SUMMARY
Yeast use the ring shaped Dam1 complex to slide down depolymerizing microtubules to move chromosomes, but current models suggest other eukaryotes do not have a sliding ring. We visualized Ndc80 and Ska complexes on microtubules by EM tomography to identify the structure of the human kinetochore-microtubule attachment. Ndc80 recruits the Ska complex so that the V-shape of the Ska dimer interacts along protofilaments. We identify a mutant of the Ndc80 tail that is deficient in Ska recruitment to kinetochores and in orienting Ska along protofilaments in vitro. This mutant Ndc80 binds microtubules with normal affinity, but is deficient in clustering along protofilaments. We propose that Ska is recruited to kinetochores by clusters of Ndc80 proteins and our structure of Ndc80 and Ska complexes on microtubules suggests a mechanism for metazoans kinetochores to couple the depolymerization of microtubules to power the movement chromosomes.
INTRODUCTION
Current models suggest that kinetochores use the energy stored in the microtubule polymer to move chromosomes to the metaphase plate and to segregate chromatids in anaphase (Powers, et al., 2009; Grishchuk, et al., 2008; Grishchuk and McIntosh, 2006; Molodtsov, et al., 2005). To do so, kinetochores must remain bound to the plus end of the microtubule polymer while it depolymerizes. As microtubules depolymerize, each protofilament first splays away from the tube to form a curved end, which subsequently loses subunits (Tran, et al., 1997). This splaying can generate force (Grishchuk, et al., 2005). Budding yeast have a ring shaped Dam1 complex that can be pushed back by the curving ends, providing a mechanism to harness the energy in a microtubule to move chromosomes (Grishchuk, et al., 2008; Wang, et al., 2007; Westermann, et al., 2006; Molodtsov, et al., 2005; Westermann, et al., 2005). However, the Dam1 complex has only been found in fungi and most metazoans have the Ska complex, which has been proposed to have similar function but does not form rings.
All eukaryotes require the Ndc80 complex to generate attachments with the microtubule plus end (McCleland, et al., 2004; McCleland, et al., 2003; DeLuca, et al., 2002; Kilmartin and Janke, 2001; Wigge and Kilmartin, 2001). Ndc80 has a microtubule attachment site at the end of a long coiled coil (Ciferri, et al., 2008; Ciferri, et al., 2005). Current models for Ndc80 function in metazoans suggest a passive model where multiple Ndc80 proteins interact with the same microtubule as it depolymerizes (Zaytsev, et al., 2015; Zaytsev, et al., 2014; Powers, et al., 2009). As Ndc80 prefers to bind the microtubule polymer rather than free tubulin subunits, the kinetochore remains attached as long as some Ndc80 remains bound to the polymer. In these models, it is unclear how the curvature of depolymerizing microtubules could be used as a power stroke.
An N-terminal Calponin homology domain (CHD) of the Ndc80 subunit (also called Hec1) directly binds the lateral sides of microtubules and this attachment is critical for the movements of chromosomes on the mitotic spindle (Tooley, et al., 2011; Ciferri, et al., 2008; Wei, et al., 2007). The binding of the CHD to microtubules is regulated by an unstructured and positively-charged 80 amino acid N-terminal tail of Ndc80 (Guimaraes, et al., 2008; Miller, et al., 2008; Cheeseman, et al., 2006). Each kinetochore has at least 7 Ndc80 complexes per microtubule attachment (Suzuki, et al., 2015; Johnston, et al., 2010; Joglekar, et al., 2008; Joglekar, et al., 2006; Emanuele, et al., 2005). Ndc80 complexes can bind along protofilaments in clusters in vitro, but current models suggest that multiple Ndc80 complexes bind a microtubule as single entities in vivo (Zaytsev, et al., 2015; Zaytsev, et al., 2014; Powers, et al., 2009). Clustering in vitro is mediated by the Ndc80 tail (Alushin, et al., 2010), which sits between CHDs of adjacent subunits (Alushin, et al., 2012). This region also directly binds microtubules, is phosphorylated on multiple sites by Aurora B, and dephosphorylation of this zone is required for proper alignment of chromosomes to the metaphase plate (Guimaraes, et al., 2008; Miller, et al., 2008; Cheeseman, et al., 2006). Phosphorylation of the tail has minimal effect on the clustering of Ndc80 molecules along microtubules in vitro (Zaytsev, et al., 2015).
The spindle and kinetochore-associated complex (Ska) is enriched on kinetochores of aligned chromosomes and has been implicated in chromosome movements and silencing of the spindle assembly checkpoint (SAC) (Sivakumar, et al., 2016; Sivakumar, et al., 2014; Schmidt, et al., 2012; Daum, et al., 2009; Welburn, et al., 2009; Hanisch, et al., 2006). It binds microtubules but its central function in mitotic exit is to recruit PP1 to kinetochores (Sivakumar, et al., 2016; Schmidt, et al., 2012; Welburn, et al., 2009). The core of the Ska complex is composed of three coiled-coil regions where the three Ska proteins come together to form a core W- or V-shaped structure (Jeyaprakash, et al., 2012). Ska directly binds microtubules and can track the plus-ends of depolymerizing microtubules (Schmidt, et al., 2012; Welburn, et al., 2009). Depletion of any member of the Ska complex results in prolonged, checkpoint dependent metaphase arrest and eventually cell death (Daum, et al., 2009). Despite lack of sequence or structural similarity, it has been suggested that the Ska complex may be functionally equivalent to the yeast Dam1 complex (Abad, et al., 2014; Gaitanos, et al., 2009; Welburn, et al., 2009).
A critical unanswered question is how the Ska complex is recruited to kinetochores. The recruitment of Ska is dependent on the Knl1-Mis12-Ndc80 (KMN) network (Chan, et al., 2012; Raaijmakers, et al., 2009; Welburn, et al., 2009; Hanisch, et al., 2006) and is regulated by Aurora B kinase (Chan, et al., 2012). Although Ndc80 and Ska have not been shown to interact in vitro, Ndc80 can increase the binding affinity of Ska to microtubules (Schmidt, et al., 2012), suggesting that they may form a complex on microtubules. Moreover, expression of a mutant Ndc80 that lacks a flexible hinge in the coiled coil region prevents Ska recruitment, but there is no evidence that Ska binds this hinged region (Zhang, et al., 2012). A recent study suggests EB1, which specifically binds polymerizing microtubules, recruits Ska to kinetochores and orients Ska across a microtubule polymer similar to the Dam1 complex in yeast (Thomas, et al., 2016).
Chromosome movements are predicated on the ability of kinetochores to remain attached to depolymerizing microtubule plus ends (Powers, et al., 2009; Grishchuk, et al., 2008; Grishchuk and McIntosh, 2006; Molodtsov, et al., 2005). Individual Ndc80 complexes can track depolymerizing ends in vitro if they are oligomerized or bound to a bead at a density high enough for multiple Ndc80 proteins to interact with a single microtubule (Powers, et al., 2009). However, individual Ndc80 complexes cannot track depolymerizing microtubule ends on their own (Schmidt, et al., 2012; Powers, et al., 2009). This has led to a passive model of depolymerization-coupled movement whereby oligomerized Ndc80 proteins can track ends because some individual complexes remain attached, while others must release when they are bound to a depolymerizing subunit. Although Ndc80 and Ska have not been shown to interact in solution, Ndc80 can increase the affinity of Ska and microtublues (Schmidt, et al., 2012). We investigated the structure of the full human Ska complex that is recruited to Taxol-stabilized microtubules by the Ndc80 by negative stain electron tomography. Ndc80 CHDs orient Ska along the straight segment of the microtubule such that both Ska microtubule binding sites interact with the same or adjacent protofilaments separated by ~5–6 tublin monomers. We provide evidence that this structure also exists in kinetochores in vivo and that Ndc80 must oligomerize to recruit Ska to kinetochores. Our data suggest mechanisms by which a complex of Ska and Ndc80 form a structure that has important implications for movements of chromosomes by kinetochores.
