Abstract
Arylamine N-acetyltransferase (NAT; E.C. 2.3.1.5) enzymes are responsible for the biotransformation of several arylamine and hydrazine drugs by acetylation. In this process, the acetyl group transferred to the acceptor substrate produces NAT deacetylation and, in consequence, it is susceptible of degradation. Sirtuins are protein deacetylases, dependent on nicotine adenine dinucleotide, which perform post-translational modifications on cytosolic proteins. To explore possible sirtuin participation in the enzymatic activity of arylamine NATs, the expression levels of NAT1, NAT2, SIRT1 and SIRT6 in peripheral blood mononuclear cells (PBMC) from healthy subjects were examined by flow cytometry and Western blot. The in situ activity of the sirtuins on NAT enzymatic activity was analyzed by HPLC, in the presence or absence of an agonist (resveratrol) and inhibitor (nicotinamide) of sirtuins. We detected a higher percentage of positive cells for NAT2 in comparison with NAT1, and higher numbers of SIRT1+ cells compared to SIRT6 in lymphocytes. In situ NAT2 activity in the presence of NAM inhibitors was higher than in the presence of its substrate, but not in the presence of resveratrol. In contrast, the activity of NAT1 was not affected by sirtuins. These results showed that NAT2 activity might be modified by sirtuins.
Abbreviations: Ac-INH, acetyl-Isoniazid; Ac-PABA, acetyl-p-aminobenzoic acid; APC, allophycocyanin; CHO, Chinese hamster ovary cells; DMEM, Dulbecco's modified Eagle's medium; E2F1, E2F transctriptios factor 1; ER81, ETS-related protein 81; FITC, fluorescein IsoTioCyanate; FOXO1, forkhead box protein O1; HeLa, adenocarcinoma epithelial cells; HPLC, high performance liquid chromatography; INH, isoniazid; NAD, nicotinamide adenine dinucleotide; NAM, nicotinamide; NAT, arylamine N-acetyltranferase; PABA, p-aminobenzoic acid; PAS, p-aminosalicilic acid; PBMC, peripheral blood mononuclear cells; PBS, phosphate-buffered saline; PGAM1, phosphoglycerate mutase 1; PGC-1α, peroxisome proliferator-activated receptor-gamma coactivator 1α; RSV, resveratrol; RUNX3, runt-related transcription factor 3; SIRT, sirtuin; SMZ, sulfamethazine; SREBP1a, sterol regulatory element-binding protein 1a; SREBP2, sterol regulatory element-binding protein 2
KEY WORDS: Arylamine N-acetyltransferase, NAT, Sirtuins, Peripheral blood mononuclear cells, Nicotinamide, Resveratrol
Graphical abstract
The present study exhibited a highly significant effect (P<0.005) on NAT2 activity in peripheral blood mononuclear cells once deacetylation function of sirtuins is inhibited by nicotinamide (NAM) generating high levels of Ac-INH which suggests a post-translational regulatory mechanism.
1. Introduction
Acetylation is the major biotransformation pathway for arylamine and hydrazine drugs with pharmacological/toxicological relevance, and it is catalyzed by arylamine N-acetyltransferases (NATs; E.C. 2.3.1.5) enzymes1., 2.. NAT1 and NAT2 are phase II cytosolic enzymes that transfer an acetyl group from acetyl CoA to a xenobiotic substrate (arylamine, aromatic, heterocyclic or hydrazine compounds)3. In spite of the high similarity at genetic structure and protein level, these two enzymes have different specificity of substrate and tissue expression. p-Aminobenzoic acid (PABA), p-aminosalicylic acid (PAS) and p-aminobenzoyl glutamic acid are specific substrates for human NAT1, and show wide tissue distribution4., 5.. Conversely, NAT2 has a more restricted distribution and expression to the liver, intestinal epithelium and colon. The known targets of this enzyme are: sulfamethazine (SMZ), isoniazid (INH), procainamide and dapsone. Moreover, other compounds function as substrates for both enzymes, e.g. 2-aminofluorene6.
NATs can be regulated at transcriptional, post-transcriptional and post-translational levels. These enzymes share three domains, the domains I and II are more conserved between NAT enzymes than domain III and have a conserved catalytic triad composed of three residues: Cysteine, Histidine and Aspartate; in their functional structure, which forms part of the active site1., 2.. Regarding the post-translational regulation of NATs, the active site Cysteine of these enzymes is acetylated in the absence of substrate, which makes it more resistant to proteasomal degradation1. In contrast, the non-acetylated form of the protein is sensitive to polyubiquitination that leads to its degradation by the proteasome7. It has been reported that acetylation of the active site cysteine (Cys68) defines the stability of NAT1. On the other hand, acetylation of NAT, like many cytosolic proteins, occurs in different amino acid residues across the entire structure, this prevents the protein from being degraded or having a greater half-life. Nevertheless, the acetyl groups have to be removed from the protein in order to maintain the equilibrium in the cell. In this mechanism of regulation some deacetylases proteins participate such as sirtuins in the specific case of lysine residues8.
The sirtuin family includes seven (SIRT1–SIRT7) deacetylase/ADP ribosyltransferase proteins that vary in cellular localization, tissue specificity, enzymatic activity and protein substrates. They are involved in post-translational modifications by deacetylation (SIRT1, 2, 3, 5 and 6) or ADP ribosylation (SIRT4, 5 and 6) and play an important regulatory role in many biological processes9. Sirtuins catalyse the deacetylation of lysine residues of target proteins using nicotinamide adenine dinucleotide (NAD+) as a cofactor and liberating nicotinamide which, in high concentrations, is able to bind to sirtuin in a non-competitive form, and by a feedback-loop mechanism, inhibit its activity10.
Due to widespread participation of sirtuins in many physiological processes such as metabolism, stress and ageing11, it has been suggested that NAT1 and NAT2 could be affected by sirtuin-regulated deacetylation. Positive modulation of SIRT1 mRNA expression was able to prevent the hepatotoxicity induced by INH and rifampicin in mice12. Since NAT is the main metabolizing enzyme of INH, it is not surprising to suggest that NAT could be a target of SIRT1 causing, therefore, a decrease in NAT activity due to an increase of the deacetylation process generated by SIRT1 overexpression. To date it is unknown whether sirtuins are able to regulate NAT activity and the consequences of this regulation on immune cells. Therefore, this field needs to be explored.
Given the fact of NATs and SIRT1 and 6, are present in the same subcellular compartment, in this study we investigated the expression and the effects of sirtuins on the in situ enzymatic activity of NAT1 and NAT2 in peripheral blood mononuclear cells (PBMC). We found higher expression and activity of NAT2 compared to NAT1 in PBMC. The results from the current study provide the first evidence of possible modulation for NAT2, but not for NAT1, by the inhibition of sirtuin activity, which might have important implications for the cellular functions of NAT2.
2. Materials and methods
2.1. Population
A total of 17 healthy subjects, ranging between 20 and 32 years of age, were recruited for this study. Nine of them were males and eight females. For this group, biochemical parameters, such as glucose and triglycerides were measured. Subjects with infectious and/or autoimmune diseases and with antibiotic therapy, alcohol consumption, tobacco or illicit drugs were excluded. The Bioethics Committee of the Autonomous University of San Luis Potosi approved this work and all participants signed a written informed consent form.
2.2. Isolation of peripheral blood mononuclear cells
Blood samples were collected in 8 mL EDTA Vacutainer tubes (BD Biosciences, CA, USA) for expression and enzymatic activity analysis. Peripheral blood mononuclear cells (PBMC) were isolated by Ficoll gradient centrifugation. Blood was diluted with an equal volume of phosphate-buffered saline (PBS) pH 7.3, overlaid on layered Ficoll-Histopaque (Sigma, St. Louis, MO, USA) and centrifuged at 2500 rpm (500×g) for 20 min at 25 °C. The PBMC layer was removed and washed twice with PBS and resuspended in Dulbecco's modified Eagle's medium (DMEM) culture medium at 2×106 cells/mL. Media were supplemented with 10% fetal calf serum, 50 U/mL penicillin and 50 μg/mL streptomycin (Sigma). Cell viability was assessed by trypan blue exclusion assay.
