method and Congo red agar method was found as 82% and 78%, respectively. The isolates exhibited high resistance toward penicillin (90%), azithromycin (60%), co-trimoxazole (60%), and ceftriaxone (40%), while all were susceptible to vancomycin and linezolid. Biofilm former isolates showed higher resistance than the non-formers.
Conclusion
Among 71 CNS isolated, S. epidermidis was the most common isolate followed by S. saprophyticus and S. haemolyticus. Biofilm formation was detected in 71.8% of the isolates. All of the methods were effective in detecting biofilm-producing CNS strains. The antimicrobial resistance was significantly higher in biofilm formers than non-formers.
Keywords: CNS, Congo red agar, bloodstream infections, foreign body-related infections, tissue culture plate
Video abstract
Introduction
Coagulase-negative staphylococci (CNS) are normal commensals of human skin and mucous membranes.1 They are typical opportunistic pathogens, especially in nosocomial settings, and have a substantial impact on human life and health. The use of implanted prosthetics or indwelling devices, which are now used invariably in modern medicine, is a major risk factor for CNS infection.2 CNS are now ranked most common infective agent in prosthetic valve infective endocarditis and third most common in native valve infective endocarditis, which demonstrates its significance in the clinical setting.3 Among various CNS, Staphylococcus epidermidis is the major cause of infections associated with catheters, surgical wounds, peritonitis, osteomyelitis, bloodstream infection, and endophthalmitis.4,5 S. saprophyticus is a common pathogen of urogenital tract infections, particularly in young, sexually active men and women, and is the second most common CNS causing human infection.6
Biofilm formation is one of the major virulence factors associated with these organisms which facilitate its adherence to and colonization in artificial materials.7 The biofilm protects CNS against the patient’s immune system and also against the action of antibiotics administered for the treatment of these infections.8 Biofilm-associated bacteria are usually less susceptible to antibiotics than planktonic bacteria; this can be explained by different mechanisms, such as the binding of antibiotics to biofilm components, reduced penetration of the antibiotic, slower growth of the microorganisms in the biofilm, high bacterial density, and altered gene expression in the bacteria present in the biofilm.9,10
Testing for the formation of biofilm is important in deciding the pathogenicity of CNS and should be routinely performed in diagnostic laboratories.11 The newer methods, such as confocal laser scanning microscopy, RNAseq, microarrays, and RT-qPCR, are expensive and difficult to perform in routine laboratories.12,13 Hence, reliable, convenient, and inexpensive methods are needed to identify CNS isolates and detect biofilm formation. These methods should be accessible to most diagnostic laboratories, particularly those located in resource-limited countries.14 In this regard, the objective of the study was to determine the prevalence of CNS among clinical isolates, characterize them up to species level, compare the three conventional methods for detection of biofilm formation, and study their antimicrobial susceptibility.
Materials and methods
Study design
A cross-sectional study was conducted at the Department of Microbiology, B.P. Koirala Institute of Health Sciences from July 1, 2014, to June 30, 2015. Among 5705 bacteria isolated during the period, we obtained 71 clinically significant CNS isolates from various clinical samples, i.e., blood, urine, pus, endotracheal tube (ETT) aspirate, and central venous catheter (CVC) tips, submitted to microbiology laboratory for routine and sensitivity testing. An attempt was made to establish significance by correlation with the clinical features and repeat culture of the specimens, whenever possible.
Isolation, identification, and characterization of CNS
All clinical samples excluding blood and urine were inoculated onto blood and MacConkey agar. The urine sample was plated on cysteine lactose electrolyte-deficient medium. All inoculated plates were incubated overnight at 35°C. Blood sample was inoculated in brain heart infusion broth and incubated overnight at 35°C before subculturing onto blood agar and MacConkey agar. Isolates which grew white opaque colonies, Gram-positive cocci in clusters on Gram staining, produced catalse, were slide and tube coagulase negative, and did not ferment mannitol were identified as CNS.15,16 Then, we characterized them up to species level using a battery of biochemical tests and antimicrobial discs following the identification model proposed by Kloos and Bannerman15 (Table 1).
Table 1.