RESULTS
Ndc80 orients the Ska complex along a microtubule protofilament
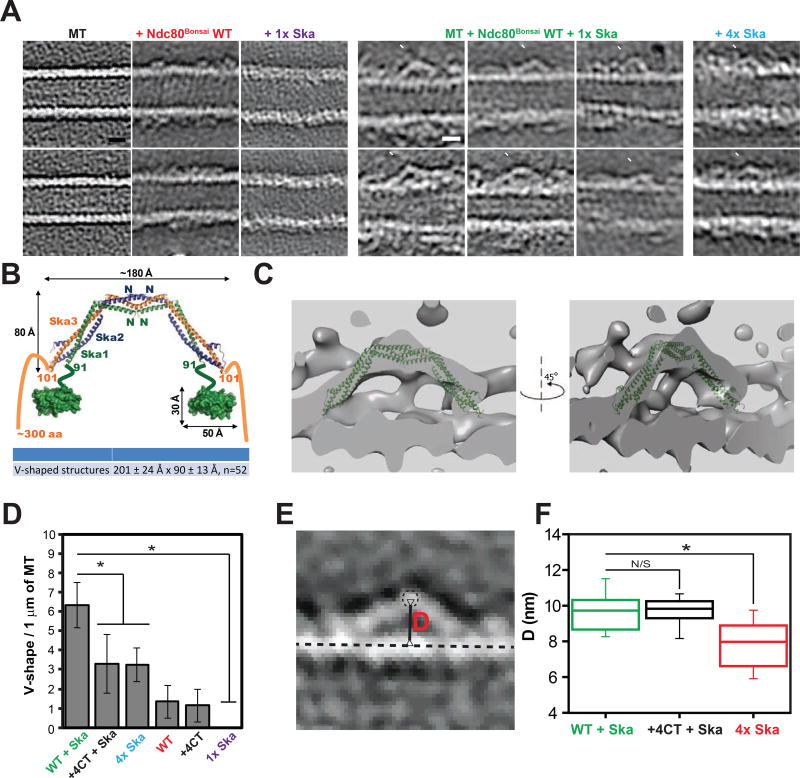
We investigated the structure of the full human Ska complex and an engineered subcomplex of the Ndc80 complex that lacks the majority of its coiled coil domains (Ndc80Bonsai) [15] on Taxol-stabilized microtubules by negative stain electron tomography (Fig. 1A). Throughout this study (unless specifically indicated) we used concentrations of Ska that were below the Kd of intrinsic microtubule binding (Kd = 2.4–5.1 µM (Schmidt, et al., 2012)) to specifically visualize the Ska complex that was recruited by Ndc80 to microtubules. V-shaped structures were detected on the microtubules that were incubated with both Ndc80Bonsai WT and Ska (Fig. 1A, Fig. S1A, Movie S2, S3). These V-shapes are highly reminiscent of the dimer unit identified in the crystal structure of the Ska core complex, which is a dimer of all three Ska subunits (180 Å × 80 Å) (Jeyaprakash, et al., 2012). Our V-shapes have a length of 201 ± 24 Å and a width of 90 ± 13 Å (Fig. 1B, Fig. S1B, n = 52 V-shapes measured), and crystal structure of the Ska core complex fits into these densities (Fig. 1C, Movie S4). Since the core structure lacks the two Ska1 C-terminal domains (CTD, ~50Å×10Å) and the Ska3 C-terminal regions that are in our structures, we suggest that the V-shapes are the correct size and shape to contain the Ska core complex and two Ska1 CTDs. We could not see any similar densities on microtubules treated only with Ska at the concentration used to visualize the complex with Ndc80Bonsai WT (1µM), however we detected some V-shaped structures on microtubules coated with Ska complex in concentrations above the Kd (4µM) (Fig. 1A). The number of those structures was significantly lower than on microtubules treated with both Ndc80 and sub-Kd concentrations of Ska, suggesting that the orientation of Ska molecules along protofilaments is dependent on the presence of Ndc80 (Fig. 1D, (+4CT will be described below)). Therefore, we suggest that the V-shapes that are recruited by Ndc80 that we observe in our tomographic reconstructions are composed of Ska complexes oriented along protofilaments on the wall of a microtubule. The V-shapes that we visualize appear to be oriented along a single microtubule-protofilament, although the resolution is not good enough to distinguish if they start and finish on adjacent protofilaments.
Figure 1. Ndc80 recruits Ska complex to microtubule protofilament.
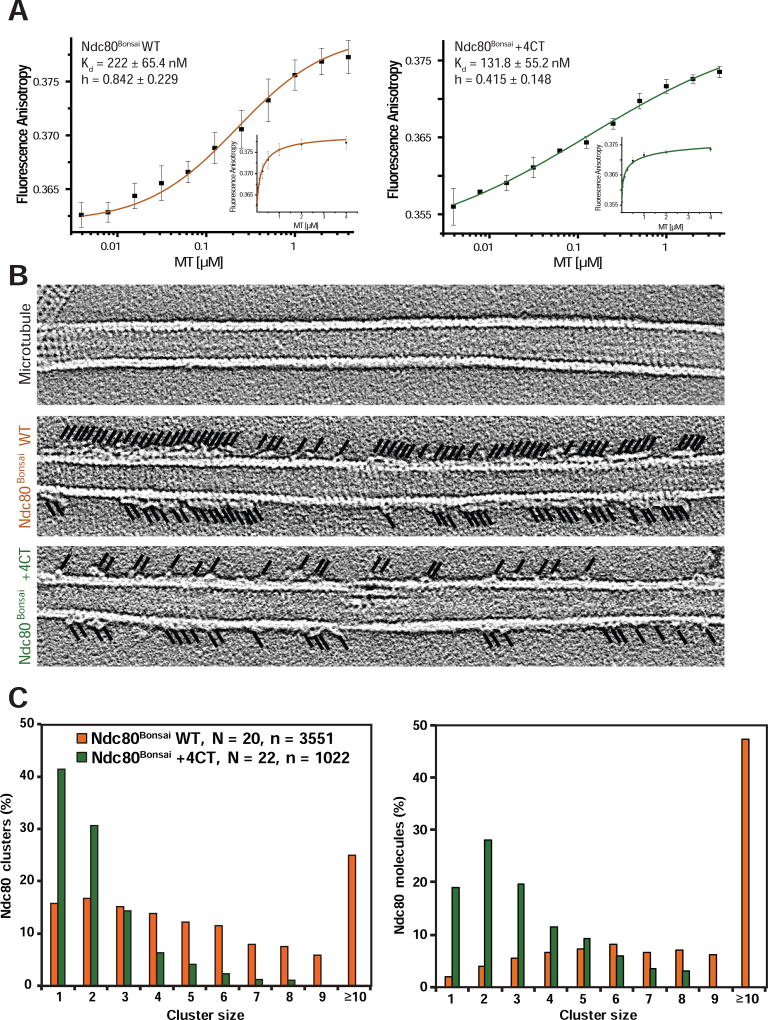
(A) Representative slices (thickness: 0.4427 nm) of tomographic reconstructions of taxol-stabilized microtubules (MT), microtubules decorated by Ndc80Bonsai WT (red, Movie S1), 1x Ska complex (purple), Ndc80Bonsai WT and 1x Ska complexes (green, 6 images), or saturating concentration of Ska (4x Ska, cyan). White arrowheads indicate positions of the tips of the V-shapes. Scale bar = 10 nm.
(B) Model of the Ska complex based on structures of the Ska complex core (PDB: 4AJ5) and Ska1 CTD (C-terminal domain = MTBD, microtubule binding domain, PDB: 4CA0). Comparison between size of the Ska core complex dimer from crystal structure and average measured size of the V-shapes identified in tomographic reconstructions of microtubules incubated with both Ndc80Bonsai WT and Ska is shown in the table below (mean ± SD). w = width; h = height; n = number of V-shapes measured.
(C) Structure of Ska core complex (PDB: 4AJ5) fitted to EM map of representative V-shape. Thickness: 2.6568 nm.
(D) Quantification of the V-shaped densities on microtubules identified in tomographic reconstructions (mean ± SD, average of 7 microtubules scored; WT = Ndc80Bonsai WT; +4CT = Ndc80Bonsai +4CT; * - p < 0.0002; Student’s t-test)
(E) Distance (D, red) between centroid of tip of V-shape (dashed line circle) and line connecting centroids of tubulin subunits within a single protofilament (dashed straight line) was measured and represented in panel E.
(F) Box and whisker plot of distribution of distances between tips of V-shapes and microtubule protofilaments measured as shown in panel D (Central line – median, Whiskers represent 10 – 90% range; n = 58, 18, 22, respectively. * - p < 0.000001; N/S – p > 0.99; Student’s t-test)
We also measured the distance (D) from the centroid of the tip of V-shapes to the attaching protofilament (as defined by the line running through centroids of tubulin monomers) (Fig. 1E, F, Fig. S1C). We observed that in presence of Ndc80 V-shapes were positioned further away from microtubule than in tomograms of high concentration of Ska and microtubules alone. This suggests Ndc80 either increases the height of Ska on the microtubule or increases the frequency that Ska binds along a single protofilament rather than spanning two or more adjacent protofilaments.
Ska is in close proximity to the calponin homology domain of Ndc80 at kinetochores
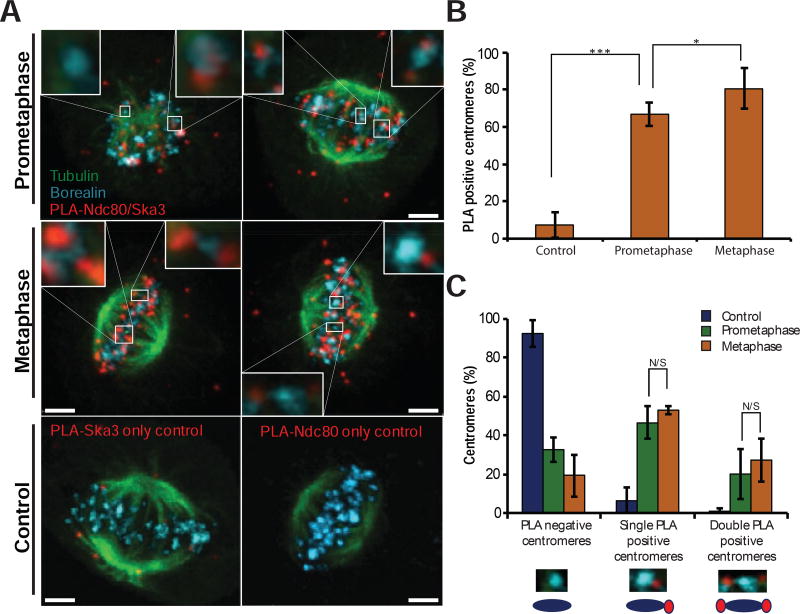
It is unclear how Ska is recruited to the kinetochore, and our tomograms suggest that Ndc80 directly recruits Ska to microtubules. We sought in vivo data to support or reject such a model. We utilized the Proximity Ligation Assay (PLA), which produces a fluorescent signal if two antigens are closer than 25 nm to each other (Soderberg, et al., 2006). We confirmed that PLA measures subkinetochore proximity by showing that antibodies recognizing a tag on the C-terminus of the Ndc80 protein generates signals when coupled with antibodies to the Spc25 subunit of the Ndc80 complex, but not when coupled with a kinetochore protein in another complex (Zw10, Fig. S2). PLA was used to measure the proximity between the Ndc80 CHD and the Ska3 protein. After the PLA reaction was performed, the cells were additionally stained with antibodies to tubulin and Borealin to identify the spindle and inner-centromeres respectively. The PLA signal was highly enriched at centromeres of mitotic cells and these signals were rarely found in control cells where one of the two primary antibodies in the PLA reaction was omitted (Fig. 2A). There were also foci of PLA signal in the cytoplasm of mitotic cells, which may represent interactions between soluble complexes. We quantified the number of Ndc80-Ska3 PLA spots adjacent to centromeres and found that Ndc80 and Ska3 appear to be in close proximity at both prometaphase and metaphase centromeres (Fig. 2B). 67% of prometaphase centromeres were associated with at least one PLA signal and this increased to 80% of centromeres during metaphase. The number of centromeres associated with two PLA signals facing opposite poles, which would be expected when both chromatids are stably attached to microtubules, increased in metaphase relative to prometaphase but the difference was not significant (Fig. 2C). These data demonstrate that Ndc80 CHD and Ska reside in close proximity at end-on attached kinetochores.