2.3. Expression of intracellular proteins by flow cytometry
The percentage of cells expressing the intracellular proteins of interest was evaluated by staining them with different combinations of monoclonal antibodies. First, intracellular staining cells were treated with the commercial Fix/Perm Buffer Kit (eBioscience, CA, USA) and then incubated with rabbit anti-NAT1 or mouse anti-NAT2 antibodies (Abcam, Cambridge, UK) for 1.5 h at 4 °C. Then, the cells were incubated with anti-rabbit APC or anti-mouse FITC secondary antibodies (eBioscience) respectively for 20 min at 4 °C in the dark. For detection of intracellular sirtuin protein levels in human lymphocytes, the cells were also permeabilized and incubated with rabbit anti-SIRT1 (Abcam) or mouse monoclonal anti-SIRT6 (Abcam) antibodies for 20 min at 4 °C, followed by incubation with anti-rabbit APC or mouse FITC secondary antibodies (eBioscience), respectively, for 20 min at 4 °C in the dark. Then, cells were fixed with 1% paraformaldehyde, and the percentage of double-positive cells was obtained in a FACSCanto II Cytometer and analyzed using FACSDiva software (Becton Dickinson, San Jose, CA, USA). The results were expressed as the percentage of positive cells.
2.4. Expression of intracellular proteins by Western blot
For protein quantification cells were washed with PBS, resuspended in lysis buffer (HEPES 100 mmol/L, 1 mol/L NaCl, 200 mmol/L MgCl2, 100 mmol/L EDTA and Triton X-100, pH 7.4) and lysed by sonication at 50% amplitude. Then, the cells were centrifuged at 12,000 rpm (2400 × g) at 4 °C for 30 min. The supernatant was used to determine protein concentrations by the Bradford method (BCA Protein Assay, Thermo Scientific; Rockford, IL, USA). For immunoblots, 50 μg of total protein of HeLa, 3T3 and PBMC samples were electrophoresed through 4%–15% SDS-polyacrylamide gel, transferred to 0.45 μm nitrocellulose membranes (Millipore, Billerica, MA, USA) on a semi-dry electro transferring unit (Trans-Blot Turbo Transfer System, Bio-Rad Laboratories) and immunoblotted using a rabbit polyclonal anti-NAT1 antibody (1: 1000), mouse polyclonal anti-NAT2 antibody (1: 500), rabbit anti-SIRT1 antibody (1:500; Abcam) or mouse monoclonal anti-SIRT6 antibody (1: 3000), and then incubated with HRP-conjugated secondary antibody for 1 h. Finally, anti-β-actin antibody (R&D) was used as a loading and transfer control. The membranes were revealed by chemiluminescence. The intensities of the bands were detected using the ChemiDoc™ XRS+ System (Bio-Rad).
2.5. In situ arylamine N-acetyltransferase assay
PBMC (2×105 cells/mL) were cultured in DMEM medium supplemented with 10% fetal calf serum, 50 U/mL penicillin and 50 μg/mL streptomycin (Sigma), and maintained at 37 °C in a humidified atmosphere of 5% CO2. Cells were treated with the sirtuin antagonist nicotinamide: 0, 30, 100, 5 and 20 mmol/L (Sigma–Aldrich) and with the sirtuin agonist resveratrol: 0, 10, 50 and 100 µmol/L (Sigma–Aldrich), for 3 h at 37 °C. After this time, the medium was replaced by fresh supplemented medium containing the specific substrate: 10, 30 and 100 μmol/L PABA or INH (Sigma–Aldrich) for each enzyme. The cells were incubated for 6, 12, 24 or 48 h. HeLa and CHO cultures were used as positive and negative controls for NAT enzymatic activity.
2.6. In situ NAT activity determination
NAT1 and NAT2 activity was determined by HPLC on supernatants of PBMC, CHO (data not shown) and HeLa cell cultures, by quantified concentrations of the substrates and metabolites of each enzyme: PABA and acetyl-PABA (Ac-PABA) for NAT1, INH and acetyl-INH (Ac-INH) for NAT2. For the extraction of PABA/Ac-PABA and INH/Ac-INH, 100 µL of supernatant was deproteinized with 30 µL of 13% acetonitrile in H2O with 0.5% acetic acid for NAT1, and 30 µL of 10% trichloroacetic acid (Sigma–Aldrich) for NAT2. Then, it was centrifuged at 11,000 rpm (2200 × g) for 10 min at 4 °C, and 20 µL of supernatant was injected into Waters HPLC chromatographic equipment, which consists of a 1525 binary pump system linked to a 717 Plus autoinjector with a 20 µL injection loop, a UV/VIS 2487 detector and Breeze software v3.2 (Waters Corporation, MA, USA). The standards used for PABA/Ac-PABA and INH/Ac-INH were of USP grade, and chromatographic solvents were HPLC grade. For the analytical separation, we used a 150 mm × 30 mm Waters X-terra RP18 column of 3.5 µm particle size, and a 30 mm × 10 mm Waters X-terra RP18 guard column of 3.5 µm particle size (Waters Corporation, MA, USA). The mobile phase for quantification of INH/Ac-INH consisted of a mixture of 2.5 mmol/L phosphate buffer and 30 mmol/L sodium heptanesulfonate with acetonitrile at an 80: 20 (v/v) ratio at pH 2.7 and a flow rate of 0.4 mL/min. For PABA/Ac-PABA, the mobile phase consisted of 50 mmol/L acetic acid and acetonitrile in a 90: 10 (v/v) ratio. The detection of each compound and its metabolite was performed at a wavelength of 270 nm for NAT1 and 266 nm for NAT2 by integrating the chromatographic peaks with Breeze software (v3.2). The method described was analytically validated according to NOM-177-SSA1-2013. The concentration of each analyte in the sample was calculated using respective calibration curves in a 0 to 30 ng/µL range. NATs activity was normalized to lysate protein concentration and under these conditions the rate of PABA acetylation was linear with respect to time and protein concentration. All enzyme reactions were performed in duplicate, in conditions in which the initial rates were linear. Enzyme activities are shown as nmol metabolite/mg protein/min13.
2.7. Statistical analysis
Statistical analysis was performed using GraphPad Prism v5.01 (GraphPad Software, Inc.). The results are expressed as arithmetic mean±the standard error of the mean (SEM) of the percentage of positive cells, and enzymatic activity of NATs is expressed as nmol metabolite/mg protein/min. We used Student's t-test to identify the differences between the treatment and control, ANOVA was used to identify the differences between the groups, and Pearson's correlation analysis was used to identify correlations between percentage sets of positive cells and activity data. Values of P<0.05 were considered to be statistically significant.
