Abstract
Peptides rich in basic residues such as lysine and arginine play important roles in biology such as bacterial defense and cell penetration. Although peptide-binding materials with high sequence-specificity have broad potential applications, the diverse functionalities of peptide side chains make their molecular recognition extremely difficult. By covalently capturing micelles of a doubly cross-linkable surfactant with solubilized peptide templates, we prepared water-soluble molecularly imprinted nanoparticles with high sequence-specificity for basic peptides. The nanoparticles interact with the side chains of lysine and arginine through hydrogen bonds strengthened by the nonpolar environment of the micelle. They have hydrophobic pockets in their core complementary to the hydrophobic side chains in size and shape. These recognition sites allowed the micelles to bind basic biological peptides strongly in water, with tens to hundreds of nanomolar in binding affinity.
Graphical Abstract
Introduction
Peptides rich in basic residues such as lysine and arginine play important roles in biology. Their cationic charge under physiological conditions help them bind negative groups on cell membranes. Strategically positioned hydrophobic side chains allow the peptides to undergo appropriate conformational changes when they bind to membranes, either lysing the cell as in antimicrobial peptides1,2 or crossing the membrane as in cell-penetrating peptides.3–7 These important features have prompted continuous research effort to understand their structure–activity relationships and develop novel therapeutic and delivery agents. Materials that can selectively bind such peptides in aqueous milieu are expected to be particularly useful in their separation, purification, sensing, and biological study.
Sequence-selective recognition of oligopeptides attracted the attention of chemists and biologists for decades.8–19 A general method to recognize peptides, however, is elusive due to the many difficult challenges involved.20,21 One difficulty comes from the highly diverse functional groups on these guest molecules. Even with many tools developed in supramolecular chemistry in the last decades, researchers do not have recognition motifs for the unique hydrophobic, hydrophilic, acidic, and basic side chains of most amino acids. Not only so, the side chains of some amino acids differ minutely: leucine and isoleucine, for example, only have the position of their methyl change by one carbon. Such subtlety demands an extremely high level of precision in an effective peptide receptor.
The second difficulty in peptide recognition is related to size. As a peptide guest gets longer, its receptor, in order to possess a complementary binding interface, necessarily needs to increase in size and complexity. One strand of DNA can bind its complementary strand with matching hydrogen-bonding code, but a concave binding interface is needed to accommodate the different side chains of peptides, implying a much larger host structure is required for peptide recognition. Even if individual recognition motifs or “codes” can be developed for each amino acids, joining the individual motifs on an appropriate scaffold in a preorganized fashion to recognize the matching peptide will take tremendous efforts, especially if the host is a discrete molecule built from the scratch.
For biological applications, the medium represents the third difficulty, as directional intermolecular forces such as hydrogen bonds are compromised severely by competition from water.22,23 Although hydrophobic interactions can be strong in water, their usage for selective molecular recognition is not as straightforward as in hydrogen-bonded systems with clearly defined donor–acceptor patterns.24
Molecular imprinting is a conceptual breakaway from the molecular construction of receptors and relies on “chemical molding”—i.e., binding, polymerization, and cross-linking—around the templates to afford the binding sites directly. Provided that appropriate functional monomers (FMs) are available for the type of templates involved and the imprinting process works well, individual design of the receptor is eliminated.25–35 This feature makes the method inherently general and potentially highly powerful. Many imprinted materials indeed have been created for peptides, showing promising results for various applications.36–41
An ideal peptide receptor should be generally applicable to peptides with varying properties and lengths. For biological applications, water-solubility is important. Their synthesis and purification should be easy, even better if achievable by nonchemists.
We recently reported our first step toward a general method for peptide recognition. Using micellar imprinting, a technique developed in our laboratory,42 we created molecularly imprinted nanoparticles (MINPs) with high affinity (as low as 20 nM in water) and exceptional selectivity for peptides 2–12 residues in length.43 Recognition was based on imprinted “hydrophobic dimples” created on the surface of the cross-linked micelle that complement the hydrophobic side chains of the templating peptide in size and shape. These “dimples” essentially encoded the MINP receptors with molecular recognition information for the peptide guests and were precise enough to distinguish leucine and isoleucine, as well as phenylalanine and tyrosine that differ by one hydroxyl.43
The previously reported method, nonetheless, mainly relied on hydrophobic interactions for the imprinting and binding. Thus, highly polar peptides including basic ones are unlikely to work well with the previous materials. Herein, we report methods to target the side chains of lysine and arginine, in addition to hydrophobic amino acids. As mentioned above, the basic and hydrophobic side chains together are critical to the antimicrobial and cell-penetrating properties of this important class of peptides. Materials that can recognize these groups should facilitate the biological study of these peptides. The strategies used in the binding also are useful to the fundamental understanding of molecular recognition in water, an important challenge in supramolecular chemistry.
Results and Discussion
Design and Synthesis
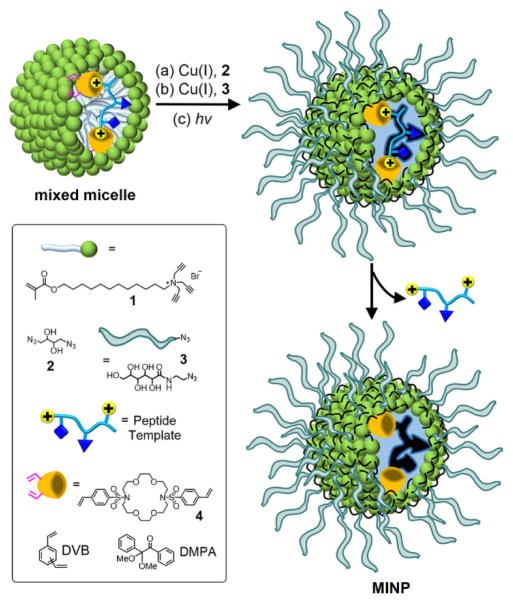
The synthesis of receptors for basic peptides was adapted from previous procedures.42–47 As shown in Scheme 1, the peptide was first solubilized by mixed micelles comprising cross-linkable surfactant 1, an equivalent amount of divinylbenzene (DVB, a free radical cross-linker), a small amount of 2,2-dimethoxy-2-phenylacetophenone (DMPA, a photoinitiator), and the appropriate FM. Although cationic materials are expected to repel basic peptides that tend to be positively charged under physiological conditions, we envisioned their usage would minimize non-specific adsorption of the peptides and thus enhance selectivity in the binding (vide infra).
SCHEME 1.
Preparation of peptide-binding MINP.
The micelles were cross-linked by diazide 2 on surface and then decorated with a layer of hydrophilic groups by sugar-derived azide 3, both using the Cu(I)-catalyzed click reaction.48–50 UV irradiation led to free radical polymerization/cross-linking of the core between the methacrylate of 1 and DVB. Repeated solvent washing removed the templates to vacate the binding sites.