Figure 2. Ndc80 interacts with Ska3 in both prometaphase and metaphase.
(A) Representative images of Ska3-Ndc80 Proximity Ligation Assay (red), with additional immunostaining of Borealin (cyan) and tubulin (green). Top – prometaphase cells, middle – metaphase cells, bottom – control metaphase cells.
(B) Centromeres in prometaphase or metaphase cells, as identified by Borealin immunostaining, were scored based on the proximity to Ska3-Ndc80 PLA signal (mean ± SD; N > 100 centromeres from four cells). Cells with only Ska3 probe were used as a control. N/S – p > 0.05, * - p < 0.05, *** - p < 0.0001. (Student’s t-test)
(C) Centromeres in prometaphase or metaphase cells, as identified by Borealin immunostaining, scored based on the number of proximal Ska3-Ndc80 PLA signals (mean ± SD; N > 100 centromeres from four cells). N/S - p > 0.05, * - p < 0.05, *** – p < 0.0001. (Student’s t-test)
A point mutation in the Ndc80 tail prevents recruitment of Ska to kinetochores in vivo and inhibits formation of V-shapes in vitro
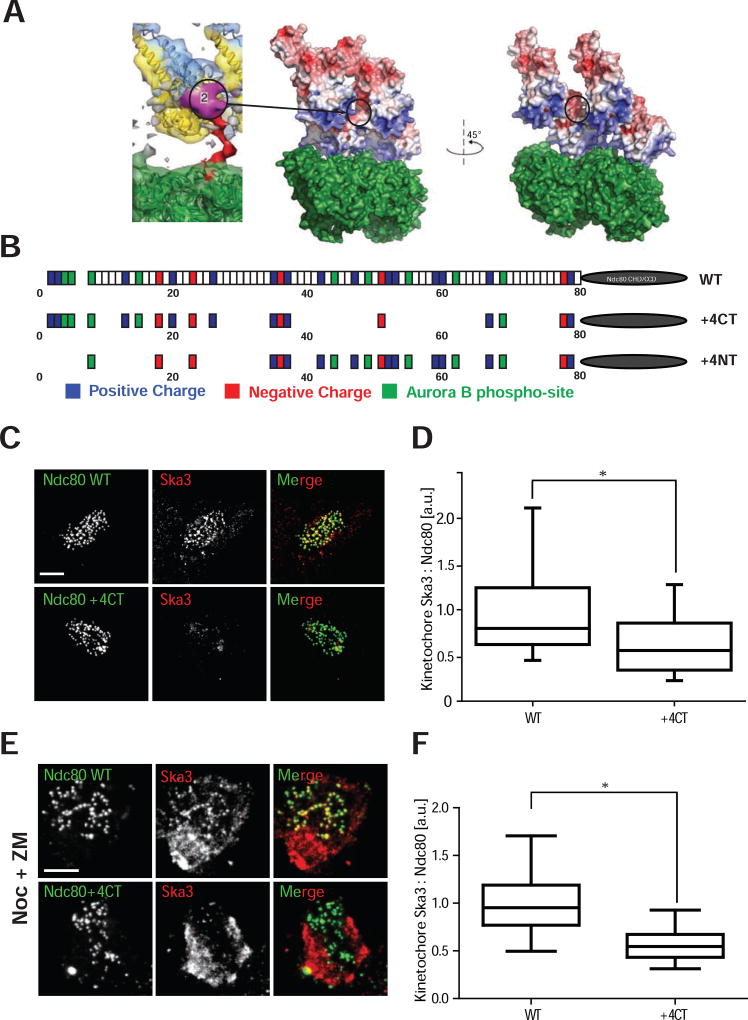
We often saw densities reminiscent of clusters of Ndc80 on a protofilament adjacent to our V-shapes in our tomograms, which suggested that Ska is recruited by a cluster of Ndc80 molecules along an end-on attached microtubule protofilament. We designed a point mutant in Ndc80 that would allow us to test whether clusters of Ndc80 recruit Ska to kinetochores in vivo. Clustering of Ndc80 is mediated by an 80 amino acid unstructured N-terminal tail of the Ndc80 protein. The amino acid residues within the tail have a net charge of +10 and this charge is essential for Ndc80 function (Tooley, et al., 2011). CryoEM densities of Ndc80Bonsai proteins on microtubules have oriented the globular domains unambiguously on microtubules (Alushin, et al., 2010). In these studies the tail can be seen as a density, but definitive assignments are not possible, consistent with the tail region being more loosely structured after microtubule binding. Instead, the tail can be seen as a density connected to the unambiguously assigned amino acid 81, which fills a groove formed by two adjacent Ndc80 CHDs (Alushin, et al., 2012). These tail densities then emerge on the back of two adjacent Ndc80 CHDs to form a large region that appears to bridge two adjacent CHDs (Fig. 3A). We examined the surface charge of the groove between adjacent CHDs and found adjacent negatively charged patches facing each other in the groove. We hypothesized that these negatively charged patches would repel adjacent subunit binding and that clustering is enabled by positive charges on the tail, which negate this repulsion. Therefore, we mutated the positively charged regions that bridge two adjacent subunits by generating the mutant Ndc80K(42, 47, 53, 59)A, R(52, 60)A (Fig. 3B). We have named this mutant +4CT because the overall charge of the unstructured tail is now +4 (instead of +10) and the charge has been removed from the C-terminal region. As a control, we generated a similar mutant (Ndc80K(2,26,35)A, R(3,13,20)A (Ndc80+4NT), Fig. 3B), which eliminates the same amount of charge on amino acids in the N-terminal part of the Ndc80 tail. Both of these mutants could be phosphorylated by Aurora B in vitro (Fig. S3A–C).
Figure 3. Ndc80+ mutant is deficient in recruitment of Ska3 to the kinetochore.
(A) Left: Purple-Localization of electron density patch attributed to the C-terminal region of Ndc80 tail. Note that it sits between two adjacent calponin homology domains (CHD) (Adapted from Alushin, et al., 2012). Right: Electrostatic surface potential of two Ndc80 CHD bound to tubulin dimer with the tail densities removed bound to a microtubule (PDB: 3IZ0) to illustrate the negatively charged surface between the Ndc80 CHD molecules. Black ellipse represents the localization of the purple patch visualized on the left panel. We mutated positively charged residues in the C-terminal region of the patch assuming that they bridged the negatively charged surfaces on the Ndc80 CHD to generate cooperative binding.
(B) Cartoon depicting the wild type Ndc80, Ndc80+ and Ndc80+ tail mutants to illustrate the change in charge that are created by the K/R to A changes in the Ndc80+ and Ndc80+ proteins. Although, it is likely that we are also affecting Aurora B phosphorylation sites since they are defined by K/RxS/T amino acids, we do not observe significant differences in in vitro phosphorylation by Aurora B between mutants and wild type protein (Fig. S3A–C). CHD = Calponin Homology Domain, CCD = coiled-coil domain. Drawing not to scale.
(C) Immunofluorescence staining of Ndc80WT and Ndc80+4CT stable cell lines after depletion of endogenous Ndc80 shows reduced levels of Ska3, on the kinetochores of Ndc80+4CT stable cell lines. Scale bar = 5 µm.
(D) Box and whisker plots representing the quantification of Ska staining intensities on kinetochores represented in (C) (N > 100 kinetochores from at least 5 cells, Whiskers – 5–95% percentile). a.u. = arbitrary units. * - p < 0.0001 (Unpaired t-test with Welch’s correction).
(E) Immunofluorescence staining of Ndc80WT and Ndc80+4CT stable cell lines treated with 3.3 µM nocodazole (Noc) and 2 µM ZM447439 (ZM) after depletion of endogenous Ndc80 shows reduced levels of Ska3 (red) on kinetochores. Scale bar = 5 µm.
