Abstract
We sequenced and characterized PMP22 (22-kD peroxisomal membrane protein) from Arabidopsis, which shares 28% to 30% amino acid identity and 55% to 57% similarity to two related mammalian peroxisomal membrane proteins, PMP22 and Mpv17. Subcellular fractionation studies confirmed that the Arabidopsis PMP22 is a genuine peroxisomal membrane protein. Biochemical analyses established that the Arabidopsis PMP22 is an integral membrane protein that is completely embedded in the lipid bilayer. In vitro import assays demonstrated that the protein is inserted into the membrane posttranslationally in the absence of ATP, but that ATP stimulates the assembly into the native state. Arabidopsis PMP22 is expressed in all organs of the mature plant and in tissue-cultured cells. Expression of PMP22 is not associated with a specific peroxisome type, as it is detected in seeds and throughout postgerminative growth as cotyledon peroxisomes undergo conversion from glyoxysomes to leaf-type peroxisomes. Although PMP22 shows increased accumulation during the growth of young seedlings, its expression is not stimulated by light.
The peroxisome is a single membrane-bound organelle present in almost all eukaryotic cells (Lazarow and Fujiki, 1985). Peroxisomes possess different metabolic functions depending not only on the type of tissue in which they are found, but also on the metabolic and developmental state of the organism (Tolbert, 1981). A common function of all peroxisomes is their ability to degrade hydrogen peroxide, thereby allowing the co-compartmentalization of metabolic pathways that produce this toxic by-product. In plants specialized peroxisomes (glyoxysomes) in cotyledons or endosperm tissues perform β-oxidation and undergo the glyoxylate cycle to convert carbon from stored fatty acids to carbohydrate during germination. In photosynthetic tissues peroxisomes are involved in the salvage of glycolate produced by photorespiration (Canvin and Salon, 1997). Such functional diversity in a single compartment is possible because the composition of peroxisomes can be modified through the uptake and assembly of the required proteins and enzymes.
The peroxisome membrane forms the interface between the organelle and the rest of the cell (Mullen and Trelease, 1996). It mediates not only the transport of metabolites, but also the import of proteins that maintain or modify the identity and function of the organelle. It also almost certainly plays a role in organelle movement and division. The peroxisomal membrane (especially that of plants) is one of the least characterized, mainly because of the difficulties in isolating pure membranes that are free from contamination by other cellular membranes. The use of yeast mutants that are defective in peroxisomal function circumvents this difficulty and has resulted in the identification of PMPs such as Pex2p (Per6) (Waterham et al., 1996), Pex3p (Pas3p) (Höhfeld et al., 1991), Pex9p (Pay2p) (Eitzen et al., 1995), Pex10p (Per8p) (Tan et al., 1995), Pex13p (Elgersma et al., 1996; Erdman and Blöbel, 1996; Gould et al., 1996), and Pex15p (Elgersma et al., 1997), all of which are required for peroxisome biogenesis, although their precise functions remain unknown.
Other proteins identified by reverse genetics in yeasts include Pex11p (Pmp27) (Erdman and Blöbel, 1995; Marshall et al., 1995), which is implicated in the control of peroxisome size; Pmp47 (McCammon et al., 1994), a putative solute transporter; and Pat1p and Pat2p (Hettema et al., 1996), which are involved in fatty acid transport into peroxisomes. Studies addressing the molecular basis of human genetic disorders that result in peroxisomal dysfunction have also yielded the identification of human PMPs such as PAF-1 (Pex2p) (Tsukamoto et al., 1991) and the adrenoleukodystrophy gene product (Mosser et al., 1993; Lombard-Platet et al., 1996). In other mammals the related PMPs, PMP22 and Mpv17, were identified by biochemical (Fujiki et al., 1982; Kaldi et al., 1993) and genetic (Weiher et al., 1990) studies. Mpv17 was implicated in the production of reactive oxygen species (Zwacka et al., 1994).
Plant homologs of these proteins have not yet been identified. In the absence of a straightforward genetic approach, most work done to date to has tried to characterize plant PMPs by searching for activities that copurify with peroxisomal membranes. Some have also attempted to purify and compare peroxisomal membranes from different plant tissues to analyze their polypeptide compositions (Corpas et al., 1995). Among the activities present in plant peroxisome membranes are those of alkaline lipase (Maeshima and Beevers, 1985), NADH:Cyt c reductase, NADH:ferricyanide reductase (Hicks and Donaldson, 1982; Fang et al., 1987; Luster and Donaldson, 1987; Struglics et al., 1993), NADH:ascorbate free-radical reductase (Bowditch and Donaldson, 1990), Mn-superoxide dismutase (Sandalio and del Rio, 1988), and an 18-kD Cyt b5 (López-Huertas et al., 1997).
Most of these activities are postulated to be involved in redox reactions associated with the reoxidation of NADH, or in defense against potentially harmful reactive oxygen species such as hydrogen peroxide and superoxide radicals produced in the matrix and membranes. In peroxisomes from pea leaves, there are two sites of superoxide generation, one in the organelle matrix, in which the generating system was identified as xanthine oxidase, and another in the peroxisomal membrane, which is dependent on NADH (del Río et al., 1992). Recently, the integral PMPs of pea leaf peroxisomes were identified using SDS-PAGE by López-Huertas et al. (1995); three of these membrane polypeptides with molecular masses of 18, 29, and 32 kD have also been characterized and demonstrated to be responsible for superoxide radical generation (López-Huertas et al., 1996, 1997). However, in plants we lack genetic information on oxygen-radical-producing PMPs. With the exception of a 31-kD membrane-associated peroxisomal ascorbate peroxidase (Yamaguchi et al., 1995; Bunkelmann and Trelease, 1996), the genes encoding these enzymic activities have not yet been identified.
Advances in the Arabidopsis Genome Project are providing a vital link between genetic and biochemical approaches in plants. To further our understanding of how plant peroxisomes are assembled, we sought to identify and characterize a PMP. We were able to use the data from the Arabidopsis sequencing project to identify a 22-kD integral membrane protein that is related to the mammalian PMP22/Mpv17 family of PMPs. In this paper we describe the characterization of this 22-kD Arabidopsis PMP.
MATERIALS AND METHODS
Materials
The Arabidopsis clone TAY089 (accession no. Z18516), derived from a cDNA library prepared from cycling cells of a cell-suspension culture of the Columbia ecotype, was obtained from the Arabidopsis Biological Research Center (Ohio State University, Columbus). We purchased all chemicals, unless otherwise specified, from either Sigma or BDH (Poole, Dorset, UK). DNA sequences were determined by automated sequencing using the DNA sequencer (model 373A-XL, Applied Biosystems) at the University of Cambridge Department of Biochemistry (Cambridge, UK).
Plasmid Constructs and Other Molecular Biological Procedures
All molecular biological procedures were performed as described by Sambrook et al. (1989) unless otherwise specified. The 0.85-kb cDNA insert of clone TAY089 flanked by EcoRI restriction enzyme sites was subcloned into the same site in pBluescript SK (Stratagene) (E10:SK). For in vitro transcription of the open reading frame, the insert was excised from E10:SK with SalI and XbaI and subcloned into pBluescript KS (Stratagene) to permit transcription from the T7 promoter. For the production of recombinant hexahistidine-PMP22, the PMP22 open reading frame was cloned into a pET16b expression vector (hexahistidine-PMP22:pET16b) to create an in-frame N-terminal hexahistidine tag. Genomic DNA from Arabidopsis was extracted from leaves using a DNA-extraction kit (PhytoPure, Scotlabs, Woburn, MA); and 10-μg aliquots at a concentration of 0.1 μg μL−1 were digested with the required restriction enzymes for 14 h. The restriction fragments were separated by agarose-gel electrophoresis and transferred to Hybond-N membranes (Amersham) by capillary blotting as described by Sambrook et al. (1989). The PMP22 open reading frame randomly labeled with [32P]α-dCTP 3000 Ci mmol−1 (Prime-It II, Stratagene) was used to probe PMP22, which contained restriction fragments in the presence of 6× SSC, 5× Denhardt's solution, 0.1% SDS, and 250 μg mL−1 denatured and fragmented herring-sperm DNA for 16 h at 65°C. The membranes were washed with 2× SSC and 0.1% (v/v) SDS for 10 min at 65°C, followed by two washes of 0.1× SSC and 0.1% (v/v) SDS for 20 min at 65°C. After the membranes were washed, they were autoradiographed.