Biochemical tests for the identification and speciation of CNS isolates
| Species | Urease | Acetoin production | Novobiocin | Polymyxin B | Slide coagulase | Tube coagulase | Alkaline phosphatase | Pyrrolidonyl arylamidase | Ornithine decarboxylase | Trehalose | Mannitol | Mannose | Xylose | Maltose | Sucrose |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Staphylococcus epidermidis | + | + | S | R | − | − | + | − | − | − | − | + | − | + | + |
| S. saprophyticus | + | + | R | S | − | − | − | − | − | + | − | − | − | + | + |
| S. haemolyticus | − | + | S | S | − | − | − | + | − | + | − | − | − | + | + |
| S. hominis | + | − | S | S | − | − | − | − | − | − | − | − | − | + | + |
| S. capitis | − | − | S | S | − | − | − | − | − | − | + | + | − | − | + |
| S. warneri | + | + | S | S | − | − | − | − | − | + | − | − | − | + | + |
| S. lugdunensis | D | + | S | D | + | − | + | + | + | − | + | − | + | + |
Notes: +, positive; −, negative; S, sensitive; R, resistant; D, differential. Adapted from Kloos WE, Bannerman TL. Update on clinical significance of coagulasenegative staphylococci. Clin Microbiol Rev. 1994;7(1):117–140. Amended with permission from American Society for Microbiology.15
Abbreviation: CNS, coagulase-negative staphylococci.
Study of biofilm formation
We studied the biofilm production of the isolates by tube adherence method, tissue culture plate (TCP) method, and Congo red agar (CRA) method.17–19 We considered TCP method as the gold standard for the detection of biofilm formation and interpreted the results accordingly.20,21 We used S. epidermidis ATCC 35984 and S. epidermidis ATCC 12228 as positive and negative control, respectively, for biofilm formation.18
Tube adherence method
We inoculated a loopful of colony suspension from an overnight culture into the trypticase soy broth (TSB) medium (HiMedia, Mumbai, India) and incubated it for 24 hours at 35°C. Then, we inverted the tubes and washed it with phosphate-buffered saline (PBS) of pH 7.3. After drying, we stained them with 0.1% crystal violet (HiMedia). We rinsed the tubes multiple times with running tap water to remove excess stains. We kept the tubes in an inverted position and observed for biofilm formation.17 The experiment was performed in triplicate.
TCP method
This method, proposed by Christensen et al, is the gold standard method for detection of biofilm formation.18 We inoculated the isolates in TSB medium (HiMedia) and incubated it for 18–24 hours at 37°C in aerobic conditions. Then, we filled the individual wells of 96-well TCPs with 100 µL of culture suspension. After overnight incubation, we tapped the plate gently for the removal of free-flowing bacteria and washed four times with PBS. We air-dried, fixed, and stained the plates with 0.1% crystal violet for 5 minutes. After that, we rinsed the plate under running tap water several times to remove excess stain. The wells which retained uniform staining of crystal violet were interpreted as biofilm formers.18
CRA method
This method was proposed by Freeman et al. We prepared the CRA by mixing 37 g brain heart infusion broth, 50 g sucrose, 0.8 g Congo red dye, and 10 g agar (all from HiMedia) in 1 L distilled water. Then, we inoculated the isolates on CRA and incubated it aerobically at 35°C for 18–24 hours. The strains which produced biofilm formed black colonies while non-forming isolates developed red colonies.19 The experiment was performed in triplicate.
Quality control
S. epidermidis ATCC 35984 was used as positive control for biofilm formation and S. epidermidis ATCC 12228 was used as negative control.18
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed by Kirby Bauer disc diffusion method following the Clinical and Laboratory Standards Institute (CLSI) guidelines against these antimicrobials: amikacin, ceftriaxone, cefoxitin, ofloxacin, penicillin, co-trimoxazole, and linezolid. Vancomycin susceptibility was tested by calculating the minimum inhibitory concentration (MIC) of vancomycin against the isolates using agar dilution method. The antimicrobial discs were selected on the basis of CLSI guidelines 2014.22
Methicillin-resistant coagulase-negative staphylococci (MRCNS) and MIC
Methicillin resistance in CNS was detected by using cefoxitin disc diffusion test (30 µg). It was further confirmed as MRCNS by calculating the MIC of oxacillin against the isolates using the agar dilution method.22,23
Quality control of antimicrobial susceptibility testing
All the antimicrobial discs were tested against S. aureus ATCC 25923 to ensure the potency of the discs.22
Data analysis
The data were entered into Microsoft Excel 2013 and analyzed by using SPSS version 16 (SPSS Inc., Chicago, IL, USA). Chi-square test was applied and p-value <0.05 was considered statistically significant.