The MINPs were characterized by previously reported procedures.42–47 The surface- and core-cross-linking were monitored by 1H NMR spectroscopy (see Supporting Information for details). The surface-cross-linking has been verified by mass spectrometry after the 1,2-diol in the cross-linked 2 was cleaved.48,49 Dynamic light scattering (DLS) afforded the size of the nanoparticles and their average molecular weights. The DLS size (~5 nm) was confirmed by transmission electron microscopy (Figure 1).
FIGURE 1.
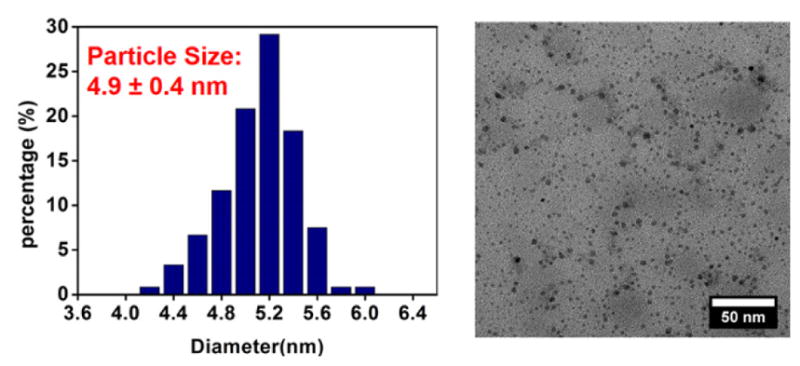
Particle size distribution from TEM imaging of MINPs.
Creating complementary hydrophobic “dimples” for the hydrophobic side chains of peptides does not require any FMs beyond the cross-linkable surfactant and DVB that polymerize around the template. As long as the double cross-linking of the micelle could “freeze” the initial peptide-binding state, the resulting MINP could bind the templating peptide afterwards. Given the exceptional selectivity in binding for leucine/isoleucine and phenylalanine/tyrosine, the polymerization and cross-linking in the confined nanospace of micelle must be extremely amenable to imprinting.43
For peptides rich in polar amino acids such as lysine and arginine, however, imprinting would not be as easy with the surfactant and DVB. First of all, these polar side chains are expected to stay on the surface of the micelle to be solvated by water. Even if they can migrate into the core of the micelle (e.g., in the amino form for lysine), the hydrophobic driving binding interactions will be quite weak. Thus, to imprint effectively, we need to have appropriate FMs to bind the polar side chains and, importantly, the binding needs to have sufficient strength in an aqueous micellar solution.51,52
As shown in Scheme 1, the FM utilized in this work was 4, prepared from the commercially available azacrown derivative.53 Its two styrenyl groups enable the FM to be covalently attached to the MINP during the core-cross-linking. Crown ether derivatives (including azacrowns) are known to bind amine,54 ammonium55,56 and guanidinium57–59 groups by various hydrogen-bonding motifs. We envisioned that the sulfonamide groups could provide additional sites for hydrogen-bonding and may be particularly suitable to bind the guanidinium side chain of arginine. Our hope was that the styrenyl groups of 4 would provide enough hydrophobicity for the FM to migrate to the hydrophobic region of the micelle where hydrogen bonds might be sufficiently strong to allow successful imprinting.
Binding Affinities
The first templates studied were tripeptide lysine-tryptophan-tryptophan (KWW) and lysine-lysine-lysine (KKK), with their structures shown below. KWW is very hydrophobic with two large indole side chains and KKK has no hydrophobic side chains. Their comparison thus could help us understand the effectiveness of FM 4 for two very different classes of peptides.
Table 1 summarizes the results of our binding studies. We included the binding data for MINPs prepared with and without FM 4 generally. We also listed the Krel values, which is the binding constant of the MINP with FM 4 relative to that without, and thus is a measure of the effectiveness of the FM in the molecular imprinting.
Table 1.
Comparison of binding for peptides by MINPs prepared with and without FM 4.a
| Entry | template | FM | −ΔG (kcal/mol) | Ka (×105 M−1) | Krel |
|---|---|---|---|---|---|
| 1 | KWW | none | 8.62 | 20.7 ± 1.7 | 1 |
| KWWb | none | 8.58 | 19.5 ± 2.2 | -- | |
| KWWc | none | 8.47 | 16.3 ± 1.8 | -- | |
| 2 | KWW | 1 equiv 4 | 8.78 | 27.5 ± 1.2 | 1.33 |
| 3 | KWW | 2 equiv 4 | 8.96 | 36.7 ± 2.4 | 1.77 |
| KWWb | 2 equiv 4 | 8.89 | 33.1 ± 1.9 | -- | |
| KWWc | 2 equiv 4 | 8.99 | 39.2 ± 3.0 | -- | |
| 4 | KWW | 3 equiv 4 | 8.94 | 35.9 ± 2.2 | 1.73 |
| 5 | KKK | none | 5.56 | 0.12 ± 0.02 | 1 |
| KKKb | none | 5.70 | 0.15 ± 0.01 | -- | |
| KKKc | none | 5.32 | 0.08 ± 0.01 | -- | |
| 6 | KKK | 2 equiv 4 | 5.74 | 0.16 ± 0.02 | 1.3 |
| 7 | KKK | 3 equiv 4 | 6.58 | 0.67 ± 0.02 | 5.6 |
| 8 | KKK | 4 equiv 4 | 7.10 | 1.59 ± 0.02 | 13 |
| KKKb | 4 equiv 4 | 7.04 | 1.45 ± 0.12 | -- | |
| KKKc | 4 equiv 4 | 7.24 | 2.04 ± 0.10 | -- | |
| 9 | KKK | 5 equiv 4 | 7.08 | 1.54 ± 0.08 | 13 |
| 10 | KKK | 6 equiv 4 | 7.08 | 1.55 ± 0.04 | 13 |
| 11 | WKW | none | 8.33 | 12.8 ± 0.6 | 1 |
| 12 | WKW | 2 equiv 4 | 8.88 | 32.1 ± 2.3 | 2.51 |
| 13 | KKW | none | 7.19 | 1.86 ± 0.05 | 1 |
| 14 | KKW | 3 equiv 4 | 7.65 | 4.08 ± 0.24 | 2.20 |
| 15 | WKK | none | 7.26 | 2.11 ± 0.07 | 1 |
| 16 | WKK | 3 equiv 4 | 7.60 | 3.70 ± 0.09 | 1.75 |
| 17 | KWK | none | 6.98 | 1.31 ± 0.05 | 1 |
| 18 | KWK | 3 equiv 4 | 7.81 | 5.29 ± 0.52 | 4.04 |
| 19 | RWW | none | 8.41 | 14.7 ± 1.0 | 1 |
| 20 | RWW | 2 equiv 4 | 9.05 | 43.1 ± 3.8 | 2.93 |
| 21 | RRW | none | 6.92 | 1.18 ± 0.04 | 1 |
| 22 | RRW | 3 equiv 4 | 8.11 | 8.77 ± 0.52 | 7.43 |
Binding constants were determined by fluorescence titrations in Millipore water for tryptophan-containing peptides and by ITC for KKK. Krel is the binding constant of the MINP with FM 4 relative to that without in Millipore water.
The binding was measured by ITC in HEPES buffer (pH 7.0).
The binding was measured by ITC in HEPES buffer (pH 4.7).
