Abstract
Using an untargeted metabolomics approach in initial (N = 99 subjects) and replication cohorts (N = 1,162), we discovered and structurally identified a plasma metabolite associated with cardiovascular disease (CVD) risks, N6,N6,N6-trimethyl-L-lysine (trimethyllysine, TML). Stable-isotope-dilution tandem mass spectrometry analyses of an independent validation cohort (N = 2,140) confirmed TML levels are independently associated with incident (3-year) major adverse cardiovascular event risks (hazards ratio [HR], 2.4; 95% CI, 1.7–3.4) and incident (5-year) mortality risk (HR, 2.9; 95% CI, 2.0–4.2). Genome-wide association studies identified several suggestive loci for TML levels, but none reached genome-wide significance; and d9(trimethyl)-TML isotope tracer studies confirmed TML can serve as a nutrient precursor for gut microbiota–dependent generation of trimethylamine (TMA) and the atherogenic metabolite trimethylamine N-oxide (TMAO). Although TML was shown to be abundant in both plant- and animal-derived foods, mouse and human fecal cultures (omnivores and vegans) showed slow conversion of TML to TMA. Furthermore, unlike chronic dietary choline, TML supplementation in mice failed to elevate plasma TMAO or heighten thrombosis potential in vivo. Thus, TML is identified as a strong predictor of incident CVD risks in subjects and to serve as a dietary precursor for gut microbiota–dependent generation of TMAO; however, TML does not appear to be a major microbial source for TMAO generation in vivo.
Keywords: Cardiology, Vascular Biology
Keywords: Atherosclerosis, Cardiovascular disease, Cholesterol
Trimethyllysine predicts incident cardiovascular disease risks in subjects and is a dietary precursor for gut microbiota-dependent generation of atherogenic metabolite TMAO.
Introduction
Untargeted metabolomics analyses of biological samples hold promise as a discovery platform for the potential identification of new metabolites and pathways linked to pathophysiological processes and disease susceptibility (1–7). For example, in previous studies we used an untargeted mass spectrometry (MS) approach to discover an association between plasma levels of the metabolite trimethylamine N-oxide (TMAO) and incident risk for cardiovascular disease (CVD) (1). Subsequent studies revealed TMAO is produced via metaorganismal pathways initiated by ingestion of trimethylamine-containing (TMA-containing) dietary nutrients, including phosphatidylcholine (1), choline (1), betaine (1), carnitine (8), and γ-butyrobetaine (9). Following ingestion, gut microbial fermentation of the TMA-containing nutrient results in TMA release, which upon delivery to the liver via the portal circulation is converted into TMAO via host enzymes (1). Further, mechanistic animal models have shown that dietary provision of TMA-containing nutrients can result in the gut microbiota–dependent generation of TMA and TMAO, and enhanced atherosclerosis and thrombosis (1, 8–11).
During the conduct of untargeted metabolomics studies, the majority of spectral features detected in biological matrices arise from metabolites of unknown structural identity (12). Consequently, unambiguous identification of the unknowns of interest requires multiple studies and diverse approaches including performance of multiple distinct case/control studies using different MS and chromatographic techniques (to narrow the focus of efforts to only those unknown analytes that appear to be reproducibly associated with the phenotype of interest), as well as synthesis of both structural isomers and isotopologues of candidate molecules (for direct comparisons with the unknown analyte). Moreover, once structural elucidation is achieved, because untargeted metabolomics studies are intrinsically only semiquantitative, replication studies using more quantitatively accurate analytical approaches are necessary to validate candidate biomarker associations with disease risks. Further, to move beyond association, mechanistic studies are necessary to explore potential causal relationships, if any, between the candidate biomarker and disease. Herein we describe our initial structural and subsequent validation and mechanistic studies identifying plasma levels of the unusual free amino acid N6,N6,N6-trimethyl-L-lysine (trimethyllysine, TML) as a strong and independent prognostic biomarker of incident CVD risks in subjects independent of traditional cardiovascular risk factors. We also show TML is a relatively abundant posttranslational modification of proteins in both animal and plants alike, is found in a wide array of animal and vegetable matter, and can serve as a nutrient precursor for gut microbiota–dependent generation of TMA and TMAO in vivo. Finally, our studies show that despite the clear structural and metabolic link between TML and gut microbial TMA/TMAO generation, the clinical association between circulating TML levels and incident CVD risks in subjects appears to be predominantly independent of TMAO, and that dietary TML does not appear to serve as a major source for microbial TMA and TMAO generation compared with choline and carnitine in both mice and humans.