Results
Isolates and identification
A total of 5705 bacteria were isolated during the study period, 71 (1.24%) of them were CNS. Among the 71 isolates, 17 were obtained from blood, 16 from urine, 13 from pus, 17 from ETT, and 8 from CVC. A total of seven species were identified. S. epidermidis (40%, n=28) was the most common species followed by S. saprophyticus (18%, n=13), S. haemolyticus (14%, n=10), and S. lugdunensis (11%, n=8; Figure 1).
Figure 1.
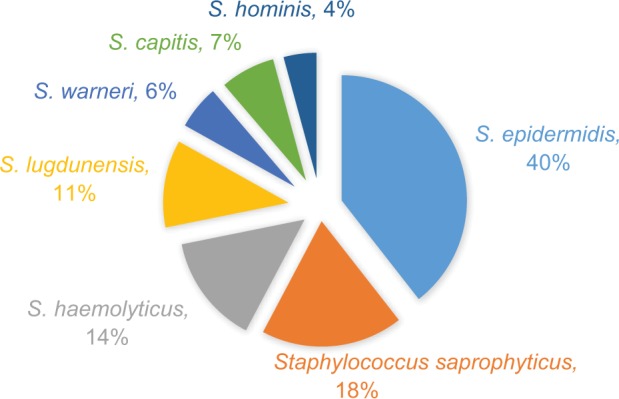
Different species of CNS isolated from various clinical samples (n=71).
Abbreviation: CNS, coagulase-negative staphylococci.
Study of biofilm production
Among the 71 CNS isolates, 51 (71.8%) were biofilm producers. With respect to the clinical sample, biofilm formation was detected among 94% (16/17) of isolates obtained from ETT, 87% (7/8) from CVC tube, 70% (12/17) from blood, 84.6% (11/13) from pus, and 31% (5/16) from the urine sample. S. epidermidis, the most common isolate, showed biofilm formation in 82% (23/28) of isolates, while S. haemolyticus showed 90% (9/10) biofilm forming ability. The least biofilm formation was observed in S. saprophyticus, i.e., 15% (2/13; Table 2).
Table 2.
Biofilm formation with respect to the species and clinical samples
| Species | Blood (n=17) |
Urine (n=16) |
Pus (n=13) |
ETT (n=17) |
CVC (n=8) |
Number of biofilm formers |
|---|---|---|---|---|---|---|
| Staphylococcus epidermidis (n=28) | 5 | 2 | 6 | 4 | 6 | 23 |
| S. saprophyticus (n=13) | 2 | 2 | ||||
| S. haemolyticus (n=10) | 2 | 1 | 2 | 4 | 9 | |
| S. lugdunensis (n=8) | 1 | 2 | 2 | 1 | 6 | |
| S. capitis (n=5) | 2 | 1 | 2 | 5 | ||
| S. warneri (n=4) | 1 | 2 | 3 | |||
| S. hominis (n=3) | 1 | 2 | 3 | |||
| Total | 12 | 5 | 11 | 16 | 7 | 51/71 |
Abbreviations: ETT, endotracheal tube; CVC, central venous catheter.
Among the 51 biofilm forming CNS isolates, tissue culture method detected biofilm production in 50 isolates, tube method in 42 isolates, and CRA method in 40 isolates. Statistical analysis was done using 2 × 2 table to calculate the sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and accuracy of CRA and tube adherence method considering TCP method as the gold standard method (Table 3). The sensitivity and accuracy of the tube adherence method (82% and 85.9%) was higher than CRA method (78% and 83%). However the specificity, PPV, and NPV were almost the same.
Table 3.
Statistical evaluation of Congo red agar and tube adherence method for detection of biofilm formation
| Methods | Sensitivity (%) | Specificity (%) | PPV (%) | NPV (%) | Accuracy (%) |
|---|---|---|---|---|---|
| Congo red agar | 78 | 95.2 | 97.5 | 64.5 | 83 |
| Tube adherence method | 82 | 95.2 | 97.6 | 64.5 | 85.9 |
Abbreviations: PPV, positive predictive value; NPV, negative predictive value.
Antimicrobial resistance
Antimicrobial susceptibility pattern of the isolates showed a variable level of resistance; 90% to penicillin, 40% to ceftriaxone, 60% to co-trimoxazole, and 60% to azithromycin. All the isolates were susceptible to vancomycin and linezolid. Forty percent of the isolates were MRCNS which was confirmed by MIC of oxacillin against the isolates.