(F) Box and whisker plot representing the quantification of the Ska staining intensities on kinetochores in cells treated with nocodazole and ZM447439 represented in (E) (N > 100 kinetochores from at least 5 cells, Whiskers – 5–95% percentile). a.u. = arbitrary units. * - p < 0.0001 (Unpaired t-test with Welch’s correction).
We depleted endogenous Ndc80 from HeLa cells and expressed a siRNA resistant Ndc80+4CT-GFP or Ndc80-GFP from an integrated loci. We then measured the levels of the endogenous Ska complex at kinetochores using antibodies to the Ska3 protein. Cells complemented with the Ndc80+4CT protein had significantly reduced levels of the Ska protein, but not another outer kinetochore protein Dsn1, relative to those rescued with the wild-type Ndc80 protein (Fig. 3C, D and Fig. S4A, B). Cells complemented with Ndc80+4NT-GFP had similar amounts of Ska as those complemented with the wild type Ndc80 protein demonstrating that the location of the positive charge on the tail, and not simply the overall charge, is critical to localize Ska (Fig. S4C, D). Aurora kinases directly phosphorylate the Ska protein to inhibit its localization to kinetochores (Chan, et al., 2012). This ensures that the Ska protein is not recruited to kinetochores before microtubule attachment. However, cells treated with nocodazole can recruit Ska complex if the cells are also treated with Aurora inhibitors (Chan, et al., 2012). The Ndc80+4CT mutant was also defective in recruiting Ska under these conditions (Fig. 3E, F). Thus, the Ndc80 tail regulates the recruitment of the Ska complex to kinetochores under all known conditions that have been shown to localize Ska.
We also visualized complexes generated by Ndc80Bonsai +4CT, Ska, and microtubules by negative stain EM tomography in vitro. The V-shape Ska-containing complexes that were easily seen with Ndc80Bonsai WT were observed significantly less often on microtubules visualized by tomography after incubation with Ndc80Bonsai +4CT and Ska (Fig. 1D). We think that Ndc80Bonsai +4CT can recruit Ska, albeit with reduced frequency to Ndc80Bonsai WT, since the increased distance from the top of the V-shape to the microtubule was similar in Ndc80Bonsai +4CT and Ndc80Bonsai WT (Fig. 1F). Together our identification of a mutant that is deficient at forming V-shapes with Ndc80 in vitro and recruitment at kinetochores provides evidence for that the tail of Ndc80 has an important function in recruiting the Ska protein to kinetochores and it suggests that V-shapes we see in vitro can form on microtubules at kinetochores.
The Ndc80+4CT phenocopies loss of Ska complex, while the Ndc80+4NT recruits Ska to kinetochores that lack “end-on” microtubule attachments
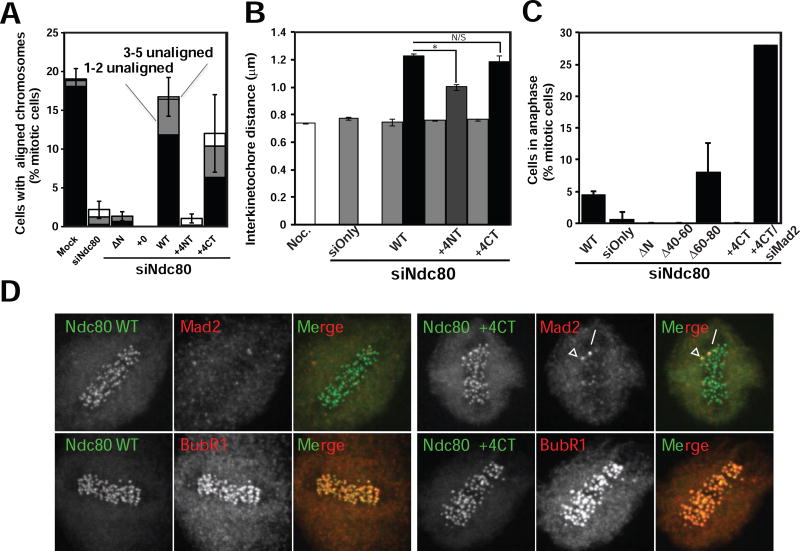
We characterized the mitotic phenotypes of HeLa cells depleted of the endogenous Ndc80 protein and rescued with the Ndc80+4CT and Ndc80+4NT mutants. The Ndc80+4CT mutant was capable of aligning chromosomes, although slightly less efficiently than cells rescued with the wild type protein (12.0 ± 1.7% of total mitotic cells) (Fig. 4A). This phenotype is similar to the reduction of Ska, which slightly delays chromosome congression (Daum, et al., 2009; Gaitanos, et al., 2009; Hanisch, et al., 2006). Strikingly, the Ndc80+4NT mutant was far less effective at aligning chromosomes than the Ndc80+4CT. Only 1.7 ± 0.8% of mitotic Ndc80+4NT cells were able to align their chromosomes (Fig. 4A), making the Ndc80+4NT alignment phenotype more comparable to the phenotype achieved by complete depletion of the Ndc80 complex or by the loss of all net positive charges from the tail (Tooley, et al., 2011). This result reinforces the concept that the Ndc80 tail is instead composed of two distinct regions and not a random stretch of positive charges (Alushin, et al., 2012).
Figure 4. Ndc80+4CT tail mutant metaphase arrest does not satisfy spindle assembly checkpoint and is Aurora dependent.
(A) Graph representing chromosome alignment among mitotic cells expressing Ndc80 mutants. Mitotic cells were scored for chromosome alignment into a metaphase plate, and the percentage of metaphase cells was plotted. Metaphase cells were further subdivided to indicate cells with all chromosomes aligned (black), cells with 1–2 unaligned chromosomes (gray) and cells with 3–5 unaligned chromosomes (white). Mitotic cells with 6 or more unaligned chromosomes are not shown in the graph. N > 100 mitotic cells counted per experiment, N = 3. Data is represented as mean ± SD (SD represents total of cells with 0–5 unaligned chromosomes).
(B) Graph representing interkinetochore distance measurements in cells expressing Ndc80 mutants. Ten sister kinetochores in at least five cells (N > 50) were identified by ACA staining between Ndc80 signals, and the distance between those sister kinetochores was measured (N=3). The distance (mean ± SD) is plotted for early prometaphase (EPM) (gray), late prometaphase (LPM) (dark gray), and metaphase (black) cells, Noc = nocodazole-treated cells (white). * - p < 0.0002. N/S - p > 0.2. (Student’s t-test)
(C) The percentage of mitotic cells in anaphase was plotted for Ndc80WT, Ndc80 knockdown and indicated Ndc80 tail mutant cells (mean ± SD). For the Ndc80+4CT mutant, cells were also co-transfected with siRNA targeting Mad2. For Ndc80 tail mutants, one hundred mitotic cells counted per experiment (n = 3). For Mad2 co-transfection with Ndc80+4CT, twenty-five cells were counted.
(D) Immunofluorescence staining of Ndc80WT and Ndc80+4CT (green) stable lines after depletion of endogenous Ndc80. Ndc80+4CT cells arrest with at least one Mad2 positive kinetochore (red, upper row) with no loss of BubR1 localization (red, lower row). Scale bar = 5 µm.
Next, we determined the ability of kinetochores with mutated Ndc80 to exert pulling forces on bound microtubules. We measured the distance between the sister kinetochores (inter-kinetochore distance) of aligned chromosomes as a readout for the strength of the kinetochore-microtubule interaction. Cells in early prometaphase have formed few productive kinetochore-microtubule interactions, and inter-kinetochore distance measurements for these cells were therefore comparable to Ndc80 knockdown cells or cells treated with high doses of nocodazole (Fig. 4B). Few cells expressing the Ndc80+4NT mutant had metaphase-aligned chromosomes, so we measured late prometaphase Ndc80+4NT cells instead. The average inter-kinetochore distance in these cells was 1.00 ± 0.02 µm, suggesting that kinetochores containing the Ndc80+4NT mutant generated reduced force. In contrast, aligned chromosomes in Ndc80+4CT metaphase cells had an average inter-kinetochore distance measurement of 1.18 ± 0.04 µm, which is similar to the average measurement for Ndc80WT metaphase chromosomes (1.23 ± 0.02 µm). We conclude that the Ndc80+4CT mutant can move chromosomes and generate full interkinetochore pulling forces, but the Ndc80+4NT loses all Ndc80 function and acts like mutants that completely lose microtubule binding activities (Tooley, et al., 2011; Miller, et al., 2008), even though it still efficiently recruits Ska complex to the kinetochores (Fig. S3C, D). This suggests that Ska recruitment is not triggered by the generation of end-on attachments.
If the Ndc80+4CT mutant is deficient in recruitment of Ska to kinetochores it should phenocopy loss of Ska. Cells depleted of the Ska complex align chromosomes to the metaphase plate but fail to silence the SAC because Ska has a second function as a protein that directly recruits PP1 to kinetochores (Sivakumar, et al., 2016; Daum, et al., 2009). Ndc80+4CT cells were capable of forming a metaphase plate and generating full tension on aligned chromosomes, however no cells expressing this mutant entered anaphase over the course of the experiment. A similar phenotype was seen when this mutated C-terminal region of the Ndc80 tail was deleted (Ndc80Δ40–60). In contrast, 4.4 ± 0.7% of Ndc80WT mitotic cells and 8.0 ± 4.6% of Ndc80Δ60–80 mitotic cells were observed in anaphase (Fig. 4C). This suggests that impairing the function of the C-terminal microtubule binding region, either by engineering point mutations or by making wholesale deletions, halts anaphase entry and phenocopies loss of Ska.
