Abstract
Nonalcoholic fatty liver disease (NAFLD), a metabolic predisposition for development of hepatocellular carcinoma (HCC), represents a disease spectrum ranging from steatosis to steatohepatitis to cirrhosis. Acox1, a rate-limiting enzyme in peroxisomal fatty acid β-oxidation, regulates metabolism, spontaneous hepatic steatosis, and hepatocellular damage over time. However, it is unknown whether Acox1 modulates inflammation relevant to NAFLD pathogenesis or if Acox1-associated metabolic and inflammatory derangements uncover and accelerate potential for NAFLD progression. Here, we show that mice with a point mutation in Acox1 (Acox1Lampe1) exhibited altered cellular metabolism, modified T cell polarization, and exacerbated immune cell inflammatory potential. Further, in context of a brief obesogenic diet stress, NAFLD progression associated with Acox1 mutation resulted in significantly accelerated and exacerbated hepatocellular damage via induction of profound histological changes in hepatocytes, hepatic inflammation, and robust upregulation of gene expression associated with HCC development. Collectively, these data demonstrate that β-oxidation links metabolism and immune responsiveness and that a better understanding of peroxisomal β-oxidation may allow for discovery of mechanisms central for NAFLD progression.
Keywords: Inflammation, Metabolism
Keywords: Cytokines, Fatty acid oxidation, Obesity
Peroxisomal beta-oxidation links metabolism, immune cells polarization, and inflammation in NAFLD pathogenesis.
Introduction
Nonalcoholic fatty liver disease (NAFLD), the world’s most common chronic liver disorder, represents a spectrum of liver diseases ranging from benign hepatic lipid accumulation (nonalcoholic steatosis; NAFL) to inflammation and fibrosis (nonalcoholic steatohepatitis; NASH) and, potentially, hepatocellular carcinoma (HCC) (1–5). However, despite NAFLD’s clinical and public health significance, the mechanisms underlying its immunopathogenesis, including those leading to HCC, remain underdefined. Thus, the discovery of novel mechanisms regulating NAFLD progression may provide novel therapeutic targets for the inhibition of NASH progression to HCC and ultimately reduce NAFLD prevalence.
Innate and adaptive immune responses are central to both NAFLD and HCC pathogenesis (4, 6). Among innate immune activators, TLR4 signaling, activated in response to endotoxin (LPS) sensing, has been associated with NAFLD progression in mice (7–9) and correlated with NAFLD severity in humans (10). Obesity-associated increased intestinal permeability and systemic endotoxemia (11) have been linked with NAFLD pathogenesis (12). Increased TLR4 activation augments hepatic and systemic cytokine production, including cytokines known to induce the differentiation and activation of Th17 cells (13). Notably, IL-17 axis activation is critical for NAFLD to NASH transition (9, 14–18).
In addition to inflammation, hepatic cellular metabolism plays a critical role in NAFLD progression. Modulation of mitochondrial function (19) and lipolytic and lipogenic pathways in hepatocytes have profound effects on the development of steatosis (20). In fact, altered mitochondrial activity, or increased de novo lipogenesis in the liver, in parallel with steatosis, promotes cellular stress, lipid peroxidation, and induction of inflammation (21). However, while inflammatory and metabolic pathways are often causally linked with NAFLD, the pathways essential for the interaction between these biological processes in NAFLD progression are not well defined.
Acyl-coenzyme A oxidase (Acox1), a rate-limiting enzyme in the peroxisomal β-oxidation pathway, is responsible for catabolism of very long–chain fatty acids. Saturated long-chain fatty acids are known to activate inflammatory and innate immune responses in immune cells (22–24). Altered Acox1 function is associated with spontaneous liver damage in humans (25–27). In fact, patients with pseudoneonatal adrenoleukodystrophy, a rare inflammatory neurodegenerative disease caused by Acox1 deficiency, exhibit hepatomegaly due to impairment of peroxisomal fatty acid β-oxidation of very long–chain fatty acids (28). Similarly, Acox1-deficient and Acox1 mutant mice exhibit spontaneous steatosis and steatohepatitis, augmented liver regeneration, and spontaneous development of HCC around 1 year of age (26, 29). These findings suggest that Acox1, in addition to its role in metabolism, may also play an important role in regulation of inflammation via peroxisomal β-oxidation — something that has not been previously interrogated, to our knowledge. Thus, we hypothesized that Acox1-dependent modulation of metabolic function and inflammatory vigor in obesity would augment induction of proinflammatory immune axes and allow for accelerated and exacerbated NAFLD pathogenesis.
Results
Acox1-dependent peroxisomal β-oxidation regulates inflammatory cytokine production and T cell polarization.
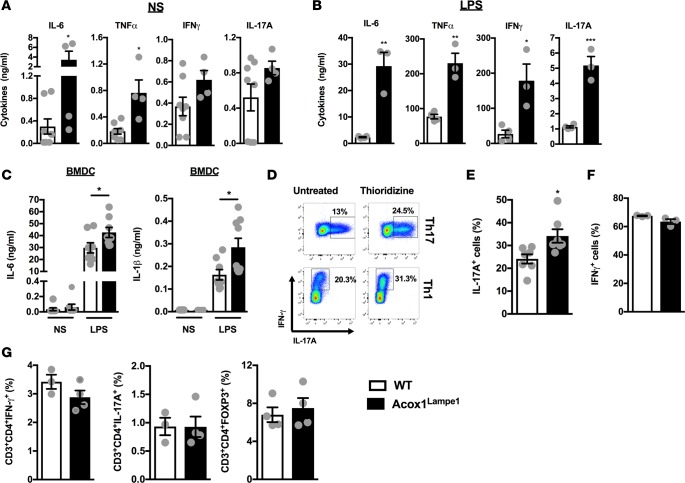
The role of Acox1 in modulation of systemic inflammatory cascades in vivo, at base line, and after inflammatory challenge has not been previously defined. Hence, we quantified in vivo basal and LPS-induced systemic cytokine production in WT and Acox1Lampe1 mice, which have a splice donor site mutation in the Acox1 gene between exons 12 and 13, resulting in partial deletion of the C-terminal acyl-CoA oxidase domain (29). Acox1Lampe1 mice, compared with WT controls, exhibited significantly increased basal serum IL-6 and TNF-α levels and trends toward increased IFN-γ and IL-17A levels (Figure 1A). Following LPS challenge, in addition to elevated IL-6 and TNF-α, the serum levels of IFN-γ and IL-17A were also significantly elevated in Acox1Lampe1 mice (Figure 1B). LPS does not directly induce IL-17 production; rather, it induces production of cytokines (e.g., IL-6, IL-1β, and IL-12), which promote IL-17 production via polarization of Th17 cells (30) and NK cell and NKT cell activation (31–33). Accordingly, Acox1Lampe1 BM-derived DCs exhibited significantly increased production of IL-6 and IL-1β, potential IL-17–inducing cytokines, after LPS stimulation (Figure 1C).
Figure 1. Acox1Lampe1 mutation promotes proinflammatory immune responsiveness.
(A and B) Twelve-week-old, chow diet–fed, Acox1Lampe1 mice and WT littermate controls were challenged i.p. with biotinylated capture Ab (20 μg/mouse) 3 hours prior to i.v. saline (NS) or LPS challenge (25 μg/mouse; ultrapure E. coli 0111:B4). Twenty-four hours later, serum was collected, and IL-6, TNF-α, IFN-γ, and IL-17A levels were quantified by IVCCA ELISA. (C) Supernatant IL-6 and IL-1β levels from BM-derived DCs (BMDC), of chow diet–fed, Acox1Lampe1 mice and WT littermate controls stimulated for 24 hours with saline (NS) or LPS (100 ng/ml). (D) Splenic naive CD4+ T cells from chow diet–fed WT mice, isolated by negative selection using magnetic beads, were cultured under Th17 and Th1 polarizing conditions with or without peroxisomal β-oxidation inhibitor (thioridizine, 1 μM) and the frequency of IL-17A– and IFN-γexpressing cells were quantified by flow cytometry. (E and F) Splenic naive CD4+ T cells from 12-week-old, chow diet–fed, Acox1Lampe1 mice and WT littermate controls, isolated by negative selection using magnetic beads, were cultured under Th17 and Th1 polarizing conditions, and the frequency of IL-17A– and IFN-γ–expressing cells was quantified by flow cytometry. (G) Frequency of total splenic Th1 (CD3+CD4+IFN-γ+IL-17A–), Th17 (CD3+CD4+IL-17A+IFN-γ–), and Treg (CD3+CD4+FOXP3+) in chow diet–fed Acox1Lampe1 mice and WT littermate controls. Data represent means ± SEM. (A, B, and E–G) Unpaired Student’s t test; *P < 0.05, **P < 0.01. (C) One-way ANOVA followed by Tukey’s correction; *P < 0.05. White bars denote WT mice; black bars denote Acox1Lampe1 mice. (A, B and D) Data combined from 2 independent experiments, n = 3–8/condition. (C and E) Data combined from 2 independent experiments, n = 7/condition. (F and G) A single experiment, n = 3–4/condition.
To test whether peroxisomal β-oxidation has a direct effect on CD4+ T cell polarization, naive WT CD4+ T cells were treated with thioridazine, a chemical inhibitor of peroxisomal β-oxidation, prior to being cultured under Th1 or Th17 polarizing conditions and stimulated with PMA/ionomycin. Notably, thioridazine treatment in vitro promoted both Th17 and Th1 polarization (Figure 1D) and reduced Treg polarization (data not shown). Hence, the impact of Acox1Lampe1 mutation on naive CD4+ T polarization was examined next. Naive CD4+ T cells from Acox1Lampe1 mice, cultured under Th17 and Th1 polarizing conditions, exhibited increased skewing toward the Th17 but not the Th1 phenotype compared with naive WT CD4+ T cells (Figure 1, E and F). However, at baseline, Acox1Lampe1 mice had similar splenic Th17, Th1, and Treg numbers (Figure 1G). These findings suggest that Acox1Lampe1 mutation does not alter basal, splenic T cell composition or total numbers — rather, it alters the T cell response to a stimulus. Overall, our findings suggest that Acox1 regulates both innate and adaptive immune responsiveness and immune cell inflammatory vigor.
Acox1-dependent peroxisomal β-oxidation regulates whole body metabolism and brown adipose tissue activity.