Recombinant Hexahistidine-PMP22 Production and Purification
The construct hexahistidine-PMP22:pET16b was transformed into Escherichia coli strain NovaBlue DE3 (Novagen, Madison, WI). Recombinant hexahistidine-PMP22 production in exponentially growing cells was induced by 500 μm isopropylthio-β-galactoside over 4 h at 37°C, and the protein was purified by its affinity to nickel-agarose (Qiagen, Chatsworth, CA) under denaturing conditions according to the manufacturer's recommendations.
Plant Material and Growth Conditions
Arabidopsis plants were germinated and grown in compost:sand (10:1, v/v) at 18°C with 8 h light d−1 for the first 4 weeks, then at 20°C with 16 h light d−1. Suspension cultures of Arabidopsis (obtained from Dr. Paul Knox, Leeds Institute for Plant Biotechnology and Agriculture) were maintained at 20°C in 1× Murashige and Skoog medium, pH 5.7, with 3% (w/v) Suc, 0.05 mg L−1 kinetin, and 0.5 mg L−1 NAA under dark and sterile conditions with constant agitation. The cells were subcultured once every 7 d by transferring 0.1× final culture volume into fresh medium as described above. For the germination time-course experiment, 0.1-g batches of Arabidopsis seeds were sterilized in 10% (v/v) domestic bleach for 30 min and washed 10 times with 1 mL of sterile distilled water. The seeds were imbibed for 1 h at room temperature in the final sterile distilled water wash and placed onto plates containing 0.2% (w/v) Phytogel (Sigma) and 0.5× Murashige and Skoog medium, pH 5.7.
Protein Isolation and Subcellular Fractionation of Plant Material
Glyoxysomes from sunflower cotyledons 3 d postimbibition were prepared according to the method described by Horng et al. (1995). For the isolation of organelles from dark-cultured Arabidopsis suspension-cultured cells, the cells were harvested by centrifugation at 1000g for 5 min at 4°C and resuspended in ice-cold 50 mm Mes-KOH, pH 6.0, 0.5 m Suc, 10 mm KCl, and 1 mm EDTA at a pellet-to-buffer ratio of 1:5. The cell clumps were loosened by homogenization (10 strokes) of a loose-fitting Teflon/glass homogenizer. The cell suspension was initially ruptured by five strokes of a 3-mL glass/glass homogenizer (no. 2 clearance, Jencons Scientific, Bridgeville, PA), followed by 20 strokes of a 1-mL glass/glass homogenizer (no. 2 clearance, Jencons Scientific).
Postnuclear supernatant was obtained by pelleting the cellular debris at 1,000g for 10 min at 4°C. For some experiments, a mixed-organelle pellet fraction was obtained by centrifuging the postnuclear supernatant at 20,000g for 30 min at 4°C. Membranes were prepared from the mixed-organelle pellet by resuspending them by homogenization first in 20 mm Hepes-KOH, pH 7.5, then with the addition of NaCl to a final concentration of 250 mm. A membrane pellet was obtained by centrifuging the organelle lysate at 100,000g for 30 min at 4°C. Continuous Suc gradients of subcellular organelles were performed in 13-mL SW40 tubes (Beckman). Gradients ranged from 0.7 to 2.1 m Suc dissolved in 50 mm Mes-KOH, pH 6.0, 10 mm KCl, and 1 mm EDTA. Typically, 5 to 10 mg (1 mL) of postnuclear supernatant was centrifuged at 200,000g for 12 h at 4°C, and the gradients were fractionated into 12 1-mL aliquots from their dense Suc end. Total protein extracts of plant material were obtained by freezing approximately 1 to 5 g of the required tissue in liquid nitrogen and grinding it into a smooth powder. This powder was immediately resuspended in 10 mL of ice-cold TCA (10%, v/v). Proteins were removed from this mixture for analysis by centrifuging aliquots at 14,000g for 15 min at 4°C.
Arabidopsis rosettes (20 g) were chopped with razor blades in 50 mL of chilled 50 mm Mes-KOH, pH 6.0, 0.5 m Suc, 10 mm KCl, and 1 mm EDTA on ice for 20 min and filtered through four layers of muslin to produce a homogenate fraction. This was centrifuged at 1,000g for 10 min at 4°C to produce a pellet and supernatant fraction. The supernatant fraction was centrifuged at 25,000g for 30 min at 4°C to produce a 25,000g pellet and a supernatant fraction.
Treatments of Subcellular Fractions
Organelles at a protein concentration of 1 mg mL−1 were washed with 0.1 m Na2CO3 (pH 11.0) as described by Fujiki and Lazaraow (1982). Fractionation of organelles with Triton X-114 was performed as described by Pryde and Phillips (1986). In protease-digestion experiments, intact organelles or their ruptured and salt-washed membranes (prepared as described above) at a protein concentration of 1 mg mL−1 were incubated in the presence of the required concentration of thermolysin in 50 mm Mes-KOH, pH 6.0, and 10 mm KCl for 30 min at 4°C. Enzymatic activities of catalase and fumarase were measured as described by Cooper and Beevers (1969), and the activity of NADH:Cyt c reductase was measured as described by Gomez and Chrispeels (1994). Protein estimations were performed using BCA reagent (Pierce) according to the manufacturer's recommendations, with BSA as the standard.
Electrophoretic Methods
Protein composition was analyzed in 15% (w/v) polyacrylamide gels (unless otherwise specified) as described by Laemmli (1970). For immunodetection, proteins separated by SDS-PAGE were transferred electrophoretically onto 0.45-μm nitrocellulose membranes (Schleicher & Schuell) in the presence of 20 mm Na2HPO4, 0.02% (w/v) SDS, and 20% (v/v) methanol using a semidry blotter (model 2117–250 Novablot, LKB-Pharmacia) for 2 h at 0.8 mA cm−2.
Immunological Methods
Nitrocellulose membranes for immunoblotting were blocked for nonspecific immunoreactivity initially with 1× TBS (20 mm Tris-HCl, pH 7.4, and 150 mm NaCl), 0.5% (v/v) Tween 20, and 0.1% (w/v) NaN3 containing 10 mg mL−1 BSA overnight and then with 1× TBS, 0.5% (v/v) Tween 20, and 0.1% (w/v) NaN3 containing 10% (w/v) defatted dried milk (Sainsbury's, London) for 6 to 8 h. Antibodies were diluted in 1× TBS, 0.05% (v/v) Tween 20, 0.1% (w/v) NaN3, and 10 mg mL−1 BSA and incubated with the blots for 16 h. Nonspecifically bound antibodies were removed with eight 15-min washes using 1× TBS and 0.1% (v/v) Tween 20. Specifically bound IgGs were detected after incubation with anti-rabbit antibodies conjugated with horseradish peroxidase (Sigma) using enhanced chemiluminescence (Amersham).
Anti-PMP22 specific antibodies were affinity purified by incubating the antisera at a dilution of 1:100 with a blocked nitrocellulose strip (0.5 × 8 cm) containing the region of migration of electrophoretically separated, purified hexahistidine-PMP22 (250 μg). Following incubation for 16 h, the strip was washed with eight 15-min washes of 1× TBS and 0.1% (v/v) Tween 20. The PMP22-specific antibodies were eluted with 0.1 m Gly, pH 2.8, for 1 min and immediately neutralized. For immunoblots, affinity-purified anti-PMP22 antibodies were used at a dilution of 1:10,000 with respect to the original antisera. Anti-glycolate oxidase antisera (Volokita and Somerville, 1989) was affinity purified by the same procedure using spinach glycolate oxidase (Sigma) and used at a dilution of 1:10,000 with respect to the original antiserum. The anti-isocitrate lyase antiserum, previously characterized by Martin and Northcote (1982), was used at a dilution of 1:100,000.