The antimicrobial susceptibility was also studied on the basis of biofilm forming ability of the isolates. Biofilm former strains showed higher resistance than the non-former isolates. Forty-two percent of biofilm formers were resistant to amikacin while the figure for biofilm non-formers was 14%, which is statistically significant (p-value <0.05). A similar type of resistance pattern was seen with most of the antimicrobials used (Table 4).
Table 4.
Antimicrobial resistance of the biofilm former isolates vs non-formers
| Antibiotics | Biofilm former (n=50) (resistance in %) |
Biofilm non-former (n=21) (resistance in %) |
p-value |
|---|---|---|---|
| Amikacin | 42 | 14 | 0.024 |
| Azithromycin | 66 | 33 | 0.011 |
| Cefoxitin | 50 | 24 | 0.041 |
| Ceftriaxone | 50 | 24 | 0.041 |
| Co-trimoxazole | 72 | 43 | 0.02 |
| Ofloxacin | 72 | 38 | 0.007 |
| Penicillin | 92 | 71 | 0.023 |
| Vancomycin | 0 | 0 | * |
| Linezolid | 0 | 0 | * |
Note:
No resistance was observed against these antimicrobials, p-value could not be calculated.
Discussion
CNS are important causative agents of implanted device-related infections, endocarditis, bloodstream infections, urinary tract infections (UTIs), ophthalmitis, and soft tissue infections.1 However, a large proportion of CNS are reported as possible laboratory contaminants without identifying their species. Since many species of CNS have their own clinical significance and are associated with biofilm production, their characterization up to species level and association of biofilm production should be prioritized in diagnostic laboratories. Therefore, simple, reliable, and inexpensive methods should be the focus in resource-constrained settings.14,24
In the present study, we isolated 71 clinically significant CNS from various clinical samples, i.e., blood (n=17), urine (n=16), ETT (n=17), CVC (n=8), and pus (n=13). Device (ETT, CVC tip) and blood samples were the major source of CNS isolates in our study. Similar results have been reported by Oliveria and Cunha Mde, who obtained 50 isolates from catheter tips and 30 from blood.13 Foreign body-related infections, also known as “device associated health care-associated infections (DA-HAIs)”, are the most significant clinical entity associated with CNS.25
We identified a total of seven CNS species. S. epidermidis (40%, n=28) was the most common species followed by S. saprophyticus (18%, n=13), S. haemolyticus (14%, n=10), S. lugdunensis (11%, n=8), S. capitis (7%, n=5), S. warneri (6%, n=4), and S. hominis (4%, n=3). The finding of our study is consistent with many other studies.24,26,27 S. epidermidis is the most common CNS species causing human infections.2,7 In contrast to the finding of our study, Jain et al isolated S. haemolyticus (58%) as the most common isolate, followed by S. epidermidis (17%).28 Another study conducted by Kashid and Raghuraman in India showed that S. haemolyticus (30%) was the most common species isolated followed by S. warneri (14%), which is in contrast to our findings.29 The finding of S. epidermidis as the most commonly isolated species in our study might be due to the fact that this organism produces biofilm which helps it to attach to surfaces, and it is the most prevalent bacterium in human skin and mucosa.2,13 The human body is colonized with CNS during the first few days after birth, with S. epidermidis, S. warneri, and S. haemolyticus as the most prevalent species.2,30 S. saprophyticus was the second most common species isolated in our study, all of which were obtained from the urine sample. Literature suggests that S. saprophyticus is the second most frequent causative microorganism of uncomplicated lower UTIs in young, sexually active women.2