3. Results
3.1. Sirtuins 1 and 6 are expressed in lymphocytes
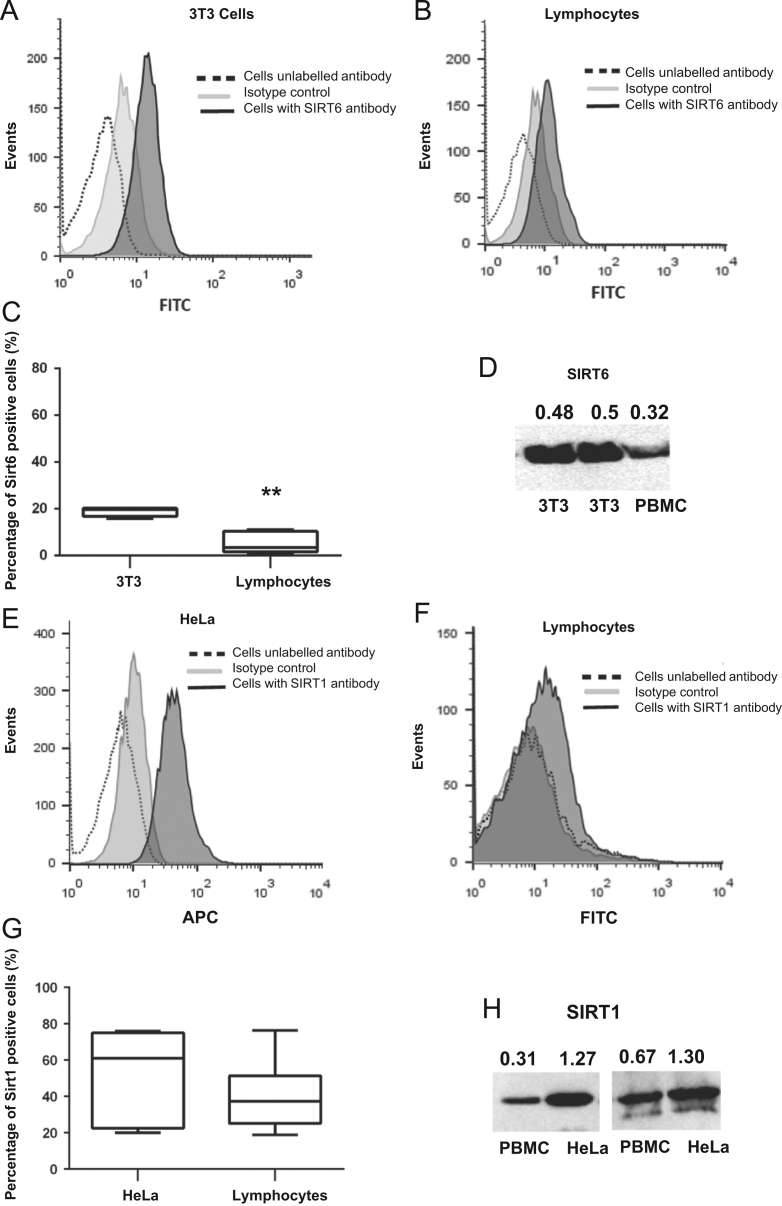
In order to determine whether SIRT6 and SIRT1 were expressed in lymphocytes, we examined sirtuin protein expression by flow cytometry (Fig. 1B and F) and Western blot (Fig. 1D and H). Fig. 1 shows representative histograms from the positive controls of SIRT6 and SIRT1 expression using 3T3 (Fig. 1A) or HeLa cells (Fig. 1E), respectively. There was a diminished level of SIRT6 positive cells in the lymphocyte gate in comparison to the positive control (Fig. 1C, P<0.001). These data were confirmed by Western blot (Fig. 1D). In contrast, due to high variability in lymphocytes and HeLa cells by flow cytometry (Fig. 1G) or Western blot (Fig. 1H), no significant changes were observed in SIRT1 expression (Fig. 1G).
Figure 1.
SIRT1 and SIRT6 expression in lymphocytes. (A) Representative histogram of 3T3 cells (positive control) or (B) Lymphocytes from healthy subjects stained with anti-SIRT6 (black) and secondary antibody FITC anti-mouse as an isotype control (grey). (C) Comparison of SIRT6 expression in 3T3 and lymphocytes. (D) Western blot of representative SIRT6 protein expression level in peripheral blood mononuclear cells (PBMC) and positive controls. (E) Representative histogram of HeLa cells (positive control) or (F) Lymphocytes from healthy subjects stained with either rabbit anti-SIRT1 (black), goat anti-rabbit APC secondary antibody as isotype control (grey) or cells in the absence of antibody (dotted line). (G) Comparison of SIRT1 expression in HeLa and lymphocytes. (H) Western blot of representative SIRT1 protein expression level in PBMC and positive controls.
3.2. Sirtuins are present in NAT-positive lymphocytes cells
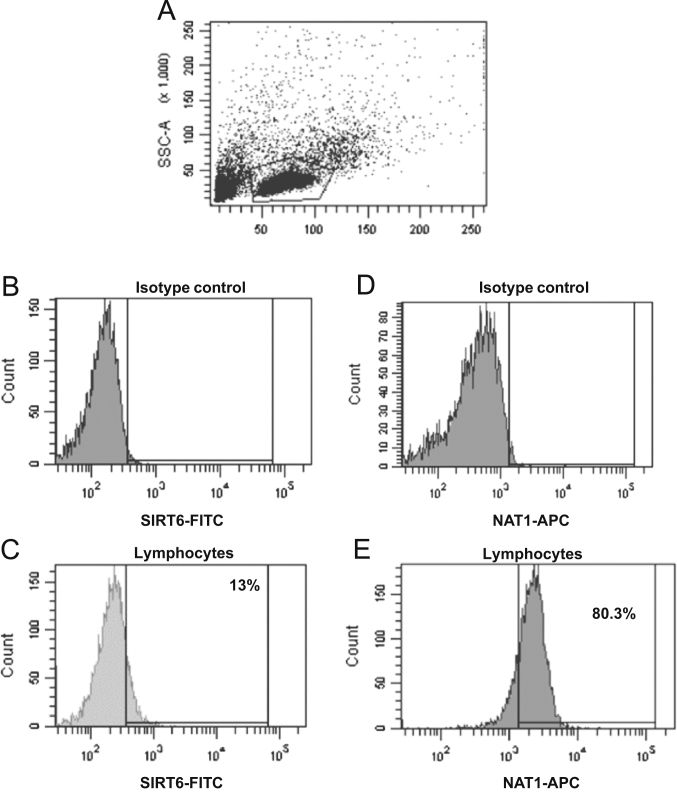