In the absence of 4, MINP(KWW), i.e., MINP prepared with KWW as the template, was found to bind its template with a binding constant of Ka = 20.7 × 105 M−1, determined by fluorescence titration and nonlinear least squares fitting of the data to a 1:1 binding isotherm. The 1:1 binding stoichiometry was confirmed additionally by the Job plot (Figure S13). The binding constant corresponds to 8.6 kcal/mol of binding free energy (−ΔG).
Addition of 4 in the MINP preparation increased the binding for KWW by <2 fold (entries 2–4). At first appearance, the small Krel seems to suggest that the FM was barely involved and inconsequential. However, when its amount was varied from 1 to 3 equiv to the template, the binding clearly responded and Krel reached the highest value at 2 equiv. Since KWW contains two amino groups, one at the N terminus and the other on the lysine side chain, our binding data suggest that a 1:1 ratio between the FM and the amino groups was optimal, a result only possible if the FM was involved in both the imprinting and binding.
The above conclusion was confirmed by MINP(KKK). Once the large hydrophobic side chains of tryptophan were removed, binding (between KKK and its MINP) became much weaker, with Ka = 0.2 × 105 M−1. This value was two orders of magnitude lower than that between KWW and MINP(KWW), consistent with the importance of hydrophobic imprinting and binding of KWW. Significantly, up to 13-fold increase in Ka was observed for KKK when the MINP contained FM 4. Interestingly, Krel reached the highest value at 4 equiv of FM for this peptide and stayed unchanged when additional FM was used (entries 6–10). Because KKK has four amino groups, the optimal ratio of FM to the amino group was thus 1:1 once again, supporting the effectiveness of FM 4 in the imprinting of lysine-containing peptides.
The above results suggest that the effectiveness of FM 4 in the imprinting of basic peptides is inversely related to the overall hydrophobicity of the template. For a peptide rich in hydrophobic side chains such as KWW, hydrophobic interactions provide sufficient driving force in binding with its MINP receptor, evident from the 8.6 kcal/mol binding free energy mentioned above. Under such conditions, additional polar interactions such as the hydrogen bonding interactions between the azacrown and the amino/ammonium groups would play a secondary role. For a peptide relatively hydrophilic, however, the polar interactions become more important, enhancing the binding by up to 13-fold in the case of KKK. It is worth noting that a 5.56 kcal/mol binding energy for its parent MINP, i.e., the MINP prepared without FM 4, was still fairly impressive, suggesting the “background” imprinting/binding was significant. In our previous work, even glycine was found to influence the imprinting and binding of peptides, despite its lack of a side chain.43 MINPs contain multiple functional groups including numerous triazole, hydroxyl, ammonium, and amide. These groups can certainly interact with the bound peptide through hydrogen bonds, van der Waals interactions, cationic–π interactions, and electrostatic interactions. Since these are less well-defined than the specific interactions between the hydrophobic “dimples” and the hydrophobic side chains or those between the azacrown and the amine/ammonium, we group them together as the background interactions.
We also studied the effects of pH on the binding of KWW and KKK, with and without FM 4 (Table 1, entries 1, 3, 5, 8). Even though the overall pH effect was relatively small, our data show that lowering pH from 7.0 to 4.7 caused a decrease of binding in the absence of the FM (entries 1 and 5) but an increase in the presence (entries 3 and 8). The results support our imprinting model. In the absence of the FM, imprinting mostly relies on the hydrophobic interactions between the peptide and the micelles. Imprinting of the lysine probably occurs in the nonpolar region of the micelle, with the side chain in the deprotonated (i.e., amino) state. During rebinding, a lower pH leads to more complete protonation of the lysine and reduces its hydrophobic interactions with the MINP. In the presence of FM 4, protonated lysine could be imprinted directly through ammonium–azacrown interactions, which are stronger than amine–azacrown intercations.54 During rebinding, more complete protonation of lysine still occurs at a lower pH but the protonated form can interact with the azacrown more strongly than the amino form. Overall, since the pH effect was rather small and the binding constants were quite similar in buffer and in water, we performed the majority of the binding in Millipore water.
Entries 11–22 of Table 1 show the effects of FM 4 on other peptides. Several interesting trends are obvious. First, the Krel value (at 1:1 FM/basic group) generally increased with the number of basic groups: up to 2.5 for two basic groups, 4.0 for three, and 13 for four. This trend supports the involvement of the azacrown in the imprinting and binding.
Second, for the peptides containing the same number of basic groups, Krel was higher when the amino groups were farther apart, e.g., 1.77 for KWW but 2.51 for WKW. For the peptides containing two lysines and one tryptophan, the same trend persisted: Krel was 2.20 for KKW and 1.75 for WKK, but 4.04 for KWK. It is not exactly clear to us why the binding depends on the distance between the basic groups. Nonetheless, to maximize the binding with its basic peptide guest, the azacrown-functionalized MINP needs to have the appropriate hydrophobic pocket to accommodate the indole ring of tryptophan. Meanwhile, the amine/ammonium group of the peptide needs to be in close proximity to the azacrown to engage in hydrogen bonds. When both tryptophan and FM 4 are quite bulky, the steric requirement might be quite demanding. When the basic groups are too close to each other, it is possible that their binding with FM 4 may cause the hydrophobic binding pocket to be less well-formed during the imprinting. It is also possible that the azacrowns may have difficulty achieving the optimal binding geometry under this situation.
Third, FM 4 was more effective for arginine (R) than for lysine (K). The binding data show that Krel was 2.93 and 7.43 with one and two arginines, but only 1.77 and 2.20 with one and two lysines, respectively. The result supports our hypothesis that the multiple hydrogen-bond acceptors (sulfonamide oxygens, ethereal oxygens, and nitrogen) of FM 4 might make it bind arginine particularly well, which has an abundance of hydrogen-bond donors.
Binding Selectivity
Having confirmed the effectiveness of FM 4 for lysine- and arginine-containing peptides, we examined its impact on the selectivity of binding, using KWW as the templating peptide. Figure 2 shows the binding constants of different peptides to MINP(KWW), normalized to Ka of KWW. The red bars show the selectivity for the MINP prepared without FM and blue bars the MINP with 2 equiv 4.
FIGURE 2.
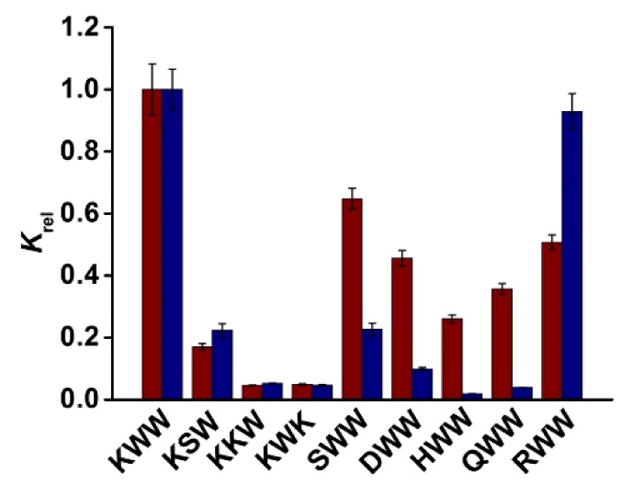
Binding selectivity of MINP(KWW) prepared without (red bars) and with FM 4 (blue bars). The binding data are reported in Table S1 in the Supporting Information.