Biofilm formation is one of the major virulence factors of CNS.6 The bacteria present inside a biofilm are protected against the action of the host immune system and antimicrobial drugs, thus permitting their survival.9 In our study, we documented that 71.8% (51/71) of the isolates were biofilm producing. This finding is consistent with the study conducted by Chokr et al, who detected biofilm formation in 73% of CNS isolates.31 In some studies, higher rates of biofilm formation have been reported. In a study conducted by Oliveria and Cunha Mde, 81% of the CNS isolates were biofilm formers.13 Similar findings have been reported by Cafiso et al (83%)32 and Soumya et al (87%).33 However, lower positive rates have been reported by Wojtyczka et al (37.5%)34 and Thilakanthy et al (39.5%).26 The higher rate of biofilm formation in our study (71.8%, n=51) may be due to the fact that most of our isolates were obtained from ETT and CVC tip, in which biofilm formation occurs invariably.2,25
We investigated biofilm production using three methods, i.e., tube method, TCP method, and CRA method. Among 51 biofilm forming isolates, tissue culture method detected the biofilm production in 50 isolates, tube method in 42 isolates, and CRA method in 40 isolates. Statistical analysis was done using a 2 × 2 table considering TCP as the gold standard method.20,21
The sensitivity, specificity, PPV, NPV, and accuracy of the tube adherence method was 82%, 95%, 97.5%, 64.5%, and 83.5%, respectively, while that of CRA method was 78%, 95.2%, 97.6%, 64.5%, and 83%, respectively. The sensitivity of the tube adherence method (82%) was slightly higher than the CRA method (78%) while specificity, PPV, NPV, and accuracy of both tube adherence method and CRA methods were similar. The finding of our study is consistent with the study by Oliveira and Cunha Mde. According to the authors, the sensitivity of the tube method was 100% as compared to 89% of CRA, but specificity of both methods was 100%.13 The finding is also supported by many other studies which report the superiority of tube adherence method as compared to CRA method for detection of biofilm formation.26,35 Although the sensitivity of CRA method is less than tube adherence method, since it is less laborious, quicker, and requires less equipment than tube adherence method for detecting slime production, it would be very useful in clinical microbiology laboratories.36
All the CNS isolates obtained from clinical samples should be characterized and the biofilm forming ability should be assessed by any of these methods in routine microbiology laboratories.
Biofilm forming bacteria are usually less susceptible to antibiotics than planktonic bacteria. The reduced penetration of the antibiotic, binding of antibiotics to biofilm components, high bacterial density, and slower bacterial growth inside biofilm could be the factors contributing to the higher antimicrobial resistance.9,10 Antimicrobial susceptibility pattern of the isolates showed a variable level of resistance. None of the isolates were resistant to vancomycin and linezolid, while 90% of the isolates were resistant to penicillin. Resistance against ceftriaxone and cefoxitin were 40%. The result of our study is similar to those of the study conducted by Jain et al. The authors showed that 94% of the CNS isolates were resistant to penicillin and none of them were resistant to vancomycin.28 The resistance pattern, when compared between the biofilm producers and non-producers, showed significant differences. The result of our study is similar to those of a study by Soumya et al.33 In contrast to our results, Hassan et al concluded that the differences in antimicrobial resistance between biofilm formers and non-formers are statistically insignificant.37 However, other studies suggest that significant differences in antimicrobial susceptibility between biofilm formers and non-formers exist.38,39 These differences could be explained by the fact that biofilm protects the bacteria from the action of antimicrobials making them resistant to most antimicrobials.2
CNS are emerging multidrug-resistant pathogens, and hence, studies on their local species distribution, antibiotic sensitivity, and prevalence of biofilm-formation are very important.
Conclusion
A total of 71 CNS were isolated from various clinical samples. S. epidermidis was the most common isolate followed by S. saprophyticus and S. haemolyticus. Biofilm formation was detected in 71.8% of the isolates. CRA, tube adherence method, and tissue culture method are all effective in detecting biofilm formation. Antimicrobial resistance is significantly higher in biofilm formers than the non-former strains.
