Abstract
Glutamate synthase (GOGAT) is a key enzyme in the assimilation of inorganic nitrogen in photosynthetic organisms. We found that, like higher plants, the facultative heterotrophic cyanobacterium Plectonema boryanum had ferredoxin (Fd)- and NADH-dependent GOGATs. The genes glsF, gltB, and gltD were cloned, and structural analyses and target mutageneses demonstrated that glsF encoded Fd-GOGAT and that gltB and gltD encoded the two subunits of NADH-GOGAT. All three mutants lacking one of the GOGAT genes were able to grow photosynthetically and heterotrophically. However, the Fd-GOGAT mutant exhibited a phenotype of marked nitrogen deficiency when grown under conditions of saturating illumination and CO2 supply. In these conditions the rate of the ammonia uptake from the culture medium was slower in the Fd-GOGAT mutant than in the wild type or in the NADH-GOGAT mutant, but no significant differences were found in the rate of the CO2 fixation-dependent O2 evolution among these strains. Our results suggest that, although both Fd- and NADH-GOGATs were operative in the cells growing in light, the contribution of Fd-GOGAT, which directly utilizes photoreducing power for the catalytic reaction, is essential for balancing photosynthetic nitrogen and carbon assimilation.
Inorganic nitrogen in the form of ammonia is assimilated into Gln and Glu through the combined actions of GS and GOGAT in all oxygenic photosynthetic organisms from cyanobacteria to higher plants (Lea et al., 1990; Flores and Herrero, 1994). GS catalyzes the ATP-dependent amination of Glu to yield Gln. GOGAT catalyzes the reductive transfer of the amide group of Gln to the keto position of 2-oxoglutarate to yield two molecules of Glu. The resulting Gln and Glu serve as nitrogen donors in the biosynthesis of various nitrogen-containing compounds (Lea et al., 1989). The GS/GOGAT pathway ultimately requires ATP and reducing power generated by photosynthesis and catabolism of carbohydrates and utilizes carbon skeletons provided from intermediates of the TCA cycle, together with the downstream metabolism of Gln and Glu. This pathway is thus involved in the integration of carbon and nitrogen assimilations.
In higher plants GOGAT occurs as two distinct forms, one that is Fd dependent (EC1.4.7.1) and one that is NADH dependent (EC 1.4.1.14); the two forms differ in their specificity for an electron donor and in their molecular architecture. cDNAs for both types of GOGAT have been cloned and characterized (Sakakibara et al., 1991; Gregerson et al., 1993), and they were found to be homologous to Escherichia coli NADPH-GOGAT (EC 1.4.1.13), which is composed of two different polypeptides, large and small subunits encoded by gltB and gltD, respectively (Oliver et al., 1987). Fd-GOGAT is an iron-sulfur flavoprotein composed of a single polypeptide that has a molecular mass of 160 kD and is similar to the large subunit of the E. coli enzyme (Sakakibara et al., 1991). NADH-GOGAT is also a single polypeptide but with two domains, the N-terminal, 160-kD and the C-terminal, 60-kD regions that are similar to the large and small subunits of the E. coli enzymes, respectively (Gregerson et al., 1993). Plant NADH-GOGAT contains the same prosthetic groups as Fd-GOGAT in the N-terminal domain and an additional iron-sulfur cluster and flavin in the C-terminal domain, which is likely to be involved in electron acceptance from NADH (Curti et al., 1995).
Only Fd-GOGAT has been reported thus far in the cyanobacteria (Rai et al., 1982; Marques et al., 1992; Navarro et al., 1995). In Synechococcus sp. PCC 6301 (Marques et al., 1992) and Synechocystis sp. PCC 6803 (Navarro et al., 1995), no pyridine nucleotide-dependent GOGAT activity was found. Two different genes for GOGAT were cloned in Synechocystis sp. PCC 6803, and both genes were found to encode Fd-dependent enzymes by using biochemical and genetic studies (Navarro et al., 1995). However, an open reading frame similar to the small subunit of E. coli NADPH-GOGAT was found in the recently published complete genomic sequence of Synechocystis sp. PCC 6803 (Kaneko et al., 1996), which suggests the presence of a gene for pyridine nucleotide-dependent GOGAT.
Different physiological roles are proposed for the two types of GOGAT in higher plants (Lam et al., 1996). Plant mutants defective in Fd-GOGAT have been identified for photorespiratory mutants in Arabidopsis (Somerville and Ogren, 1980) and barley (Kendall et al., 1986). In these mutants ammonia derived from photorespiration cannot be recaptured efficiently, and NADH-GOGAT expressed constitutively at a low level in leaves seems to have no compensatory function in photorespiration. Under conditions in which photorespiration was suppressed (high CO2 or low O2), the Fd-GOGAT mutants grew normally, which suggests that the assimilation of the primary ammonia derived from nitrate reduction can be achieved only by NADH-GOGAT. The second gene for Fd-GOGAT has been cloned in Arabidopsis and is expressed preferentially in roots. This isoenzyme of Fd-GOGAT was proposed to be mainly involved in nitrogen assimilation in roots (Coshigano et al., 1998). Recently, NADH-GOGAT was shown to be localized in the vascular parenchyma in rice, which is indicative of a role for mobilization of nitrogen compounds through the vascular system (Hayakawa et al., 1994). No mutants lacking NADH-GOGAT have been obtained; therefore, the real physiological role of NADH-GOGAT remains conjectural.
In the course of our study of GOGATs from the filamentous cyanobacterium Plectonema boryanum, which is able to grow under photoautotrophic and heterotrophic conditions, we detected significant activities of Fd- and NADH-GOGATs in contrast to reports for another species of cyanobacterium. The purpose of this work was to investigate the GOGAT genes and their mutants with the expectation of clarifying the structures and physiological functions of these two distinct GOGATs in a single species.