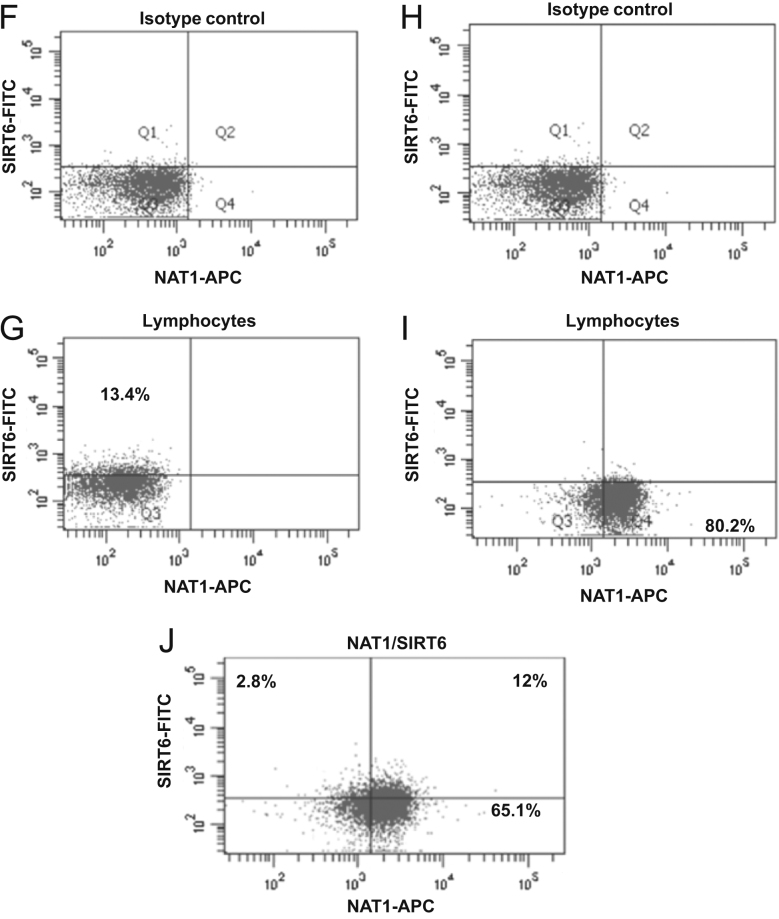
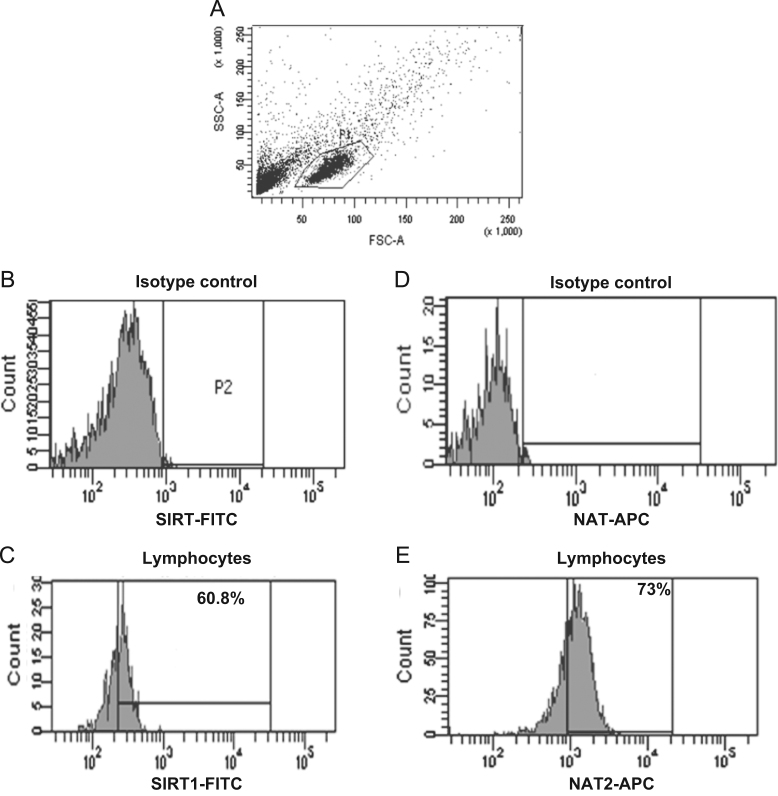
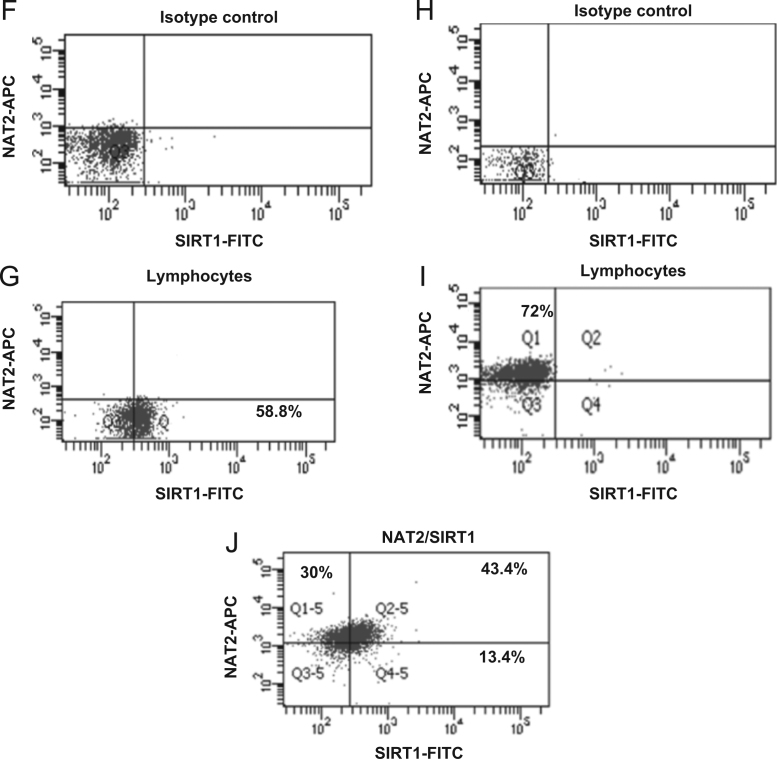
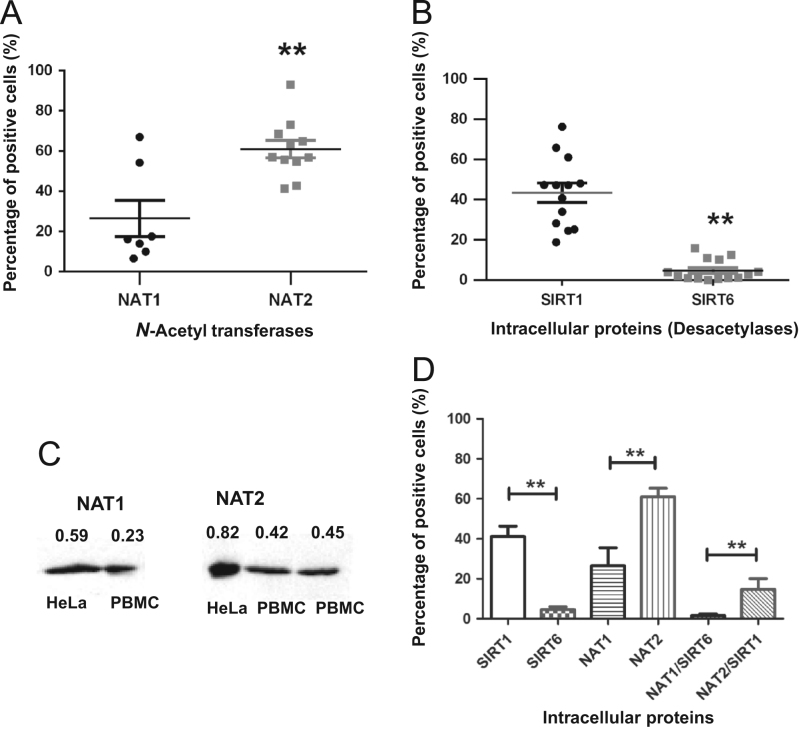
An indirect immunofluorescence staining assay was performed to identify the expression of intracellular NAT proteins in lymphocytes (Figure 2, Figure 3). The conditions used were previously described14 and confirmed by Western blot. These later assays included several experimental conditions to demonstrate the specificity of the anti-NAT1 and anti-NAT2 antibody using CHO and HeLa cells as negative and positive controls (Data not shown). Figs. 2A and 3A show the dot plots corresponding to the size (FCS) and granularity (SSC) of the lymphocyte population and isotype controls used (Fig. 2B, D and 3B, D). The histograms and dot-plot for the percentage of positive cells for NAT1 (Fig. 2E and I) and NAT2 (Fig. 3E and I) or SIRT6 (Fig. 2C and G) and SIRT1 (Fig. 3C and G) are shown. Subsequently, to verify whether sirtuins are expressed simultaneously with NAT enzymes; an indirect double staining, blocked with 0.01% BSA between each primary antibody, was designed. The dot plots show the percentage of double-positive cells for NAT1+SIRT6+ (Fig. 2J) and NAT2+SIRT1+ (Fig. 3J). When we analysed whether any of the NAT enzymes was predominant in the lymphocytes, we found that NAT2 was significantly increased in lymphocytes in comparison with NAT1 (Fig. 4A, P<0.05). These data were confirmed by Western blot where higher expression of NAT2 was found than of NAT1 on lysate of PBMC (Fig. 4C). Similarly, SIRT1 showed higher levels in comparison with SIRT6 in lymphocytes (Fig. 4B, P<0.0001). Then, a higher level of positive cells for NAT2+SIRT1+ versus NAT1+SIRT6+ was detected in lymphocytes from healthy subjects (Fig. 4D).
Figure 2.