In Vitro Import Reactions
In vitro import assays were performed as described by Behari and Baker (1993) with modifications according to Horng et al. (1995). Glyoxysomes were isolated from cotyledons of sunflower 3 d postgermination by separation of a postnuclear supernatant in a Nycodenz step gradient. Glyoxysomes harvested from the gradient were diluted and concentrated by centrifugation before resuspension in 25 mm Mes-KOH, pH 6.0, 0.5 m Suc, 10 mm KCl, and 1 mm MgCl2 at a protein concentration of 1.0 to 1.5 mg mL−1. Organelle integrity was assessed by the latency of malate synthase activity. Radiolabeled PMP22 was prepared by in vitro transcription and translation in wheat germ lysate, as previously described by Behari and Baker (1993). Import assays contained 200 μg of glyoxysome fraction, 15 μL of translation product, 2.4 mm ATP, 0.26 mg mL−1 creatine kinase, 32 mm creatine phosphate, 0.34 mg mL−1 cold Met, 25 mm Mes-KOH, pH 6.0, 0.5 m Suc, 10 mm KCl, and 1 mm MgCl2 in a final volume of 200 μL. For carbonate extraction the incubations were carried out in 400 μL and contained 400 μg of glyoxysomes and 30 μL of translation. For minus-ATP controls, ATP and the ATP-regenerating system were omitted and both glyoxysomes and translation product were pretreated with apyrase. Import reactions were incubated for 15 min at 26°C and terminated by chilling on ice. Protease-treated reactions were incubated with 0.1 mg mL−1 (final concentration) thermolysin on ice for 30 min. EDTA was added to 25 mm final concentration and the organelles were re-isolated by centrifugation through a 0.7 m Suc cushion. Samples to be Triton-treated were re-isolated through a Suc cushion, and the organelle pellet was resuspended in 25 mm Mes-KOH, pH 6.0, 0.5 m Suc, 10 mm KCl, 1 mm MgCl2, 250 mm NaCl, 1% (v/v) Triton X-100, and 0.1 mg mL−1 thermolysin and digested for 30 min on ice. Organelle pellets were subjected to SDS-PAGE and radioactivity was detected by exposure to a phosphor-imaging cassette.
RESULTS
Identification and Characterization of Arabidopsis PMP22 cDNA
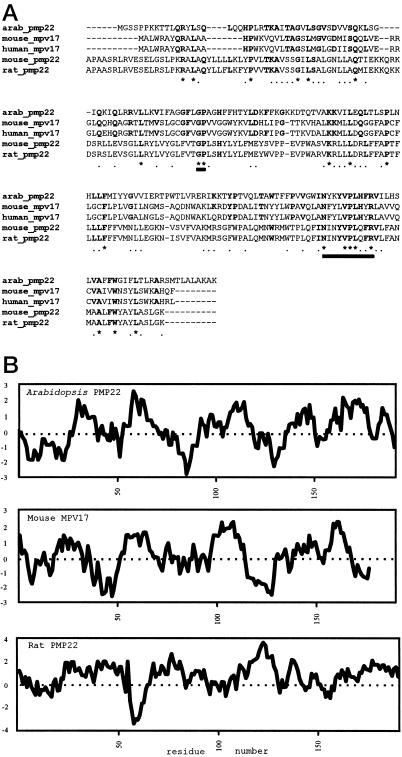
To identify putative PMPs we screened sequence databases generated by the Arabidopsis Sequencing Project for genes similar to known PMPs from other organisms using the BLAST algorithm (Altschul et al., 1990). With this procedure we identified the cDNA TAY089 (accession no. Z18516) encoding a 0.85-kb sequence with similarity to the 5′ end of mammalian PMP22 genes. The complete sequence of the cDNA, shown in Figure 1A, revealed an open reading frame encoding for a 190-amino acid protein with a calculated molecular mass of 21,687 D. As shown in Table I, the alignment of all known PMP22 polypeptides reveals two distinct but related mammalian families, the Mpv17s and the PMP22s, with the Arabidopsis sequence showing 55% amino acid sequence similarity (30% identity) to the mouse and human Mpv17 proteins, and 57% sequence similarity (28% identity) to the rat and mouse PMP22 proteins. Regions of identity at amino acids 77, 78 (-GP-), 151, 155, 156, 157, and 160 (-N-xxx-VPL-xx-R-) of the Arabidopsis polypeptide appear to be signatures of all subgroups of the PMP22 family (Fig. 1A). Another region of the Arabidopsis polypeptide, from amino acids 91 to 109 and encompassing the di-Lys at positions 92 and 93, shows weak similarity to the PMP-targeting signal postulated by Dyer et al. (1996) and Elgersma et al. (1997). Further structural similarities between the Arabidopsis polypeptide and the mammalian PMP22 sequences (especially Mpv17) are apparent on comparison of their hydropathy profiles, as shown in Figure 1B. We therefore refer to this protein as Arabidopsis PMP22.
Figure 1.
A, Clustal W alignment of the deduced amino acid sequences of Arabidopsis PMP22, mouse Mpv17 (P19258), human Mpv17 (P39210), mouse PMP22 (P42925), and rat PMP22 (Q07066). The thick underlining indicates the conserved regions mentioned in the text. B, Arabidopsis PMP22, mouse Mpv17, and rat PMP22 according to the algorithm of Kyte and Doolittle (1982) with an amino acid window of 19. Hydrophobic residues are depicted as positive and hydrophilic residues as negative.
Table I.
Amino acid sequence comparison of PMP22 related proteins
| % Similarity | Arabidopsis | Mouse MPV17 | Human MPV17 | Mouse PMP22 | Rat PMP22 |
|---|---|---|---|---|---|
| % Identity | |||||
| Arabidopsis | – | 55 | 55 | 57 | 57 |
| Mouse MPV17 | 30 | – | 97 | 52 | 54 |
| Human MPV17 | 29 | 92 | – | 52 | 50 |
| Mouse PMP22 | 28 | 28 | 26 | – | 97 |
| Rat PMP22 | 28 | 28 | 26 | 95 | – |
Amino acid sequence similarities and identities between Mpv17 and PMP22 proteins from Arabidopsis and mammals. Data derived from BESTFIT (Genetics Computer Group) by pairwise comparison of polypeptide sequences.
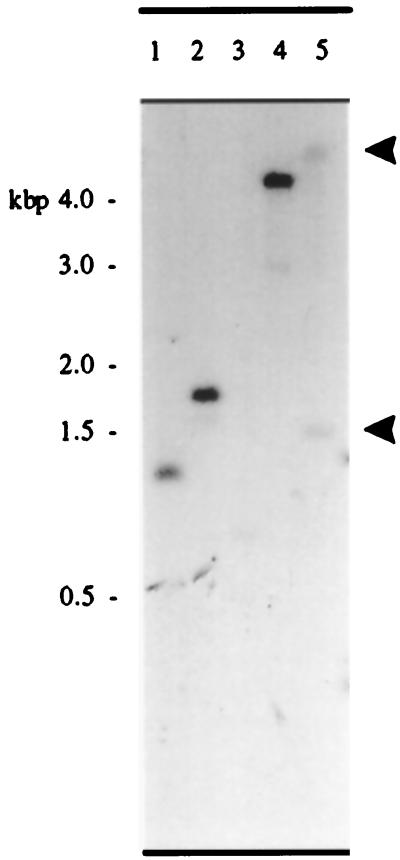
Figure 2 shows the results of Southern analysis of Arabidopsis genomic DNA probed with PMP22. The result suggests that the cDNA is the product of a single-copy gene, as genomic DNA samples digested with the restriction enzymes HindIII, EcoRI, and PstI, each contained a single fragment that hybridized to the Arabidopsis PMP22 probe sequence at high stringency (Fig. 2, lanes 1, 2, and 3). DNA digested with PvuII, which has a single site within the cDNA, gave two fragments of 6.0 and 1.7 kb (indicated by arrows in Fig. 2, lane 5). The gene encoding PMP22 is located on chromosome IV at 19.3 centimorgans within BAC T26N6 (accession no. AF076243). The start codon is located at 50,654 and the stop codon at 49,343 in the BAC sequence. The gene consists of seven exons: exon 1, 50,654 to 50,576; exon 2, 50,351 to 50,255; exon 3, 50,153 to 50,047; exon 4, 49,963 to 49,888; exon 5, 49,801 to 49,735; exon 6, 49,636 to 49,550; and exon 7, 49,451 to 49,393.
Figure 2.
Southern blot showing 10 μg of Arabidopsis genomic DNA digested with HindIII (lane 1), EcoRI (lane 2), PstI (lane 3), XbaI (lane 4), and PvuII (lane 5) probed with the [32P]PMP22 open reading frame, washed under stringent conditions, and detected by autoradiography.