In the absence of the azacrown FM, MINP(KWW) easily distinguished KSW, KKW, and KWK. Thus, replacing any one of the two tryptophans with a smaller, polar side chain (of serine and lysine) drastically weakened the binding. However, replacing the lysine of KWW with serine (S), aspartic acid (D), histidine (H), glutamine (Q), and arginine (R) did not weaken the binding as much, affording 26–65% of the binding constant. The parent MINP, therefore, was excellent at distinguishing the hydrophobic side chains but not very good at polar ones.
For the MINP(KWW) prepared with 2 equiv FM 4, selectivity generally improved. Its ability to recognize the hydrophobic side chains, to our delight, was maintained, as KSW, KKW, and KWK continued to show low binding. Meanwhile, the selectivity for SWW, DWW, HWW (with histidine also as a basic amino acid), and QWW all improved in comparison to the parent MINP, sometimes dramatically. The improved binding selectivity for these polar amino acids supports lysine being engaged in specific interactions with the aza-crown-functionalized MINP, in line with our design hypothesis and the binding affinities discussed in the previous section.
For the azacrown-functionalized MINP, the notable exception to the improved selectivity was RWW. Whereas the parent MINP(KWW) was able to distinguish RWW with a modest selectivity (ca. 1:0.5 or 2:1), the azacrown-functionalized MINP(KWW) showed essentially the same binding for KWW and RWW.
This pattern of lysine/arginine selectivity was confirmed by additional studies (Table 2). The binding selectivity in this table is shown by Krel, the binding constant of a nontemplating peptide over that of the template for the same MINP receptor. The smaller the Krel, the more selective the peptide is. In the absence of FM 4, MINP(KKW) containing two lysines once again afforded reasonable selectivity for RRW (with Krel = 0.44 or ~2.3:1 selectivity). Meanwhile, MINP(RRW) could not distinguish KKW at all: if anything, the Krel value (1.09) suggested reversed selectivity, meaning that the receptor bound the nontemplating peptide KKW slightly more strongly than its own template.
Table 2.
Differentiation of lysine (K) and arginine acid (R).a
| Entry | Host | Guest | FM 4 | Ka (×105 M−1) | K rel |
|---|---|---|---|---|---|
| 1 | MINP(KKW) | KKW | none | 1.86 ± 0.05 | |
| 2 | MINP(KKW) | RRW | none | 0.81 ± 0.02 | 0.44 |
| 3 | MINP(RRW) | RRW | none | 1.18 ± 0.04 | |
| 4 | MINP(RRW) | KKW | none | 1.27 ± 0.02 | 1.09 |
| 5 | MINP(KKW) | KKW | 3 equiv | 4.08 ± 0.24 | |
| 6 | MINP(KKW) | RRW | 3 equiv | 4.57 ± 0.13 | 1.12 |
| 7 | MINP(RRW) | RRW | 3 equiv | 8.77 ± 0.52 | |
| 8 | MINP(RRW) | KKW | 3 equiv | 3.32 ± 0.17 | 0.38 |
The Ka values were determined by fluorescence titrations in Millipore water. Krel is the binding constant of a guest relative to that of the templating peptide for a particular MINP.
In the presence of FM 4, the completely opposite trend was observed. MINP(RRW) distinguished KKW fairly easily, with Krel = 0.38 or ~2.6:1 selectivity. MINP(KKW), however, bound KKW and RRW both very well. The slightly reversed selectivity (Krel = 1.12) was observed once again.
These results indicate that, for the two basic amino acids, there is a preference for lysine by the parent MINPs and a preference for arginine by the azacrown-functionalized MINPs. The preferences can be understood from the different types of interactions involved in the two types of imprinted receptors.
In the absence of the azacrown, imprinting relies strongly on hydrophobic interactions. Lysine can deprotonate easily on the side chain due to weaker basicity of the amino group in comparison to guanidine. Once deprotonated, the side chain can insert itself into the micelle, which is beneficial to the hydrophobic imprinting. Arginine, however, has to stay on the surface of the micelle due to its charged nature, not contributing to the hydrophobic imprinting as much. The explanation is supported by the binding affinity data: MINP(RRW) bound its template with Ka = 1.18 × 105 M−1 but MINP(KKW) bound its template more strongly, with Ka = 1.86 × 105 M−1 (Table 2, entries 1 and 3). During rebinding, RRW has difficulty fitting in the binding site inside MINP(KKW) generated for KKW, due to the highly charged nature of the arginine side chain. KKW, on the other hand, could in its protonated form “pretend” to be RRW and stay on the surface of MINP(RRW), exposed to water, just like arginine does.
Once the micelle contains FM 4, the azacrown can interact with arginine better than lysine, because of its stronger and more numerous hydrogen-bond donors. Indeed, MINP(RRW) now has better binding affinity for its template than MINP(KKW) for its own (Table 2, entries 5 and 7). The stronger guanidinium–azacrown interactions not only can explain the selectivity of MINP(RRW) for KKW (entry 8) but also the reversed selectivity of RMINP(KKW) for RRW (entry 6). This is because, when KKW is used as the template, the lysine– azacrown interactions were strong enough to place the azacrown at the right place—also evident from our binding data in Table 1. Since arginine is quite close to lysine in length, the guanidinium side chain should be able to reach the azacrown binding site. The stronger arginine–azacrown interactions apparently can more than compensate for any imperfection in binding geometry and make the nontemplating RRW bind to MINP(KKW) more strongly than the templating KKW.
The binding selectivity so far was determined from the different binding constants of various peptides for the same MINP. To further confirm the selectivity, we added MINP(KWW) to an equimolar mixture of four peptides (KWW, RWW, SWW, and KKW). The peptide mixtures contained the template and three other representative peptides from Figure 2. The amounts of peptides adsorbed by the MINPs were determined by LCMS after ultracentrifugation to precipitate the MINPs and washing the MINPs with methanol (see the Experimental Section for details). The peptide binding assay essentially represents a solid-phase extraction study. The percent yields of recovery were then plotted against the amounts of MINP used in the extraction (Figures 3 and 4).
FIGURE 3.