MATERIALS AND METHODS
Culture Conditions and Growth Measurements
Plectonema boryanum IAM-M101 strain dg5 (Fujita et al., 1996) and its derivative strains obtained in this study were cultivated in BG11 medium (Rippka et al., 1979) with 20 mm Hepes-NaOH, pH 7.5; 10 mm NH4Cl was added to the medium instead of 17.6 mm NaNO3 when necessary. For gene-disrupted mutants with a kanamycin-resistant cassette, the above media were supplemented with 15 μg/mL kanamycin. Liquid cultures were bubbled with 2% (v/v) CO2 in air under continuous illumination provided by fluorescent lamps; photon flux densities were controlled in the range of about 30 to 170 μE m−2 s−1 for experimental purposes. Growth was monitored by turbidity with a Klett-Summerson photoelectric calorimeter equipped with KS66 filter (640–700 nm; Manostat, New York) or by wet weight of cells. To weigh cells, 10-mL aliquots were removed from the bottles and filtered onto 0.45-μm membrane filters by centrifugation. For plate cultures the liquid media were added with 1.5% (w/v) agar.
Enzyme Extraction, Chromatography, and Assay
P. boryanum cells were harvested by centrifugation at 5,000g for 10 min and suspended in 50 mm potassium phosphate buffer, pH 7.5, 1 mm EDTA, and 10 mm β-mercaptoethanol (buffer A) at a ratio of 2.5 mL g−1. The suspended cells were thoroughly disrupted by sonication (model 350 sonifier, Branson Ultrasonics, Danbury, CT) with the addition of 1 mm PMSF and centrifuged at 100,000g for 1 h at 4°C. The supernatant was applied onto a Resource-Q column (1 mL) and eluted with a linear gradient of NaCl from 0 to 0.5 m in buffer A with a fast protein liquid chromatography system (Pharmacia).
The GOGAT activities were determined by Glu formation according to the method of Martin et al. (1982). For the enzyme assay the total cell extracts were pretreated by being passed through a Nap-10 column (Pharmacia) equilibrated with buffer A, and the fractions obtained in the ion chromatography were used directly. The assay mixture for Fd-GOGAT contained in a final volume of 180 μL: 10 μmol of potassium phosphate buffer (pH 7.4), 1 μmol of l-Glu, 1 μmol 2-oxoglutarate, 8 nmol or 2 nmol of P. boryanum Fd, and an appropriate amount of enzyme. The reaction was started by adding 20 μL of 100 mm sodium dithionite and, after incubation for an appropriate time at 30°C, was stopped by boiling for 3 min. For NADPH- and NADH-GOGATs, 20 μL of 10 mm NADPH and NADH, respectively, was added in place of sodium dithionite to the same assay mixture as above except Fd was omitted.
Construction and Screening of a Genomic Library of P. boryanum, Subcloning, and Sequencing
Genomic DNA was prepared from P. boryanum cells as described previously (Fujita et al., 1998) and was partially digested with Sau3AI. DNA fragments were size-fractionated, and those ranging from 9 to 22 kb were ligated into λ-DASH II vector (Stratagene). Recombinant phage DNAs were packaged with Gigapack II Gold packaging extract (Stratagene).
A pair of degenerated primers, 5′-CC(ACGT)CA(TC)CA(TC)GA(TC)AT(TC)TA-3′ and 5′-CC(ACGT)CC(AGCT)AT(AG)TA(CT)TC(AG)CA-3′, was used for amplification of a certain region of the GOGAT gene in P. boryanum. These sequences were derived from the two conserved regions among known GOGATs: Pro-1097 to Tyr-1102 and Cys-1494 to Gly-1500, according to the numbering in the amino acid sequence of maize Fd-GOGAT (Sakakibara et al., 1991). The total genomic DNA was digested with BglII and used as a template for PCR. PCR amplification was carried out with the following programs: one cycle of 95°C for 2 min, 55°C for 1 min, and 72°C for 2 min; and then 24 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min. The amplified DNA fragments were subcloned into pUC19 and sequenced. The fragment was labeled with a random-prime kit (Amersham) with [α-32P]dCTP and used as a probe for the screening of the genomic library. Insert DNAs of positive plaques were mapped, and appropriate restriction fragments were subcloned into pBluescript II SK+ (Stratagene). A set of nested deletion clones was made for DNA sequencing. The nucleotide sequences of these fragments were determined using the dye-terminator method with a DNA sequencer (model 373A, Applied Biosystems).
Genetyx software (version 8.0, Software DC, Tokyo, Japan) was used for computer analysis of nucleotide and deduced amino acid sequences. Phylogenetic trees were inferred using the UPGMA program included in this package.
Insertional Inactivation of Genes and Southern Analysis
To disrupt gltB, gltD, and glsF, the three GOGAT genes cloned in this study, both ends of a kanamycin-resistant (neo) gene were connected with the 5′ and 3′ regions of the GOGAT genes, which served as platforms for homologous recombination with the chromosomal region of each GOGAT gene. The neo gene cartridge was excised from pUCK121, pUCK122 (Fujita et al., 1996), and pYFC10 (Fujita et al., 1992) by digestion with SmaI, EcoRI, and XbaI, respectively. The cartridges with three different restriction sites were conveniently used for insertion into the GOGAT genes, the EcoRV site of gltB, the EcoRI site of gltD, and the NheI site of glsF (for details, see Figs. 2 and 3). Plasmids carrying these chimeric genes were linearized and introduced into P. boryanum cells by electroporation as described previously (Fujita et al., 1992), and transformants were selected on a kanamycin-containing BG11 plate. The gene disruptions were confirmed by Southern analysis according to a method described previously (Fujita et al., 1996).