Arabidopsis PMP22 Is Ubiquitously Expressed
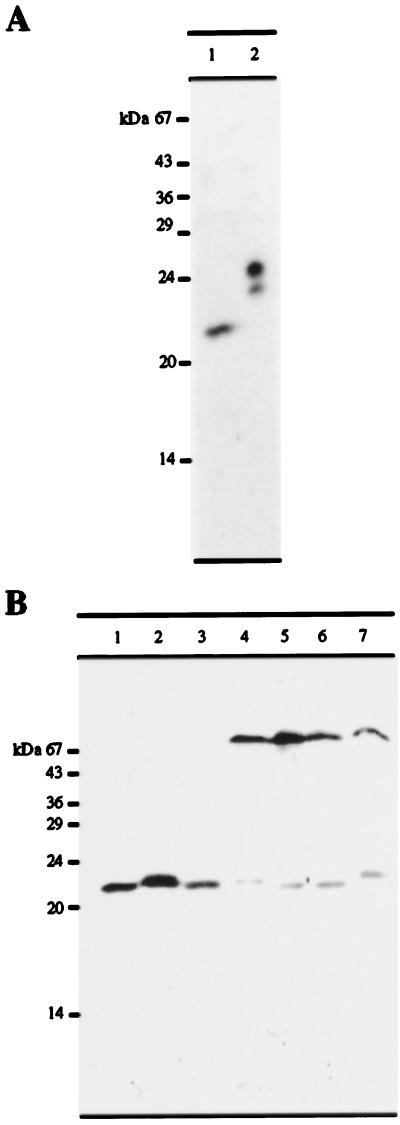
The Arabidopsis PMP22 open reading frame was cloned into the pET16b expression vector with an N-terminal hexahistidine tag and expressed in E. coli. The resulting fusion protein, which has a calculated molecular mass of 24,172 D (due to the presence of 6 His residues and 18 additional residues at the amino terminus of the protein, which were introduced as a result of cloning the PMP22 open reading frame into the pET16b expression vector) was purified on nickel-agarose under denaturing conditions and used to produce rabbit polyclonal antibodies. The anti-PMP22 antisera was affinity purified using hexahistidine-Arabidopsis PMP22 (see Methods). Because the PMP22 cDNA clone was isolated from a cell-suspension culture library, we tested the antibody using a total protein extract from a dark-grown Arabidopsis cell-suspension culture (Fig. 3A, lane 1) and purified recombinant hexahistidine PMP22 as a positive control (Fig. 3A, lane 2). The affinity-purified antibodies bound to a 22-kD protein (in good agreement with the calculated molecular mass of 21,687 D) and to hexahistidine-PMP22, which, as expected, migrated at approximately 24 kD.
Figure 3.
A, Immunoblot showing 150 μg of total protein extract from dark-grown Arabidopsis cell-suspension culture (lane 1) and 0.25 μg of recombinant purified hexahistidine-PMP22 (lane 2) probed with affinity-purified anti-PMP22 antibodies and detected by enhanced chemiluminescence. B, Equal amounts of protein (150 μg) extracted from various organs of Arabidopsis plants and from dark-grown tissue-cultured cells were separated by SDS-PAGE and probed with affinity-purified anti-PMP22 antibodies. Lane 1, Dark-grown tissue-culture cells; lane 2, flowers; lane 3, siliques; lane 4, stems; lane 5, upper leaves; lane 6, basal leaves; and lane 7, roots.
To determine whether PMP22 was expressed in whole plants, equal amounts of total protein from organs of 12-week-old flowering Arabidopsis plants (see Methods for growth conditions) were immunoblotted with the affinity-purified anti-PMP22 antibody. Figure 3B shows that PMP22 is present in all Arabidopsis tissues, with the highest enrichment in flowers and green siliques. There appeared to be a tissue-dependent difference in the electrophoretic mobility of PMP22, with a larger isoform detected in flowers, stems, and roots. Although the anti-PMP22 was affinity purified using recombinant hexahistidine-PMP22 and was monospecific when tested against whole-cell extracts of suspension-cultured cells (Fig. 3, lanes 1), there was an additional and persistent cross-reaction with a protein of approximately 70 kD in green tissues and roots (Fig. 3B, lanes 4–7) that was absent in flowers and siliques (Fig. 3B, lanes 2 and 3). Because it was not possible to remove the cross-reaction with this protein by affinity purification of the anti-PMP22 IgGs, it may share epitopes with PMP22.
Localization of PMP22 to Peroxisomes by Subcellular Fractionation and Suc-Density-Gradient Centrifugation
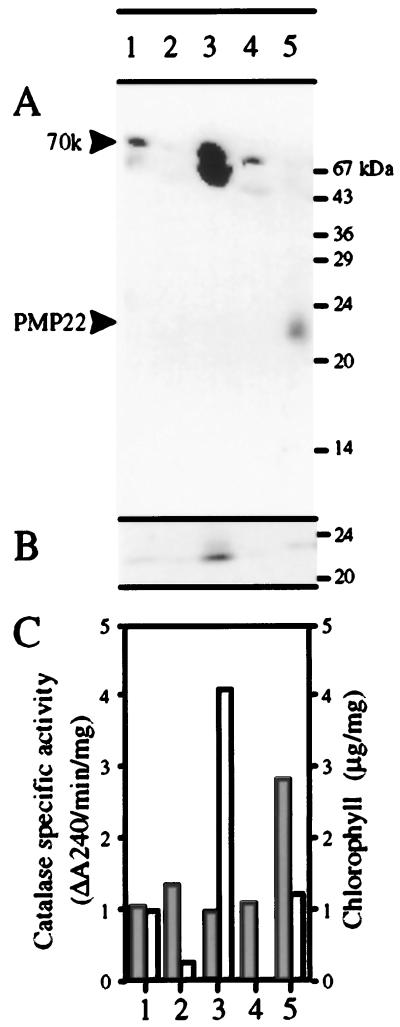
Arabidopsis rosette leaves were fractionated by differential centrifugation. A homogenate fraction was centrifuged to produce a 1,000g pellet and supernatant. The 1,000g supernatant was further centrifuged to produce a 25,000g pellet and supernatant. The 1,000g pellet would be expected to be enriched in nuclei and chloroplasts, whereas the 25,000g pellet would be expected to be enriched in mitochondria and peroxisomes. The 25,000g supernatant contained soluble proteins and light membranes. Equal amounts of protein (120 μg) from each fraction were separated by SDS-PAGE and subjected to blotting with affinity-purified anti-PMP22 and antibodies against the 23-kD subunit of the PSII oxygen-evolving complex (a peripheral membrane protein of the thylakoid membrane). Figure 4A shows that the 70-kD protein is detectable in the homogenate and greatly enriched in the 1,000g pellet. A trace was detectable in the 25,000g supernatant, but nothing was detectable in the 25,000g pellet. Because the 70-kD protein behaved as a soluble protein (data not shown), a low level of the antigen in the 25,000g supernatant due to the release of soluble proteins from ruptured organelles was to be expected. The 23-kD protein (Fig. 4B) and chlorophyll (Fig. 4C) showed a similar distribution, being predominantly in the 1,000g pellet. However, both of these markers are associated with the thylakoid membrane, so they were also present at a low level in the 25,000g pellet due to the presence of fragments from the thylakoid membrane released from ruptured chloroplasts. In contrast, PMP22 could not be detected in the homogenate, in the 1000g supernatant, or in the pellet (because of its low abundance), but was readily detectable in the 25,000g pellet (Fig. 4A). The specific activity of catalase (a peroxisomal marker) showed a similar distribution (Fig. 4C), being most enriched in the 25,000g pellet. These results are consistent with a peroxisomal localization for PMP22 but not for the 70-kD protein, which may be a soluble plastidial protein.
Figure 4.
Subcellular localization of Arabidopsis PMP22 in leaf tissue by differential centrifugation. Arabidopsis leaves were fractionated as described in Methods. Equal amounts of protein (120 μg) were separated by SDS-PAGE, transferred to nitrocellulose, and probed with affinity-purified antibodies against PMP22 (A) or the 23-kD protein of the PSII oxygen-evolving complex (B). C, Catalase activity, a peroxisomal matrix marker (shaded bars) and chlorophyll (white bars) were measured in each fraction. Catalase recovery was 102% and chlorophyll recovery was 61%. Lane 1, Homogenate; lane 2, 1,000g supernatant; lane 3, 1,000g pellet; lane 4, 25,000g supernatant; lane 5, 25,000g pellet.