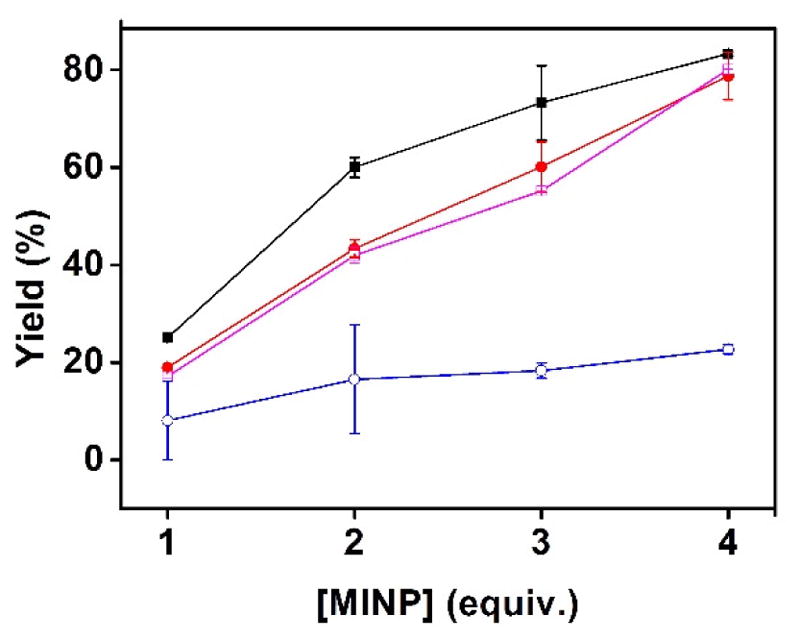
Extraction yields of different peptides (■: KWW;
 : RWW;
: RWW;
 : SWW;
: SWW;
 : KKW) by MINP(KWW) prepared without FM 4. [All peptides in the mixture] = 1.0 μM. [MINP used for extraction] = 1–4 μM.
: KKW) by MINP(KWW) prepared without FM 4. [All peptides in the mixture] = 1.0 μM. [MINP used for extraction] = 1–4 μM.
FIGURE 4.
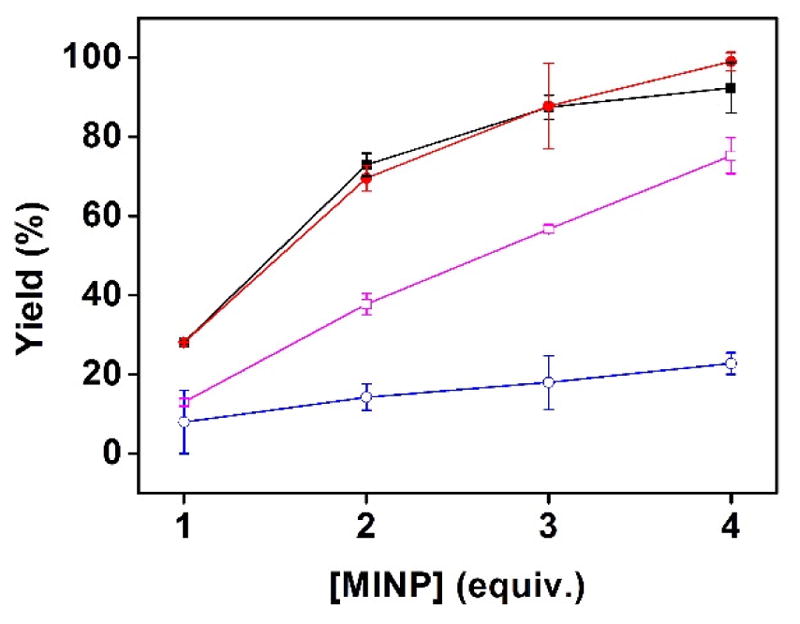
Extraction yields of different peptides (■: KWW;
 : RWW;
: RWW;
 : SWW;
: SWW;
 : KKW) by MINP(KWW) prepared with 2 equiv FM 4. [All peptides in the mixture] = 1.0 μM. [MINP used for extraction] = 1–4 μM.
: KKW) by MINP(KWW) prepared with 2 equiv FM 4. [All peptides in the mixture] = 1.0 μM. [MINP used for extraction] = 1–4 μM.
In general, the extraction yields were in line with the binding constants of the different peptides. For example, KKW showed the weakest binding for MINP(KWW) among the four peptides, whether the MINP contained FM 4 or not (Figure 2). During the extractions, its adsorption by the MINP was also the lowest, whether the MINP contained FM 4 or not (Figures 3 and 4). MINP(KWW) without the azacrown FM were able to distinguish SWW and RWW from its template in Figure 2. The extraction yields of SWW and RWW by MINP(KWW) were also lower than that of KWW in Figure 3.
On the other hand, MINP(KWW) with FM 4 had difficulty distinguishing RWW but distinguished SWW quite well (Figure 2). The extraction yields showed exactly the same trend (Figure 4), suggesting the binding selectivity was maintained in mixtures of peptides. These results, obtained from a single extraction, bode well for the usage of MINPs in chromatographic separation of peptides.
Binding of Biological Basic Peptides
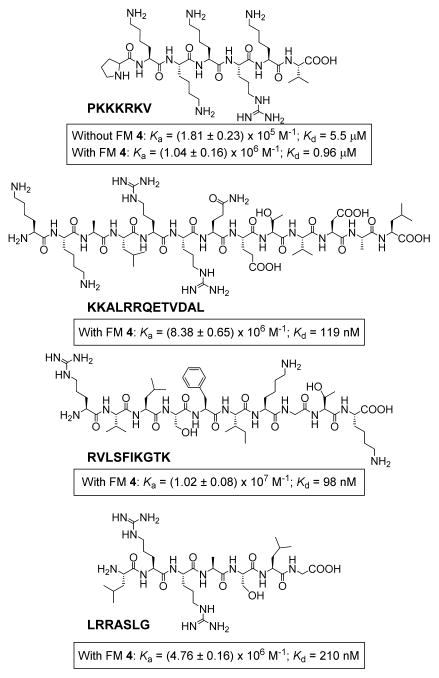
Figure 5 shows the binding data for several biological peptides, all containing a significant number of lysine/arginine. PKKKRKV is a nuclear localization signal peptide very useful in delivery.60 Without FM 4, its MINP receptor gave a dissociation constant (Kd) of 5.5 μM and the azacrown FM was able to lower the Kd to 0.96 μM. The other three peptides are KKALRRQETVDAL (a selective substrate for Ca2+/calmodulin-dependent protein kinase), RVLSFIKGTK (an immune epitope for Influenza A virus), LRRASLG (a phosphate receptor peptide). With slightly more hydrophobic residues and/or a longer chain length, they could be bound by the azacrown-functionalized MINP receptor with affinities in the range of 100–200 nM in water.
FIGURE 5.
Structures of biological peptides examined in this study and the binding constants (Ka) and dissociation constants (Kd) obtained.
Conclusions
A general method to recognize oligopeptides in a sequence-selective manner has been an unsolved problem in supramolecular and bioorganic chemistry for decades. With the micellar imprinting technique, we can prepare and purify artificial peptide receptors in 2 days if all the starting surfactants, FMs, and cross-linkers are available. The receptors can distinguish peptides based on their hydrophobic side chains43 and, as shown in this work, recognize basic amino acids (lysine and arginine) when appropriate functional monomers are included.
Importantly, the azacrown-derived FM 4 generally improved both the binding affinity and selectivity of the imprinted micelles (Table 1, Figure 2). Although two reversed selectivities were observed in our binding studies (Table 2), it is fortunate that the parent MNP was more selective for lysine and the azacrown-functionalized MINP more selective for arginine. Thus, selectivity for these two particular amino acids could be controlled by whether or not to include the azacrown in the formation. According to our previous study, all residues contribute to the binding affinity and selectivity, either by primary or secondary (i.e., background) interactions.43 As long as two peptide differ in multiple places including in chain length, their distinction is quite likely because every mismatch contributes to selectivity.