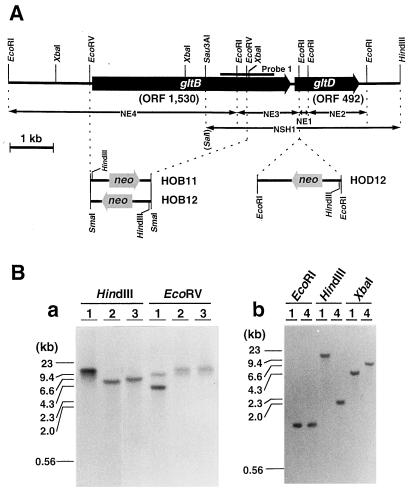
Figure 2.
Physical map of NADH-GOGAT gene (A) and Southern analysis of the genome of gltB- and gltD-disrupted mutants (B). A, The two open reading frames ORF 1530 and ORF 492, named gltB and gltD, respectively, are contained in a 9036-bp DNA fragment cloned from the P. boryanum genome. Various subcloned fragments (NE1–NE4 and NSH1) indicated with double-headed arrows were used for the sequence determination. The position of the amplified fragment (probe 1) by PCR using a pair of degenerated primers as described in Methods is shown with a heavy bar. The SalI site at the end of NSH1 was derived from a linker sequence of the cloning vector. The position and direction of the neo gene cassette inserted between two EcoRV sites of gltB or into EcoRI site of gltD are shown below the physical map. The accession number for this genomic sequence is D85230. B, Genomic DNAs (1 μg of each) from the wild type (lanes 1), gltB-disrupted mutants (HOB11, lanes 2; HOB12, lanes 3), and the gltD-disrupted mutant (HOD12, lanes 4) were digested with the restriction enzymes indicated, size-fractionated by agarose gel electrophoresis, and probed with NE4 (a) and NE1 (b). Sizes of marker DNA fragments are shown on the left of each panel.
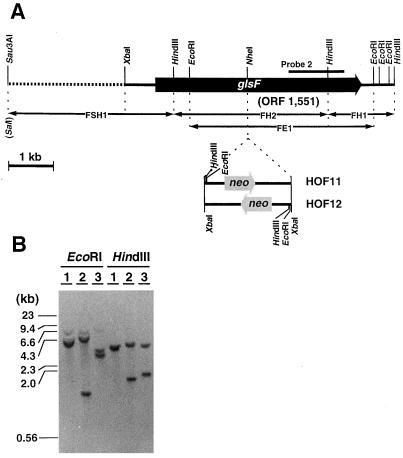
Figure 3.
Physical map of the Fd-GOGAT gene (A) and Southern analysis of the genome of the glsF-disrupted mutant (B). A, The open reading frame ORF 1551 (glsF) is contained in a 8.5-kb DNA region of the P. boryanum genome. The position of the amplified fragment (probe 2) by PCR is shown with a heavy bar. Various subcloned fragments (FH1, FH2, FSH1, and FE1) indicated with double-headed arrows were used for the sequence analysis, and a region of 6063 bp from XbaI to HindIII was determined. The region indicated by the dotted line was not sequenced. The SalI site at the end of FSH1 was derived from a linker sequence of the λ-cloning vector. The position and direction of neo gene cassette inserted into NheI site of glsF is shown below the physical map. The accession number for this genomic sequence is D85735. B, Genomic DNAs (1 μg of each) from wild type (lanes 1) and glsF disruptants (HOF11, lanes 2; HOF12, lanes 3) were digested separately with EcoRI and HindIII, size-fractionated by agarose gel electrophoresis, and probed with the FH2 fragment.
Determination of Phycobiliprotein and Chlorophyll Content
Contents of phycobiliproteins and chlorophyll were determined according to a previously published method (Tandeau de Marsac and Houmand, 1988). P. boryanum cells were suspended in 20 mm sodium acetate buffer, pH 5.5, and were disrupted by sonication. The extracts were treated with 1% (w/v) streptomycin for 30 min at 4°C, and the membrane fractions and insoluble materials were removed by centrifugation at 10,000g for 10 min. The amounts of phycocyanin and allophycocyanin in the supernatant were calculated from A620 and A650. Methanolic extracts of cells were measured at A665 to determine the chlorophyll content.
Measurements of O2 Evolution and Ammonia Uptake
Cells growing in a log phase were harvested and suspended in BG11 without NaNO3. After NaHCO3 was added to the cell suspension to a final concentration of 10 mm, the rates of oxygen evolution were determined with an oxygen electrode (DWI, Hansatech, King's Lynn, UK) at 30°C under light intensities from 10 to 210 μE m−2 s−1. For the measurement of ammonia uptake, the cells were cultured in a new BG11 medium containing 4 mm NH4Cl, and the concentrations of ammonia in the culture medium were monitored using an ammonia-selected ion electrode (5002A-10C, Horiba, Tokyo, Japan).
RESULTS
Two Different GOGATs in P. boryanum Cells
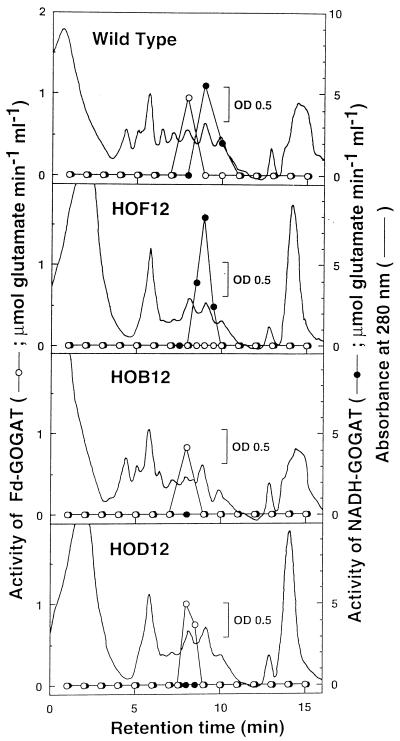
To examine which type(s) of GOGAT is present in P. boryanum, we measured GOGAT activities in the crude extract of the cyanobacterial cells using Fd, NADH, and NADPH as electron donors (Table I). Activities of Fd- and NADH-GOGATs were detected at comparable levels. No significant activity of NADPH-GOGAT was found. The total proteins in the cell extract were separated by an anion-exchange chromatography, and the activities of Fd- and NADH-GOGATs were eluted in different fractions (Fig. 1), which suggests that the two activities are attributable to distinct enzymes.