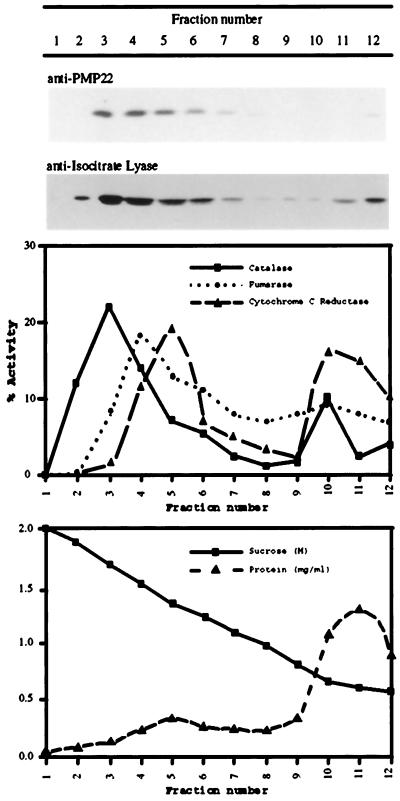
To provide more definitive evidence for the location of PMP22, organelles were separated by Suc-density-gradient centrifugation. Because of the size and abundance of chloroplasts it was not possible to obtain clean separation between chloroplast and peroxisomal fractions in Suc gradients from leaf tissue. Therefore, suspension-cultured cells were chosen for the Suc-gradient experiments because sufficient quantities of material could readily be obtained. PMP22 is abundant in these cells, which also lack the 70-kD cross-reacting protein. A postnuclear supernatant prepared from dark-grown tissue-cultured cells was separated in a 0.7 to 2.1 m continuous Suc gradient. The gradient was fractionated and the distribution of organelles was determined by the enzymatic activities of the marker enzymes catalase (for peroxisomes), fumarase (for mitochondria), and NADH:Cyt c reductase (for the ER), as well as with the immunoreactivity of isocitrate lyase. Figure 5 is an immunoblot of equal volume loadings for each fraction, and shows that Arabidopsis PMP22 was localized mainly in fractions 3 and 4 (1.80 and 1.65 m Suc), where the peroxisomal markers catalase and isocitrate lyase were also detected. The fairly broad distribution of these markers is typical for peroxisomal proteins and reflects the heterogeneity of the peroxisomal compartment (Lüers et al., 1993). Fumarase and NADH:Cyt c reductase were localized in fractions 4 and 5 (1.5 and 1.35 m Suc, respectively).
Figure 5.
Subcellular localization of PMP22 in tissue culture cells by Suc-density-gradient centrifugation. A postnuclear supernatant prepared from dark-grown Arabidopsis suspension-cultured cells was separated on a 0.7 to 2.1 m continuous Suc-density gradient. Fractions were assayed for catalase (a peroxisomal marker), fumarase (a mitochondrial marker), NADH:Cyt c reductase (an ER marker), and protein. The recoveries of the enzyme activities relative to the postnuclear supernatant were: catalase, 76%; fumarase, 96%; and NADH Cyt c reductase, 91%. Equal volumes of the Suc-gradient fractions were separated by SDS-PAGE and probed with affinity-purified anti-PMP22 antibodies and anti-castor bean isocitrate lyase antibodies.
Membrane Localization of Arabidopsis PMP22
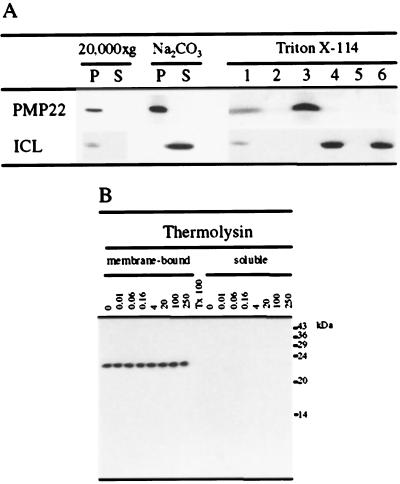
The presence of significant hydrophobic regions in the Arabidopsis PMP22 and its sequence similarity to other PMP22s that are known to be integral membrane proteins indicated its probable membrane association. To determine the nature of this membrane association, a 20,000g organelle pellet obtained from dark-cultured Arabidopsis suspension-cultured cells enriched in PMP22 and isocitrate lyase was treated with 0.1 m Na2CO3 (pH 11.0) to lyse the organelles and to remove peripheral and lumenal proteins. As shown in Figure 6A, Arabidopsis PMP22 remained in the membrane pellet fraction, whereas isocitrate lyase, which is a peroxisomal matrix protein, was extracted into the supernatant (Fig. 6A).
Figure 6.
A, PMP22 is resistant to extraction by Na2CO3 and partitions in the detergent phase of Triton X-114. A 20,000g organelle pellet (P) and the corresponding supernatant (S) were prepared from dark-grown tissue-cultured cells and analyzed for PMP22 and isocitrate lyase (ICL) by SDS-PAGE and immunoblotting (20,000g). The 20,000g pellet fraction enriched in PMP22 was treated with 0.1 m Na2CO3 (pH 11.0) and centrifuged at 100,000g to produce a membrane pellet and supernatant fraction. A postnuclear supernatant prepared from dark-grown tissue-cultured cells was fractionated using 2% (v/v) Triton X-114 phase separation, and the various fractions were analyzed by SDS-PAGE and immunoblotting with antibodies against isocitrate lyase and affinity-purified anti-PMP22 antibodies. Lane 1, Postnuclear supernatant; lane 2, Triton X-114-insoluble pellet; lane 3, detergent phase; lane 4, aqueous phase; lane 5, “glycoprotein”-rich pellet; and lane 6, postaqueous supernatant. In all lanes 150 μg of protein was analyzed. B, Accessibility of Arabidopsis PMP22 to protease. The peroxisome-enriched 20,000g pellet fraction was subjected to hypotonic lysis followed by a wash with 250 mm NaCl. Salt-washed membranes (250 μg) were incubated with the protease thermolysin at the indicated concentrations (see Methods). After the incubation the protease was inhibited and the membrane-bound and protease-solubilized peptides were separated by ultracentrifugation. Triton X-100 was included in a duplicate incubation containing 100 μg mL−1 thermolysin, which was not subjected to ultracentrifugation before analysis by SDS-PAGE and immunoblotting. All samples were analyzed by SDS-PAGE and immunoblotting with affinity-purified anti-PMP22 antibodies.
Fractionation of biological membranes using the phase-separation properties of the nonionic detergent Triton X-114 provides a method of determining the hydrophobic nature of membrane-associated proteins (Bordier, 1981; Pryde and Phillips, 1986). Although all membrane-associated proteins are extracted by Triton X-114, only integral membrane proteins with significant hydrophobic regions partition into a “detergent” phase; those that are peripherally associated and/or glycosylated are enriched in the “aqueous” phase. A postnuclear supernatant prepared from dark-grown Arabidopsis suspension-cultured cells was treated with Triton X-114, and the resulting detergent-soluble fraction was phase-separated according to the method of Pryde and Phillips (1986). Arabidopsis PMP22 was observed to partition only into the Triton X-114 detergent phase (Fig. 6A), as expected for an integral membrane protein and unlike the soluble matrix protein isocitrate lyase, which partitioned into the aqueous phase.
To investigate the topology and the extent of integration of Arabidopsis PMP22 in the peroxisomal membrane, both intact organelles (not shown) and lysed membranes derived from them were treated with the protease thermolysin at a range of concentrations. Following protease treatment, membrane-bound and protease-solubilized peptides were separated by ultracentrifugation and analyzed for Arabidopsis PMP22 immunoreactivity. Figure 6B shows that Arabidopsis PMP22 remained completely protease resistant even when lysed membranes prewashed with 250 mm NaCl were treated with high protease concentrations (Fig. 6B). Protease resistance is dependent on the presence of a phospholipid bilayer, because the inclusion of 1% (v/v) Triton X-100 during the protease treatment resulted in the complete digestion of Arabidopsis PMP22. Similar results were obtained with the protease trysin (data not shown).