SIRT6+, NAT1+ and NAT1+SIRT6+ expression in lymphocytes. Peripheral blood mononuclear cells (PBMC) were stained as described in Section materials and Methods. (A) Dot plots of FCS and SSC where the lymphocyte gate was selected. (B) Histogram of isotype control coupled to FITC in the lymphocyte gate (P1). (C) Histogram of anti-SIRT6 (primary antibody) and FITC anti-mouse (secondary antibody) in the lymphocyte gate (P1). (D) Histogram of isotype control coupled to APC in the lymphocyte gate (P1). (E) Histogram of anti-NAT1 (primary antibody) and APC anti-rabbit (secondary antibody) in the lymphocyte gate. (F) Dot plot of isotype control-FITC in the lymphocyte gate. (G) Dot plot of anti-SIRT6 (primary antibody) and FITC anti-mouse (secondary antibody) in the lymphocyte gate. (H) Dot plot of isotype control-APC in the lymphocyte gate. (I) Dot plot of anti-NAT1 (primary antibody) and APC anti-rabbit (secondary antibody) in the lymphocyte gate. (J) Representative dot plot of double staining with anti-SIRT6-FITC and anti-NAT1-APC in the lymphocyte gate.
Figure 3.
SIRT1+, NAT2+ and NAT2+SIRT1+ expression in lymphocytes. Peripheral blood mononuclear cells (PBMC) were stained as described in Section materials and methods. (A) Dot plots of FCS and SSC where the lymphocyte gate was selected. (B) Histogram of isotype control coupled to FITC in the lymphocyte gate (P1). (C) Histogram of anti-SIRT1 (primary antibody) and FITC anti-mouse (secondary antibody) in the lymphocyte gate (P1). D) Histogram of isotype control coupled to APC in the lymphocyte gate (P1). E) Histogram of anti-NAT2 (primary antibody) and APC anti-mouse (secondary antibody) in the lymphocyte gate. F) Dot plot of isotype control-FITC in the lymphocyte gate. G) Dot plot of anti-SIRT1 (primary antibody) and FITC anti-mouse (secondary antibody) in the lymphocyte gate. H) Dot plot of isotype control-APC in the lymphocyte gate. I) Dot plot of anti-NAT2 (primary antibody) and APC anti-mouse (secondary antibody) in the lymphocyte gate. J) Representative dot plot of double staining with anti-SIRT1-FITC and anti-NAT2-APC in the lymphocyte gate.
Figure 4.
Double-positive cells for intracellular N-acetyl transferase and sirtuin proteins. (A) Comparison of NAT1 and NAT2 expression in lymphocytes. (B) Comparison of SIRT1 and SIRT6 expression in lymphocytes. (C) Western blot of representative NAT1 and NAT2 protein expression levels in PBMC and positive controls. (D) Percentage of protein expression of intracellular arylamine N-acetyl transferases and sirtuins in lymphocytes (**P>0.01).
3.3. In situ NAT activity in cultures of mononuclear cells
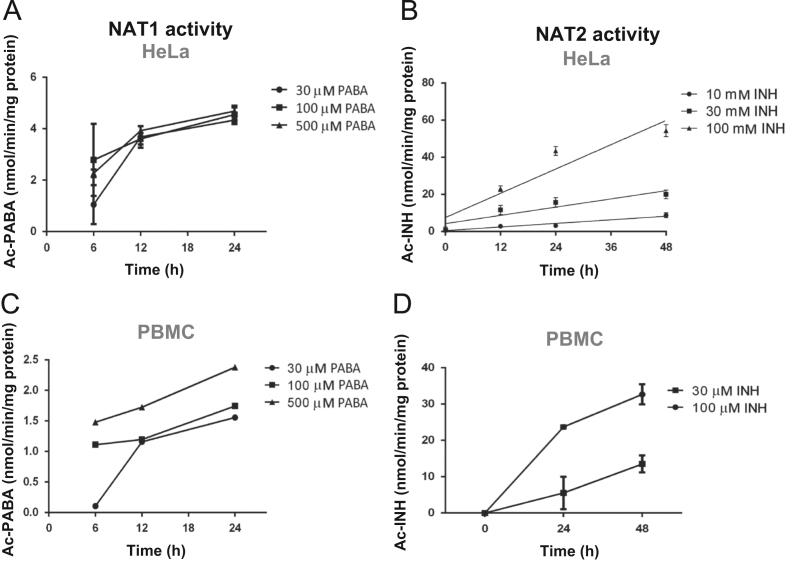
To evaluate the enzymatic activity of both NAT proteins, it was necessary to design and validate the method. Using the HPLC technique, we quantified the metabolites PABA and Ac-PABA for NAT1, and INH and Ac-INH for NAT2 in cells that constitutively expressed such proteins in mononuclear cells. Cultures of human PBMC and HeLa cells were conducted to investigate the effect of PABA and INH on NAT1 and NAT2 in situ enzymatic activity, respectively as described by Hein et al.15 The cells were cultured in DMEM for up to 24 h in the absence or presence of PABA, and for up to 48 h in the absence or presence of INH; then, the enzymatic activity was assessed. The NAT1 and NAT2 activity (measured as the N-acetylation of PABA or INH) was determined at concentrations ranging between 30 and 500 µmol/L of PABA (Fig. 5A and C), and concentrations between 10 and 100 µmol/L of INH for HeLa cells (positive control) and PBMC (Fig. 5B and D). Cultured cells showed no loss of NAT1 or NAT2 activity at the culture time tested.
Figure 5.
Dose-response and kinetic curves for the detection of acetyl-p-aminobenzoic acid (Ac-PABA) and acetyl-isoniazid (Ac-INH) in peripheral blood mononuclear cells (PBMC) and HeLa cells. HeLa cells (A), (B) and PBMC (C), (D) were incubated in the presence of different concentrations of PABA (A), (C) or INH (B), (D). Both metabolites were quantified by HPLC as described in Section materials and methods. Each point represents the mean of duplicate assays performed in four cultures. M, mol/L.
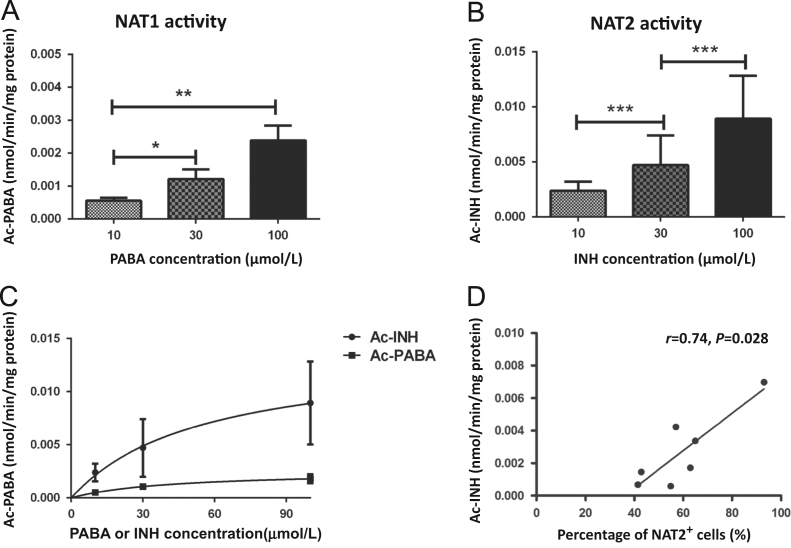
Different PABA and INH concentrations were used to evaluate the NAT1 and NAT2 activity in PBMC after 24 h of incubation (Fig. 6A and B). We observed a dose-dependent activity due to significantly increased levels of the metabolite of NAT1 (nmol Ac-PABA/min/mg protein) and NAT2 (nmol Ac-INH/min/mg protein) with 10, 30 and 100 µmol/L INH after 24 h of incubation. However, the enzymatic activity observed in NAT1 culture did not show the same proportion as with the NAT2 culture, i.e. although both were dependent on the concentration of the substrate (Fig. 6C). In addition, the NAT2 activity showed a positive correlation associated with NAT2 expression in lymphocytes from healthy volunteers (Fig. 6D).