Table I.
Fd (40 μm)-, NADH (1 mm)-, and NADPH (1 mm)-dependent GOGAT activities in crude extracts of the control strain and GOGAT-disrupted strains of P. boryanum
| Strain | Fd | NADH | NADPH |
|---|---|---|---|
| nmol Glu min−1 mg−1 protein | |||
| YFD1 (control)a | 45 ± 15 | 42 ± 11 | <1b |
| HOF12 (glsF mutant) | <1 | 48 ± 10 | <1 |
| HOB12 (gltB mutant) | 44 ± 7 | <1 | <1 |
| HOD12 (gltD mutant) | 44 ± 5 | <1 | <1 |
Control strain with the kanamycin cartridge inserted in a neutral site of the genome.
<1, Negligible values (below the detection limit).
Figure 1.
Separation of Fd- and NADH-GOGAT by anion-exchange column chromatography. The crude extracts of wild-type, HOF12, HOB12, and HOD12 strains of P. boryanum were applied to a Resource-Q column (0.64 × 3 cm) and eluted with a linear gradient of NaCl at a flow rate of 1.0 mL/min as described in Methods. Elution profiles of Fd- and NADH-GOGAT were monitored by assay of their activities using 10 μm Fd and 1 mm NADH as electron donors, respectively.
Cloning and Targeted Mutagenesis of gltB and gltD Genes Encoding Two Subunits of NADH-GOGAT
A 1.2-kb DNA fragment (probe 1) was amplified from the genomic DNA of P. boryanum by PCR with a pair of mixed primers as described in Methods. The nucleotide sequence of the amplified fragment was homologous to the expected region of the GOGAT genes. A genomic library of P. boryanum was then screened by plaque hybridization with the 32P-labeled probe 1, and six positive clones (λ-N1–λ-N6) were selected. Insert DNAs of the clones were mapped by digestion with various restriction enzymes, followed by Southern analysis with the same probe. All clones overlapped each other and contained a common 1.4-kb EcoRI fragment hybridized with the probe (data not shown). Because the region of the genomic DNA covered by λ-N1 and λ-N4 was the longest among various combinations of the six clones, two clones were chosen for further analyses. Four EcoRI fragments (NE1–NE4) from λ-N4 and a SalI/HindIII fragment (NSH1) from λ-N1 were subcloned and sequenced (Fig. 2). The complete nucleotide sequence of a 9036-bp region from the EcoRI to the HindIII site was determined, and two open reading frames encoding polypeptides of 1530 amino acids (ORF 1, 530, gltB) and 492 amino acids (ORF 492, gltD) 106 bp apart were found. Two genes, gltB and gltD, which showed significant homologies to the genes for the large and small subunits of Escherichia coli NADPH-GOGAT (Oliver et al., 1987), respectively, seemed to encode the NADH-GOGAT of P. boryanum.
We tried to isolate gltB- and gltD-disrupted mutants to confirm the above assignment of the genes. As shown in Figure 2, gltB was disrupted by replacing its internal 3.6-kb EcoRV region with a neo cassette, and two mutant strains, HOB11 and HOB12, were selected. A gltD-disrupted mutant, HOD12, was also obtained by the insertion of the neo cassette into the EcoRI site located downstream of the putative initiation codon of gltD. The insertions of the cassette into the expected regions were confirmed by Southern analyses of genomic DNAs of each mutant (Fig. 2). Neither HOB12 nor HOD12 had significant NADH-GOGAT activity in their total cell extracts, whereas both mutants retained the original activity of Fd-GOGAT (Table I). Furthermore, when the total cell extracts of HOB12 and HOD12 were chromatographed, only the peak for Fd-GOGAT activity was observed in the fraction corresponding to that of the wild-type strain (Fig. 1). These results showed that two polypeptides encoded by gltB and gltD are the large and small subunits of NADH-GOGAT, respectively.
Further Cloning and Targeted Mutagenesis of the glsF Gene, Which Encodes Fd-GOGAT
As described above, the gltB-disrupted mutant retained the activity of Fd-GOGAT at the wild-type level, and this led us to clone further the gene encoding Fd-GOGAT. In the above PCR with the genomic DNA from the wild-type cells as a template, the 1.2-kb fragment corresponding to gltB was predominantly amplified. Therefore, we carried out PCR using the genomic DNA from HOB12, in which the annealing sites of the primers within gltB had been lost, as a template. A 1.2-kb DNA fragment (probe 2) was obtained and used to screen the genomic library. Three positive clones (λ-F1–λ-F3) were selected; all of them had common restriction fragments hybridizable with probe 2, a 4.1-kb EcoRI fragment (FE1), and 1.5- and 3.5-kb HindIII fragments (FH1 and FH2, respectively) (data not shown). Multiple sequence analysis of these subfragments and inspection of overlapping regions revealed the complete nucleotide sequence of a 6063-bp region from the XbaI to the HindIII site (Fig. 3). There was an open reading frame encoding 1551 amino acids (ORF 1551), which shared significant homology with maize glsF encoding Fd-GOGAT (Sakakibara et al., 1991). ORF 1551 was disrupted by insertion of the neo cassette into the NheI site within its coding region (Fig. 3). The disrupted mutant, HOF12, lost its Fd-GOGAT activity completely (Table I); only the peak for the activity of NADH-GOGAT was detected on a chromatogram of the total cell extract (Fig. 1). These results confirmed the identification of ORF 1551 as glsF, which encodes Fd-GOGAT.