Acknowledgments
We would like to thank Dr Prajwal Pyakurel for his help with the biostatistics. We would also like to acknowledge all the faculty members and laboratory staff of the Department of Microbiology for their continuous support during the study period.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Rupp ME, Fey PD. Staphylococci and other coagulase-negative Staphylococci. In: Mandell GL, Benett JE, Dolin R, editors. Principle & Practice of infectious diseases. 7th ed. Philadelphia: Churchill Livingstone Elsevier; 2010. pp. 2579–2581. [Google Scholar]
- 2.Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27(4):870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murdoch DR, Corey GR, Hoen B, et al. Clinical presentation, etiology, and outcome of infective endocarditis in the 21st century: the International Collaboration on Endocarditis-Prospective Cohort Study. Arch Intern Med. 2009;169(5):463–473. doi: 10.1001/archinternmed.2008.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Gara JP, Humphreys H. Staphylococcus epidermidis biofilms: importance and implications. J Med Microbiol. 2001;50(7):582–587. doi: 10.1099/0022-1317-50-7-582. [DOI] [PubMed] [Google Scholar]
- 5.Upadhyayula S, Kambalapalli M, Asmar BI. Staphylococcus epidermidis urinary tract infection in an infant. Case Rep Infect Dis. 2012;2012:983153. doi: 10.1155/2012/983153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Eiff C, Proctor RA, Peters G. Coagulase-negative staphylococci. Pathogens have major role in nosocomial infections. Postgrad Med. 2001;110(4):63–64. 69–70, 73–76. [PubMed] [Google Scholar]
- 7.Otto M. Virulence factors of the coagulase-negative staphylococci. Front Biosci. 2004;9:841–863. doi: 10.2741/1295. [DOI] [PubMed] [Google Scholar]
- 8.Klingenberg C, Aarag E, Ronnestad A, et al. Coagulase-negative staphylococcal sepsis in neonates. Association between antibiotic resistance, biofilm formation and the host inflammatory response. Pediatr Infect Dis J. 2005;24(9):817–822. doi: 10.1097/01.inf.0000176735.20008.cd. [DOI] [PubMed] [Google Scholar]
- 9.Pinheiro L, Brito CI, Pereira VC, Oliveira A, Camargo CH, Cunha Mde L. Reduced susceptibility to vancomycin and biofilm formation in methicillin-resistant Staphylococcus epidermidis isolated from blood cultures. Mem Inst Oswaldo Cruz. 2014;109(7):871–878. doi: 10.1590/0074-0276140120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358(9276):135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 11.Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74(2):470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franklin MJ, Chang C, Akiyama T, Bothner B. New technologies for studying biofilms. Microbiol Spectr. 2015;3(4) doi: 10.1128/microbiolspec.MB-0016-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliveira A, Cunha Mde L. Comparison of methods for the detection of biofilm production in coagulase-negative staphylococci. BMC Res Notes. 2010;3:260. doi: 10.1186/1756-0500-3-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goyal R, Singh NP, Kumar A, et al. Simple and economical method for speciation and resistotyping of clinically significant coagulase negative staphylococci. Indian J Med Microbiol. 2006;24(3):201–204. [PubMed] [Google Scholar]
- 15.Kloos WE, Bannerman TL. Update on clinical significance of coagulase-negative staphylococci. Clin Microbiol Rev. 1994;7(1):117–140. doi: 10.1128/cmr.7.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloos WE, Schleifer KH. Simplified scheme for routine identification of human Staphylococcus species. J Clin Microbiol. 1975;1(1):82–88. doi: 10.1128/jcm.1.1.82-88.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christensen GD, Simpson WA, Bisno AL, Beachey EH. Adherence of slime-producing strains of Staphylococcus epidermidis to smooth surfaces. Infect Immun. 1982;37(1):318–326. doi: 10.1128/iai.37.1.318-326.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Christensen GD, Simpson WA, Younger JJ, et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J Clin Microbiol. 1985;22(6):996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freeman DJ, Falkiner FR, Keane CT. New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol. 1989;42(8):872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mathur T, Singhal S, Khan S, Upadhyay DJ, Fatma T, Rattan A. Detection of biofilm formation among the clinical isolates of Staphylococci: an evaluation of three different screening methods. Indian J Med Microbiol. 2006;24(1):25–29. doi: 10.4103/0255-0857.19890. [DOI] [PubMed] [Google Scholar]
- 21.Jain A, Agarwal A. Biofilm production, a marker of pathogenic potential of colonizing and commensal staphylococci. J Microbiol Methods. 2009;76(1):88–92. doi: 10.1016/j.mimet.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 22.Clinical and Laboratory Standards Institute . CLSI document M100-S24. Performance Standards for Antimicrobial Susceptibility Testing. 24th ed. Wayne, PA: CLSI; 2014. [Google Scholar]