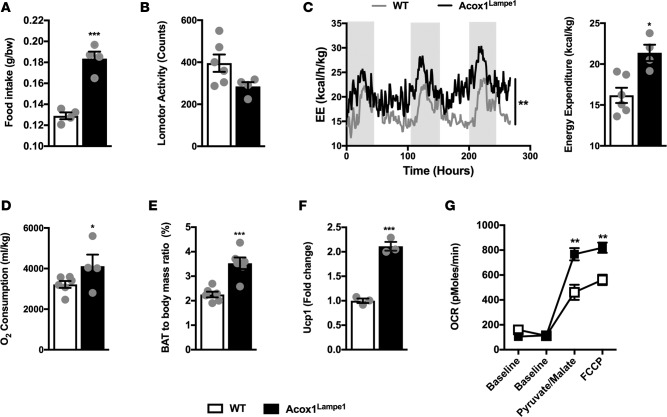
While the Acox1Lampe1 mutation is associated with protection from weight gain (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.93626DS1), whether Acox1Lampe1 mutation modulates whole body metabolism has not been defined (29). Notably, despite increased food intake (Figure 2A) and similar locomotor activity (Figure 2B), Acox1Lampe1 mice displayed significantly increased energy expenditure (Figure 2C) and oxygen consumption (Figure 2D) compared with WT controls. These results suggested a potential increase in brown adipose tissue–driven (BAT-driven) metabolic activity. In fact, subsequent analyses revealed that Acox1Lampe1 mice exhibited a significant increase in BAT/body mass ratio (Figure 2E) and expression of uncoupling protein 1 (Ucp1), a key mediator of energy expenditure (34) (Figure 2F). Further, this correlated with increased mitochondrial respiration in the BAT (Figure 2G). Specifically, mitochondria from AcoxLampe1 BAT displayed increased oxygen consumption rate (OCR) after supplementation with pyruvate and malate and after uncoupling of mitochondrial respiration using carbonyl cyanide p-trifluoromethoxyphenylhydrazone (FCCP) (Figure 2G). Of interest, during periods of increased energy expenditure, triglycerides (TGs) stored in the white adipose tissue (WAT) are degraded and released as fatty acids, via the process of lipolysis, to be utilized by BAT (35). Congruent with this observation, WAT mRNA expression of Lipe, Pparg, and Pnpla2, genes associated with lipolysis, was increased in Acox1Lampe1 mice compared with WT controls (Supplemental Figure 1B). In sum, these differences suggest that Acox1Lampe1 mutation augments whole body energy expenditure and BAT activity.
Figure 2. Acox1Lampe1 mutation modulates whole body metabolism and brown adipose tissue activity.
(A–D) Analysis of factors contributing to differential weight gain. Twelve-week-old, chow diet–fed, Acox1Lampe1 mice and WT littermate controls were placed in metabolic chambers, and (A) food intake, (B) total daily locomotor activity, (C) total daily energy expenditure (EE), and (D) total daily oxygen consumption were quantified. (E–G) Analysis of brown adipose tissue (BAT) function. Twelve-week-old, chow diet–fed, Acox1Lampe1 mice and WT littermate controls were utilized for analysis of (E) BAT/body mass ratio, (F) BAT Ucp1 mRNA expression levels, and (G) BAT mitochondrial oxygen consumption rate (OCR). Data represent means ± SEM. (A–G) Unpaired Student’s t test; *P < 0.05, **P < 0.01, ***P < 0.001. (A–F) White bars denote WT mice; black bars denote Acox1Lampe1 mice. (C) Shaded regions denote night cycle; clear regions denote day cycle. (G) White squares denote WT mice; black squares denote Acox1Lampe1 mice. (A) Representative of 3 individual experiments, n = 4/condition. (B–G) Data combined from 2 independent experiments, n = 3-6/condition.
Acox1-dependent peroxisomal β-oxidation regulates hepatic mitochondrial activity and inflammation.
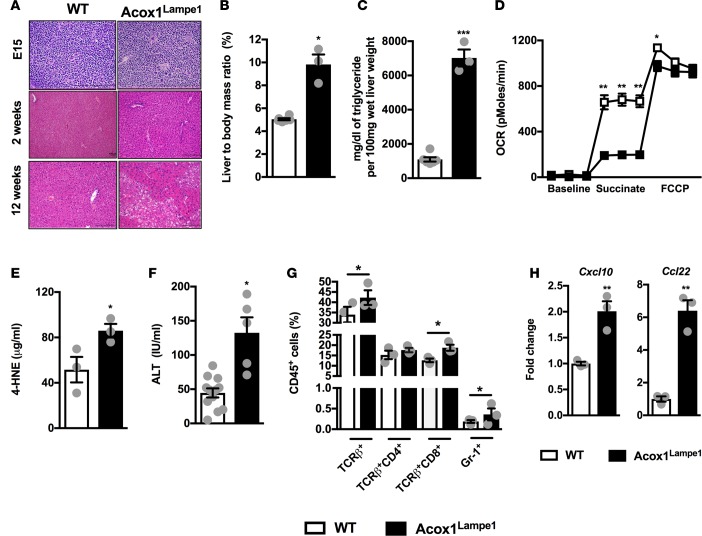
Given that the liver plays a major role in numerous metabolic processes and that hepatic mitochondrial function is linked with hepatocellular damage (19), we next evaluated whether hepatic metabolism was altered in Acox1Lampe1 mice. Initial analysis of the liver indicated that Acox1Lampe1 mice displayed normal embryonic and neonatal liver histology, with evidence of hepatic steatosis at 12 weeks of age (Figure 3A). At this time, Acox1Lampe1 mice also exhibited an increased liver/body mass ratio and augmented hepatic TG accumulation (Figure 3, B and C). At the cellular level, unlike in BAT, mitochondria from Acox1Lampe1 hepatocytes displayed decreased OCR in response to succinate and FCCP — both responses indicative of impaired mitochondrial function (Figure 3D). This reduction in mitochondrial function inversely correlated with increased hepatic lipid peroxidation and levels of hepatocellular damage, as measured by 4-hydroxynonenal (4-HNE) and alanine transaminase (ALT) levels, respectively (Figure 3, E and F).
Figure 3. Acox1Lampe1 mutation alters hepatic mitochondrial activity, hepatic inflammation, and hepatocellular damage.
(A) Representative liver histology (H&E staining; 20×) of chow diet–fed Acox1Lampe1 mice and WT littermate controls. E15; 2 weeks of age; and 12 weeks of age were analyzed. Representative hepatic H&E staining. (B–H) Characterization of liver phenotype and function in 12-week-old, chow diet–fed, Acox1Lampe1 mice and WT littermate controls. (B) Liver/body mass ratio. (C) Hepatic triglyceride (TG) levels. (D) Hepatic mitochondrial OCR. (E) Hepatic 4-hydroxynonenal (4-HNE) levels. (F) Serum alanine transaminase (ALT) levels. (G) Total hepatic immune (CD45+) cell infiltration determined by flow cytometry. (H) Hepatic chemokine mRNA expression of Cxcl10 and Ccl22. Data represent means ± SEM. (B–H) Unpaired Student’s t test; *P < 0.05, **P < 0.01, ***P < 0.001. White bars denote WT mice; black bars denote Acox1Lampe1 mice. (B–D) Representative of 3 individual experiments, n = 3/condition. (E, G, H) A single experiment, n = 3/condition. (F) Data combined from 2 independent experiments, n = 5–12/condition.
Congruent with exacerbated hepatocellular damage and dysregulated hepatic mitochondrial metabolism, Acox1Lampe1 mice, compared with WT controls, had increased hepatic T cell and neutrophil infiltration (Figure 3G) that correlated with increased hepatic mRNA expression of chemokines Cxcl10 and Ccl22 (Figure 3H), previously associated with T cell infiltration and IL-17 axis activation in NAFLD (16). Of note, hepatic T cells from Acox1Lampe1 mice had augmented capacity to produce IL-17A but not IFN-γ (Supplemental Figure 2, A–D). In contrast, Acox1Lampe1 mice displayed a significant decrease in the presence of hepatic Tregs (Supplemental Figure 2E). Notably, reduced hepatic Treg numbers and polarization was not observed in the spleen (Figure 1G and data not shown), possibly suggesting that the Acox1-driven hepatic milieu is critical for skewed hepatic T cell infiltration.
Short-term obesogenic-diet challenge accelerates and exacerbates Acox1-dependent peroxisomal β-oxidation–driven hepatocellular damage and systemic inflammation.
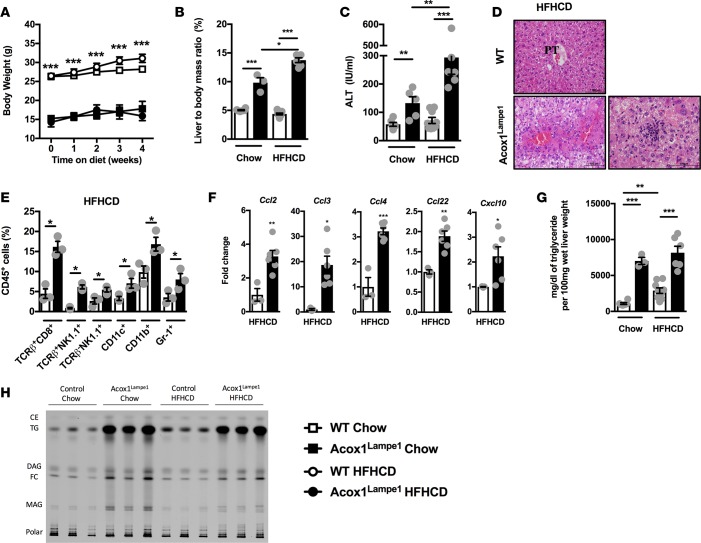
At baseline, Acox1Lampe1 mice exhibited significantly altered energy expenditure, hepatic metabolism and systemic and hepatic inflammation. All of these parameters are also thought to play a significant role in obesogenic diet–induced NAFLD pathogenesis. When fed a short–term (4 weeks) high-fat high-carbohydrate diet (HFHCD), Acox1Lampe1 mice exhibited increased food consumption in proportion to their body weight (Supplemental Figure 3A) but were protected from weight gain as compared with WT controls (Figure 4A). As expected, the lack of weight gain in Acox1Lampe1 mice correlated with significantly decreased adiposity (Supplemental Figure 3, B–D). However, despite the protection from HFHCD-driven adiposity and weight gain, Acox1Lampe1 mice fed HFHCD had significantly increased liver/body mass ratio and hepatocellular damage compared with WT controls (Figure 4, B and C). Importantly, short-term HFHCD challenge, compared with chow diet, exacerbated liver weight and hepatocellular damage only in Acox1Lampe1 mice — not WT controls (Figure 4, B and C). Further, in Acox1Lampe1 livers, secondary insult with HFHCD accelerated hepatocellular injury, which correlated with augmented steatosis, lobular inflammation (Figure 4D), and expression of genes associated with increase ER stress (Supplemental Figure 4).
Figure 4. Short-term obesogenic-diet challenge accelerates and exacerbates Acox1Lampe1 mutation–driven hepatocellular damage and systemic inflammation.