To determine if this cell cycle delay was caused by an active SAC signal, we performed a knockdown and rescue experiment of Ndc80 and concurrently depleted the SAC protein Mad2. 28% of Ndc80+4CT cells depleted of Mad2 were observed in anaphase, in sharp contrast to the absence of anaphase figures in Ndc80+4CT cells with functional SAC (Fig. 4C). When we examined the kinetochore localization of Mad2 in metaphase Ndc80+4CT cells, we found that Mad2 was lost from most kinetochores similarly to Ndc80WT controls. However, we almost always observed one kinetochore with robust Mad1 staining. This is consistent with either an inability to silence the SAC or the loss of stable microtubule attachments in a small population of cells (Fig. 4D). We also found similar levels of BubR1 at the kinetochores of metaphase Ndc80+4CT or Ndc80WT complemented cells. We conclude that the failure of our Ndc80+4CT mutant to progress into anaphase was due to active SAC signaling. Together, these data argue that the Ndc80+4CT mutant phenocopies loss of the Ska protein, which is consistent with Ndc80-dependent recruitment of Ska to kinetochores.
Ndc80+4CT binds microtubules with wild-type affinity but is deficient in clustering along microtubule protofilaments
Structural analysis suggests that the C-terminal region of the Ndc80 unstructured tail has two functions. First it interacts with the E-hooks of tubulin and second it bridges two adjacent Ndc80 proteins on microtubules (Alushin, et al., 2012). To determine if either or both of these activities are affected in the Ndc80+4CT mutant we characterized the biophysical properties of Ndc80+4CT using Ndc80Bonsai +4CT expressed in E.coli. We first measured the interaction of Ndc80Bonsai +4CT with Taxol-stabilized microtubules by fluorescence anisotropy. The interaction between Ndc80Bonsai +4CT and microtubules had a higher binding affinity than Ndc80Bonsai WT, with a Kd of 131.8 ± 55.2 nM (Fig. 5A). This distinguishes Ndc80+4CT mutant from other reported phospho-mimetic mutants in the C-terminal region of the Ndc80 tail which demonstrate reduced affinity for microtubules (Zaytsev, et al., 2015). These measurements complement our observation that Ndc80+4CT cells are capable of forming stable kinetochore-microtubule attachments and generating pulling forces during metaphase (Fig. 4). Strikingly, the Hill coefficient was noticeably reduced between Ndc80Bonsai WT (h = 0.842 ± 0.229) and Ndc80Bonsai +4CT (h = 0.415 ± 0.148). In fact, this change in slope of the binding curve suggests that Ndc80Bonsai +4CT binds with negative cooperativity to microtubules. We suggest that negative cooperativity arises since the Ndc80+4CT proteins cannot negate the repulsive forces generated by the negative patches that line the groove between adjacent subunits (Figure 3A). Therefore, if an Ndc80+4CT that interacts with a tubulin subunit next to a second Ndc80+4CT that this is less likely to be a productive binding event than Ndc80 WT
Figure 5. Ndc80+4CT tail mutant is deficient in clustering on microtubules.
(A) Fluorescence anisotropy measurements of fluorescently labeled Ndc80Bonsai WT (left panel, orange) or Ndc80Bonsai +4CT (right panel, dark green) incubated with increasing concentrations of taxol-stabilized microtubules, plotted on log10 scale (mean ± SD). Hill equation was used for fitment of the data. Small graphs represent the data in the linear scale. N = 3.
(B) Representative projections of 5 consecutive Z-sections (1.107 nm) of the tomographic reconstructions show Ndc80Bonsai +4CT form smaller clusters than Ndc80Bonsai WT. Black lines indicate the positions of the Ndc80 molecules.
(C) Quantification of cluster sizes represented in (B). N, number of quantified microtubules, n, total number of Ndc80Bonsai molecules quantified.
Ndc80Bonsai +4NT bound microtubules with lower affinity than Ndc80Bonsai WT (Kd = 345 ± 47 nM), but it did not affect the hill coefficient in our binding assays (Fig. S5A). Thus, the charge on the N-terminal part of the tail is required for binding microtubules in vitro and in vivo.
Structural studies show that the Ndc80 complex oligomerizes along microtubule protofilaments in vitro (Alushin, et al., 2012; Alushin, et al., 2010; Ciferri, et al., 2008). Our model to explain the negative cooperativity of Ndc80Bonsai +4CT binding to microtubules predicts that Ndc80Bonsai +4CT should have reduced clustering. We visualized the binding of Ndc80Bonsai by negative stain electron tomography to directly measure the binding of adjacent Ndc80 proteins along protofilaments. The formation of clusters was significantly decreased in the Ndc80Bonsai +4CT mutant compared to Ndc80Bonsai WT (Fig. 5B, C, Fig. S5B). We additionally observed a significantly reduced amount of V-shaped structures on microtubules incubated with Ska and Ndc80Bonsai +4CT mutant, as compared to Ndc80Bonsai WT (Fig. 1C). This suggests that the clustering of Ndc80 along microtubule protofilament is important for Ndc80-Ska complex formation.
DISCUSSION
Here we provide important insight into three critical processes that are required for mitotic progression. First, we identify the molecular steps that result in recruitment of Ska to kinetochores. Second, we identify the structure of a novel Ndc80-Ska complex on microtubules, which may explain how the curvature of a depolymerizing microtubule is used as a power-stroke to move chromosomes. Third, we provide the evidence that the clustering of Ndc80 proteins on microtubules, a phenomenon established in vitro, also exists and functions in vivo. These conclusions provide new models of kinetochore function and provide important insights into the mechanisms of SAC silencing and chromosome segregation during anaphase.
The mechanisms governing recruitment of Ska to kinetochores have proven elusive. Low levels of Ska can be measured at prometaphase kinetochores and its levels are highest in metaphase, when robust kinetochore-microtubule attachments are present. However, reduced levels of Ska can also localize to kinetochores in cells treated with nocodazole, when no kinetochore-microtubule attachments are present. These seemingly incongruent observations can be explained if Ska recognizes clustered Ndc80 proteins, or if Ska recognizes the Ndc80 unstructured tail which is exposed upon microtubule binding.
There are at least five experiments that support our model of Ska recruitment to kinetochore by direct binding to the N-terminus of the Ndc80 complex which contains the microtubule binding region. First, using PLA we show that Ska and the CHD of Ndc80 are in close proximity on microtubules in vivo. Second, Ndc80Bonsai, which lacks most of the coiled coil regions of the Ndc80 complex, orients Ska along microtubule protofilaments. Third, we have characterized mutants of the Ndc80 tail region that is deficient in Ska recruitment to microtubules in vitro and to kinetochores in vivo. Fourth and fifth, our model is consistent with published experiments showing that Ska enables the plus-end tracking of individual Ndc80 complexes on depolymerizing microtubules (Schmidt, et al., 2012), and the requirement of the Ndc80 complex to recruit Ska (Chan, et al., 2012; Raaijmakers, et al., 2009; Welburn, et al., 2009; Hanisch, et al., 2006).
Our tomographic reconstructions demonstrate that Ndc80 can orient Ska along a microtubule protofilament, while it was recently shown that EB1 orients Ska across multiple protofilaments to partially encircle the microtubule (Thomas, et al., 2016). We were not able to clearly visualize Ska lying across multiple protofilaments by EM-tomography of either Ska alone or the protein recruited by Ndc80. We do not understand the reason for this discrepancy. EB1 is asymmetrically recruited to sister kinetochores because it binds polymerizing microtubules that are on the anti-poleward sister (Tirnauer, et al., 2002). Thus, it is possible that Ska is bound along protofilaments on the poleward sister by interaction with Ndc80 to pull chromosomes, while EB1 on the anti-poleward sister binds Ska across the microtubule to allow microtubules to lengthen.
Functions of the Ndc80 unstructured tail
The N-terminal tail of Ndc80 modulates Ndc80-microtubule interactions, where it acts to decrease the koff of individual Ndc80 proteins (Zaytsev, et al., 2015; Alushin, et al., 2012; Cheeseman, et al., 2006; DeLuca, et al., 2006). This activity is countered by Aurora phosphorylation, which increases the koff to reduce binding affinity. Our studies suggest that the Ndc80 tail has an additional function in recruiting Ska. A similar conclusion was made in the accompanying paper in C. elegans, demonstrating that this function is highly conserved (REF needed). We also demonstrate that the tail enables clustering of Ndc80 along microtubule protofilaments as suggested by a previous structural study (Alushin, et al., 2012). We propose that the key event to recruit Ska is a tail-dependent clustering of Ndc80 proteins along microtubules, which forms a structure that is recognized by Ska. However, it is also possible that the C-terminal region of the Ndc80 tail controls both clustering and binding of Ska.
Cells depleted of Ska or cells expressing Ndc80+4CT arrest in metaphase and fail to silence the SAC. Ska recruits PP1 to kinetochores to enable progression to anaphase (Sivakumar, et al., 2016). Our data place Ska and presumably Ska/PP1 at the plus tip of the microtubule in close proximity to the microtubule binding sites of Ndc80, where PP1 would be in a position to dephosphorylate the Ndc80 tail and other nearby substrates. We suggest that this event would enhance binding of the Ndc80 tail to microtubules and drive the maturation of kinetochore-microtubule attachments, ultimately silencing the SAC signal. We have not shown that PP1 can participate in formation of a V-shaped complex, which is an important future experiment.