- 23.Clinical and Laboratory Standards Institute . CLSI document no M07-A10 Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. 10th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2015. [Google Scholar]
- 24.Usha MG, Shwetha DC, Vishwanath G. Speciation of coagulase negative Staphylococcal isolates from clinically significant specimens and their antibiogram. Indian J Pathol Microbiol. 2013;56(3):258–260. doi: 10.4103/0377-4929.120383. [DOI] [PubMed] [Google Scholar]
- 25.von Eiff C, Jansen B, Kohnen W, Becker K. Infections associated with medical devices: pathogenesis, management and prophylaxis. Drugs. 2005;65(2):179–214. doi: 10.2165/00003495-200565020-00003. [DOI] [PubMed] [Google Scholar]
- 26.Thilakavathy P, Priyan RM, Jagatheeswari PA, et al. Evaluation of Ica gene in comparison with phenotypic methods for detection of biofilm production by coagulase negative staphylococci in a tertiary care hospital. J Clin Diagn Res. 2015;9(8):DC16–DC19. doi: 10.7860/JCDR/2015/11725.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al Tayyar IA, Al-Zoubi MS, Hussein E, Khudairat S, Sarosiekf K. Prevalence and antimicrobial susceptibility pattern of coagulase-negative staphylococci (CoNS) isolated from clinical specimens in Northern of Jordan. Iran J Microbiol. 2015;7(6):294–301. [PMC free article] [PubMed] [Google Scholar]
- 28.Jain A, Agarwal J, Bansal S. Prevalence of methicillin-resistant, coagulase-negative staphylococci in neonatal intensive care units: findings from a tertiary care hospital in India. J Med Microbiol. 2004;53(Pt 9):941–944. doi: 10.1099/jmm.0.45565-0. [DOI] [PubMed] [Google Scholar]
- 29.Kashid RA, Raghuraman K. Speciation and antimicrobial susceptibility of coagulase negative staphylococci, isolated from the anterior nares of health care workers, in a tertiary care hospital in South India, with special reference to methicillin resistance. Int J Contemporary Med Res. 2016;3(8):2329–2333. [Google Scholar]
- 30.Bjorkqvist M, Liljedahl M, Zimmermann J, Schollin J, Soderquist B. Colonization pattern of coagulase-negative staphylococci in preterm neonates and the relation to bacteremia. Eur J Clin Microbiol Infect Dis. 2010;29(9):1085–1093. doi: 10.1007/s10096-010-0966-3. [DOI] [PubMed] [Google Scholar]
- 31.Chokr A, Watier D, Eleaume H, et al. Correlation between biofilm formation and production of polysaccharide intercellular adhesin in clinical isolates of coagulase-negative staphylococci. Int J Med Microbiol. 2006;296(6):381–388. doi: 10.1016/j.ijmm.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 32.Cafiso V, Bertuccio T, Santagati M, et al. Presence of the ica operon in clinical isolates of Staphylococcus epidermidis and its role in biofilm production. Clin Microbiol Infect. 2004;10(12):1081–1088. doi: 10.1111/j.1469-0691.2004.01024.x. [DOI] [PubMed] [Google Scholar]
- 33.Soumya KR, Philip S, Sugathan S, Mathew J, Radhakrishnan EK. Virulence factors associated with coagulase negative staphylococci isolated from human infections. 3 Biotech. 2017;7(2):140. doi: 10.1007/s13205-017-0753-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wojtyczka RD, Orlewska K, Kepa M, et al. Biofilm formation and antimicrobial susceptibility of Staphylococcus epidermidis strains from a hospital environment. Int J Environ Res Public Health. 2014;11(5):4619–4633. doi: 10.3390/ijerph110504619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruzicka F, Hola V, Votava M, et al. Biofilm detection and the clinical significance of Staphylococcus epidermidis isolates. Folia Microbiol (Praha) 2004;49(5):596–600. doi: 10.1007/BF02931540. [DOI] [PubMed] [Google Scholar]
- 36.Garcia P, Benitez R, Lam M, et al. Coagulase-negative staphylococci: clinical, microbiological and molecular features to predict true bacteraemia. J Med Microbiol. 2004;53(Pt 1):67–72. doi: 10.1099/jmm.0.04994-0. [DOI] [PubMed] [Google Scholar]
- 37.Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis. 2011;15(4):305–311. [PubMed] [Google Scholar]
- 38.Qi L, Li H, Zhang C, et al. Relationship between antibiotic resistance, biofilm formation, and biofilm-specific resistance in Acinetobacter baumannii. Front Microbiol. 2016;7:483. doi: 10.3389/fmicb.2016.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerca N, Martins S, Cerca F, et al. Comparative assessment of antibiotic susceptibility of coagulase-negative staphylococci in biofilm versus planktonic culture as assessed by bacterial enumeration or rapid XTT colorimetry. J Antimicrob Chemother. 2005;56(2):331–336. doi: 10.1093/jac/dki217. [DOI] [PMC free article] [PubMed] [Google Scholar]