Growth Experiments on NADH-GOGAT- and Fd-GOGAT-Deficient Mutant Cells
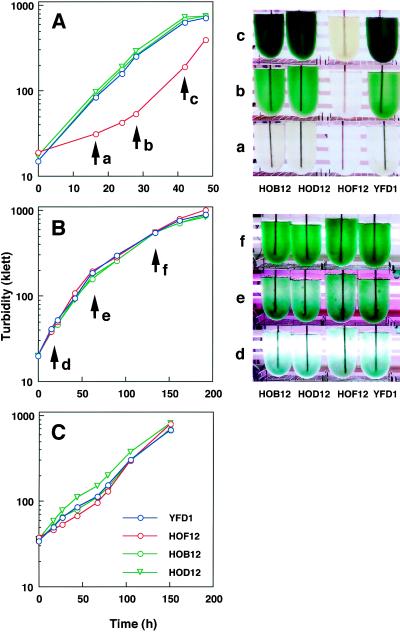
Growth analysis was undertaken using gltB-, gltD-, and glsF-disrupted mutants (HOB12, HOD12, and HOF12, respectively) and the control strain (YFD1). All were cultivated photoautotrophically in a medium containing 10 mm NH4Cl as a nitrogen source with aeration of 2% CO2 in air under two different light intensities of 25 and 160 μE m−2 s−1. Under the more intense light condition, the cells of HOF12 developed a yellow color markedly different from those of HOB12, HOD12, and YFD1, and their growth curve, based on turbidity of the culture, showed a sigmoidal shape, whereas those of HOB12, HOD12, and YFD1 showed normal hyperbolic shapes (Fig. 4A). The sigmoidal growth curve was due to the yellow coloration, because the actual growth rate of HOF12 (doubling time, 9 h), which was measured on a cell-fresh-weight basis, was not severely reduced compared with that of the control strain (doubling time, 7 h) (data not shown). When these strains were grown under the lower light intensity (Fig. 4B) or under conditons in which the 2% CO2 supplement was omitted from the aeration gas (Fig. 4C), all of them grew in the same manner (doubling times of 17 or 32 h, respectively). Therefore, the yellow coloration of HOF12 appeared when this mutant was grown under conditions for efficient carbon assimilation (high CO2 and high light). The same phenotype of HOF12 was also observed when the cells were grown in culture medium containing nitrate instead of ammonia (data not shown). Under heterotrophic conditions in the dark, no remarkable phenotype was observed in any mutant strain (data not shown).
Figure 4.
Growth curves and tonalities of the control (YFD1), Fd-GOGAT-deficient (HOF12), and NADH-GOGAT-deficient (HOB12 and HOD12) strains of P. boryanum under different photoautotrophic conditions. The four strains were cultivated in a modified BG11 medium containing 10 mm NH4Cl under high light (165 μE m−2 s−1) (A) and low light (25 μE m−2 s−1) (B) with aeration with a supplement of 2% CO2 or under high light (165 μE m−2 s−1) with aeration without the CO2 supplement (C). Growth was monitored by turbidity in Klett units. Culture bottles of the four strains were photographed at different growth stages as indicated by arrows labeled a, b, and c in A and d, e, and f in B.
Imbalance of Nitrogen and Carbon Assimilation in the Fd-GOGAT-Deficient Mutant
The results described above suggested that considerable changes in the contents of the cellular pigments and proteins were induced by the deficiency of Fd-GOGAT. This is clearly demonstrated in Table II. The contents of chlorophyll and phycobiliproteins were decreased in HOF12 by 2.5- to 3.3-fold from those in HOB12 and YFD1.
Table II.
Chlorophyll and phycobiliprotein content in the control strain and GOGAT disruptants of P. boryanum cultured under the high light condition (165 μE m−2 s−1) with aeration by 2% CO2
| Strain | Chlorophyll | Phycocyanin | Allophycocyanin |
|---|---|---|---|
| μg mg−1 protein | |||
| YFD1 (control) | 73 | 265 ± 20 | 275 ± 16 |
| HOF12 (glsF mutant) | 28 | 80 ± 7 | 111 ± 9 |
| HOB12 (gltB mutant) | 71 | 273 ± 9 | 285 ± 12 |
| HOD12 (gltD mutant) | 67 | 277 ± 14 | 289 ± 13 |
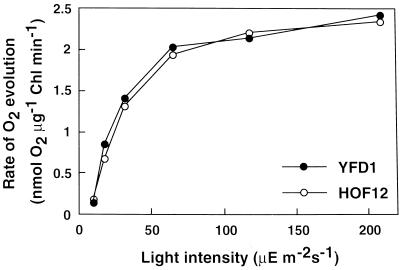
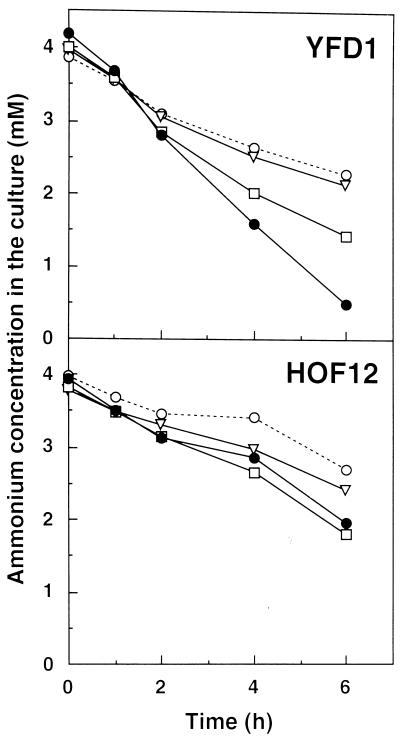
The capacities for carbon and nitrogen assimilation were compared between HOF12 and YFD1 by measuring the rates of the O2 evolution and ammonium uptake from the culture medium. As shown in Figure 5, the rates of O2 evolution increased similarly up to saturating levels of approximately 2.5 nmol μg−1 chlorophyll min−1 with increasing light intensities in both strains. On the other hand, the two strains differed significantly in their capacity for ammonia utilization (Fig. 6): The rate of ammonia uptake in the control strain increased in response to the increasing light intensities, whereas no such increase in response was found in HOF12. The increase in the rate of the ammonium uptake was dependent on the availability of CO2, because removal of the supplemental CO2 from the aeration gas resulted in a decrease in the rate.