Integration of Arabidopsis PMP22 into Peroxisomes in Vitro
As an independent method of determining the intracellular location of Arabidopsis PMP22, the specificity of its targeting and insertion into peroxisomes in vitro was addressed. Transcripts were translated in vitro with wheat germ lysate in the presence of [35S]-l-Met and incubated with isolated intact sunflower glyoxysomes, a stage-specific type of peroxisome whose purity and integrity was established by Behari and Baker (1993) and Horng et al. (1995). As established in the previous section, native Arabidopsis PMP22 is deeply buried in the membrane so that it is resistant to proteolysis by thermolysin and cannot be extracted with 0.1 m Na2CO3, pH 11.0. This provided the means of assessing the correct insertion of PMP22 into peroxisomal membranes in vitro.
After the incubation of intact peroxisomes with radiolabeled, in vitro-translated Arabidopsis PMP22 in the presence of ATP and an ATP-regeneration system, the peroxisomes were incubated with thermolysin and then re-isolated through a 0.7 m Suc cushion. To differentiate between correctly integrated and possible membrane-associated protease-resistant aggregates of PMP22, the re-isolated peroxisomes were washed with 0.1 m Na2CO3 (pH 11.0). As shown in Figure 7, most of the radiolabeled Arabidopsis PMP22 bound to peroxisomes (Fig. 7, lane 2), and was resistant to carbonate washing (Fig. 7, lane 10). Protease treatment removed surface-bound PMP22 and left intact those molecules that were correctly inserted (Fig. 7, lane 3). These molecules were also resistant to carbonate extraction (Fig. 7, lane 12). Insertion was dependent on the presence of the peroxisome membrane, because pretreatment with 1% (v/v) Triton X-100 prior to protease treatment had abolished protease resistance (Fig. 7, lane 4).
Figure 7.
Insertion of Arabidopsis PMP22 into isolated peroxisomes. Radiolabeled PMP22 was prepared by in vitro transcription and translation in wheat germ lysate and incubated with peroxisomes isolated from 3-d postimbibition sunflower cotyledons (see Methods). Lane 1, 40% of the translation product added to the other reactions; lane 2, import reaction carried out in the presence of the ATP-regeneration system; lane 3, import in the presence of the ATP-regeneration system followed by protease treatment; lane 4, same as lane 3 but in buffer containing 1% Triton X-100 and 250 mm NaCl before the addition of protease; lane 5, import in the absence of ATP and the ATP-regenerating system; lane 6, import in the absence of ATP and the ATP-regenerating system followed by treatment with protease; lane 7, same as lane 3 but no peroxisomes were added to the import assay; lane 8, same as lane 2 but glyoxysomes were replaced with 50 μg of washed red blood cells from calf ascites; lane 9, same as lane 8 but treated with protease; lanes 10 to 15, Na2CO3 (pH 11.0) treated pellets and supernatants derived from import reactions (lane 10 is the pellet and lane 11 the supernatant of the import reaction in lane 2, lane 12 is the pellet and lane 13 the supernatant of the import reaction in lane 3; and lane 14 is the pellet and lane 15 the supernatant of the import reaction in lane 5).
If peroxisomes were omitted from the import assay, (Fig. 7, lane 7) or replaced with washed red blood cells from calf ascites (Fig. 7, lanes 8 and 9), Arabidopsis PMP22 was not re-isolated through the 0.7 m Suc cushion, demonstrating that the re-isolation and insertion is dependent upon the presence of the correct membrane. Red blood cells were used as an “irrelevant membrane” control in these experiments; because of the fragility and heterogeneity of peroxisomes it is extremely difficulty to prepare other organelles that are totally free from peroxisome membrane contamination (Fig. 5; Behari and Baker, 1993).
When organelles and the translation product were pretreated with apyrase to remove any nucleoside triphosphates and the ATP-regeneration system was omitted from the import reaction, the level of insertion was substantially reduced (Fig. 7, compare lanes 3 and 6). In the absence of ATP, slightly more PMP22 bound to peroxisomes (Fig. 7, compare lanes 2 and 5), and it was also resistant to carbonate extraction (Fig. 7, lane 14).
Developmental Expression of PMP22 in Arabidopsis Seedlings
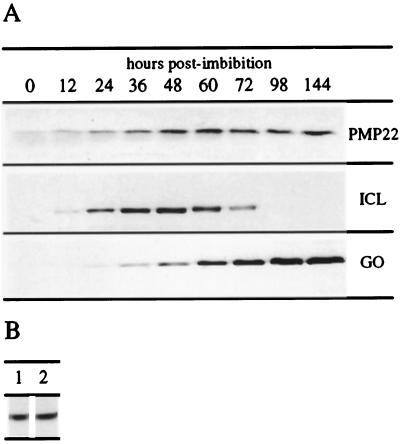
To begin to understand the function of Arabidopsis PMP22, we investigated its expression pattern in seedlings and in suspension-cultured cells. During germination glyoxysomes are important organelles, taking part in the mobilization of stored lipids for the provision of the carbohydrate required for postgerminative growth. By the time stored lipid reserves begin to diminish, the cotyledons break out of the soil and become photosynthetic. As this occurs, peroxisomes change metabolic function from fatty acid oxidation to salvaging glycolate, a by-product of photorespiration. As a consequence, the levels of isocitrate lyase decrease and the levels of glycolate oxidase increase. To investigate whether PMP22 is involved in such stage-specific roles, the levels of PMP22 in Arabidopsis seedlings of different ages were compared with levels of isocitrate lyase and glycolate oxidase. Total protein was extracted from equal numbers of Arabidopsis seedlings at different time points after imbibition and then immunoblotted with affinity-purified anti-PMP22, affinity-purified anti-glycolate oxidase, and anti-isocitrate lyase antibodies. Figure 8A shows that PMP22, which is present at low levels in seeds, progressively accumulated on a per-seedling basis to reach a steady level by 48 h postimbibition. The level of isocitrate lyase was, as expected, temporally dependent, with maximum protein levels at 48 h. The level of glycolate oxidase increased steadily from 36 h on, as did the level of chlorophyll (data not shown). Greening was observed between 48 and 72 h by measuring total chlorophyll levels.
Figure 8.
A, Expression of PMP22 during seedling development. Total protein was extracted from an equal number (based on dry weight at sowing) of Arabidopsis seedlings at the indicated number of hours postimbibition. Protein samples were separated by SDS-PAGE and probed with affinity-purified anti-PMP22 antibodies, anti-castor bean isocitrate lyase antibodies (ICL), or affinity-purified anti-spinach glycolate oxidase antibodies (GO). B, Effects of light on expression of PMP22 in tissue cultures. Arabidopsis suspension-cultured cells were grown in either continuous light (lane 1) or total dark (lane 2), subcultured, and cultured for a further 5 d under the same conditions. Total protein was extracted from these samples and subjected to SDS-PAGE and immunoblotting with affinity-purified anti-PMP22 antibodies.
To establish whether PMP22 expression was light dependent, total protein was extracted from the same growth stage (5 d postsubculture) of constant-dark- and constant-light-cultured Arabidopsis suspension-cultured cells (these cells turned green upon exposure to light). Immunoblotting equivalent amounts of total protein (compared on the basis of equal packed-cell volume) revealed that the levels of PMP22 were the same in both dark and light conditions (Fig. 8B).
DISCUSSION
We have identified and characterized at the molecular level a plant PMP, Arabidopsis PMP22, which is approximately 55% similar (30% identical) to its mammalian counterparts, which form two subgroups, Mpv17 and PMP22. Although Mpv17 from human and mouse are 92% identical at the amino acid level, and rat and mouse PMP22 are 95% identical, the identity between Mpv17 and PMP22 from mouse is only 28%. Thus, the two different subgroups from the same organism are no more similar to each other than they are to Arabidopsis PMP22. However, comparison of the hydropathy profiles shows that Arabidopsis PMP22 is more similar to Mpv17 than to PMP22. The Arabidopsis PMP22 is 190 amino acids long and is the product of a single-copy gene.
Arabidopsis PMP22 was localized to peroxisomes by subcellular fractionation and immunoblotting and by co-sedimenting in Suc gradients with catalase and the peroxisomal marker protein isocitrate lyase. As predicted by its hydropathy plot, Arabidopsis PMP22 behaves as an integral PMP: it is not extracted by Na2CO3 (pH 11.0) and it fractionates into the Triton X-114 detergent phase. Despite the prediction by the hydropathy profile that significant hydrophilic domains flank the transmembrane regions, these were not accessible to proteases such as thermolysin and trypsin even if salt-washed membranes were treated with high concentrations of protease. This implies that Arabidopsis PMP22 is either deeply buried in the membrane or that it may form homo- or hetero-complexes in the membrane such that the hydrophilic domains are not accessible to protease. Protease protection by the membrane was completely abolished in the presence of Triton X-100.