Experimental Section
Syntheses of compound 1–3 have been reported.42
Compound 4
To a solution of diaza-18-crown-6 (131 mg, 0.5 mmol) in DCM (6 mL), 4-vinylbenzenesulfonyl chloride (405 mg, 2 mmol) in DCM (4 mL) and TEA (278 μL, 2 mmol) were added dropwise at 0 °C gradually. The mixture was allowed to warm to room temperature and stirred overnight. The organic solution was extracted with water (3 × 10 mL), dried over anhydrous sodium sulfate and concentrated by rotary evaporation. The residue was purified by column chromatography over silica gel using 1: 200 methanol/DCM as eluent to afford compound 4 as white powder (201 mg, 67 %). 1H NMR (600 MHz, DMSO-d6) δ 7.77 (d, J = 8.0 Hz, 2H), 7.68 (d, J = 8.0 Hz, 2H), 6.82 (dd, J1 = 17.6 Hz, J2 = 10.9 Hz, 1H), 6.01 (d, J = 17.6 Hz, 1H), 5.75 (d, J = 1.3 Hz, 1H), 5.45 (d, J = 10.9 Hz, 1H), 3.52 (t, J = 5.9 Hz, 4H), 3.45 (s, 4H), 3.30 (t, J = 6.0 Hz, 4H) ppm. 13C NMR (100 MHz, d6-DMSO) δ 141.7, 138.42 135.7, 127.7, 127.3, 118.3, 70.3, 70.1, 49.0. ESI-HRMS calcd for C28H38N2O8S2 (m/z): [M + H]+, 595.2142; found, 595.2147.
Typical procedure for the synthesis of MINPs.42
To a micellar solution of compound 1 (9.3 mg, 0.02 mmol) in H2O (2.0 mL), divinylbenzene (DVB, 2.8 μL, 0.02 mmol), compound 4 in H2O (10 μL of a 14.1 mg/mL in H2O, 0.0004 mmol), and 2,2-dimethoxy-2-phenylacetophenone (DMPA,10 μL of a 12.8 mg/mL solution in DMSO, 0.0005 mmol) were added. The mixture was subjected to ultrasonication for 10 min before compound 2 (4.1 mg, 0.024 mmol), CuCl2 (10 μL of a 6.7 mg/mL solution in H2O, 0.0005 mmol), and sodium ascorbate (10 μL of a 99 mg/mL solution in H2O, 0.005 mmol) were added. After the reaction mixture was stirred slowly at room temperature for 12 h, compound 3 (10.6 mg, 0.04 mmol), CuCl2 (10 μL of a 6.7 mg/mL solution in H2O, 0.0005 mmol), and sodium ascorbate (10 μL of a 99 mg/mL solution in H2O, 0.005 mmol) were added. After being stirred for another 6 h at room temperature, the reaction mixture was transferred to a glass vial, purged with nitrogen for 15 min, sealed with a rubber stopper, and irradiated in a Rayonet reactor for 12 h. The reaction mixture was poured into acetone (8 mL). The precipitate was collected by centrifugation and washed with a mixture of acetone/water (5 mL/1 mL) three times, followed by methanol/acetic acid (5 mL/0.1 mL) three times. The product was dried in air to afford MINP(4). Yields generally were >80%.
ITC Titration
ITC was performed using a MicroCal VP-ITC Microcalorimeter with Origin 7 software and VPViewer2000 (GE Healthcare, Northampton, MA). The determination of binding constants by ITC followed standard procedures.61–63 In general, a solution of an appropriate guest in Millipore water was injected in equal steps into 1.43 mL of the corresponding MINP in the same solution. The top panel shows the raw calorimetric data. The area under each peak represents the amount of heat generated at each ejection and is plotted against the molar ratio of the MINP to the guest. The smooth solid line is the best fit of the experimental data to the sequential binding of N binding site on the MINP. The heat of dilution for the guest, obtained by titration carried out beyond the saturation point, was subtracted from the heat released during the binding. Binding parameters were auto-generated after curve fitting using Microcal Origin 7.
Solid-Phase Extraction of Peptides
Four 1.0 mL solutions of peptide mixtures were prepared in Millipore water containing KWW, RWW, SWW and KKW each at 100 μM. MINP(KWW) was added to the four solutions, at 100, 200, 300, and 400 μM, respectively. The solutions were diluted to 100 mL by Millipore water and stirred at room temperature overnight.64 For the determination of the amounts of peptides adsorbed by the MINPs, the solutions were centrifuged at 42,000 g for 2 min to precipitate the MINPs. The MINP precipitates were washed with methanol (3 × 10 mL) and the combined organic phase was concentrated under vacuum to dryness for each sample. The residue in each sample was dissolved in 0.95 mL of Millipore water, followed by the addition of 50 μL of 1.0 mM L-Tryptophan aqueous solution as the internal standard. The amounts of the peptides in the solutions were analyzed by an Agilent 1200 Series Binary VWD HPLC system coupled to an Agilent 6540 UHD Accurate Mass Q-TOF mass spectrometry detector.
Supplementary Material
Acknowledgments
We thank NIGMS (R01GM113883) for supporting this research.
Footnotes
Author Contributions
All authors have given approval to the final version of the manuscript.
Notes
Any additional relevant notes should be placed here.