Figure 6.
NAT1 and NAT2 enzymatic activity in mononuclear cell cultures. Peripheral blood mononuclear cells (PBMC) were cultured with 10, 30 and 100 µmol/L of PABA (A) or INH (B) for 24 h. (C) Comparison of enzymatic activity between NAT1 (■) and NAT2 (•). Levels of Ac-PABA and Ac-INH from each culture were evaluated by HPLC as described in Section materials and methods. Each point represents the mean of duplicate assays of cultured cells. (D) Correlation between the percentage of NAT2 positive cells and NAT2 activity measured as acetyl-isoniazid (Ac-INH). A Pearson correlation analysis was performed for each pair; the trend line, correlation coefficient (r), and significance (P) are shown for the plot (*P<0.05; **P<0.01; ***P<0.001).
3.4. NAT2 activity is increased by inhibition of sirtuins
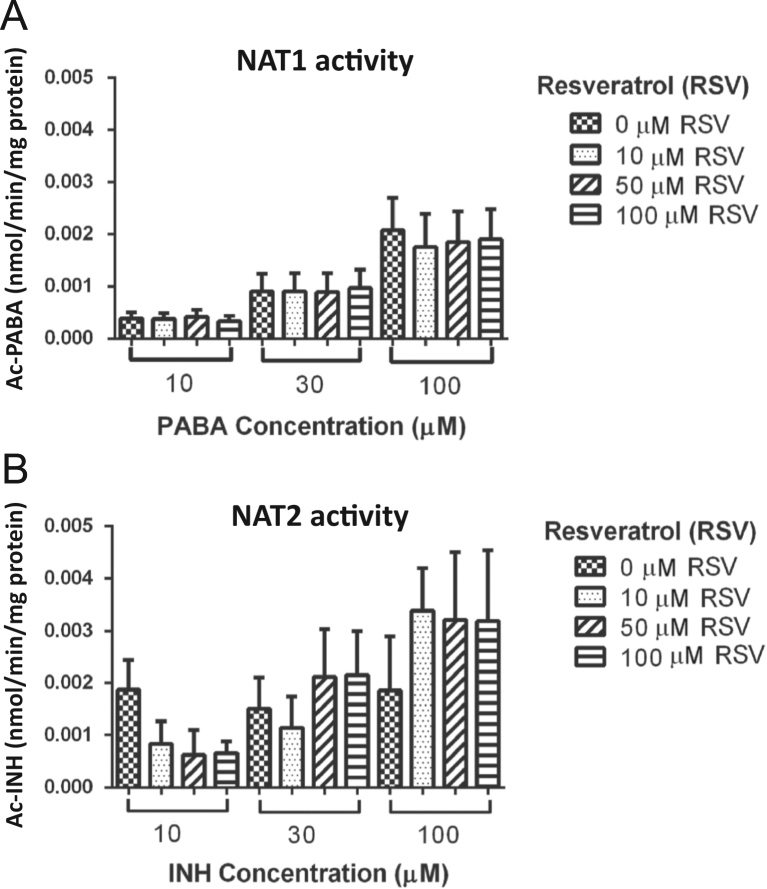
The possible effect of sirtuins on NAT activity in PBMC was examined as nmol Ac-PABA/min/mg protein for NAT1 and nmol Ac-INH/min/mg protein for NAT2 in the presence and absence of an agonist (resveratrol, RSV) and an inhibitor (nicotinamide, NAM) of sirtuins. To determine the exposure time, the cells were cultured with and without RSV or NAM at 37 °C for 3, 6 and 12 h. Cell viability was measured by MTT assay (Sigma–Aldrich). An increase in the time of incubation longer to than 6 h for NAM and 3 h for RSV caused the death of 25% of the cells (data not shown). Therefore, the exposure time used in the treatments was 3 h. PBMC were cultivated in DMEM in the presence and absence of RSV (0, 10, 50 and 100 µmol/L) for 3 h; subsequently, the medium was replaced with fresh medium containing different concentrations of PABA and INH (10, 30 and 100 µmol/L) and the activity was evaluated. In the latter conditions we found that the activity of NAT1 (Fig. 7A) and NAT2 (Fig. 7B) in cell culture did not change when cells were stimulated with different concentrations of RSV, compared to cells in culture medium.
Figure 7.
Effect of an agonist of sirtuins on NAT1 and NAT2 activity. Peripheral blood mononuclear cells (PBMC) were cultured with different concentrations of a sirtuin agonist (resveratrol, RSV: 0, 10, 50 and 100 µmol/L) for 3 h and, subsequently, with p-aminobenzoic acid (PABA) or isoniazid (INH) were added. NAT1 (A) and NAT2 (B) activity expressed in nmol of metabolite/min/mg of protein was determined by HPLC. PABA or INH concentrations = 10, 30 and 100 µmol/L. M, mol/L.
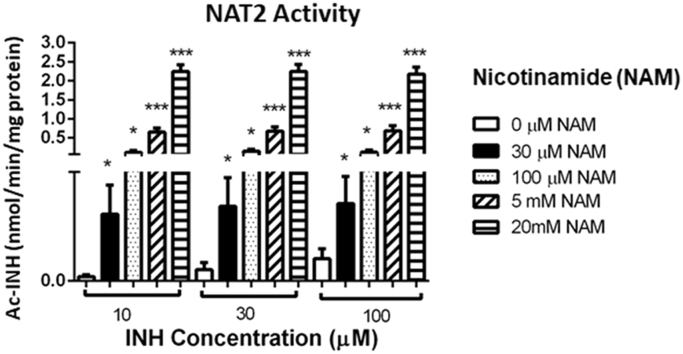
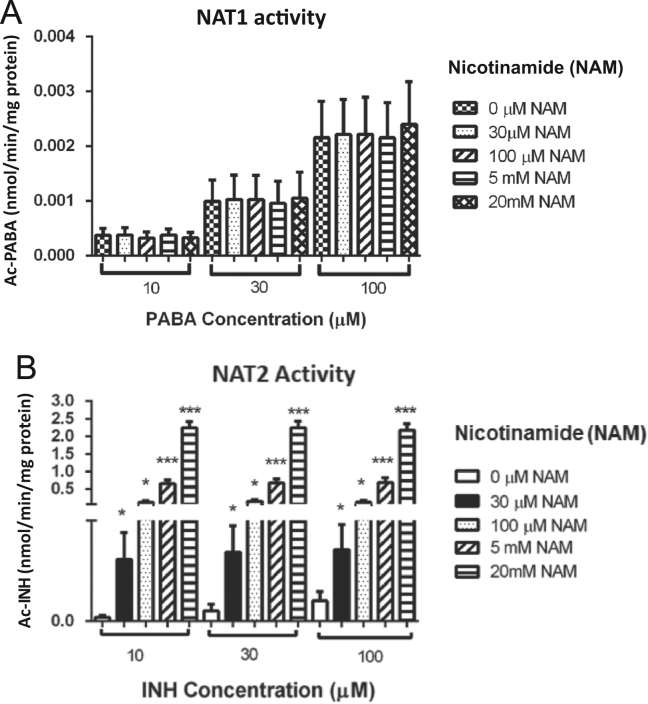
Since sirtuins have a deacetylase capacity and, in consequence, can alter the metabolizing activity of NATs, it was analyzed whether sirtuin inhibition by increased concentrations of NAM metabolite could have an effect on NAT1 or NAT2 activity, measuring Ac-PABA and Ac-INH, respectively. Mononuclear cell cultures with 10, 30 and 100 µmol/L of INH indicated that NAT2 metabolizing capacity increased significantly (P<0.05) when sirtuins were inhibited with NAM compared with those not treated for 24 h (Fig. 8B). In contrast, we did not observe any effect on NAT1 activity when sirtuins were inhibited with different concentrations of NAM for 24 h (Fig. 8A).