Eight-week-old Acox1Lampe1 mice and WT littermate controls were fed a HFHCD or chow diet for 4 weeks. (A) Weight gain. (B) Liver/body mass ratio. (C) Serum ALT levels. (D) Representative liver histology (H&E staining; 20x). (E) Total hepatic immune (CD45+) cell infiltration determined by flow cytometry. (F) Hepatic chemokine mRNA expression of Ccl2, Ccl3, Ccl4, Ccl22, and Cxcl10. (G) Hepatic TG levels. (H) Hepatic lipid content measured by thin layer chromatography and visualized by primuline staining. Cholesterol ester (CE), triglyceride (TG), diacylglycerol (DAG), free cholesterol (FC), monoacylglycerol (MAG), phospholipids (polar). Data represent means ± SEM. (A, E and F) Unpaired Student’s t test; *P < 0.05, **P < 0.01, ***P < 0.01. (B, C, and G) One-way ANOVA followed by Tukey’s correction; *P < 0.05, **P < 0.01, ***P < 0.01. (A) White squares denote WT mice fed chow diet; white circles denote WT mice fed HFHCD; black squares denote Acox1Lampe1 mice fed chow diet; black circles denote Acox1Lampe1 mice fed HFHCD. (B, C, and E–G) White bars denote WT mice fed HFHCD; black bars denote Acox1Lampe1 mice fed HFHCD. (A) Representative of 2 individual experiments, n = 3/condition. (B, C, F and G) Data combined from 2 independent experiments, n = 6–9/condition. (E and H) A single experiment, n = 3/condition.
We next examined if the increased hepatocellular damage and histological changes observed in HFHCD-fed Acox1Lampe1 mice were also associated with augmented hepatic inflammation. Short-term HFHCD feeding resulted in a robust increase in numbers of hepatic CD8+ T cells, NKT cells, NK cells, DCs, myeloid cells, and neutrophils in Acox1Lampe1 mice compared with WT littermate controls (Figure 4E). Increased hepatic immune cell infiltration further correlated with exacerbated hepatic expression of macrophage and T cell–recruiting chemokines, Ccl2, Ccl3, Ccl4, Ccl22, and Cxcl10 (Figure 4F), and ezymes associated with ROS production (Supplemental Figure 5), known to play a role in NAFLD pathogenesis (36).
As Acox1 plays an important role in regulation of lipid processing, and as lipid accumulation is central to NAFLD progression, hepatic accumulation of specific lipid species known to play a role in NAFLD progression was analyzed. Notably, toxic lipid accumulation is associated with both the induction of steatosis and the conversion of NAFL to NASH (37). Quantification of hepatic TG levels revealed that, despite a significant difference between Acox1Lampe1 mice and WT controls, HFHCD — compared with chow diet — did not additively impact hepatic TG accumulation in Acox1Lampe1 mice (Figure 4G). Additional quantification of hepatic lipid species by chromatography revealed that Acox1Lampe1 mice, in comparison with WT controls, displayed a robust hepatic accumulation of diacylglycerol (DAG), free cholesterol (FC), monoacylglycerol (MAG), and TG at baseline — lipids all known to promote NAFLD pathogenesis (37). However, despite significantly exacerbated hepatocellular damage and inflammatory capacity, HFHCD feeding of Acox1Lampe1 mice did not exhibit exacerbated toxic lipid accumulation in the liver (Figure 4H). Further, Acox1Lampe1 mice fed HFHCD displayed similar hepatic Srebp1c mRNA expression but decreased hepatic Ppara mRNA expression — genes associated with fatty acid oxidation (FAO; Supplemental Figure 6). In sum, these findings suggest that both lipid metabolism and inflammation play a significant, synergistic role in hepatocellular damage and that Acox1Lampe1 mutation likely works via augmentation of pathways alternate to Ppara mRNA expression. However, whether Acox1 function in hepatocytes, or in immune cells, is the main driver of the observed phenotype is underdefined.
Hematopoietic cell Acox1-dependent peroxisomal β-oxidation contributes to exacerbated hepatocellular damage and systemic inflammation.
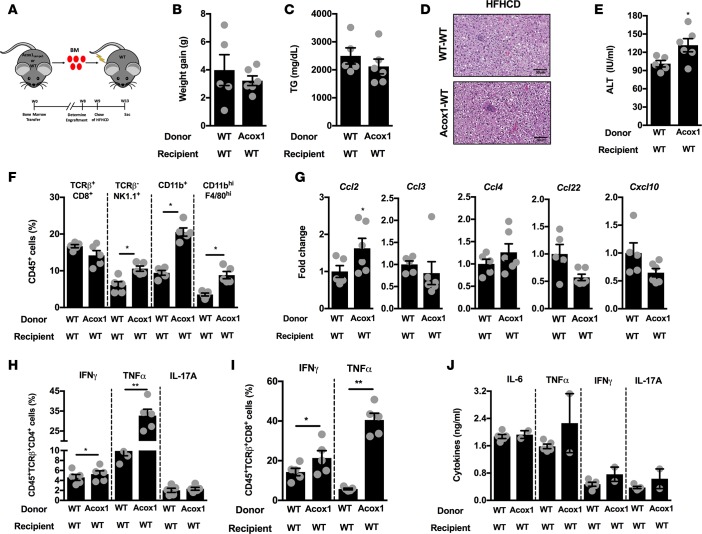
To define whether hematopoietic or nonhematopoietic Acox1 expression is the key contributor to exacerbated inflammation and hepatic disease, BM transfer experiments were performed (Figure 5A). WT or Acox1Lampe1 BM was transferred into irradiated WT mice, and following successful immunological reconstitution, mice were fed a HFHCD for 4 weeks. Acox1Lampe1 mutation in the hematopoietic compartment did not alter HFHCD-driven weight gain (Figure 5B), but — in contrast to full body mutation — it negatively impacted food intake (Supplemental Figure 7A). As expected, the similarity in body weight gain correlated with comparable hepatic TG accumulation and histological manifestations (Figure 5, C and D). In contrast, the Acox1Lampe1 mutation in the hematopoietic compartment was sufficient to exacerbate hepatocellular damage (Figure 5E). However, the level of hepatocellular damage was blunted compared with total body Acox1Lampe1 mice fed a HFHCD (Supplemental Figure 7B). Further, Acox1Lampe1 mutation in the hematopoietic compartment resulted in a robust increase in the hepatic numbers of NK cells, myeloid cells, and macrophages (Figure 5F), which correlated with a modest increase in hepatic expression of macrophage-recruiting chemokines, Ccl2, Ccl3, Ccl4, Ccl22, and Cxcl10 (Figure 5G), known to play a role in NAFLD pathogenesis (36). PMA/ionomycin stimulation of hepatic infiltrating immune cells isolated from HFHCD-fed WT mice that received Acox1Lampe1 BM revealed increased IFN-γ, TNF-α, and IL-17A production by CD4+ and CD8+ T cells (Figure 5, H and I), compared with CD4+ and CD8+ T cells isolated from WT mice that received WT BM. Transfer of Acox1Lampe1 BM did not impact systemic proinflammatory cytokine levels (Figure 5J). These data support observations depicted in Supplemental Figure 1 and suggest that baseline differences in food intake and systemic inflammatory phenotype seen in the total body Acox1Lampe1 mice arise from a nonhematopoietic compartment. However, these data also indicated that Acox1Lampe1-dependent peroxisomal β-oxidation in the hematopoietic compartment impacts immune cell responsiveness, which is, at least in part, responsible for the observed hepatic immune cell infiltration and augmented hepatocellular damage.
Figure 5. Acox1Lampe1 mutation in hematopoietic cells contributes to exacerbated hepatocellular damage and systemic inflammation during short-term obesogenic-diet challenge.
Eight- ton 10-week-old WT mice were lethally irradiated and subsequently reconstituted with BM from WT or Acox1Lampe1 mice. Following successful immunological reconstitution, mice were fed a HFHCD for 4 weeks. (A) Schematic representation. (B) Weight gain. (C) Hepatic TG levels. (D) Representative liver histology (H&E staining; 20×). (E) Serum ALT levels. (F) Total hepatic immune (CD45+) cell infiltration determined by flow cytometry. (G) Hepatic chemokine mRNA expression of Ccl2, Ccl3, Ccl4, Ccl22, and Cxcl10. (H) Frequency of hepatic CD45+TCRB+CD4+ T cells producing IFN-γ, TNF-α, and IL-17A. (I) Frequency of hepatic CD45+TCRB+CD8+ T cells producing IFN-γ and TNF-α. (J) Mice were challenged i.p. with biotinylated capture Ab (20 μg/mouse) for 3 hours, and serum IL-6, TNF-α, IFN-γ, and IL-17A levels were quantified by IVCCA ELISA. Data represent means ± SEM. (B–J) Unpaired Student’s t test; *P < 0.05, **P < 0.01. A single experiment, n = 5–6/condition.
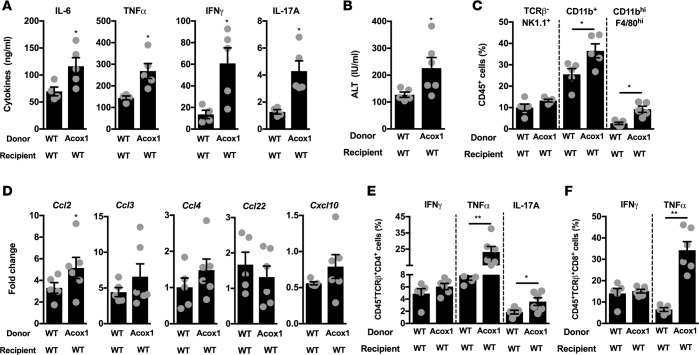
Inflammatory mediators play a role in delivering a “second hit” during NAFL to NASH transition (38), and our data (Figure 1) suggest that Acox1Lampe1 mice exhibit significantly increased proinflammatory cytokine production following inflammatory challenge. Hence, we next examined the contribution of hematopoietic Acox1Lampe1 mutation on immune responsiveness following inflammatory challenge. Notably, as metabolic endotexemia (LPS levels) directly correlates with obesity and NAFLD progression, we employed LPS challenge as a measurement of inducible inflammatory responses in the context of altered peroxisomal β-oxidation. Specifically, following LPS challenge of HFHCD-fed mice, Acox1Lampe1 mutation in the hematopoietic compartment resulted in significantly increased systemic proinflammatory cytokine levels (e.g., IL-6, IFN-γ, TNF-α, and IL-17A) compared with WT controls (Figure 6A). In fact, the increase in proinflammatory cytokine levels in response to LPS challenge was comparable between hematopoietic and total body Acox1Lampe1 (Figure 1B). The robust exacerbation of inflammatory vigor in mice with hematopoietic Acox1Lampe1 mutation positively correlated with exacerbated hepatocellular damage (Figure 6B), significantly increasing frequency of hepatic myeloid cells (Figure 6C) and hepatic expression of immune cell–recruiting chemokines, Ccl2, Ccl3, Ccl4, Ccl22 and Cxcl10, (Figure 6D) known to play a role in NAFLD pathogenesis (36). Further, under such experimental conditions, hematopoietic Acox1Lampe1 mutation significantly augmented cytokine production (e.g., IFN-γ, TNF-α, and IL-17A) by hepatic CD4+ and CD8+ T cells (Figure 6, E and F). These data suggest that Acox1 expression in immune cells contributes to the phenotype seen in total body Acox1Lampe1 mice. However, the lack of a significant increase in potentially toxic hepatic lipids, which may render hepatocytes susceptible to parallel/sequential inflammatory insult, due to normal Acox1 expression in hepatocytes may contribute to the cumulatively blunted liver phenotype. In sum, these findings suggest that both lipid metabolism and inflammation play a significant, synergistic role in hepatocellular damage and that Acox1 function may represent a critical link between the 2 biological processes.