Supporting Information. Experimental details, ITC and fluorescence titration curves, and additional data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 2.Coates AR, Hu Y. Targeting non-multiplying organisms as a way to develop novel antimicrobials. Trends Pharmacol Sci. 2008;29:143–150. doi: 10.1016/j.tips.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Lindgren M, Hällbrink M, Prochiantz A, Langel Ü. Cell-penetrating peptides. Trends Pharmacol Sci. 2000;21:99–103. doi: 10.1016/s0165-6147(00)01447-4. [DOI] [PubMed] [Google Scholar]
- 4.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. Cell-penetrating Peptides: A REEVALUATION OF THE MECHANISM OF CELLULAR UPTAKE. J Biol Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 5.Zorko M, Langel Ü. Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Adv Drug Deliv Rev. 2005;57:529–545. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Heitz F, Morris MC, Divita G. Twenty years of cell-penetrating peptides: from molecular mechanisms to therapeutics. Br J Pharmacol. 2009;157:195–206. doi: 10.1111/j.1476-5381.2009.00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fonseca SB, Pereira MP, Kelley SO. Recent advances in the use of cell-penetrating peptides for medical and biological applications. Adv Drug Deliv Rev. 2009;61:953–964. doi: 10.1016/j.addr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Hong JI, Namgoong SK, Bernardi A, Still WC. Highly Selective Binding of Simple Peptides by a C3 Macrotricyclic Receptor. J Am Chem Soc. 1991;113:5111–5112. [Google Scholar]
- 9.Breslow R, Yang Z, Ching R, Trojandt G, Odobel F. Sequence Selective Binding of Peptides by Artificial Receptors in Aqueous Solution. J Am Chem Soc. 1998;120:3536–3537. [Google Scholar]
- 10.Hossain MA, Schneider HJ. Sequence-Selective Evaluation of Peptide Side-Chain Interaction. New Artificial Receptors for Selective Recognition in Water. J Am Chem Soc. 1998;120:11208–11209. [Google Scholar]
- 11.Yamamura H, Rekharsky MV, Ishihara Y, Kawai M, Inoue Y. Factors controlling the complex architecture of native and modified cyclodextrins with dipeptide (Z-Glu-Tyr) studied by microcalorimetry and NMR spectroscopy: critical effects of peripheral bis-trimethylamination and cavity size. J Am Chem Soc. 2004;126:14224–14233. doi: 10.1021/ja046612r. [DOI] [PubMed] [Google Scholar]
- 12.Tashiro S, Tominaga M, Kawano M, Therrien B, Ozeki T, Fujita M. Sequence-selective recognition of peptides within the single binding pocket of a self-assembled coordination cage. J Am Chem Soc. 2005;127:4546–4547. doi: 10.1021/ja044782y. [DOI] [PubMed] [Google Scholar]
- 13.Wright AT, Anslyn EV, McDevitt JT. A differential array of metalated synthetic receptors for the analysis of tripeptide mixtures. J Am Chem Soc. 2005;127:17405–17411. doi: 10.1021/ja055696g. [DOI] [PubMed] [Google Scholar]
- 14.Schmuck C, Wich P. Sequence-dependent stereoselectivity in the binding of tetrapeptides in water by a flexible artificial receptor. Angew Chem Int Ed. 2006;45:4277–4281. doi: 10.1002/anie.200601046. [DOI] [PubMed] [Google Scholar]
- 15.Reczek JJ, Kennedy AA, Halbert BT, Urbach AR. Multivalent Recognition of Peptides by Modular Self-Assembled Receptors. J Am Chem Soc. 2009;131:2408–2415. doi: 10.1021/ja808936y. [DOI] [PubMed] [Google Scholar]
- 16.Niebling S, Kuchelmeister HY, Schmuck C, Schlucker S. Quantitative label-free monitoring of peptide recognition by artificial receptors: a comparative FT-IR and UV resonance Raman spectroscopic study. Chem Sci. 2012;3:3371–3377. [Google Scholar]
- 17.Smith LC, Leach DG, Blaylock BE, Ali OA, Urbach AR. Sequence-Specific, Nanomolar Peptide Binding via Cucurbit[8]uril-Induced Folding and Inclusion of Neighboring Side Chains. J Am Chem Soc. 2015;137:3663–3669. doi: 10.1021/jacs.5b00718. [DOI] [PubMed] [Google Scholar]
- 18.Faggi E, Vicent C, Luis SV, Alfonso I. Stereoselective recognition of the Ac-Glu-Tyr-OH dipeptide by pseudopeptidic cages. Org Biomol Chem. 2015;13:11721–11731. doi: 10.1039/c5ob01889g. [DOI] [PubMed] [Google Scholar]
- 19.Sonzini S, Marcozzi A, Gubeli RJ, van der Walle CF, Ravn P, Herrmann A, Scherman OA. High Affinity Recognition of a Selected Amino Acid Epitope within a Protein by Cucurbit[8]uril Complexation. Angew Chem Int Ed. 2016;55:14000–14004. doi: 10.1002/anie.201606763. [DOI] [PubMed] [Google Scholar]
- 20.Maity D, Schmuck C. Synthetic Receptors for Biomolecules: Design Principles and Applications. The Royal Society of Chemistry; 2015. Chapter 8 Synthetic Receptors for Amino Acids and Peptides; pp. 326–368. [Google Scholar]
- 21.Peczuh MW, Hamilton AD. Peptide and Protein Recognition by Designed Molecules. Chem Rev. 2000;100:2479–2494. doi: 10.1021/cr9900026. [DOI] [PubMed] [Google Scholar]
- 22.Oshovsky GV, Reinhoudt DN, Verboom W. Supramolecular Chemistry in Water. Angew Chem Int Ed. 2007;46:2366–2393. doi: 10.1002/anie.200602815. [DOI] [PubMed] [Google Scholar]
- 23.Kataev EA, Müller C. Recent advances in molecular recognition in water: artificial receptors and supramolecular catalysis. Tetrahedron. 2014;70:137–167. [Google Scholar]
- 24.Prins LJ, Reinhoudt DN, Timmerman P. Noncovalent Synthesis Using Hydrogen Bonding. Angew Chem Int Ed. 2001;40:2382–2426. doi: 10.1002/1521-3773(20010702)40:13<2382::aid-anie2382>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 25.Wulff G. Molecular Imprinting in Cross-Linked Materials with the Aid of Molecular Templates— A Way towards Artificial Antibodies. Angew Chem Int Ed Engl. 1995;34:1812–1832. [Google Scholar]
- 26.Wulff G. Enzyme-like Catalysis by Molecularly Imprinted Polymers. Chem Rev. 2001;102:1–28. doi: 10.1021/cr980039a. [DOI] [PubMed] [Google Scholar]
- 27.Haupt K, Mosbach K. Molecularly Imprinted Polymers and Their Use in Biomimetic Sensors. Chem Rev. 2000;100:2495–2504. doi: 10.1021/cr990099w. [DOI] [PubMed] [Google Scholar]
- 28.Ye L, Mosbach K. Molecular Imprinting: Synthetic Materials As Substitutes for Biological Antibodies and Receptors. Chem Mater. 2008;20:859–868. [Google Scholar]
- 29.Shea KJ. Molecular imprinting of synthetic network polymers: the de novo synthesis of macromolecular binding and catalytic sites. Trends Polym Sci. 1994;2:166–173. [Google Scholar]
- 30.Sellergren B. Molecularly imprinted polymers: man-made mimics of antibodies and their applications in analytical chemistry. Elsevier; Amsterdam: 2001. [Google Scholar]
- 31.Komiyama M. Molecular imprinting: from fundamentals to applications. Wiley-VCH; Weinheim: 2003. [Google Scholar]
- 32.Yan M, Ramström O. Molecularly imprinted materials: science and technology. Marcel Dekker; New York: 2005. [Google Scholar]