Figure 8.
Effect of an inhibitor of sirtuins on NAT1 and NAT2 activity. Peripheral blood mononuclear cells (C) from healthy subjects were cultured with different concentrations of a sirtuin inhibitor (nicotinamide, NAM: 0, 30, 100 µmol/L, 5 and 20 mmol/L) for 3 h and, subsequently, the substrate of each enzyme with p-aminobenzoic acid (PABA) or isoniazid (INH) were added. NAT1 (A) and NAT2 (B) activity expressed in nmol of metabolite/min/mg of protein was determined by HPLC. PABA or INH concentrations = 10, 30 and 100 µmol/L. *P<0.05; ***P<0.001. M, mol/L.
4. Discussion
In order to evaluate the possible role of arylamine N-acetyltransferases NAT1 and NAT2 on immune cells, the expression and activity of these enzymes in PBMC from healthy subjects were studied and evaluated. We found a higher NAT2 expression in lymphocytes, confirming the results previously reported by our research group14. However, in this study we observed a higher variability in the percentage of NAT2 positive cells between the studied subjects. This high variability in the level of NAT2 in these cells could be attributed to polymorphisms affecting their expression. NATs polymorphisms (rapid, intermediate and slow phenotypes) which explain the variability in NAT2 expression are under investigation (unpublished results). On the other hand, NAT1 is a protein that in a conventional form has been determined by Western blot in different tissues and cellular types5., 16., 17.. Today, studies about the presence of NAT1 in mononuclear cells are reported as the relative expression of protein in individuals with different genotypes3. Although NAT1 appears to be present in almost all examined tissues, it was important to evaluate the percentage of lymphocytes expressing this protein, as reported with NAT2 by flow cytometry. Interestingly, our results show NAT1 expression of approximately 20% and it is noticeable that, when we compared the expression of these proteins, NAT2 levels were higher than NAT1 in lymphocytes. In this regard, the worldwide consensus indicates that NAT2 presents a lower distribution than NAT1 in the organism. However, our data indicate in the case of these type of blood cells, the percentage of NAT2 positive cells is higher than NAT1+ cells in our study group. In addition, it is important to consider the influence of certain NAT1 and NAT2 polymorphism in the expression of these enzymes. Therefore, it would be important to determine in subsequent studies whether these expression levels of NAT1 and NAT2 are similar in PBMC or whether are altered in some pathological conditions. These assays are undergoing in our group of work.
NAT1 and NAT2 activity was quantified through the measurement of their metabolite levels by HPLC as previously described13., 18.. Our results showed that NAT activity was dependent on substrate concentration for both enzymes (NAT1 and NAT2) in a range from 10 to 100 µmol/L; in addition, for the first time, we found higher concentrations of Ac-INH in comparison to Ac-PABA, indicating that NAT2 would has a greater metabolizing function than NAT1 in PBMC. Given that the results were measured on supernatants of PBMC cultures and they were not from the supernatant of lysate of cells, these low levels of product formation were similar to others previously reported19. NAT1 and NAT2 acetylate their substrates by ‘ping pong bi-bi’ reactions and in the NATs acetylated state, they behave as stable proteins; conversely, their deacetylation generates fast polyubiquitination and subsequent protein degradation3. In this regard, reversible acetylation is a regulation mechanism for the enzyme activity of several proteins. HDAC enzymes, like sirtuins, are responsible of carrying out this mechanism, through the removal of the acetyl group and, therefore, they generate unstable proteins9.
Specifically, SIRT1 and SIRT6 are cytoplasmic enzymes with a deacetylase function that are found in the same environment together with NATs and could have an effect on these proteins; since it has been reported that sirtuins target different substrates such as proteins involved in metabolism. Therefore, in this study, we analyzed the expression profile of the cytoplasmic sirtuins 1 and 6 in lymphocytes, since to date there are no reports of its detection by flow cytometry. Our results show for the first time that SIRT6 is expressed at levels below 15% in lymphocytes, while for SIRT1 there is a greater variability in its expression levels, which are above 42%. This means that the percentage of SIRT1-positive cells is higher than the percentage of SIRT6-positive cells. It could be interesting to evaluate the presence of these proteins in subpopulations of lymphocytes and explore their function in relevant processes such as proliferation or cytokine production.
Acetylation is a crucial step for NAT proteins in order to induce their function and/or activity on xenobiotic compounds and drugs. Since we observed that there is a co-expression of NAT2 and SIRT1, and NAT1 and SIRT6 in lymphocytes, we prompted us to explore the regulation mechanism that sirtuin exert on NATs. Sirtuins are deacetylases that target proteins such as FOXO1, PGAM1, p53, p73, RUNX3, SREB1a, SREBP2, E2F1 and ER8120., 21., 22. among others. The deacetylated-state of proteins carried out by sirtuins, generates the instability of their substrates and, in consequence, an alteration in their activity. All sirtuin enzymes are dependent on oxidized NAD+ and there are different conditions that can alter the levels of this cofactor, affecting the biosynthesis and activity of sirtuins23 such as age, weight or percentage of adipose tissue11., 22., 24.. It has been reported that by suppressing SIRT1 activity with NAM, there is an increase in the acetylation of PGC-1α; on the contrary, when concentrations of 50 μmol/L of RSV, a sirtuin agonist, were used, the acetylation of PGC-1α decreased25. In our work, we investigated the possibility that sirtuins are regulators of NAT1 and NAT2 through the deacetylation mechanism that affects the enzymatic activity of NATs. The concentrations of RSV and NAM for the activation and inhibition of sirtuins were used at around 100 µmol/L for RSV and 20 mmol/L for NAM given that numerous studies have shown that higher concentrations of resveratrol or NAM induce apoptosis in a variety of cells26. Our results showed no significant loss of NAT1 and NAT2 activity with any concentration of RSV evaluated. Therefore, the capacity of sirtuins to adjust NATs downward require more new studies to determine why RSV did not alter the NAT activity in the conditions of our study. Recently, a number of non-polyphenolic synthetic SIRT1-activating compounds (SRT series) have been described, which have no structural similarity to RSV and are 1000 times more potent than RSV. These compounds could be used for future applications to increase the capacity to monitor the status of sirtuin activity in a cellular environment27.
On the other hand, NAM, an inhibitor of sirtuins, modified NAT2 activity and generated an important increase, which was not observed in the NAT1 activity. This suggests that when sirtuins are inhibited, they prevent NAT2 deacetylation; causing, therefore, a shift in the equilibrium of the NAT2 proteins towards a stable acetylated state that is capable of performing a better and faster metabolizing function on its substrates. Regarding to this, we carried out an in silico analysis in order to search for acetylation sites within NAT2 protein. Using the PHOSIDA database (http://www.phosida.com), we were able to predict the next acetylated sites: K13, K100, K188, K243, K272; which can be target of sirtuins.
Although several studies have reported the activation and inhibition of sirtuins with agonists and inhibitors of sirtuins, such as RSV and NAM, the selectivity and sensitivity to individual isoforms within the human SIRT1-7 group is not equal26., 27.. To date, there are more reports describing the action that these compounds exert on SIRT1 than on SIRT6, consequently, the possible post-transcriptional regulation of NAT2 evaluated in this study could be related to SIRT1. Moreover, our investigation did not make it possible to identify the member of the sirtuin family responsible for the possible effect on NAT2. Our results are the first to be reported on this type of post-transcriptional regulation; however, further studies are necessary in order to determine whether these proteins are linked or which sirtuins are involved in this regulatory mechanism.