Figure 6. Acox1Lampe1 mutation in hematopoietic cells alters immune responsiveness to secondary inflammatory challenge during short-term obesogenic-diet challenge.
Eight- to 10-week-old WT mice were lethally irradiated and subsequently reconstituted with BM from WT or Acox1Lampe1 mice. Following successful immunological reconstitution, mice were fed a HFHCD for 4 weeks and challenged with LPS. (A) Mice were challenged i.p. with biotinylated capture Ab (20 μg/mouse) 3 hours prior to i.v. LPS challenge (25 μg/mouse; ultrapure E. coli 0111:B4). Twenty-four hours later, serum was collected, and IL-6, TNF-α, IFN-γ, and IL-17A levels were quantified by IVCCA ELISA. (B) Serum ALT levels. (C) Total hepatic immune (CD45+) cell infiltration determined by flow cytometry. (D) Hepatic chemokine mRNA expression of Ccl2, Ccl3, Ccl4, Ccl22, and Cxcl10. (E) Frequency of hepatic CD45+TCRB+CD4+ T cells producing IFN-γ, TNF-α, and IL-17A. (F) Frequency of hepatic CD45+TCRB+CD8+ T cells producing IFN-γ and TNF-α. Data represent means ± SEM. (A–F) Unpaired Student’s t test; *P < 0.05, **P < 0.01. A single experiment, n = 5–6/condition.
Coupling of altered Acox1-dependent peroxisomal β-oxidation and obesogenic diet stress amplifies biological pathways that correlate with HCC development.
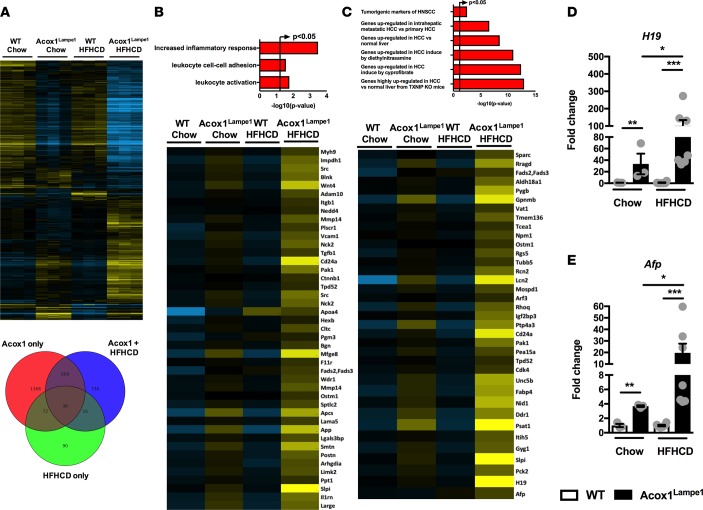
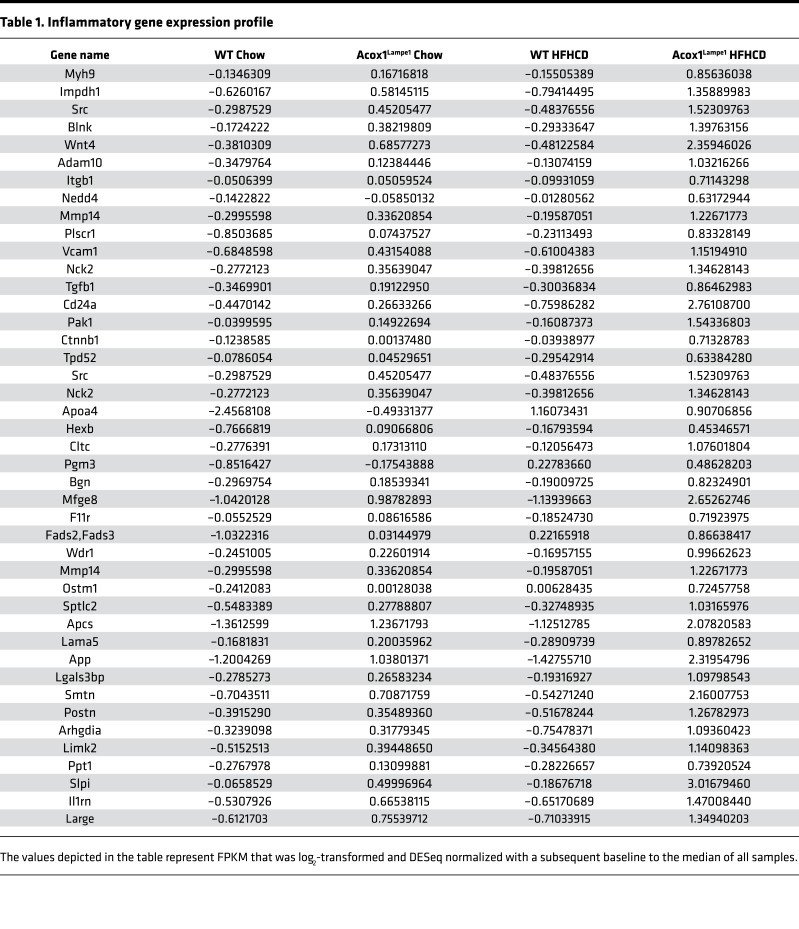
To assess the preclinical relevance of exacerbated and accelerated hepatocellular damage and histological changes observed in AcoxLampe1 mice fed HFHCD, an unbiased analysis of hepatic gene expression was performed. Both genetic (Acox1Lampe1 vs. WT) and dietary (chow vs. HFHCD) perturbations drove differential hepatic gene expression (Figure 7A). In fact, the combination of Acox1Lampe1 mice and HFHCD feeding resulted in the highest number of significant, differentially expressed hepatic genes compared with WT chow-fed mice (Figure 7A). Comprehensive analysis of gene expression profiles, central to biological processes known to be associated with NAFLD progression, revealed that HFHCD feeding of Acox1Lampe1 mice led to a significant increase in expression of genes associated with inflammatory responses, leukocyte activation, and leukocyte adhesion (Figure 7B and Table 1). These findings are supported by our observed increase in hepatic immune cell infiltration and chemokine expression (Figure 4, E and F).
Figure 7. Coupling of Acox1Lampe1 mutation with obesogenic diet stress amplifies biological pathways associated with HCC development.
Eight-week-old Acox1Lampe1 mice and WT littermate controls were fed a HFHCD or chow diet for 4 weeks. (A) Genes upregulated at least 2-fold in the liver of mice fed chow or HFHCD, as determined by RNA-seq. (B and C) Upregulated gene expression pathways determined from RNA-seq analysis. (D and E) Hepatic mRNA expression of genes associated with HCC development: H19 and Afp. Data represent means ± SEM. One-way ANOVA with Tukey’s correction; *P < 0.05, **P < 0.01, ***P < 0.001. White bars denote WT mice; black bars denote Acox1Lampe1 mice. (A–C) A single experiment, n = 3/condition. (D and E) Data combined from 2 independent experiments, n = 3–6/condition.
Table 1. Inflammatory gene expression profile.
Chronic inflammation and mitochondrial dysfunction have been shown to promote tumorigenesis (39, 40). In fact, NAFLD and its related inflammation are associated with increased susceptibility for HCC development in humans (41, 42). However, mouse models of NAFLD have largely failed to encompass the full spectrum of human HCC development. Previous studies have shown that Acox1Lampe1 mice spontaneously develop HCC after 1 year of age (29). Thus, we hypothesized that short-term HFHCD feeding coupled with whole body Acox1Lampe1 mutation would allow for accelerated and exacerbated expression of genes associated with HCC development.
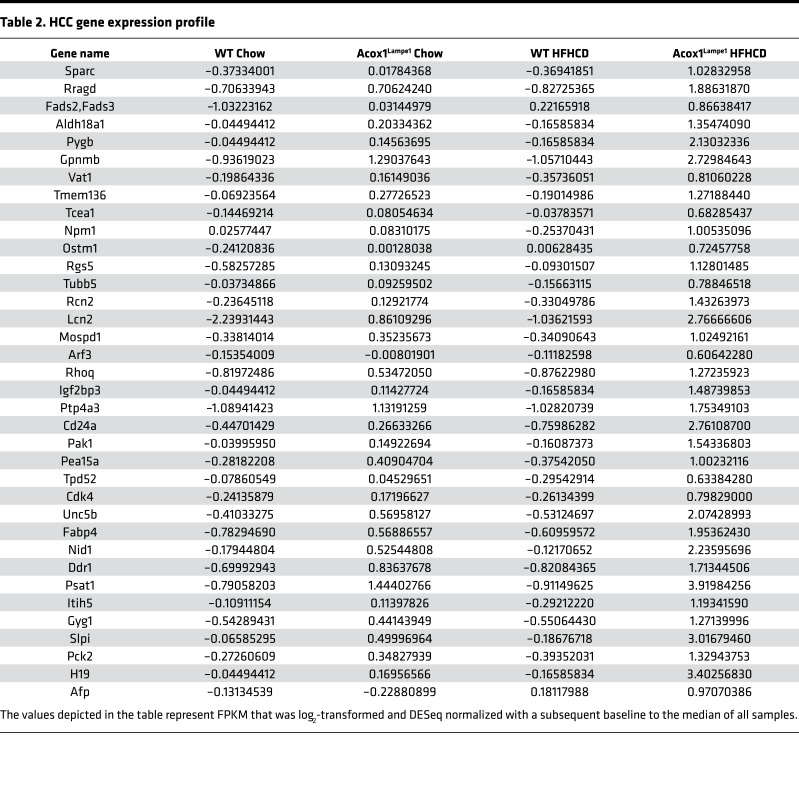
Although 4 weeks of HFHCD did not induce full-blown HCC, such short-term obesogenic diet feeding allowed for the robust induction of multiple genes associated with HCC. In fact, the expression of over 35 genes associated with HCC development in both murine models and humans were dramatically increased in young Acox1Lampe1 mice fed short-term HFHCD (Figure 7C and Table 2) — a finding further validated by quantitative PCR (qPCR) expression of selected genes (e.g., H19, Afp, Igfb1, Fabp5, and Spp1) (Figure 7, D and E, and Supplemental Figure 8). Notably, hepatic levels of H19, an oncogene associated with HCC development, and α-fetoprotein (Afp), a biomarker used for the diagnosis of human HCC (43) were expressed at significantly higher levels only in young Acox1Lampe1 mice fed a short-term HFHCD (Figure 7, D and E). These data demonstrate that, in addition to exacerbated inflammatory potential and systemic and hepatic inflammation, Acox1Lampe1 mice challenged with short-term HFHCD stress exhibit significant hepatic gene expression profiles associated with the induction of HCC.