The protease resistance of native Arabidopsis PMP22 and its inextractability with Na2CO3 (pH 11.0) allowed us to investigate the manner of its insertion into the peroxisomal membrane. As established for PMP70 (Imanaka et al., 1996), PAF-1, and rat PMP22 (Just and Diestelkötter, 1996), Arabidopsis PMP22 inserted into peroxisomal membranes posttranslationally. There was no binding or insertion observed if glyoxysomes were replaced by an irrelevant membrane (red blood cells), demonstrating that insertion was not due to nonspecific partitioning of a hydrophobic protein into a membrane. We observed that in contrast to results obtained with rat PMP22, but similar to results with PMP70 and PAF-1, the insertion of Arabidopsis PMP22 into peroxisomal membranes was stimulated by the presence of 2.4 mm ATP with an ATP-regeneration system.
In the absence of ATP and the ATP-regeneration system, binding of Arabidopsis PMP22 to the surface of glyoxysomes was not affected, and this binding was carbonate-insensitive, suggesting partial insertion into the bilayer. However, the level of the native protease-resistant conformation was reduced by 50%. This suggests that after binding/insertion of Arabidopsis PMP22 to and/or into the peroxisomal membrane by an ATP-independent mechanism, ATP is required at the membrane for rearrangement into the native conformation. However, because this process was not completely abolished in the absence of ATP, it may also suggest that the membrane protein insertion/assembly machinery of the isolated peroxisomes could remain “primed” for the import of membrane proteins, e.g. with ATP tightly bound. These results are in contrast to those obtained for the matrix proteins isocitrate lyase and glycolate oxidase, in which import into the matrix is dependent upon added ATP (Behari and Baker, 1993; Horng et al., 1995).
The expression of Arabidopsis PMP22 is not light dependent, because it is expressed at similar levels in both dark- and light-grown suspension-cultured cells. After imbibition the levels of PMP22 per seedling steadily increased to a constant level by the onset of greening. Thus, the requirement for PMP22 was not associated with a specific peroxisome type and may represent a universal component of the peroxisome membrane. The steady-state level of PMP22 differs between tissues—it is most abundant in open flowers and siliques, with lesser amounts in stems, leaves, and roots. We observed a tissue-specific alteration in the mobility of PMP22 (<0.5 kD), with a larger isoform detected in flowers, stems, and roots. We are currently investigating the molecular basis of this heterogeneity.
The mouse Mpv17 protein has been shown to generate reactive oxygen species (Zwacka et al., 1994), but it is not known if this is its principal function or if it is a by-product of some other reaction catalyzed by the protein. Although, like Arabidopsis PMP22, Mpv17 appears to be expressed in all tissues, its inactivation by retroviral insertion principally affects the kidney, leading to glomerosclerosis and nephrotic syndrome (Weiher et al., 1990). Plant peroxisomes are significant generators of active oxygen species in both the membranes and the matrix; they contain defense mechanisms in the form of catalase, superoxide dismutase, and ascorbate peroxidase. It will be interesting to discover if the loss of PMP22 has phenotypic effects on any particular organ, or if it results in altered sensitivity to growth conditions that stimulate free radical production in plant peroxisomes. Such information may help to shed light on the function of PMP22 in both mammals and plants.
ACKNOWLEDGMENTS
We thank Dr Claude Kaplan for identifying the TAY089 expressed sequence tag as a possible PMP22 ortholog and the Arabidopsis Biological Resource Center for dispatching the expressed sequence tag clone. We are grateful to Prof. Colin Robinson (University of Warwick, UK) for antibodies to the 23-kD subunit of PSII and to Prof. L.A. del Río and Dr. L.M. Sandalio for helpful comments on the text.
The accession number for the Arabidopsis PMP22 sequence reported here is AJ006053.
Abbreviation:
- PMP
peroxisomal membrane protein
Footnotes
This work was supported by the Biotechnology and Biology Research Council, UK (grant no. 24/C06847), and by the Leverhulme Trust, UK (grant no. F/122/AW to A.B.).
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Behari R, Baker A. The carboxy terminus of isocitrate lyase is not essential for import into glyoxysomes in an in vitro system. J Biol Chem. 1993;268:7315–7322. [PubMed] [Google Scholar]
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- Bowditch MI, Donaldson RP. Ascorbate free-radical reduction by glyoxysomal membranes. Plant Physiol. 1990;94:531–537. doi: 10.1104/pp.94.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunkelmann J, Trelease RN. Ascorbate peroxidase. A prominent membrane protein in oil seed glyoxysomes. Plant Physiol. 1996;110:589–598. doi: 10.1104/pp.110.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canvin DT, Salon C. Photorespiration and CO2-concentrating mechanisms. In: Dennis DT, Turpin DH, Lefebvre DD, Layzell DB, editors. Plant Metabolism, Ed 2. Addison Wesley. Harlow, UK: Longman; 1997. pp. 314–340. [Google Scholar]
- Cooper TG, Beevers H. Mitochondria and glyoxysomes from castor bean endosperm. J Biol Chem. 1969;244:3514–3520. [PubMed] [Google Scholar]
- Corpas FJ, Bunkelman J, Trelease RN. Identification and immunocytochemical characterization of a family of peroxisome membrane proteins (PMPs) in oilseed glyoxysomes. Eur J Cell Biol. 1995;65:280–290. [PubMed] [Google Scholar]
- del Río LA, Sandalio LM, Palma JM, Bueno P, Corpas FJ. Metabolism of oxygen radicals in peroxisomes and cellular implications. Free Radical Biol Med. 1992;13:557–580. doi: 10.1016/0891-5849(92)90150-f. [DOI] [PubMed] [Google Scholar]
- Dyer JM, McNew JA, Goodman JM. The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J Cell Biol. 1996;133:269–280. doi: 10.1083/jcb.133.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen GA, Aitchinson JD, Szilard RK, Veenhuis M, Nuttley WM, Rachubinski RA. The Yarrowia lipolytica gene PAY2 encodes a 42-kD peroxisomal integral membrane protein essential for matrix protein import and peroxisome enlargement but not for peroxisome proliferation. J Biol Chem. 1995;270:1429–1436. doi: 10.1074/jbc.270.3.1429. [DOI] [PubMed] [Google Scholar]
- Elgersma Y, Kwast L, Klein A, Voorn-Brouwer T, van den Berg M, Metzig B, America T, Tabak HF, Distel B. The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex 5p, a mobile receptor for the import of PTS-1 containing proteins. J Cell Biol. 1996;135:97–109. doi: 10.1083/jcb.135.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgersma Y, Kwast L, van den Berg M, Snyder WB, Distel B, Subramani S, Tabak HF. Overexpression of Pex15p, phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S. cerevisiae, causes proliferation of the ER membrane. EMBO J. 1997;16:7326–7341. doi: 10.1093/emboj/16.24.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R, Blöbel G. Giant peroxisomes in oleic acid induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J Cell Biol. 1995;128:509–523. doi: 10.1083/jcb.128.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann R, Blöbel G. Identification of PEX13p, a peroxisomal membrane receptor for the PTS1 recognition factor. J Cell Biol. 1996;135:111–121. doi: 10.1083/jcb.135.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang TK, Donaldson RP, Vigil EL. Electron transport in purified glyoxysomal membranes from castor bean endosperm. Planta. 1987;172:1–13. doi: 10.1007/BF00403023. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Fowler S, Shio H, Hubbard A, Lazaraow PB. Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes, comparison with ER and mitochondrial membrane. J Cell Biol. 1982;93:103–110. doi: 10.1083/jcb.93.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki Y, Lazaraow PB. Isolation of intracellular membranes by means of sodium carbonate treatment: application to ER. J Cell Biol. 1982;93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez L, Chrispeels MJ. Complementation of an Arabidopsis thaliana mutant that lacks complex Asn-linked glycans with the human cDNA encoding N-acetylglucosaminyltransferase I. Proc Natl Acad Sci USA. 1994;91:1829–1833. doi: 10.1073/pnas.91.5.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Kalish JE, Morrell JC, Bjorkman J, Urquhart AJ, Crane DI. PEX13p is a SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTS1 receptor. J Cell Biol. 1996;135:85–95. doi: 10.1083/jcb.135.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, van Roermund CWT, Distel B, van den Berg M, Viela C, Rodrigues-Pousada C, Wanders RJA, Tabak HF. The ABC transporter proteins PAT-1 and PAT-2 are required for the import of long chain fatty acids into peroxisomes of Saccharomyces cerevisiae. EMBO J. 1996;15:3813–3822. [PMC free article] [PubMed] [Google Scholar]