Figure 5.
The rate of CO2-dependent oxygen evolution as a function of light intensity in the control (YFD1) and Fd-GOGAT-deficient (HOF12) strains of P. boryanum. The cyanobacterial cells were suspended in nitrate-free BG11 medium. After addition of 10 mm NaHCO3 to the medium, oxygen evolution was measured under increasing light intensities from 10 to 210 μE m−2 s−1.
Figure 6.
Time course of ammonia consumption in control (YFD1) and Fd-GOGAT-deficient (HOF12) strains of P. boryanum under different photoautotrophic conditions. Cyanobacterial cells at the exponential growth stage were inoculated into a BG11 medium containing 4 mm NH4Cl at a density of 200 Klett and cultured for an additional 6 h under one of four conditions: 165 μE m−2 s−1 (•), 75 μE m−2 s−1 (□), 25 μE m−2 s−1 with aeration with 2% CO2 supplement (▿), or 165 μE m−2 s−1 with aeration without the CO2 supplement (○). The concentrations of ammonium ion in the medium were measured at the indicated culture times.
DISCUSSION
We present definitive evidence that, like higher plants, the filamentous cyanobacterium P. boryanum has both Fd- and NADH-GOGATs. Fd-GOGAT is encoded by glsF and NADH-GOGAT is encoded by gltB and gltD. The overall structures of P. boryanum Fd- and NADH-GOGATs are similar to those of higher-plant GOGATs, except that P. boryanum NADH-GOGAT consists of two subunits with molecular masses of 160 and 60 kD. NADH-GOGATs from alfalfa (Gregerson et al., 1993) and rice (accession no. AB001916) are known to be single polypeptides with molecular masses of approximately 220 kD. The large and small subunits of P. boryanum NADH-GOGAT correspond to the N- and C-terminal domains of the higher-plant NADH-GOGAT, respectively.
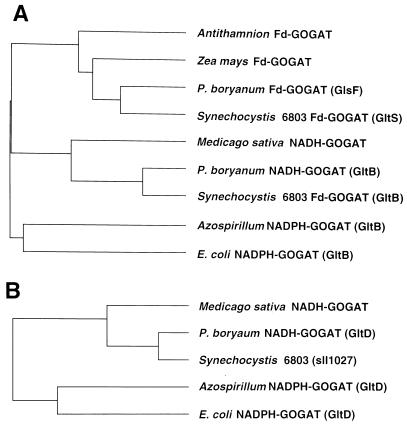
In a previous report of gltB and gltS, the GOGAT genes of the unicellular cyanobacterium Synechocystis sp. PCC 6803 (Navarro et al., 1995), the authors postulated that the two genes encoded distinct Fd-GOGATs because mutant strains with either gltB or gltS inactivated still retained a significant activity of Fd-GOGAT. However, the complete genomic sequence of this cyanobacterium contained an open reading frame, sll1027, which was homologous to gltD of P. boryanum NADH-GOGAT, at a site far from gltB (Kaneko et al., 1996). We constructed phylogenetic trees of GOGATs from representative organisms (Fig. 7). Synechocystis sp. PCC 6803 GltS and GltB proteins are closely related to P. boryanum GlsF protein in the branch for Fd-GOGAT and GltB protein in the branch of NADH-GOGAT, respectively. The protein encoded by sll1027 fits into the group of small subunits of NAD(P)H-GOGATs. Therefore, the previous assignment of gltB of Synechocystis sp. PCC6083 as the gene for Fd-GOGAT needs to be reexamined.
Figure 7.
Phylogenetic trees of Fd- and NAD(P)H-GOGATs. A, Tree for Fd-GOGAT and the large subunit (or the N-terminal domain) of NAD(P)H-GOGAT; B, tree for the small subunit (or the C-terminal domain) of NAD(P)H-GOGAT. The trees were constructed with the UPGMA program as described in Methods. Included sequences are from Antithamnion sp. Fd-GOGAT (Valentin et al., 1993), maize Fd-GOGAT (Sakakibara et al., 1991), P. boryanum Fd-GOGAT (GlsF) and NADH-GOGAT (GltB and GltD) (this study), alfalfa NADH-GOGAT (Gregerson et al., 1993), Synechocystis sp. PCC 6803 Fd-GOGATs (GltS and GltB) (Navarro et al., 1995), Synechocystis sp. PCC 6803 sll1027 (Kaneko et al., 1996), Azospirillum brasilense NADPH-GOGAT (GltB and GltD) (Pelanda et al., 1993), and E. coli NADPH-GOGAT (GltB and GltD) (Oliver et al., 1987).
For the first time to our knowledge, we were able to inactivate each of the P. boryanum Fd- and NADH-GOGAT genes in a single species, and the analysis of the resultant mutants has enabled us to examine the physiological roles of the two distinct GOGATs. In higher plants Fd-GOGAT is widely known to function in photorespiratory metabolism. In the Fd-GOGAT-deficient mutants of Arabidopsis (Somerville and Ogren, 1980) and barley (Kendall et al., 1986), NADH-GOGAT present at the normal wild-type level contributes to the primary nitrogen assimilation sufficient for growth of the plants under nonphotorespiratory conditions. In their metabolism of photorespiration, cyanobacteria differ from higher terrestrial plants. It is questionable whether the photorespiratory glycolate pathway operates actively in cyanobacteria (Colman, 1989), because cyanobacterial cells have the capacity to accumulate inorganic carbon inside the cell by actively taking up CO2 and HCO3− from their environment (Aizawa and Miyachi, 1986) and because Rubisco may be protected from O2 inhibition by sequestration in carboxysomes (McKay et al., 1992).