Table 2. HCC gene expression profile.
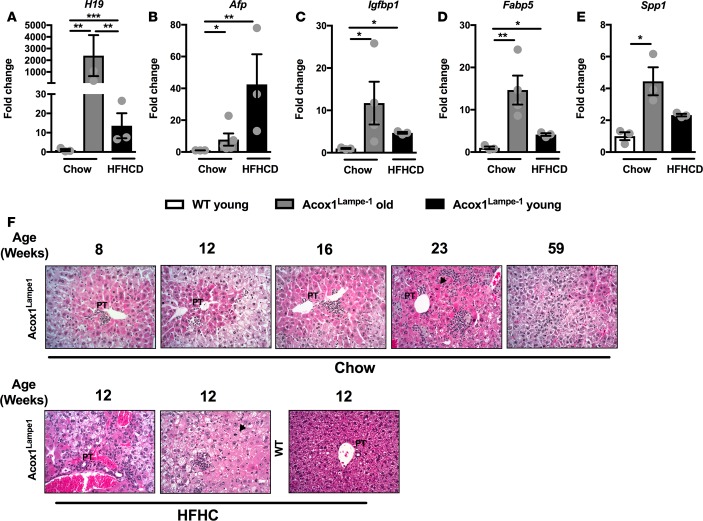
We next compared the amplitude of hepatic inflammation and HCC-related gene expression between short-term HFHCD diet stress in young, Acox1Lampe1 mice and aged (1-year-old) Acox1Lampe1 mice fed a chow diet. This approach allowed us to determine whether similar pathways were upregulated under these independent conditions and whether amplitude of gene expression correlated with the observed development of tumors in aged mice. Similar to HFHCD Acox1Lampe1 young mice, tumor-bearing, 1-year-old, Acox1Lampe1 mice exhibited increased hepatic chemokine expression (e.g., Ccl2, Ccl3, Ccl4, Ccl22, and Cxcl10; Supplemental Figure 9). Further, although HFHCD feeding robustly induced expression of genes considered markers of HCC (e.g., H19, Afp, Igfbp1, Fabp5, and Spp1) in young Acox1Lampe1 mice, the expression of such genes was predominantly higher in chow-fed, aged Acox1Lampe1 mice (Figure 8, A–E) — mice that develop hepatic tumors. The temporal evolution of histopathological changes that occur in the liver at different time points demonstrated that Acox1Lampe1 mice display steatosis, increasing inflammation and liver cell injury as they age, with development of hepatocellular tumors at 59 weeks of age (Figure 8F, upper panel). Indeed, at 12 weeks, HFHCD-fed Acox1Lampe1 mice displayed accelerated liver cell damage and steatosis, equivalent to a 23-week-old Acox1Lampe1 mouse on a regular diet (Figure 8F, lower panel). Thus, utility of young Acox1Lampe1 mice under obesogenic diet stress may allow for accelerated analysis of pathways underlying the interplay between metabolism and inflammation in obesity-driven NAFLD to HCC transition. Notably, such an approach may allow for faster, more efficacious future discovery of mechanisms that regulate NAFLD progression and pathogenesis.
Figure 8. Coupling of Acox1Lampe1 mutation with obesogenic diet stress accelerates hepatic injury and expression of genes associated with HCC development.
Eight-week-old Acox1Lampe1 mice and WT littermate controls were fed a HFHCD or chow diet for 4 weeks (young) and compared with Acox1Lampe1 were fed chow diet for 59 weeks (old). (A–E) Hepatic mRNA expression of genes associated with HCC development; H19, Afp, Igfbp1, Fabp5, and Spp1. (F) Upper panel: Representative liver histology (H&E staining; 20×) of the temporal evolution (8, 12, 16, 23, and 59 weeks of age) and histopathological changes in chow-fed Acox1Lampe1 mice. Acox1Lampe1 mice display steatosis, increasing inflammation, and liver cell injury over time with development of hepatocellular tumors at 59 weeks of age. Lower panel: Representative liver histology (H&E staining; 20×) of 12-week-old (young) Acox1Lampe1 and WT mice fed a HFHCD. Acox1Lampe1 mice display steatosis, inflammation, and liver cell injury after 4 weeks of HFHCD feeding. PT depicts portal tract; arrowhead depicts mitotic hepatocytes. Data represent mean ± SEM. One-way ANOVA with Tukey’s correction; *P < 0.05, **P < 0.01, ***P < 0.001. White bars denote WT mice fed chow diet; gray bars denote old Acox1Lampe1 mice fed chow diet; black bars denote young Acox1Lampe1 mice fed HFHCD. A single experiment, n = 3/condition.
Discussion
Here, we demonstrate that Acox1 regulates both cellular and whole body metabolism, immune cell polarization, and inflammation and that the interplay between these biological processes correlates with increased hepatocellular damage and impaired hepatic mitochondrial function. Notably, increased hepatic inflammation observed at baseline is exacerbated by HFHCD-driven metabolic burden and linked to augmented expression of HCC-related genes. In sum, our findings suggest that Acox1Lampe1 mice coupled with obesogenic diet stress provide a relevant experimental model, encompassing both metabolism and inflammation, for induction of accelerated and exacerbated NAFLD progression and induction of HCC-related genes.
Our data demonstrate an important role for peroxisomal β-oxidation in regulation of whole body energy expenditure and end-organ mitochondrial function. In the context of Acox1Lampe1 mutation, the unmetabolized substrates of this enzyme (very long–chain fatty acyl-CoAs), function as endogenous ligands for the lipid-sensing nuclear receptor Ppara to upregulate mitochondrial, peroxisomal, and microsomal FAO systems (44–46). Thus, the sustained increase in FAO, resulting from Acox1 deficiency, could influence Ppara function and regulate energy metabolism and body weight via regulation of Ucp1 activation in BAT (47). However, despite the possible advantages of Acox1 deficiency in terms of weight gain, compounded adverse effects prevail in the context of end-organ sequelae and, specifically, liver pathology (48). Acox1Lampe1 mice exhibited similar expression levels of Ppara after short-term HFHCD feeding compared with chow-fed counterparts. These data suggest that other pathways, in addition to Ppara, may be involved in the induction of genes associated with HCC in this model.
While we have demonstrated a role for Acox1 in immune function, the underlying mechanisms of Acox1-dependent modulation of inflammation require further exploration. Specifically, a role for Acox1 in inflammation has only been previously described in human fibroblasts from a patient with a mutation in the Acox1 gene (25). Acox1 deficiency in these cells was associated with significant alterations in the inflammatory response, leading to the activation of the IL-1 pathway and the induction of both IL-6 and IL-8 cytokines (25). Importantly, both IL-1β and IL-6 promote T cell polarization (49). In fact, our data demonstrated that Acox1 function particularly and profoundly impacted Th17 cell polarization in vivo and in vitro, and it impacted the ability of Th17 cells for multicytokine production, including IFN-γ and IL-17A secretion. Notably, systemic LPS levels, IL-17 levels, and IFN-γ levels correlate with NAFLD progression (12, 14, 16, 50, 51), and Th17 cells have been shown to play an important pathogenic role in NAFLD-to-HCC progression (52). The fact that we observed an increase in IFN-γ and IL-17 using chemical inhibition of peroxisomal β-oxidation suggested an intrinsic CD4+ T helper bias. Notably, however, Acox1Lampe1 mutation in naive CD4+ T cells, compared with WT T cells, selectively promoted higher Th17 polarization compared with WT naive CD4+ T cells. These data suggest that (i) thioridizine may have off-target effects; (ii) the increased Th1 responses in Acox1 mice may not be T cell intrinsic and may be associated with increased proinflammatory potential of antigen-presenting cells; and (iii) the source of high levels of IFN-γ in the Acox1Lampe1 mice could also be from other immune cells, including NK and CD8+ T cells. While, the role of fatty acid oxidation (FAO) through the mitochondrial β-oxidation pathway has previously been shown to play a role in Th17 cell polarization, our study shows for the first time to our knowledge an important role for peroxisomal β-oxidation in Th17 cell polarization. Nonetheless, the molecular mechanisms that link functional Th17 cell differentiation to FAO remain poorly defined. FAO has been shown to be one of the main pathways to fuel oxidative phosphorylation, which in turn fuels glycolysis, fast proliferative response, and proinflammatory cytokine production by memory CD8+ T cells (53). These studies indicate that the proper regulation of intracellular lipid metabolism is critical for T cell responses. Although future studies utilizing cell-specific deletion of Acox1 are needed, our current findings suggest that better understanding of peroxisomal β-oxidation in Th17 polarization may lead to the identification of novel mechanisms by which metabolism and inflammation are coregulated and contribute to NAFLD progression.
Of note, while our work demonstrates that peroxisomal β-oxidation regulates immune responses, whether immune responses regulate NAFLD-associated metabolic processes remains to be fully elucidated. Interestingly, the role of IL-17 in NAFLD may not be limited simply to inflammation. In fact, IL-17 has been shown to alter metabolic processes, including adipogenesis via activation of Kruppel-like factors (54). Further, we have previously demonstrated that IL-17RA–deficient mice exhibit significantly increased steatosis (14). Therefore, whether the IL-17 axis regulates peroxisomal β-oxidation and subsequent hepatic lipid accumulation, and the mechanisms underlying this potential effect, is an important question moving forward.