Our findings demonstrated that lymphocytes from peripheral venous blood express NAT1 and NAT2 proteins and sirtuins, with low concentration levels of SIRT6 and higher levels of SIRT1. We provide strong evidence that high levels of Ac-INH generated by inhibition with NAM could be an effect of the mechanism of regulation that sirtuins are exerting on NAT2. i.e. once deacetylation function of sirtuins is inhibited, they are not able to remove the acetyl groups from NAT2, making this protein more stable and, therefore, increasing its activity. We hypothesize that an interaction between sirtuins and NAT2, does exist which mediates NAT2 regulation. However, it is necessary to carry out further investigation in order to explore whether such interaction is direct and functional, and to determine whether the predicted lysine sites are target of sirtuins. Moreover, future studies are aimed at determining whether SIRT1 is directly involved in post-transcriptional regulation of NATs in different subpopulations of lymphocytes and their effect, by identifying this previously unrecognized function of SIRT1 in NAT2. Our study has further expanded the range of biological functions aimed at by this regulation so that this should provide a basis for future studies aimed at further delineation of disorders linked to SIRT1 dysfunction.
Acknowledgements
This study was supported by grants 255781 and 248950 (to Portales-Perez Diana Patricia) from CONACYT, Mexico. Turiján-Espinoza Eneida was the recipient of a scholarship (592537) from CONACYT, Mexico.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
Contributor Information
Eneida Turiján-Espinoza, Email: eneida.turijan.espinoza@gmail.com.
Diana Portales-Pérez, Email: dportale@uaslp.mx.
References
- 1.Sinclair J.C., Sandy J., Degoda R., Sim E., Noble M.E. Structure of arylamine N-acetyltransferase reveals a protease-like catalytic triad. Nat Struct Biol. 2000;7:560–564. doi: 10.1038/76783. [DOI] [PubMed] [Google Scholar]
- 2.Kubiak X., Dairou J., Dupret J.M., Rodrigues-Lima F. Crystal structure of arylamine N-acetyltransferases: insights into the mechanisms of action and substrate selectivity. Drug Metab Toxicol. 2013;9:349–362. doi: 10.1517/17425255.2013.742505. [DOI] [PubMed] [Google Scholar]
- 3.Butcher N.J., Arulpragasam M.R., Minchin R.F. Proteasomal degradation of N-acetyltransferase 1 is prevented by acetylation of the active site cysteine: a mechanism for the slow acetylator phenotype and substrate-dependent down-regulation. J Biol Chem. 2004;279:22131–22137. doi: 10.1074/jbc.M312858200. [DOI] [PubMed] [Google Scholar]
- 4.Zhou X.T., Ma Z.G., Dong D., Wu B.J. Arylamine N-acetyltransferases: a structural perspective. Br J Pharmacol. 2013;169:748–760. doi: 10.1111/bph.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sim E., Walters K., Boukouvala S. Arylamine N-acetyltransferases: from structure to function. Drug Metab Rev. 2008;40:479–510. doi: 10.1080/03602530802186603. [DOI] [PubMed] [Google Scholar]
- 6.Butcher N.J., Tiang J., Minchin R.F. Regulation of arylamine N-acetyltransferases. Curr Drug Metab. 2008;9:498–504. doi: 10.2174/138920008784892128. [DOI] [PubMed] [Google Scholar]
- 7.Caron C., Boyault C., Khochbin S. Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. BioEssays. 2005;27:408–415. doi: 10.1002/bies.20210. [DOI] [PubMed] [Google Scholar]
- 8.Yuan H., Rossetto D., Mellert H., Dang W.W., Srinivasan M.J., Hodawadekar S. MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 2011;31:58–70. doi: 10.1038/emboj.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Houtkooper R.H., Pirinen E., Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saunders L.R., Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489–5504. doi: 10.1038/sj.onc.1210616. [DOI] [PubMed] [Google Scholar]
- 11.Moschen A.R., Wieser V., Gerner R.R., Bichler A., Enrich B., Moser P. Adipose tissue and liver expression of SIRT1, 3, and 6 increase after extensive weight loss in morbid obesity. J Hepatol. 2013;59:1315–1322. doi: 10.1016/j.jhep.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 12.Nicoletti N.F., Rodrigues-Junior V., Santos Jr, AA, Leite C.E., Dias A.C.O., Batista Jr., EL Protective effects of resveratrol on hepatotoxicity induced by isoniazid and rifampicin via SIRT1 modulation. J Nat Prod. 2014;77:2190–2195. doi: 10.1021/np5003143. [DOI] [PubMed] [Google Scholar]
- 13.Queen M. Wiley; New York: 2002. Measuring the activity of arylamine N-acetyltransferase (NAT). Current protocols in toxicology. [DOI] [PubMed] [Google Scholar]
- 14.Salazar-González R.A., Gómez R., Romano-Moreno S., Medellín-Garibay S., Núñez-Ruíz A., Magaña-Aquino M. Expression of NAT2 in immune system cells and the relation of NAT2 gene polymorphisms in the anti-tuberculosis therapy in Mexican mestizo population. Mol Biol Rep. 2014;41:7833–7843. doi: 10.1007/s11033-014-3677-5. [DOI] [PubMed] [Google Scholar]
- 15.Doll M.A., Hein D.W. Genetic heterogeneity among slow acetylator N-acetyltransferase 2 phenotypes in cryopreserved human hepatocytes. Arch Toxicol. 2017;91:2655–2661. doi: 10.1007/s00204-017-1988-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paterson S., Sin K.L., Tiang J.M., Minchin R.F., Butcher N.J. Histone deacetylase inhibitors increase human arylamine N-acetyltransferase-1 expression in human tumor cells. Drug Metab Dispos. 2010;39:77–82. doi: 10.1124/dmd.110.036202. [DOI] [PubMed] [Google Scholar]
- 17.Tiang J.M., Butcher N.J., Minchin R.F. Effects of human arylamine N-acetyltransferase I knockdown in triple-negative breast cancer cell lines. Cancer Med. 2015;4:565–574. doi: 10.1002/cam4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung J.G., Wang H.H., Tsou M.F., Hsieh S.E., Lo H.H., Yen Y.S. Evidence for arylamine N-acetyltransferase activity in the bacterium. Helicobacter pylori. Toxicol Lett. 1997;91:63–71. doi: 10.1016/s0378-4274(97)03870-8. [DOI] [PubMed] [Google Scholar]
- 19.Doll M.A., Salazar-gonzález R.A., Bodduluri S., Hein D.W. Arylamine N-acetyltransferase 2 genotype-dependent N-acetylation of isoniazid in cryopreserved human hepatocytes. Acta Pharm Sin B. 2017;7:517–522. doi: 10.1016/j.apsb.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang C.G., Chen L.H., Hou X.H., Li Z.Y., Kabra N., Ma Y.H. Interactions between E2F1 and SIRT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8:1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 21.Yamakuchi M., Lowenstein C.J. MiR-34, SIRT1, and p53: the feedback loop. Cell Cycle. 2009;8:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- 22.Hori Y.S., Kuno A., Hosoda R., Horio Y. Regulation of FOXOs and p53 by SIRT1 modulators under oxidative stress. PLoS One. 2013;8:e73875. doi: 10.1371/journal.pone.0073875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong Y., Guan K.L. Mechanistic insights into the regulation of metabolic enzymes by acetylation. J Cell Biol. 2012;198:155–164. doi: 10.1083/jcb.201202056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Kreutzenberg S.V., Ceolotto G., Papparella I., Bortoluzzi A., Semplicini A., Man C.D. Downregulation of the longevity-associated protein sirtuin 1 in insulin resistance and metabolic syndrome: potential biochemical mechanisms. Diabetes. 2010;59:1006–1015. doi: 10.2337/db09-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higashida K., Kim S.H., Jung S.R., Asaka M., Holloszy J.O., Han D.H. Effects of resveratrol and SIRT1 on PGC-1α activity and mitochondrial biogenesis: a reevaluation. PLoS Biol. 2013;11:e1001603. doi: 10.1371/journal.pbio.1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villalba J.M., Alcaín F.J. Sirtuin activators and inhibitors. Biofactors. 2012;38:349–359. doi: 10.1002/biof.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haigis M.C., Sinclair D.A. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]