With these concerns in mind, we measured the growth, ammonia uptake, and CO2-dependent O2 evolution of the P. boryanum GOGAT mutants under different CO2 and light conditions. The remarkable phenotype of nitrogen deficiency was observed only in the Fd-GOGAT mutant grown under high CO2 and high light intensity (Fig. 4; Table II). This defective phenotype is caused by the inability to assimilate inorganic nitrogen and carbon in a coordinated fashion. In the control strain, the rate of ammonia uptake increased in response to increasing light intensity, and this light-driven uptake of ammonia was suppressed by removal of the supplemental CO2 in the culture medium (Fig. 6). The suppression of the nitrate utilization by low availability of CO2 was also previously reported with Synechococcus sp. strain PCC6301 (Flores et al., 1983). These data suggest that the nitrogen and carbon metabolisms interact with each other to balance these elements in cyanobacterial cells. In contrast, the Fd-GOGAT mutant shows no significant increase in the rate of the light-driven ammonia uptake under high CO2 (Fig. 6), whereas O2 evolution increases in a light-dependent manner similar to that of the control strain (Fig. 5).
Under the lower light condition, where photosynthetic carbon assimilation is limited, all strains grew similarly and the Fd-GOGAT mutant did not suffer from nitrogen deficiency (Fig. 4). It is noteworthy that when the Fd-GOGAT mutant was grown photomixotrophically with a supplement of 2% Glc in the culture medium, the mutant showed the nitrogen-deficiency phenotype even under the lower light condition (data not shown). These phenotypic characteristics indicate that NADH-GOGAT present at the wild-type level (Table I) is able to support nitrogen assimilation to a certain extent, but this contribution is not sufficient for the great demand for nitrogen assimilation by the cells growing rapidly with sufficient recent photosynthate of CO2 or exogeneously added carbohydrate.
We have not yet found any defective phenotype for the NADH-GOGAT mutant. The mutant grows normally in darkness and under conditions of anaerobic nitrogen fixation (data not shown). Therefore, nitrogen assimilation can be supported solely by Fd-GOGAT under conditions in which photoreducing power is not available, suggesting that a system for the donation of electrons from NADPH to Fd is operative, as it is in the roots of higher plants (Matsumura et al., 1997). We have not succeeded in generating a double mutant with both genes inactivated.
In conclusion, Fd- and NADH-GOGATs are not essential in P. boryanum and have some overlapping roles in primary nitrogen assimilation under growth conditions irrespective of photoautotrophy and heterotrophy. The contribution by Fd-GOGAT to the photoassimilation of nitrogen becomes dominant to that by NADH-GOGAT with increasing availability of carbohydrates. The Fd-GOGAT mutant thus becomes unable to maintain the cellular contents of nitrogenous compounds at the wild-type level in response to carbohydrate sufficiency. In Arabidopsis two Fd-GOGAT genes, Glu1 and Glu2, were recently identified, and the mutants lacking Glu1 fail to increase chlorophyll accumulation when exposed to high CO2 and inorganic nitrogen concentrations (Coshigano et al., 1998). It is probable that higher-plant Fd-GOGAT, which is essential for photorespiration, also plays a role in the primary nitrogen assimilation in leaves. The Fd-GOGAT mutant of P. boryanum with the phenotype of conditional nitrogen deficiency will be useful for further study of the in vivo roles of the GOGATs in balancing nitrogen demands in cells grown under various physiological conditions.
Abbreviations:
- GOGAT
Glu synthase
- GS
Gln synthetase
Footnotes
This work was supported in part by a Grant-in-Aid for Scientific Research (no. 10640630) and by a Grant-in-Aid for Research on Priority Areas (nos. 09274101 and 09274103) from the Ministry of Education, Science and Culture of Japan.