We also demonstrate that Acox1Lampe1 mice fed HFHCD develop accelerated and exacerbated hepatocellular damage — something that uncoupled lipid toxicity from inflammation during obesogenic diet feeding. Although the hepatic accumulation of toxic lipids in Acox1Lampe1 mice was dependent on Acox1 function, it was found to be independent of dietary intake. Further, to define whether Acox1 expression by hematopoietic or nonhematopoietic cells was central to the exacerbated inflammation and hepatic disease, BM transfer experiments were performed. WT mice reconstituted with Acox1Lampe1 BM fed a HFHCD displayed augmented inflammation and hepatocellular damage. However, the severity of hepatocellular damage was blunted in comparison with the levels observed in nonirradiated, total body Acox1Lampe1 mice fed a HFHCD. These data suggest that hematopoietic Acox1 expression plays an important role in the inflammatory phenotype seen in Acox1Lampe1 mice. However, these data also suggest that such effects alone are not sufficient to drive differential development of hepatic steatosis or fully alter the severity of hepatocellular damage. This effect could be partially due to the short duration of HFHCD feeding (4 weeks) — something that precludes robust hepatic triglyceride accumulation in WT animals as compared with Acox1Lampe1 mice.
Specifically, increased hepatic triglyceride synthesis and storage represents the “first hit” in NAFLD and renders the hepatocytes susceptible to parallel/sequential inflammatory hits, potential cell death, and disease progression. Endotoxemia, oxidative stress, cytokine production, and other proinflammatory mediators have all been shown to play roles in delivering a “second hit” during the transition from simple steatosis to NASH (38). As hepatic inflammation is one of the most distinguished features of NASH, hepatic immune responses may also play critical roles in the pathogenesis of NASH. In fact, exogenous administration of an inflammatory stimuli to WT mice reconstituted with Acox1Lampe1 BM and fed a HFHCD exacerbated hepatocellular damage, as compared with WT mice reconstituted with WT BM. These data suggest that, in mice with impaired hematopoietic peroxisomal β-oxidation (e.g., Acox1Lampe1 mice), the intrinsic immune cell inflammatory potential might further contribute to the “second hit” stress–mediated damage and NAFLD progression to HCC. In fact, the utility of our model may allow for definition of the metabolic processes that regulate inflammatory potential and the complex interactions between inflammation and lipid deposition/toxicity in NAFLD progression. However, specific experiments involving Acox1–conditional KO mice will be necessary for determination of the critical cell types and pathways regulating peroxisomal β-oxidation in NAFLD progression.
Taken together, our data provide potentially novel insights into the interactions of metabolism and inflammation in the pathogenesis of NAFLD progression. Overall, our findings suggest that coupling of genetic and dietary stress significantly augments inflammatory vigor, accelerates and exacerbates NAFLD, and promotes induction of HCC-associated genes — something possibly dependent on exacerbated immune activation and impaired fatty acid β-oxidation. Additionally, the introduction of short-term HFHCD feeding in the context of Acox1 deficiency induces expression of HCC genes that normally take more than a year to develop in the absence of dietary challenge.
Methods
Mice.
Acox1Lampe1 mice on a C57BL/6 background were generated as previously described (29) and were matched for nicotinamide nucleotide transhydrogenase (Nnt) mutation status (55). Animals were housed in a specific pathogen-free animal facility at CCHMC and handled in high-efficiency particulate-filtered laminar flow hoods with free access to food and water. For all studies, littermate WT and Acox1Lampe1 heterozygous or homozygous mice derived from Acox1Lampe1 heterozygous parents were utilized.
Dietary modulation.
Mice were fed either regular chow (fat 13.5% kcal, carbohydrate 59% kcal, protein 27.5% kcal; LabDiet 5010) or a HFHCD (fat 58% kcal, carbohydrate 25.5% kcal, protein 16.4% kcal; Research Diets, catalog D12331; plus carbohydrates in drinking water [55 mM sucrose + 128 mM fructose]; refs. 14, 56). For all studies, body weight and food consumption were quantified weekly.
BM transfer.
BM chimeric mice were generated using 8-week-old WT recipient mice. Briefly, recipient mice were conditioned by whole body irradiation and were reconstituted via tail vein injection of 1 × 106 BM cells derived from femurs of WT or Acox1Lampe1 mice. Peripheral blood chimerism was assessed by flow cytometric analysis 8 weeks after BM reconstitution. Following successful BM reconstitution, mice were placed on chow or HFHCD as described above.
Hepatic function and phenotyping.
Hepatic TGs were quantified using Triglyceride Reagent and Triglyceride Standards (Pointe Scientific). Serum ALT levels were quantified using ALT Reagent and Catatrol I and II (Catachem). Lipid peroxidation was quantified using 4-HNE ELISA reagents (Cell Biolabs) as previously described (16). For histology, liver tissue was fixed in 10% buffered formalin, stained with H&E, and evaluated by a board-certified pathologist. Hepatic immune infiltration was quantified by flow cytometry of single cell suspensions following hepatic digestion as previously described (14, 16). All antibodies used were from eBioscience.
Quantification of cytokine levels.
For quantification of in vivo cytokine production, in vivo cytokine capture assay (IVCCA) was employed as previously described. Briefly, biotinylated capture antibodies to IL-6 (MP5-32C11, catalog 36-7062-85), TNF-α (TN3, catalog 36-7423-85), IFN-γ (R4-6A2, catalog 36-7312-85), and IL-17A (eBio17B7, catalog 13-7177-85) (all from eBioscience) were injected i.p., and terminal serum collection was performed 24 hours later (9, 14, 16, 57, 58). Further, for quantification of LPS-driven cytokine production in vivo, mice were injected i.p. 3 hours prior to LPS (25 μg/mouse) challenge, and serum cytokine levels were determined 24 hours later. For in vitro cytokine production, IL-6 and IL-1β ELISAs (BD Biosciences) were utilized according to the manufacturer’s instruction.
T cell polarization.
Naive CD4+ T cells were isolated from the spleen and cultured using anti-CD3/CD28 (catalog 14-0031-82 and catalog 14-0281-81, respectively; 1 μg/ml and 0.5 μg/ml, respectively), in DMEM supplemented with FBS, nonessential amino acids, and vitamins. Polarization of naive CD4+ T cell to Th17, Th1, or Tregs was achieved via supplementation of culture media for 6 days. Th17 skewing was induced by supplementation of culture media with TGFB (1 ng/ml), IL-6 (10 ng/ml), IL-23 (20 ng/ml), and IL-1β (1 ng/ml). Th1 skewing was induced by supplementation of culture media with IL-12 (20 ng/ml). Treg skewing was induced by supplementation of culture media with TGFB (10 ng/ml) and IL-2 (100 U/ml). All recombinant proteins and antibodies were from eBioscience.
Mitochondrial isolation and quantification of substrate utilization.
Mitochondria was isolated from intrascapular BAT and the left lateral liver lobe. Mitochondrial protein concentration was determined with a bicinchoninic acid protein assay kit (BCA; Thermo Fisher Scientific) according to manufacturer’s instructions. An XF Analyzer (Seahorse Bioscience) was used to measure bioenergetics in mitochondria isolated from BAT and liver. Briefly, XF24 extracellular flux assay cartridge (Seahorse Bioscience) was hydrated overnight at 37oC according to manufacturer’s instruction. Isolated mitochondria (6 μg per well for BAT and 10 μg per well for liver) were plated in MAS-1 buffer supplemented with 70 mM sucrose, 220 mM D-mannitol (MilliporeSigma), 5 mM potassium dihydrogen phosphate (Thermo Fisher Scientific), 5 mM magnesium chloride (MilliporeSigma), 2 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 1 mM EDTA (MilliporeSigma) and 0.2% fatty free acid BSA (MP Biologicals, pH 7.4) on a polyethylenimine (MilliporeSigma) precoated XF Cell Culture Microplate, centrifuged at 200 g for 12 minutes at 4oC, and incubated at 37oC in a non-CO2 incubator for 10 minutes. Liver mitochondria were plated on an uncoated plate, centrifuged at 2,000 g for 20 minutes at 4oC, and incubated at 37oC in a non-CO2 incubator for 10 minutes. For BAT, 3 mM sodium pyruvate/3 mM malate (MilliporeSigma), and 4 μM FCCP (MilliporeSigma) were sequentially injected, and mitochondrial OCR was quantified. For liver, 5 mM succinate (MilliporeSigma) and 4 μM FCCP were sequentially injected, and OCR was quantified.
Analysis of mouse metabolic phenotypes.
Assessment of energy intake, energy expenditure, and home-cage activity were performed using an indirect calorimetric system (TSE PhenoMaster, TSE Systems). Data for energy expenditure were analyzed using analysis of covariance (ANCOVA) with body weight as covariant as previously described (59, 60). Briefly, mice were placed in calorimetric chambers that recorded oxygen consumption, as well as locomotor activity through use beam breaks.
Flow cytometry.
Single cell suspensions were derived from hepatic and splenic homogenates. To determine immune cell populations, cells were labeled with monoclonal antibodies (all from eBioscience) at room temperature. For cytokine production, total single cells were stimulated for 4 hours with PMA (50 ng/ml; MilliporeSigma) and Ionomycin (1 μg/ml; Calbiochem) in presence of Brefeldin A (10 μg/ml; MilliporeSigma). Subsequently, data were collected using a LSR Fortessa (BD Biosciences) and analyzed using FlowJo X software (vX0.7). Briefly, cells were stained with Live/Dead stain (Zombie UV Dye, BioLegend, catalog 423107) and with directly conjugated monoclonal antibodies to CD45 (clone 104, BioLegend, catalog 109845), CD11b (clone M1/70, catalog 12-0112-81), F4/80 (clone BM8, catalog 17-4801-82), Gr1 (clone RB6-8C5, catalog 56-5931-82), NK1.1 (clone PK136, catalog 25-5941-82), TCRB (clone H57-597, catalog 47-5961-82), CD4 (clone GK1.5, catalog 11-0042-85), CD8 (clone 53-6.7, catalog 48-0081-82), IL-17A (clone 17B7, catalog45-7177-82), IFN-γ (clone XMG1.2, catalog 25-7311-82 ), and FOXP3 (clone FJK-16s, catalog 17-5773-82) (all from eBioscience).
RNA sequencing and gene expression quantification.
Gene expression was determined by running 50 bp single-end reads (~20 million reads per sample). All analysis was performed in Cufflinks 1.3.0, TopHat 1.4.0, and GeneSpring 7.0. Single-end 50-bp reads were aligned to the mouse genome (mm9) in TopHat v1.4.0. Following alignment, transcripts were quantified following the protocol described (61). Functional enrichments of differentially expressed genes were assessed through ToppGene, with a P value cutoff of 0.05. All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE89626).
Tri-one dimensional thin layer chromatography (TOD-TLC) analysis of neutral lipids from liver.