- 33.Alexander C, Andersson HS, Andersson LI, Ansell RJ, Kirsch N, Nicholls IA, O’Mahony J, Whitcombe MJ. Molecular imprinting science and technology: a survey of the literature for the years up to and including 2003. J Mol Recognit. 2006;19:106–180. doi: 10.1002/jmr.760. [DOI] [PubMed] [Google Scholar]
- 34.Sellergren B, Hall AJ. Molecularly Imprinted Polymers. In: Steed JW, Gale PA, editors. Supramolecular Chemistry: From Molecules to Nanomaterials. Wiley; 2012. Online. [Google Scholar]
- 35.Haupt K, Ayela C. Molecular Imprinting. Springer; Heidelberg ; New York: 2012. [Google Scholar]
- 36.Klein JU, Whitcombe MJ, Mulholland F, Vulfson EN. Template-mediated synthesis of a polymeric receptor specific to amino acid sequences. Angew Chem Int Ed. 1999;38:2057–2060. doi: 10.1002/(SICI)1521-3773(19990712)38:13/14<2057::AID-ANIE2057>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 37.Nishino H, Huang CS, Shea KJ. Selective protein capture by epitope imprinting. Angew Chem Int Ed. 2006;45:2392–2396. doi: 10.1002/anie.200503760. [DOI] [PubMed] [Google Scholar]
- 38.Hoshino Y, Kodama T, Okahata Y, Shea KJ. Peptide Imprinted Polymer Nanoparticles: A Plastic Antibody. J Am Chem Soc. 2008;130:15242–15243. doi: 10.1021/ja8062875. [DOI] [PubMed] [Google Scholar]
- 39.Hoshino Y, Koide H, Urakami T, Kanazawa H, Kodama T, Oku N, Shea KJ. Recognition, Neutralization, and Clearance of Target Peptides in the Bloodstream of Living Mice by Molecularly Imprinted Polymer Nanoparticles: A Plastic Antibody. J Am Chem Soc. 2010;132:6644–6645. doi: 10.1021/ja102148f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee S, König B. Molecular Imprinting of Luminescent Vesicles. J Am Chem Soc. 2013;135:2967–2970. doi: 10.1021/ja4001568. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Y, Deng C, Liu S, Wu J, Chen Z, Li C, Lu W. Active Targeting of Tumors through Conformational Epitope Imprinting. Angew Chem Int Ed. 2015;54:5157–5160. doi: 10.1002/anie.201412114. [DOI] [PubMed] [Google Scholar]
- 42.Awino JK, Zhao Y. Protein-Mimetic, Molecularly Imprinted Nanoparticles for Selective Binding of Bile Salt Derivatives in Water. J Am Chem Soc. 2013;135:12552–12555. doi: 10.1021/ja406089c. [DOI] [PubMed] [Google Scholar]
- 43.Awino JK, Gunasekara RW, Zhao Y. Sequence-Selective Binding of Oligopeptides in Water through Hydrophobic Coding. J Am Chem Soc. 2017;139:2188–2191. doi: 10.1021/jacs.6b12949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Awino JK, Zhao Y. Molecularly Imprinted Nanoparticles as Tailor-Made Sensors for Small Fluorescent Molecules. Chem Commun. 2014;50:5752–5755. doi: 10.1039/c4cc01516a. [DOI] [PubMed] [Google Scholar]
- 45.Awino JK, Zhao Y. Water-Soluble Molecularly Imprinted Nanoparticles (MINPs) with Tailored, Functionalized, Modifiable Binding Pockets. Chem-Eur J. 2015;21:655–661. doi: 10.1002/chem.201404919. [DOI] [PubMed] [Google Scholar]
- 46.Awino JK, Hu L, Zhao Y. Molecularly Responsive Binding through Co-occupation of Binding Space: A Lock–Key Story. Org Lett. 2016;18:1650–1653. doi: 10.1021/acs.orglett.6b00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Awino JK, Zhao Y. Polymeric Nanoparticle Receptors as Synthetic Antibodies for Nonsteroidal Anti-Inflammatory Drugs (NSAIDs) ACS Biomater Sci Eng. 2015;1:425–430. doi: 10.1021/acsbiomaterials.5b00042. [DOI] [PubMed] [Google Scholar]
- 48.Zhang S, Zhao Y. Facile Synthesis of Multivalent Water-Soluble Organic Nanoparticles via “Surface Clicking” of Alkynylated Surfactant Micelles. Macromolecules. 2010;43:4020–4022. [Google Scholar]
- 49.Li X, Zhao Y. Protection/Deprotection of Surface Activity and Its Applications in the Controlled Release of Liposomal Contents. Langmuir. 2012;28:4152–4159. doi: 10.1021/la2050702. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Y. Surface-Cross-Linked Micelles as Multifunctionalized Organic Nanoparticles for Controlled Release, Light Harvesting, and Catalysis. Langmuir. 2016;32:5703–5713. doi: 10.1021/acs.langmuir.6b01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Awino JK, Gunasekara RW, Zhao Y. Selective Recognition of d-Aldohexoses in Water by Boronic Acid-Functionalized, Molecularly Imprinted Cross-Linked Micelles. J Am Chem Soc. 2016;138:9759–9762. doi: 10.1021/jacs.6b04613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gunasekara RW, Zhao Y. A General Method for Selective Recognition of Monosaccharides and Oligosaccharides in Water. J Am Chem Soc. 2017;139:829–835. doi: 10.1021/jacs.6b10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gatto VJ, Arnold KA, Viscariello AM, Miller SR, Morgan CR, Gokel GW. Syntheses and binding properties of bibrachial lariat ethers (BiBLEs): survey of synthetic methods and cation selectivities. J Org Chem. 1986;51:5373–5384. [Google Scholar]
- 54.Buschmann HJ, Mutihac L. Complex formation between the macrocyclic ligand 18-crown-6 and unprotonated amines in methanol. Thermochim Acta. 1994;237:203–206. [Google Scholar]
- 55.Izatt RM, Pawlak K, Bradshaw JS, Bruening RL. Thermodynamic and kinetic data for macrocycle interactions with cations and anions. Chem Rev. 1991;91:1721–2085. [Google Scholar]
- 56.Izatt RM, Bradshaw JS, Nielsen SA, Lamb JD, Christensen JJ, Sen D. Thermodynamic and kinetic data for cation-macrocycle interaction. Chem Rev. 1985;85:271–339. [Google Scholar]
- 57.Watson WH, Galloy J, Grossie DA, Voegtle F, Mueller WM. Host-guest complex chemistry. Structures of 18-crown-6 and diaza-18-crown-6 with neutral molecules. J Org Chem. 1984;49:347–353. [Google Scholar]
- 58.Buschmann HJ, Dong H, Mutihac L, Schollmeyer E. Complex formation between crown ethers and urea and some guanidinium derivatives in methanol. J Therm Anal Cal. 1999;57:487–491. [Google Scholar]
- 59.Gawley RE, Pinet S, Cardona CM, Datta PK, Ren T, Guida WC, Nydick J, Leblanc RM. Chemosensors for the Marine Toxin Saxitoxin. J Am Chem Soc. 2002;124:13448–13453. doi: 10.1021/ja027507p. [DOI] [PubMed] [Google Scholar]
- 60.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 61.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid Measurement of Binding Constants and Heats of Binding Using a New Titration Calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 62.Jelesarov I, Bosshard HR. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J Mol Recognit. 1999;12:3–18. doi: 10.1002/(SICI)1099-1352(199901/02)12:1<3::AID-JMR441>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 63.Velazquez-Campoy A, Leavitt SA, Freire E. Characterization of protein-protein interactions by isothermal titration calorimetry. Methods Mol Biol. 2004;261:35–54. doi: 10.1385/1-59259-762-9:035. [DOI] [PubMed] [Google Scholar]
- 64.The extractions were performed at micromolar concentrations because of the strong binding affinities of the MINP for the peptides.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.