LITERATURE CITED
- Aizawa K, Miyachi S. Carbonic anhydrase and CO2 concentrating mechanism in microalga and cyanobacteria. FEMS Microbiol Rev. 1986;39:215–233. [Google Scholar]
- Colman B. Photosynthetic carbon assimilation and the suppression of photorespiration in the cyanobacteria. Aquat Bot. 1989;34:211–231. [Google Scholar]
- Coshigano KT, Melo-Oliveita R, Lim J, Coruzzi M. Arabidopsis gls mutants and distinct Fd-GOGAT genes: implications for photorespiration and primary nitrogen assimilation. Plant Cell. 1998;10:741–752. doi: 10.1105/tpc.10.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curti B, Vanoni MA, Verzotti E, Zanetti G. Glutamate synthase: a complex iron-sulphur flavoprotein. Biochem Soc Trans. 1995;24:95–99. doi: 10.1042/bst0240095. [DOI] [PubMed] [Google Scholar]
- Flores E, Herrero A. Assimilatory nitrogen metabolism and its regulation. In: Bryant DA, editor. The Molecular Biology of Cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 487–517. [Google Scholar]
- Flores E, Romero JM, Guerrero MG, Losada M. Biochim Biophys Acta. 1983;725:529–532. [Google Scholar]
- Fujita Y, Takagi H, Hase T. Plant Cell Physiol. 1996;37:313–323. doi: 10.1093/oxfordjournals.pcp.a028948. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Takagi H, Hase T. Cloning of the gene encoding a protochlorophyllide reductase: the physiological significance of the co-existence of light-dependent and -independent protochlorophyllide reduction systems in the cyanobacterium Plectonema boryanum. Plant Cell Physiol. 1998;39:177–185. doi: 10.1093/oxfordjournals.pcp.a029355. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Takahashi Y, Chuganji M, Matsubara H. The nifH-like (frxC) gene is involved in the biosynthesis of chlorophyll in the filamentous cyanobacterium Plectonema boryanum. Plant Cell Physiol. 1992;33:81–92. [PubMed] [Google Scholar]
- Gregerson RG, Miller SS, Twary SN, Gantt JS, Vance CP. Molecular characterization of NADH-dependent glutamate synthase from alfalfa nodule. Plant Cell. 1993;5:215–226. doi: 10.1105/tpc.5.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T, Nakamura T, Hattori F, Mae T, Ojima K, Yamaya T. Cellular localization of NADH-dependent glutamate synthase protein in vascular bundles of unexpanded leaf blades and young grains in rice plants. Planta. 1994;193:455–460. [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sakamoto S and others. Sequence analysis of the genome of the unicellular cyanobacterium Synechosystis sp. strain PCC6893. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Kendall AC, Wallsgrove RM, Hall NP, Turner JC, Lea PJ. Carbon and nitrogen metabolism in barley (Hordeum vulgare) mutants lacking ferredoxin dependent glutamate synthase activity. Planta. 1986;168:316–323. doi: 10.1007/BF00392355. [DOI] [PubMed] [Google Scholar]
- Lam H-M, Coschigano KT, Oliveira IC, Melo-Oliveira R, Coruzzi GM. The molecular-genetics of nitrogen assimilation into amino acids in higher plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:569–593. doi: 10.1146/annurev.arplant.47.1.569. [DOI] [PubMed] [Google Scholar]
- Lea PJ, Blackwell RD, Murray AJS, Joy KW (1989) The use of mutants lacking glutamine synthetase and glutamate synthase to study their role in plant nitrogen metabolism. In JE Poulton, JT Romeo, EE Conn, eds, Recent Advances in Phytochemistry. Plenum Press, New York, pp 157–189
- Lea PJ, Robinson SA, Stewart GR. The enzymology and metabolism of glutamine, glutamate, and asparagine. In: Miflin BJ, Lea PJ, editors. The Biochemistry of Plants, Vol 16. San Diego, CA: Academic Press; 1990. pp. 121–159. [Google Scholar]
- Marqués S, Florencio FJ, Candau P (1992) Purification and characterization of the ferredoxin-glutamate synthase from the unicellular cyanobacterium Synechococcus sp. PCC 6301. Eur J Biochem 296: 69–77 [DOI] [PubMed]
- Martin F, Suzuki A, Hirel B. A new high-performance liquid chromatography assay for glutamine synthetase and glutamate synthase in plant tissues. Anal Biochem. 1982;125:24–29. doi: 10.1016/0003-2697(82)90378-5. [DOI] [PubMed] [Google Scholar]
- Matsumura T, Sakakibara H, Nakano R, Kimata Y, Sugiyama T, Hase T. A nitrate-inducible ferredoxin in maize roots. Genomic organization and differential expression of two nonphotosynthetic ferredoxin isoproteins. Plant Physiol. 1997;114:653–660. doi: 10.1104/pp.114.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay RML, Gibbs SP, Espie GS. Effect of dissolved inorganic carbon on the expression of carboxysomes, localization of Rubisco and the mode of inorganic carbon transport in cells of the cyanobacterium Synechococcus UTEX 625. Arch Microbiol. 1992;159:21–29. [Google Scholar]
- Navarro F, Chávez S, Candau P, Florencio FJ (1995) Existence of two ferredoxin-glutamate synthases in the cyanobacterium Synechocystis sp. PCC 6803: isolation and insertional inactivation of gltB and gltS genes. Plant Mol Biol 27: 753–767 [DOI] [PubMed]
- Oliver G, Gosset G, Sanchez-Pescador R, Lozoya E, Ku LM, Flores N, Becerril B, Valle F, Boliver F. Determination of the nucleotide sequence for the glutamate synthase structural genes of Escherichia coli K12. Gene. 1987;60:1–11. doi: 10.1016/0378-1119(87)90207-1. [DOI] [PubMed] [Google Scholar]
- Pelanda R, Vanoni MA, Perego M, Piubelli L, Galizzi A, Curti B, Zanetti G. Glutamate synthase genes of the diazotroph Azospillum brasilense: cloning, sequencing, and analysis of functional domains. J Biol Chem. 1993;268:3099–3106. [PubMed] [Google Scholar]
- Rai AN, Rowell P, Stewart WDP. Glutamate synthase activity of heterocysts and vegetative cells of the cyanobacterium Anabaena variabilis Kutz. J Gen Microbiol. 1982;128:2203–2205. [Google Scholar]
- Ripkka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. Genetic assignments, strain histories, and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- Sakakibara H, Watanabe M, Hase T, Sugiyama T. Molecular cloning and characterization of complementary DNA encoding for ferredoxin-dependent glutamate synthase in maize leaf. J Biol Chem. 1991;266:2028–2035. [PubMed] [Google Scholar]
- Somerville CR, Ogren WL. Inhibition of photosynthesis in Arabidopsis mutants lacking leaf glutamate synthase activity. Nature. 1980;286:257–259. [Google Scholar]
- Tandeau de Marsac N, Houmard J. Complementary chromatic adaptation: physiological conditions and action spectra. Methods Enzymol. 1988;167:318–328. [Google Scholar]
- Valentin K, Kostrzewa M, Zetsche K. Glutamate synthase is plastid-encoded in a red alga: implications for the evolution of glutamate synthases. Plant Mol Biol. 1993;23:77–85. doi: 10.1007/BF00021421. [DOI] [PubMed] [Google Scholar]