Acox1Lampe1 and WT control liver tissue (0.1 mg) from mice fed chow and HFHCD were homogenized in PBS, and cellular lipids were extracted into chloroform/methanol. Subsequently, lipid samples were evaporated using a nitrogen stream and a 52oC water bath and loaded onto high-performance thin layer chromatography plates precoated with silica gel 60 (Thermo Fisher Scientific). Plates were washed with chloroform and methanol to remove any contaminants, dried overnight at 120oC, and developed in a solvent system modified from a previous report (62). Briefly, plates were initially developed in a solvent mixture of chloroform/methanol/acetic acid (90:10:1) up to 3 cm of a 10-centimeter plate and then dried and placed in a second solvent of hexane/diethyl ether/acetone (60:40:5) up to 8 cm of the plate. Next, plates were dried again and placed in a third solvent of hexane/diethyl ether (97:3) up to 9.5 cm of the plate. Finally, plates were placed in the final solvent of pure hexane, and the final solvant was run to the top of the plate. Bands were visualized by spraying with a 0.05% solution of primuline in acetone and water (80:20) and detected as ultraviolet spots at 366 nm on a Typhoon 9500 molecular imager (62).
qPCR.
Tissue samples were homogenized in TRIzol. RNA was extracted, reverse transcribed to complementary DNA (cDNA), and subjected to qPCR analysis (Light Cycler 480 II; Roche Diagnostics) as previously described (14, 16). The primer sequences, all from Invitrogen are as follows: Ccl2 forward, 5′ - AGATGCAGTTAACGCCCCAC - 3′, and reverse, 5′ - TGTCTGGACCCATTCCTTCTTG - 3′; Ccl3 forward, 5′ - ACCATGACACTCTGCAACCAAG - 3′, and reverse, 5′ - TTGGAGTCAGCGCAGATCTG - 3′; Ccl4 forward, 5′ - TCTCTCCTCTTGCTCGTGGC - 3′, and reverse, 5′ - GAATACCACAGCTGGCTTGGA - 3′; Cxcl10 forward, 5′ - CCTATGGCCCTCATTCTCAC - 3′, and reverse, 5′ - CGTCATTTTCTGCCTCATCC - 3′; Ccl22 forward, 5′ - TGGAGTAGCTTCTTCACCCA - 3′, and reverse, 5′ - TCTGGACCTCAAAATCCTGC - 3′; Ucp1 forward, 5′ - TCAGCTGTTCAAAGCACACA - 3′, and reverse, 5′ - GTACCAAGCTGTGCGATGTC - 3′; Igfbp1 forward, 5′ - TCCTCTGTCATCTCTGGGCT - 3′, and reverse, 5′ - TGTGTACCAGAACCTGCTGC - 3′; Fabp5 forward, 5′ - TGTTGTTGCCATCACACGTA - 3′, and reverse, 5′ - AGGATCTCGAAGGGAAGTGG - 3′; Spp1 forward, 5′ - ATTTGCTTTTGCCTGTTTGG - 3′, and reverse, 5′ - GGACACCTCACTGAATGGCT - 3′; H19 forward, 5′ - CAACATCCCACCCACCGTAA - 3′, and reverse, 5′ - GCTCACCAAGAAGGCTGGAT - 3′; Afp forward, 5′ - TCTTTCCACTCCACTTTGGC - 3′, and reverse, 5′ - GGCGATGGGTGTTTAGAAAG - 3′; Lipe forward, 5′ - TCTCGTTGCGTTTGTAGTGC - 3′, and reverse, 5′ - ACGCTACACAAAGGCTGCTT - 3′; Pnpla2 forward, 5′ - GTTGAAGGAGGGATGCAGAG - 3′, and reverse, 5′ - GCCACTCACATCTACGGAGC - 3′; Srebp1c forward, 5′ - CTGTCTCACCCCCAGCATAG - 3′, and reverse, 5′ - GATGTGCGAACTGGACACAG - 3′; Ppara forward, 5′ - CATGGGGAGAGAGGACAGA - 3′, and reverse 5′ - AGTTCGGGAACAAGACGTTG - 3′; and Bactin forward, 5′ - GGCCCAGAGCAAGAGAGGTA - 3′, and reverse, 5′ - GGTTGGCCTTAGGTTTCAGG - 3′.
Statistics.
For statistical analysis, a normal distribution was assumed and parametric tests were employed. A 2-tailed student’s t test was used when the comparison was between 2 groups, while a 1-way ANOVA with Tukey’s post hoc test to assess differences between specific groups was employed for 3 or more groups. Statistical analysis was completed using Prism 5a (GraphPad Software Inc.). All values are represented as means ± SEM. A P value less than 0.05 was considered significant. For determination of sample size, an online statistical tool was utilized (https://www.dssresearch.com/KnowledgeCenter/toolkitcalculators/statisticalpowercalculators.aspx).
Study approval.
All studies were performed in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and approved by the CCHMC Institutional Animal Care and Use Committee (IACUC).
Author contributions
MEMF, DAG, TES, RS, MC, RM, KL, AS, JPB, and SD participated in data generation. RS, RK, CS, AS, SPH, JPB, BJA, KH, and SD participated in analysis and interpretation of data. MEMF, DAG, KH, and SD participated in the conception and design of the study, obtained the funding, and wrote the manuscript. All authors have reviewed the manuscript and approve the final version.
Supplementary Material
Acknowledgments
This study was supported, in part, by NIH R01DK099222 (to SD); R01DK099222-02S1 (associated with SD and MEMF); AHA 17POST33650045 (to MEMF); NIH T32AI118697 (associated with DAG); and PHS grant P30 DK078392 Pathology of the Digestive Disease Research Core Center at CCHMC (associated with SD). We thank Calvin C. Chan, Matthew J. Lawson, and Vishakha Sharma for technical assistance.
Version 1. 03/22/2018
Electronic publication
Funding Statement
This work was supported in part by NIH R01DK099222 (to S.D) and R01DK099222-02S1 (to M.E.M.F); NIH T32AI118697 (to D.A.G.); and PHS Grant P30 DK078392 Pathology of the Digestive Disease Research Core Center at CCHMC.
Footnotes
DAG’s present address is: La Jolla Institute for Allergy and Immunology, La Jolla, CA, USA.
MC’s present address is: Divisions of Neonatology and Developmental Biology, David Geffen School of Medicine at UCLA, Mattel Children’s Hospital UCLA, CA, USA.
AS’s present address is: Department of Biology, Miami University, OH, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: JCI Insight. 2018;3(6):e93626. https://doi.org/10.1172/jci.insight.93626.
Contributor Information
Rachel Sheridan, Email: Rachel.Sheridan@cchmc.org.
Rebekah Karns, Email: Rebekah.Karns@cchmc.org.
Monica Cappelletti, Email: MCappelletti@mednet.ucla.edu.
Kristin Lampe, Email: kristin.Lampe@cchmc.org.
Rajib Mukherjee, Email: rajib.mukherjee@cchmc.org.
Christian Sina, Email: christian.sina@uk-sh.de.
Anthony Sallese, Email: sallesaf@miamioh.edu.
Kasper Hoebe, Email: kasper.hoebe@cchmc.org.
Senad Divanovic, Email: senad.divanovic@cchmc.org.
References
- 1.Schenk S, Saberi M, Olefsky JM. Insulin sensitivity: modulation by nutrients and inflammation. J Clin Invest. 2008;118(9):2992–3002. doi: 10.1172/JCI34260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strissel KJ, et al. Adipocyte death, adipose tissue remodeling, and obesity complications. Diabetes. 2007;56(12):2910–2918. doi: 10.2337/db07-0767. [DOI] [PubMed] [Google Scholar]
- 3.Weisberg SP, McCann D, Desai M, Rosenbaum M, Leibel RL, Ferrante AW. Obesity is associated with macrophage accumulation in adipose tissue. J Clin Invest. 2003;112(12):1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tiniakos DG, Vos MB, Brunt EM. Nonalcoholic fatty liver disease: pathology and pathogenesis. Annu Rev Pathol. 2010;5:145–171. doi: 10.1146/annurev-pathol-121808-102132. [DOI] [PubMed] [Google Scholar]
- 5.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332(6037):1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter-Kent C, Zein NN, Feldstein AE. Cytokines in the pathogenesis of fatty liver and disease progression to steatohepatitis: implications for treatment. Am J Gastroenterol. 2008;103(4):1036–1042. doi: 10.1111/j.1572-0241.2007.01709.x. [DOI] [PubMed] [Google Scholar]
- 7.Jia L, et al. Hepatocyte Toll-like receptor 4 regulates obesity-induced inflammation and insulin resistance. Nat Commun. 2014;5:3878. doi: 10.1038/ncomms4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L, et al. Nuclear factor high-mobility group box1 mediating the activation of Toll-like receptor 4 signaling in hepatocytes in the early stage of nonalcoholic fatty liver disease in mice. Hepatology. 2011;54(5):1620–1630. doi: 10.1002/hep.24552. [DOI] [PubMed] [Google Scholar]
- 9.Giles DA, et al. Thermoneutral housing exacerbates nonalcoholic fatty liver disease in mice and allows for sex-independent disease modeling. Nat Med. 2017;23(7):829–838. doi: 10.1038/nm.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharifnia T, et al. Hepatic TLR4 signaling in obese NAFLD. Am J Physiol Gastrointest Liver Physiol. 2015;309(4):G270–G278. doi: 10.1152/ajpgi.00304.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miele L, et al. Increased intestinal permeability and tight junction alterations in nonalcoholic fatty liver disease. Hepatology. 2009;49(6):1877–1887. doi: 10.1002/hep.22848. [DOI] [PubMed] [Google Scholar]
- 12.Mehal WZ. The Gordian Knot of dysbiosis, obesity and NAFLD. Nat Rev Gastroenterol Hepatol. 2013;10(11):637–644. doi: 10.1038/nrgastro.2013.146. [DOI] [PubMed] [Google Scholar]
- 13.Mills KH. TLR-dependent T cell activation in autoimmunity. Nat Rev Immunol. 2011;11(12):807–822. doi: 10.1038/nri3095. [DOI] [PubMed] [Google Scholar]
- 14.Harley IT, et al. IL-17 signaling accelerates the progression of nonalcoholic fatty liver disease in mice. Hepatology. 2014;59(5):1830–1839. doi: 10.1002/hep.26746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giles DA, Moreno-Fernandez ME, Divanovic S. IL-17 Axis Driven Inflammation in Non-Alcoholic Fatty Liver Disease Progression. Curr Drug Targets. 2015;16(12):1315–1323. doi: 10.2174/1389450116666150531153627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giles DA, et al. Regulation of Inflammation by IL-17A and IL-17F Modulates Non-Alcoholic Fatty Liver Disease Pathogenesis. PLoS ONE. 2016;11(2):e0149783. doi: 10.1371/journal.pone.0149783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu FM, et al. IL-17 induces AKT-dependent IL-6/JAK2/STAT3 activation and tumor progression in hepatocellular carcinoma. Mol Cancer. 2011;10:150. doi: 10.1186/1476-4598-10-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao R, et al. High expression of IL-17 and IL-17RE associate with poor prognosis of hepatocellular carcinoma. J Exp Clin Cancer Res. 2013;32:3. doi: 10.1186/1756-9966-32-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schröder T, et al. Mitochondrial gene polymorphisms alter hepatic cellular energy metabolism and aggravate diet-induced non-alcoholic steatohepatitis. Mol Metab. 2016;5(4):283–295. doi: 10.1016/j.molmet.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Postic C, Girard J. The role of the lipogenic pathway in the development of hepatic steatosis. Diabetes Metab. 2008;34(6 Pt 2):643–648. doi: 10.1016/S1262-3636(08)74599-3. [DOI] [PubMed] [Google Scholar]