Insight into “end-on” kinetochore-microtubule attachment and depolymerization-coupled movement of chromosomes
Our findings provide clues to connect the Ska structure with its function. It was unclear why the coiled coils of the Ska core complex formed a V-shape. Our data orient the core of Ska lengthwise along a microtubule protofilaments and we suggest that the V-shape allows Ska to bind clusters of Ndc80 complexes. It was unclear how the ability of Ska to bind both straight and curved microtubules contributed to its function(s) (Abad, et al., 2014; Jeyaprakash, et al., 2012; Schmidt, et al., 2012). Our structure orients the two microtubule binding regions of the Ska1 C-terminal domain in two distinct positions suggesting that if a depolymerizing microtubule end encounters Ska then one microtubule binding domain could interact with the curved protofilament, while the other would sit behind a cluster of Ndc80 proteins on the straight region of the polymer.
Current models, known as unbiased diffusion or the molecular lawn, suggest that the Ndc80 complex acts passively in eukaryotes. It is theorized that each kinetochore contains >7 Ndc80s and each Ndc80 protein binds microtubules independently to form a molecular lawn. Kinetochores maintain attachment with depolymerizing microtubules because some Ndc80 molecules are still associated with straight regions of microtubule, while others dissociate from depolymerizing subunits. Kinetochore remains attached to the microtubule as long as dissociated Ndc80 proteins are able to re-bind to the straight protofilaments. Our data do not disprove this model, and it is possible that the clustering of Ndc80 complexes is transient and functions to recruit Ska/PP1 to dephosphorylate substrates allowing the end-on attachments to mature.
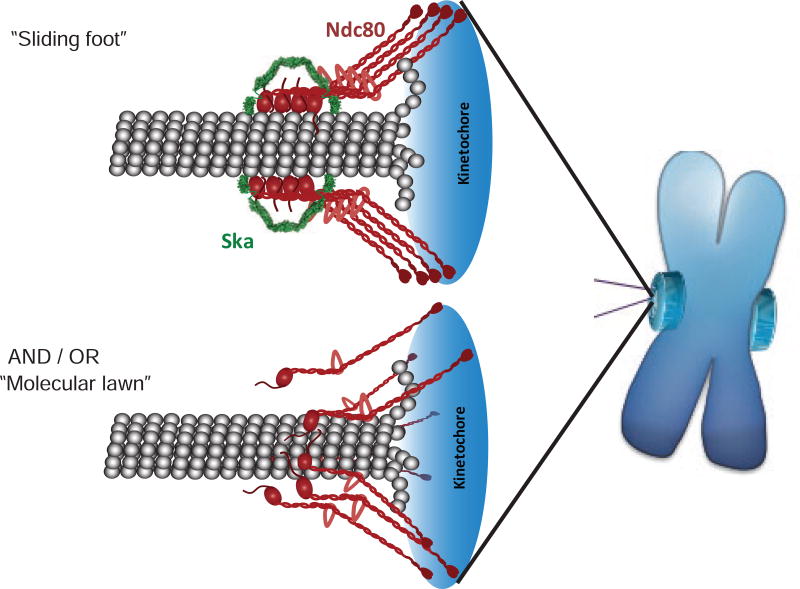
In contrast, many fungi have evolved the Dam1 complex that can form a ring around microtubules at kinetochores (Wang, et al., 2007; Westermann, et al., 2006; Westermann, et al., 2005). This ring creates a topologically constrained interaction with the polymer and enables sliding activities in a manner similar to the sliding clamps of DNA polymerases (Kong, et al., 1992; Stukenberg, et al., 1991). Elegant biophysical experiments have shown that the curvature of the depolymerizing microtubule produces force that pushes the Dam1 rings toward the minus end (Grishchuk, et al., 2008; Grishchuk and McIntosh, 2006; Westermann, et al., 2006). Our data suggest, but do not prove, that the Ndc80-Ska complex could similarly couple the curvature of depolymerizing microtubules into a pushing force and thus act similarly to the Dam1 clamp, albeit without encircling the microtubule. The V-shapes we observed suggest a speculative “sliding foot” model that can drive future experiments (Fig. 6). We propose that clusters of Ndc80 allow the complex to remain tightly bound to the microtubule protofilament, while the complex is pushed backwards. While there is a single interaction between the “toe” of the Ndc80 CHD and the tubulin surface (Tooley, et al., 2011; Alushin, et al., 2010), there are also at least two important interactions between the Ndc80 complex and tubulin that are built from unstructured tails and are required for the Ndc80 complex to interact with microtubules (Alushin, et al., 2012; Tooley, et al., 2011; Miller, et al., 2008). Clustering of 4 Ndc80 subunits would build an attachment with 12 distinct microtubule attachment points, 8 of which would be made from unstructured regions of proteins allowing Ndc80 to maintain an interaction with tubulin as it slides between adjacent tubulin subunits.
Figure 6. Schematic models summarizing potential modes of mature kinetochore-microtubule attachment.
“Sliding foot” model – Ska complex is localized and oriented along protofilament of the depolymerizing microtubule end by clusters of Ndc80. This mechanism likely allows for utilization of energy released by curving protofilaments.
“Molecular lawn” model – current model for passive binding of multiple individual Ndc80 molecules to depolymerizing microtubule end.
We propose that the “molecular lawn” represents an intermediate to forming the “sliding foot” and that either can move chromosomes. Cells depleted of the Ska complex still congress chromosomes to the metaphase plate (Sivakumar, et al., 2014; Daum, et al., 2009). We can phenocopy this result using mutant of the Ndc80 tail that is deficient in Ska recruitment. Therefore, the available data suggests that chromosomes move without a sliding foot and we suggest this is accomplished by a molecular lawn model of microtubule binding or by the motor CENP-E. However, Ska is at all kinetochores that make mature attachments and it remains bound during anaphase, so we propose that the sliding foot is the mature coupler of depolymerization energy into chromosome movement.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, P. Todd Stukenberg (pts7h@virginia.edu)
Experimental Model and Subject Details
Cell culture
HeLa cells (ATCC) or HeLa T-Rex (ThermoFisher Scientific) were maintained in Dulbecco’s Modified Eagle’s Medium (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) in a humidified incubator at 37 °C with 5% CO2.
Method Details
Electron microscopy and tomography
Preparation of negatively stained samples was done essentially as previously described (Alushin, et al., 2012). For experiment depicted in Fig. 1A, B EM grids were incubated with 2 µM taxol-stabilized microtubules in BRB80 supplemented with 10 µM taxol for 30 seconds, then with 2 µM Ndc80Bonsai WT or Ndc80Bonsai +4CT and 1 µM of the Ska complex for 2 min, followed by staining with 2% uranyl formate. 1 µM and 4 µM of the Ska complex was used for 1x Ska and 4x Ska samples, respectively. Tilt series from −60 to 60 degrees were collected semi-automatically by SerialEM software package (Mastronarde, D.N. 2005) with Tecnai F20 operating at 120 kV, at nominal magnification of 50,000x and 1.2 or 2.5 µm underfocus. IMOD software packages (Kremer, et al., 1996) were used for tomographic reconstructions: following alignment, tilt series were binned by two and filtered before first inversion of the ‘contrast transfer function’. Tomographic reconstructions were calculated using back projection algorithm. For experiment depicted in Fig. 5B and Fig. S5B EM grids were incubated with 2 µM taxol-stabilized microtubules for 30 seconds in BRB80 supplemented with 10 µM taxol, then with 3.3 µM Ndc80Bonsai WT or Ndc80Bonsai +4CT for 2 min, followed by brief wash in BRB80 and staining with 2% uranyl formate. Tilt series from −60 to 60 degrees were then collected manually and processed using IMOD software packages, with no subsequent binning and filtering of the acquired micrographs. Analysis was performed only on microtubules that were not significantly flattened or deformed during grid preparation.
Immunofluorescence
For most of the immunofluorescence experiments, coverslips were co-fixed and extracted in PHEM buffer containing 2% paraformaldehyde and 0.5% Triton X-100 for 20 min at room temperature. Antibodies used were anti-Ndc80 9G3 (1:500 [vol/vol]; GTX70268, GeneTex), anti-GFP (1:500 [vol/vol]; ab1218, Abcam), anti-ACA (1:500 [vol/vol]; 15–234-0001, Antibodies Incorporated), anti-tubulin (1:500 [vol/vol]; DM1α, NeoMarkers), anti-BubR1 (1:500 [vol/vol]), anti-Mad2 (1:100 [vol/vol]; a kind gift from Gary Gorbsky), anti-Ska3 (1.5 µg/mL; a kind gift from Gary Gorbsky), anti-Dsn1 (1:1000 [vol/vol]) and FITC conjugated anti-tubulin (1:500 [vol/vol]; DM1α, Sigma). DAPI staining (1:20000 of a 5 mg/ml stock; Invitrogen) was used to visualize DNA. The images from Fig. 3C and Fig. 4D were collected using a 100x lens on a Deltavision microscope (Applied Precision) and deconvoluted z-projections are shown. Images from Fig. 2, Fig. 3, Fig. S2 and Fig. S4 were captured using Zeiss Axiovert 200 inverted microscope with a Perkin Elmer confocal attachment, a krypton/argon laser and AOTF control for detecting fluorescence at 488, 568, and 647 nm, using 63x lens. Images were captured with a Hamamatsu digital CCD camera. Image acquisition and processing was done using Volocity 6.3 imaging software (Perkin Elmer). Quantification of the fluorescence intensities was done by semi-automated detection of kinetochore volumes using ACA mask in Volocity 6.3 imaging software. Each kinetochore signal was divided by its volume, then normalized to equivalent Ndc80 signal following background subtraction. Inter-kinetochore distance measurements were quantified using softWoRX imaging software.