- Hicks DB, Donaldson RP. Electron transport in glyoxysomal membranes. Arch Biochem Biophys. 1982;215:280–288. doi: 10.1016/0003-9861(82)90306-x. [DOI] [PubMed] [Google Scholar]
- Höhfeld J, Veenhuis M, Kunau WH. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J Cell Biol. 1991;114:1167–1178. doi: 10.1083/jcb.114.6.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng J-T, Behari R, Burke C-A, Baker A. Investigation of the energy requirement and targeting signal for the import of glycolate oxidase into glyoxysomes. Eur J Biochem. 1995;230:157–163. doi: 10.1111/j.1432-1033.1995.tb20546.x. [DOI] [PubMed] [Google Scholar]
- Imanaka T, Shiina Y, Takano T, Hashimato T, Osumi T. Insertion of the 70-kD peroxisomal membrane protein into peroxisomal membranes in vivo and in vitro. J Biol Chem. 1996;271:3706–3713. doi: 10.1074/jbc.271.7.3706. [DOI] [PubMed] [Google Scholar]
- Just WW, Diestelkötter P. Protein insertion into the peroxisomal membrane. Ann NY Acad Sci. 1996;804:60–75. doi: 10.1111/j.1749-6632.1996.tb18608.x. [DOI] [PubMed] [Google Scholar]
- Kaldi K, Diestelkötter P, Stenbeck G, Auerbach S, Jäkle U, Mägert H-J, Wieland FT, Just WW. Membrane topology of the 22-kD integral peroxisomal membrane. FEBS Lett. 1993;315:217–222. doi: 10.1016/0014-5793(93)81167-x. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazarow RB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–550. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Lombard-Platet G, Savary S, Sarde C-O, Mandel J-L, Chimini G. A close relative of the adrenoleukodystrophy (ALD) gene codes for a peroxisomal protein with a specific expression pattern. Proc Natl Acad Sci. 1996;93:1265–1269. doi: 10.1073/pnas.93.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Huertas E, Sandalio LM, del Río LA. Integral membrane polypeptides off pea leaf peroxisomes: characterization and response to plant stress. Plant Physiol Biochem. 1995;33:295–302. [Google Scholar]
- López-Huertas E, Sandalio LM, del Río LA. Superoxide generation in plant peroxisomal membranes: characterization of the redox proteins involved. Biochem Soc Trans. 1996;24:195S. doi: 10.1042/bst024195s. [DOI] [PubMed] [Google Scholar]
- López-Huertas E, Sandalio LM, Gomez M, del Río LA. Superoxide radical generation in peroxisomal membranes: evidence for the participation of the 18-kD integral membrane polypeptide. Free Radical Res. 1997;26:497–506. doi: 10.3109/10715769709097820. [DOI] [PubMed] [Google Scholar]
- Lüers G, Hashimoto T, Fahimi HD, Völkl A. Biogenesis of peroxisomes: isolation and characterization of two distinct peroxisomal populations from normal and regenerating rat liver. J Cell Biol. 1993;121:1271–2180. doi: 10.1083/jcb.121.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luster DG, Donaldson RP. Orientation of electron transport activities in the membrane of intact glyoxysomes isolated from castor bean endosperm. Plant Physiol. 1987;85:796–800. doi: 10.1104/pp.85.3.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeshima M, Beevers H. Purification and properties of glyoxysomal lipase from castor bean. Plant Physiol. 1985;79:489–493. doi: 10.1104/pp.79.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall PA, Krimkevich YI, Lark RH, Dyer JM, Veenhuis M, Goodman JM. PMP27 promotes peroxisomal proliferation. J Cell Biol. 1995;129:345–355. doi: 10.1083/jcb.129.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Northcote DH. The action of exogenous GA3 on isocitrate lyase mRNA in germinating castor bean seeds. Planta. 1982;154:174–183. doi: 10.1007/BF00387913. [DOI] [PubMed] [Google Scholar]
- McCammon MT, Dowds CA, Orth K, Moomaw CR, Slaughter CA, Goodman JM. Sorting of peroxisomal membrane protein PMP47 from Candida boidinii into peroxisomal membranes of Saccharomyces cerevisiae. J Biol Chem. 1994;265:20098–20105. [PubMed] [Google Scholar]
- Mosser J, Douar AM, Sarde CO, Kioschis P, Feil R, Moser H, Poustka AM, Mandel JL, Aubourg P. Putative X-linked adrenoleukodystrophy gene shares unexpected homology with ABC transporters. Nature. 1993;361:726–730. doi: 10.1038/361726a0. [DOI] [PubMed] [Google Scholar]
- Mullen RT, Trelease RN. Biogenesis and membrane properties of peroxisomes: does the boundary membrane serve and protect? Trends Plant Sci. 1996;1:389–394. [Google Scholar]
- Pryde JG, Phillips JH. Fractionation of membrane-proteins by temperature-induced phase-separation in Triton X-114: application to subcellular fractionation of the adrenal-medulla. Biochem J. 1986;233:525–533. doi: 10.1042/bj2330525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sandalio LM, del Río LA. Intraorganellar distribution of superoxide dismutase in plant peroxisomes (glyoxysomes and leaf peroxisomes) Plant Physiol. 1988;88:1215–1218. doi: 10.1104/pp.88.4.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struglics A, Fredlund KM, Rasmusson AG, Møller IM. The presence of a short redox chain in the membranes of intact potato tuber peroxisomes and the association of malate dehydrogenase with the peroxisome membrane. Physiol Plant. 1993;88:19–28. [Google Scholar]
- Tan X, Waterham HR, Veenhuis M, Cregg JM. The Hansenula polymorpha PER8 gene encodes a novel peroxisomal integral membrane protein involved in proliferation. J Cell Biol. 1995;128:307–319. doi: 10.1083/jcb.128.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolbert NE. Metabolic pathways in glyoxysomes and peroxisomes. Annu Rev Biochem. 1981;50:133–157. doi: 10.1146/annurev.bi.50.070181.001025. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Miura S, Fujiki Y. Restoration by a 35K membrane protein of peroxisome assembly in a peroxisome-deficient mammalian cell mutant. Nature. 1991;350:77–81. doi: 10.1038/350077a0. [DOI] [PubMed] [Google Scholar]
- Volokita M, Somerville C. The primary structure of spinach glycolate oxidase deduced from the DNA sequence of a cDNA clone. J Biol Chem. 1989;262:15825–15828. [PubMed] [Google Scholar]
- Waterham HR, Devries Y, Russell KA, Xie WQ, Veenhuis M, Cregg JM. The Pichia pastoris PER6 gene-product is a peroxisomal integral membrane protein essential for peroxisome biogenesis and has sequence similarity to the Zellweger-syndrome protein PAF-1. Mol Cell Biol. 1996;16:2527–2536. doi: 10.1128/mcb.16.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiher H, Noda T, Gray DA, Sharpe AH, Jaenisch R. Transgenic mouse model of kidney disease: insertional inactivation of ubiquitously expressed gene leads to nephrotic syndrome. Cell. 1990;62:425–434. doi: 10.1016/0092-8674(90)90008-3. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Mori H, Nishimura M. A novel isoenzyme of ascorbate peroxidase localized on glyoxysomal and leaf peroxisomal membranes in pumpkin. Plant Cell Physiol. 1995;36:1157–1162. doi: 10.1093/oxfordjournals.pcp.a078862. [DOI] [PubMed] [Google Scholar]
- Zwacka RM, Reuter A, Pfaff E, Moll J, Gorgas K, Karasawa M, Weiher H. The glomerulosclerosis gene Mpv17 encodes a peroxisomal protein producing reactive oxygen species. EMBO J. 1994;13:5129–5134. doi: 10.1002/j.1460-2075.1994.tb06842.x. [DOI] [PMC free article] [PubMed] [Google Scholar]