- 21.Berson A, et al. Steatohepatitis-inducing drugs cause mitochondrial dysfunction and lipid peroxidation in rat hepatocytes. Gastroenterology. 1998;114(4):764–774. doi: 10.1016/S0016-5085(98)70590-6. [DOI] [PubMed] [Google Scholar]
- 22.Averill MM, Bornfeldt KE. Lipids versus glucose in inflammation and the pathogenesis of macrovascular disease in diabetes. Curr Diab Rep. 2009;9(1):18–25. doi: 10.1007/s11892-009-0005-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laine PS, et al. Palmitic acid induces IP-10 expression in human macrophages via NF-kappaB activation. Biochem Biophys Res Commun. 2007;358(1):150–155. doi: 10.1016/j.bbrc.2007.04.092. [DOI] [PubMed] [Google Scholar]
- 24.de Lima-Salgado TM, Alba-Loureiro TC, do Nascimento CS, Nunes MT, Curi R. Molecular mechanisms by which saturated fatty acids modulate TNF-α expression in mouse macrophage lineage. Cell Biochem Biophys. 2011;59(2):89–97. doi: 10.1007/s12013-010-9117-9. [DOI] [PubMed] [Google Scholar]
- 25.El Hajj HI, et al. The inflammatory response in acyl-CoA oxidase 1 deficiency (pseudoneonatal adrenoleukodystrophy) Endocrinology. 2012;153(6):2568–2575. doi: 10.1210/en.2012-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meyer K, Jia Y, Cao WQ, Kashireddy P, Rao MS. Expression of peroxisome proliferator-activated receptor alpha, and PPARalpha regulated genes in spontaneously developed hepatocellular carcinomas in fatty acyl-CoA oxidase null mice. Int J Oncol. 2002;21(6):1175–1180. [PubMed] [Google Scholar]
- 27.Stauber AJ, et al. Constitutive expression of peroxisome proliferator-activated receptor alpha-regulated genes in dwarf mice. Mol Pharmacol. 2005;67(3):681–694. doi: 10.1124/mol.104.007278. [DOI] [PubMed] [Google Scholar]
- 28.Poll-The BT, et al. A new peroxisomal disorder with enlarged peroxisomes and a specific deficiency of acyl-CoA oxidase (pseudo-neonatal adrenoleukodystrophy) Am J Hum Genet. 1988;42(3):422–434. [PMC free article] [PubMed] [Google Scholar]
- 29.Sheridan R, et al. Lampe1: an ENU-germline mutation causing spontaneous hepatosteatosis identified through targeted exon-enrichment and next-generation sequencing. PLoS One. 2011;6(7):e21979. doi: 10.1371/journal.pone.0021979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Passos ST, Silver JS, O’Hara AC, Sehy D, Stumhofer JS, Hunter CA. IL-6 promotes NK cell production of IL-17 during toxoplasmosis. J Immunol. 2010;184(4):1776–1783. doi: 10.4049/jimmunol.0901843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodier MR, Londei M. Lipopolysaccharide stimulates the proliferation of human CD56+CD3- NK cells: a regulatory role of monocytes and IL-10. J Immunol. 2000;165(1):139–147. doi: 10.4049/jimmunol.165.1.139. [DOI] [PubMed] [Google Scholar]
- 33.Gerosa F, Baldani-Guerra B, Nisii C, Marchesini V, Carra G, Trinchieri G. Reciprocal activating interaction between natural killer cells and dendritic cells. J Exp Med. 2002;195(3):327–333. doi: 10.1084/jem.20010938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.González-Muniesa P, Milagro FI, Campión J, Martínez JA. Reduction in energy efficiency induced by expression of the uncoupling protein, UCP1, in mouse liver mitochondria. Int J Mol Med. 2006;17(4):591–597. [PubMed] [Google Scholar]
- 35.Townsend KL, Tseng YH. Brown fat fuel utilization and thermogenesis. Trends Endocrinol Metab. 2014;25(4):168–177. doi: 10.1016/j.tem.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wenfeng Z, Yakun W, Di M, Jianping G, Chuanxin W, Chun H. Kupffer cells: increasingly significant role in nonalcoholic fatty liver disease. Ann Hepatol. 2014;13(5):489–495. [PubMed] [Google Scholar]
- 37.Kawano Y, Nishiumi S, Saito M, Yano Y, Azuma T, Yoshida M. Identification of Lipid Species Linked to the Progression of Non-Alcoholic Fatty Liver Disease. Curr Drug Targets. 2015;16(12):1293–1300. doi: 10.2174/1389450116666150408103318. [DOI] [PubMed] [Google Scholar]
- 38.Marra F, Lotersztajn S. Pathophysiology of NASH: perspectives for a targeted treatment. Curr Pharm Des. 2013;19(29):5250–5269. doi: 10.2174/13816128113199990344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Philip M, Rowley DA, Schreiber H. Inflammation as a tumor promoter in cancer induction. Semin Cancer Biol. 2004;14(6):433–439. doi: 10.1016/j.semcancer.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bishayee A. The role of inflammation and liver cancer. Adv Exp Med Biol. 2014;816:401–435. doi: 10.1007/978-3-0348-0837-8_16. [DOI] [PubMed] [Google Scholar]
- 42.Day CP. From fat to inflammation. Gastroenterology. 2006;130(1):207–210. doi: 10.1053/j.gastro.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 43.Zhu K, Dai Z, Zhou J. Biomarkers for hepatocellular carcinoma: progression in early diagnosis, prognosis, and personalized therapy. Biomark Res. 2013;1(1):10. doi: 10.1186/2050-7771-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fan CY, Pan J, Usuda N, Yeldandi AV, Rao MS, Reddy JK. Steatohepatitis, spontaneous peroxisome proliferation and liver tumors in mice lacking peroxisomal fatty acyl-CoA oxidase. Implications for peroxisome proliferator-activated receptor alpha natural ligand metabolism. J Biol Chem. 1998;273(25):15639–15645. doi: 10.1074/jbc.273.25.15639. [DOI] [PubMed] [Google Scholar]
- 45.Reddy JK. Nonalcoholic steatosis and steatohepatitis. III. Peroxisomal beta-oxidation, PPAR alpha, and steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2001;281(6):G1333–G1339. doi: 10.1152/ajpgi.2001.281.6.G1333. [DOI] [PubMed] [Google Scholar]
- 46.Pyper SR, Viswakarma N, Yu S, Reddy JK. PPARalpha: energy combustion, hypolipidemia, inflammation and cancer. Nucl Recept Signal. 2010;8:e002. doi: 10.1621/nrs.08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cypess AM, Kahn CR. Brown fat as a therapy for obesity and diabetes. Curr Opin Endocrinol Diabetes Obes. 2010;17(2):143–149. doi: 10.1097/MED.0b013e328337a81f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang J, et al. Sustained activation of PPARα by endogenous ligands increases hepatic fatty acid oxidation and prevents obesity in ob/ob mice. FASEB J. 2012;26(2):628–638. doi: 10.1096/fj.11-194019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gaublomme JT, et al. Single-Cell Genomics Unveils Critical Regulators of Th17 Cell Pathogenicity. Cell. 2015;163(6):1400–1412. doi: 10.1016/j.cell.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertola A, et al. Hepatic expression patterns of inflammatory and immune response genes associated with obesity and NASH in morbidly obese patients. PLoS One. 2010;5(10):e13577. doi: 10.1371/journal.pone.0013577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vonghia L, et al. Peripheral and Hepatic Vein Cytokine Levels in Correlation with Non-Alcoholic Fatty Liver Disease (NAFLD)-Related Metabolic, Histological, and Haemodynamic Features. PLoS One. 2015;10(11):e0143380. doi: 10.1371/journal.pone.0143380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomes AL, et al. Metabolic Inflammation-Associated IL-17A Causes Non-alcoholic Steatohepatitis and Hepatocellular Carcinoma. Cancer Cell. 2016;30(1):161–175. doi: 10.1016/j.ccell.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 53.van der Windt GJ, et al. CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability. Proc Natl Acad Sci USA. 2013;110(35):14336–14341. doi: 10.1073/pnas.1221740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahmed M, Gaffen SL. IL-17 inhibits adipogenesis in part via C/EBPα, PPARγ and Krüppel-like factors. Cytokine. 2013;61(3):898–905. doi: 10.1016/j.cyto.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang TT, et al. Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxide dismutase. Hum Mol Genet. 2006;15(7):1187–1194. doi: 10.1093/hmg/ddl034. [DOI] [PubMed] [Google Scholar]
- 56.Kohli R, et al. High-fructose, medium chain trans fat diet induces liver fibrosis and elevates plasma coenzyme Q9 in a novel murine model of obesity and nonalcoholic steatohepatitis. Hepatology. 2010;52(3):934–944. doi: 10.1002/hep.23797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finkelman F, Morris S, Orekhova T, Sehy D. The in vivo cytokine capture assay for measurement of cytokine production in the mouse. Curr Protoc Immunol. 2003;Chapter 6:Unit 6.28. doi: 10.1002/0471142735.im0628s54. [DOI] [PubMed] [Google Scholar]
- 58.Divanovic S, et al. Opposing biological functions of tryptophan catabolizing enzymes during intracellular infection. J Infect Dis. 2012;205(1):152–161. doi: 10.1093/infdis/jir621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tschöp MH, et al. A guide to analysis of mouse energy metabolism. Nat Methods. 2011;9(1):57–63. doi: 10.1038/nmeth.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stemmer K, et al. Thermoneutral housing is a critical factor for immune function and diet-induced obesity in C57BL/6 nude mice. Int J Obes (Lond) 2015;39(5):791–797. doi: 10.1038/ijo.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7(3):562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.White T, Bursten S, Federighi D, Lewis RA, Nudelman E. High-resolution separation and quantification of neutral lipid and phospholipid species in mammalian cells and sera by multi-one-dimensional thin-layer chromatography. Anal Biochem. 1998;258(1):109–117. doi: 10.1006/abio.1997.2545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.