Protein purification
Expressions in BL21 (DE3) cells of both GST-Ska3 and untagged Ska2/Ska1 (kind gifts from Iain M. Cheeseman) were induced by addition of 0.1 M imidazole, and continued for 4hrs at 20 °C. Cells were subsequently pelleted and lysed in PBS, pH 7.4, supplemented with 250 mM NaCl and 1 mM DTT (Wash Buffer), and cell debris was removed by centrifugation. GST-Ska3 was immobilized on Glutathione Agarose Resin (Gold Biotechnology), briefly washed with Wash Buffer, and incubated with cell lysate containing untagged Ska2/Ska1 for 1 hr at 4 °C. After subsequent wash with Wash Buffer the Ska complex was eluted with Wash Buffer supplemented with 15 mM L-Glutathione Reduced and gel filtered on Superdex 200 column 10/300 GL size-exclusion column (GE Healthcare Life Sciences), previously equilibrated to 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM DTT.
For fluorescence anisotropy and kinase assay Ndc80Bonsai constructs were expressed, lysed and pre-cleared similarly to GST-Ska3. Cell lysate was then incubated with Glutathione Agarose Resin (Gold Biotechnology) followed by extensive wash with PBS supplemented with 250 mM NaCl and 1 mM DTT (Wash Buffer). Untagged proteins were eluted by overnight incubation with HRV 3C protease and gel filtered on Superdex 200 column 10/300 GL size-exclusion column (GE Healthcare Life Sciences), previously equilibrated to 50 mM Tris, pH 7.4, 150 mM NaCl, 1 mM DTT.
For electron tomography experiments Ndc80Bonsai constructs were purified similarly as described above, with following modifications. After incubation of the cell lysate with the Glutathione Agarose Resin (Gold Biotechnology) proteins were washed with Wash Buffer, eluted with Wash Buffer supplemented with 15 mM reduced L-glutathione, and flash frozen in liquid nitrogen for storage in −80 °C. Prior to grid preparation, samples were thawed and subjected to gel filtration on Superdex 200 10/300 GL size-exclusion column in (20 mM Tris pH 7.2, 100 mM KCl, 1 mM DTT). Protein were then re-bound to glutathione resin and washed into BRB80 + 1 mM DTT. Untagged Ndc80Bonsai was eluted by overnight cleavage of GST-tag by HRV 3C Protease. Samples were then precleared at 240,000 × g for 20 min at 4 °C, warmed to room temperature and centrifuged in table-top centrifuge at 17,000 × g for 10 min prior to EM grid preparation.
Knockdown and rescue of Ndc80
For synchronization, cells were seeded in media containing 2mM thymidine for 24 hr, released into fresh media for 12 hr, arrested again in 2mM thymidine for 12 hr, released for 8–12 hr, and fixed for immunofluorescence with 2% PFA in PHEM buffer + 0.5% Triton X-100 or ice-cold methanol. For knock-down experiments in stable T-Rex HeLa cell lines lines (Fig. 3, Fig. 4D and Fig. S4), transfection of Ndc80 siRNA (see Key Resource Table for sequence, 75 nM final concentration) was done at first thymidine release and second thymidine block using RNAiMAX (Life Technologies), according to manufacturer’s description. For experiment depicted in Fig. 3E 3.3 µM Nocodazol (Sigma-Aldrich) and 2 µM ZM 447439 (R&D Systems) were added two hours before fixation. For knockdown and replacement experiments (Fig. 4A–C), HeLa cells were co-transfected at the first thymidine release with siRNA oligos (75 nM for Ndc80, 20 nM for Mad2) and 100 ng rescue plasmid using Lipofectamine 2000 (Life Technologies). Cells were transfected a second time with siRNA oligos at the second thymidine block using RNAiMAX (Life Technologies). For mock and siRNA only controls, an empty pEGFP vector was used as the rescue plasmid. For transient transfection Ndc80 tail experiments, 75 nM GAPD siRNA oligos (Thermo Scientific) were included as mock controls.
Creation of stable cell lines
To create a stable cell line expressing the Ndc80WT, Ndc80+4CT and Ndc80+4NT tail mutant, appropriate sequences were cloned into the pCDNA5/FRT plasmid (Invitrogen) using flanking Not1 restriction sites. The plasmid was co-transfected into T-Rex HeLa cells (Invitrogen) with the pOG44 plasmid (Invitrogen) and the cells were cultured in DMEM + 10% FBS (Gibco) supplemented with hygromycin B (Invitrogen) for 14 days. At the end of the selection period, remaining cells were pooled and used for subsequent experiments.
Proximity Ligation Assay
After fixation of HeLa cells PLA was performed using Duolink In-Situ PLA probe anti-Mouse MINUS and anti-Rabbit PLUS and Duolink II Detection Reagents Orange, according to manufacturer’s instructions (Sigma-Aldrich), using the provided blocking solution and antibody diluent. Samples were incubated with primary antibodies overnight at 4°C. Following primary antibodies were used for the assay: anti-Ska3 (gift from Gary Gorbsky, rabbit polyclonal), anti-Ndc80 9G3 (GTX70268, GeneTex), FITC conjugated mouse monoclonal anti-tubulin (Sigma-Aldrich), and Cy5 conjugated anti-Borealin (Stukenberg Lab, rabbit polyclonal). Quantification of PLA-positive centromeres was done in Volocity 6.3 software (PerkinElmer) in 3D Opacity mode, by semi-automatic detection of the centromeric volumes and manual scoring of surrounding PLA signals.
Fluorescence Anisotropy
Ndc80Bonsai WT, Ndc80Bonsai +4CT and Ndc80Bonsai +4NT were labeled with Oregon Green 488 maleimide (ThermoFisher Scientific) according to manufacturer instructions and the un-reacted dye was removed by size exclusion chromatography. Efficiency of labeling was estimated to be between 1.5 to 2 dye molecules per Ndc80Bonsai molecule. Increasing concentrations of taxol stabilized MT were incubated at room temperature overnight with 50 nM fluorescently labeled Ndc80Bonsai, then 35 µL of mixture was loaded in triplicates into 384-well plate (Greiner Bio-One) and fluorescence anisotropy measurements were done in PHERAstar FS plate reader (BMG Labtech). The anisotropy data were fitted using Hill equation by OriginPro 7.5 software (Built-in non-linear fitting function Hill1, y = Start + (End – Start) * xn/(kn + xn)).
Kinase Assay
Aurora B-Incenp790–856 (Banerjee et al., 2014) was diluted to 0.03 mg/ml in Kinase Buffer (20 mM Tris, pH 7.5, 25 mM KCl, 1 mM MgCl2, 1 mM DTT) supplemented with 1 mM ATP/10 µM γ-[32P] ATP. Following mixture was added in 1:10 v/v ratio to 1 µM Ndc80Bonsai WT, Ndc80Bonsai +4CT or Ndc80Bonsai +4NT, previously dialyzed to Kinase Buffer. 20µL samples were taken from the reaction mixture at appropriate time points and added to 5 µL 5xSample Buffer to quench the reaction. Samples were further subjected to SDS-PAGE, followed by Coomassie staining. Gels were dehydrated on Whatman paper and exposed to Phosphor Screen overnight. Phosphor Screens were scanned on Storm 860, Molecular Dynamics, and resulting images were quantified using ImageJ software. Amount of incorporated PO4 was calculated based on initial concentrations of ATP and γ-[32P] ATP and standard curves obtained by serial dilutions of the reaction mixture.
Quantification and statistical analysis
Statistical parameters and tests used for analysis are reported in Figures and Figure Legends. Statistical analysis was performed by GraphPad Prism (GraphPad). p < 0.05 was considered to be statistically significant.
Supplementary Material
Movie S1 (Related to Fig. 1). Serial slices of tomographic reconstruction of microtubules coated with Ndc80Bonsai WT.
Movies S2 (Related to Fig. 1). Serial slices of tomographic reconstruction of microtubules coated with Ndc80Bonsai WT and Ska complex showing representative V-shape.
Movies S3 (Related to Fig. 1). Serial slices of tomographic reconstruction of microtubules coated with Ndc80Bonsai WT and Ska complex showing representative V-shape.
Movie S4. (Related to Fig. 1) Movie showing structure of Ska core complex (PDB: 4AJ5) fitted to representative V-shape.
Highlights.
The Ndc80 complex orients the Ska complex along microtubule protofilaments
The Ndc80 unstructured tail recruits the Ska complex to kinetochore
Structural view of a putative metazoan kinetochore microtubule attachment
Clusters of Ndc80, which are seen on microtubules in vitro, may function in vivo
Acknowledgments
The authors wish to thanks Eva Nogales and Greg Alushin for help with tomography and the original observation of the V-shapes on microtubules. We thank Iain Cheeseman and Andrea Musacchio for plasmids and Dan Burke, Gary Gorbsky, Bob Nakamoto and Zygmunt Derewenda for helpful discussions about the work. DRM was supported by T32 GM008136. The work was supported by GM081576 and GM112508.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
P.Ł.J. performed experiments in figure 1, 3, 5, S1, S3, S4 and S5. P.Ł.J., K.A.S. and O.P. designed and/or performed electron tomography. C.A.K. performed Proximity Ligation Assay, which was quantified by P.Ł.J. P.Ł.J., J.T., T.W. and D.R.M. performed experiments depicted in Fig. 3, 4 and S4. P.Ł.J. and P.T.S. designed the study and wrote the manuscript.
References
- Abad MA, Medina B, Santamaria A, Zou J, Plasberg-Hill C, Madhumalar A, Jayachandran U, Redli PM, Rappsilber J, Nigg EA, et al. Structural basis for microtubule recognition by the human kinetochore Ska complex. Nat Commun. 2014;5:2964. doi: 10.1038/ncomms3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- Alushin GM, Musinipally V, Matson D, Tooley J, Stukenberg PT, Nogales E. Multimodal microtubule binding by the Ndc80 kinetochore complex. Nat Struct Mol Biol. 2012;19:1161–7. doi: 10.1038/nsmb.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Ramey VH, Pasqualato S, Ball DA, Grigorieff N, Musacchio A, Nogales E. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–10. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee B, Kestner CA, Stukenberg PT. EB1 enables spindle microtubules to regulate centromeric recruitment of Aurora B. J Cell Biol. 2014;204:947–63. doi: 10.1083/jcb.201307119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J Cell Biol. 2012;196:563–71. doi: 10.1083/jcb.201109001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–97. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Ciferri C, De Luca J, Monzani S, Ferrari KJ, Ristic D, Wyman C, Stark H, Kilmartin J, Salmon ED, Musacchio A. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005;280:29088–95. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–39. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum JR, Wren JD, Daniel JJ, Sivakumar S, McAvoy JN, Potapova TA, Gorbsky GJ. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr Biol. 2009;19:1467–72. doi: 10.1016/j.cub.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, Cimini D, Musacchio A, Salmon ED. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–82. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- DeLuca JG, Moree B, Hickey JM, Kilmartin JV, Salmon ED. hNuf2 inhibition blocks stable kinetochore-microtubule attachment and induces mitotic cell death in HeLa cells. J Cell Biol. 2002;159:549–55. doi: 10.1083/jcb.200208159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca KF, Lens SM, DeLuca JG. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci. 2011;124:622–34. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, McCleland ML, Satinover DL, Stukenberg PT. Measuring the Stoichiometry and Physical Interactions between Components Elucidates the Architecture of the Vertebrate Kinetochore. Mol Biol Cell. 2005;16:4882–4892. doi: 10.1091/mbc.E05-03-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos TN, Santamaria A, Jeyaprakash AA, Wang B, Conti E, Nigg EA. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. Embo J. 2009;28:1442–52. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, Efremov AK, Volkov VA, Spiridonov IS, Gudimchuk N, Westermann S, Drubin D, Barnes G, McIntosh JR, Ataullakhanov FI. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc Natl Acad Sci U S A. 2008;105:15423–8. doi: 10.1073/pnas.0807859105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, McIntosh JR. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. Embo J. 2006;25:4888–96. doi: 10.1038/sj.emboj.7601353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–8. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–84. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. Embo J. 2006;25:5504–15. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyaprakash AA, Santamaria A, Jayachandran U, Chan YW, Benda C, Nigg EA, Conti E. Structural and functional organization of the ska complex, a key component of the kinetochore-microtubule interface. Mol Cell. 2012;46:274–86. doi: 10.1016/j.molcel.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, Bloom KS, Salmon ED. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–5. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Salmon ED, Bloom KS. Counting kinetochore protein numbers in budding yeast using genetically encoded fluorescent proteins. Methods Cell Biol. 2008;85:127–51. doi: 10.1016/S0091-679X(08)85007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston K, Joglekar A, Hori T, Suzuki A, Fukagawa T, Salmon ED. Vertebrate kinetochore protein architecture: protein copy number. J Cell Biol. 2010;189:937–43. doi: 10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Janke C. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. Journal of Cell Biology. 2001;152:349–60. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong XP, Onrust R, O’Donnell M, Kuriyan J. Three-dimensional structure of the beta subunit of E. coli DNA polymerase III holoenzyme: a sliding DNA clamp. Cell. 1992;69:425–37. doi: 10.1016/0092-8674(92)90445-i. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–6. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- McCleland ML, Gardner RD, Kallio MJ, Daum JR, Gorbsky GJ, Burke DJ, Stukenberg PT. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–14. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland ML, Kallio MJ, Barrett-Wilt GA, Kestner CA, Shabanowitz J, Hunt DF, Gorbsky GJ, Stukenberg PT. The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs, which are required to establish and maintain kinetochore-microtubule attachment. Curr Biol. 2004;14:131–7. doi: 10.1016/j.cub.2003.12.058. [DOI] [PubMed] [Google Scholar]
- Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr Biol. 2008;18:1785–91. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molodtsov MI, Grishchuk EL, Efremov AK, McIntosh JR, Ataullakhanov FI. Force production by depolymerizing microtubules: a theoretical study. Proc Natl Acad Sci U S A. 2005;102:4353–8. doi: 10.1073/pnas.0501142102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF chimera—A visualization system for exploratory research and analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Powers AF, Franck AD, Gestaut DR, Cooper J, Gracyzk B, Wei RR, Wordeman L, Davis TN, Asbury CL. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–75. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci. 2009;122:2436–45. doi: 10.1242/jcs.051912. [DOI] [PubMed] [Google Scholar]
- Schmidt JC, Arthanari H, Boeszoermenyi A, Dashkevich NM, Wilson-Kubalek EM, Monnier N, Markus M, Oberer M, Milligan RA, Bathe M, et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev Cell. 2012;23:968–80. doi: 10.1016/j.devcel.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S, Daum JR, Tipton AR, Rankin S, Gorbsky GJ. The Spindle and kinetochore-associated (Ska) complex enhances binding of the Anaphase-Promoting Complex/Cyclosome (APC/C) to chromosomes and promotes mitotic exit. Mol Biol Cell. 2014 doi: 10.1091/mbc.E13-07-0421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar S, Janczyk PL, Qu Q, Brautigam CA, Stukenberg PT, Yu H, Gorbsky GJ. The human SKA complex drives the metaphase-anaphase cell cycle transition by recruiting protein phosphatase 1 to kinetochores. Elife. 2016;5 doi: 10.7554/eLife.12902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, et al. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat Methods. 2006;3:995–1000. doi: 10.1038/nmeth947. [DOI] [PubMed] [Google Scholar]
- Stukenberg PT, Studwell PS, O’Donnell M. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. Journal of Biological Chemistry. 1991;266:21681–6. [PubMed] [Google Scholar]
- Suzuki A, Badger BL, Salmon ED. A quantitative description of Ndc80 complex linkage to human kinetochores. Nat Commun. 2015;6:8161. doi: 10.1038/ncomms9161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas GE, Bandopadhyay K, Sutradhar S, Renjith MR, Singh P, Gireesh KK, Simon S, Badarudeen B, Gupta H, Banerjee M, et al. EB1 regulates attachment of Ska1 with microtubules by forming extended structures on the microtubule lattice. Nat Commun. 2016;7:11665. doi: 10.1038/ncomms11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, Canman JC, Salmon ED, Mitchison TJ. EB1 Targets to Kinetochores with Attached, Polymerizing Microtubules. Mol Biol Cell. 2002;13:4308–16. doi: 10.1091/mbc.E02-04-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley JG, Miller SA, Stukenberg PT. The Ndc80 Complex Employs a Tripartite Attachment Point to Couple Microtubule Depolymerization to Chromosome Movement. Mol Biol Cell. 2011 doi: 10.1091/mbc.E10-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Joshi P, Salmon ED. How tubulin subunits are lost from the shortening ends of microtubules. J Struct Biol. 1997;118:107–18. [Google Scholar]
- Wang HW, Ramey VH, Westermann S, Leschziner AE, Welburn JP, Nakajima Y, Drubin DG, Barnes G, Nogales E. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat Struct Mol Biol. 2007;14:721–6. doi: 10.1038/nsmb1274. [DOI] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–9. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Welburn JP, Grishchuk EL, Backer CB, Wilson-Kubalek EM, Yates JR, 3rd, Cheeseman IM. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–85. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–90. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–9. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kilmartin JV. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol. 2001;152:349–60. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev AV, Mick JE, Maslennikov E, Nikashin B, DeLuca JG, Grishchuk EL. Multisite phosphorylation of the NDC80 complex gradually tunes its microtubule-binding affinity. Mol Biol Cell. 2015;26:1829–44. doi: 10.1091/mbc.E14-11-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaytsev AV, Sundin LJ, DeLuca KF, Grishchuk EL, DeLuca JG. Accurate phosphoregulation of kinetochore-microtubule affinity requires unconstrained molecular interactions. J Cell Biol. 2014;206:45–59. doi: 10.1083/jcb.201312107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Kelstrup CD, Hu XW, Kaas Hansen MJ, Singleton MR, Olsen JV, Nilsson J. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J Cell Sci. 2012;125:3243–53. doi: 10.1242/jcs.104208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1 (Related to Fig. 1). Serial slices of tomographic reconstruction of microtubules coated with Ndc80Bonsai WT.
Movies S2 (Related to Fig. 1). Serial slices of tomographic reconstruction of microtubules coated with Ndc80Bonsai WT and Ska complex showing representative V-shape.
Movies S3 (Related to Fig. 1). Serial slices of tomographic reconstruction of microtubules coated with Ndc80Bonsai WT and Ska complex showing representative V-shape.
Movie S4. (Related to Fig. 1) Movie showing structure of Ska core complex (PDB: 4AJ5) fitted to representative V-shape.