Results
Discovery and structural identification of TML as a plasma metabolite associated with CVD risks.
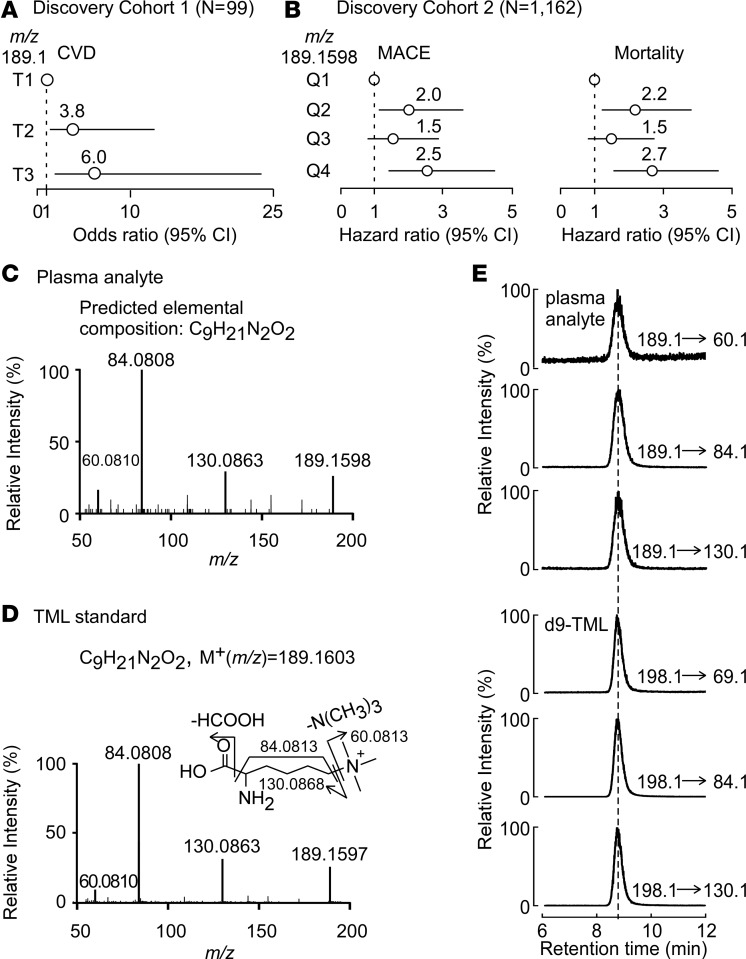
Our studies that ultimately led to the discovery of a link between TMAO and CVD used an initial case/control design (N = 49/50 cases/controls, discovery cohort 1, Table 1) and untargeted MS as a discovery platform (1). These studies also disclosed multiple plasma analytes beyond TMAO whose levels were associated with CVD based on ion intensities in the semiquantitative untargeted MS experiment. One analyte with an unknown structure (mass-to-charge ratio [m/z] of 189.1) appeared to be reproducibly associated with risks for both CVD (tertile 3 versus tertile 1, odds ratio [OR], 6.0; 95% CI, 1.7–21.0; P = 0.005) and coronary artery disease (CAD) (OR, 6.0; 95% CI, 1.7–21.0; P = 0.005) (Figure 1A). To confirm these initial findings, we performed additional untargeted metabolomics studies in a nonoverlapping cohort of sequential consenting subjects (N = 1,162) undergoing elective diagnostic cardiac evaluation using an alternative (high-resolution) MS-based approach, as described in Methods. Baseline characteristics of this cohort (discovery cohort 2) are shown in Table 2. Again, an association between a plasma metabolite with high mass accuracy m/z 189.1598 and incident CVD risks was observed. As compared with subjects in the lowest quartile of analyte (m/z 189.1598) levels, patients in the highest quartile (Q4) demonstrated a significant increased risk of both incident (3 years) major adverse cardiac events (MACE: myocardial infarction, stroke, or death) (hazards ratio [HR], 2.5; 95% CI, 1.4–4.5; P = 0.001), and incident (5-year) mortality risk (HR, 2.7; 95% CI, 1.6–4.6; P < 0.001). Following adjustments for traditional CVD risk factors and C-reactive protein levels, elevated plasma levels of the analyte (m/z 189.1598) remained a significant predictor of risks of incident (3-year) MACE (adjusted HR, 2.4; 95% CI, 1.4–4.2; P = 0.003) and mortality (5-year; adjusted HR, 2.2; 95% CI, 1.3–3.9; P = 0.005) (Figure 1B).
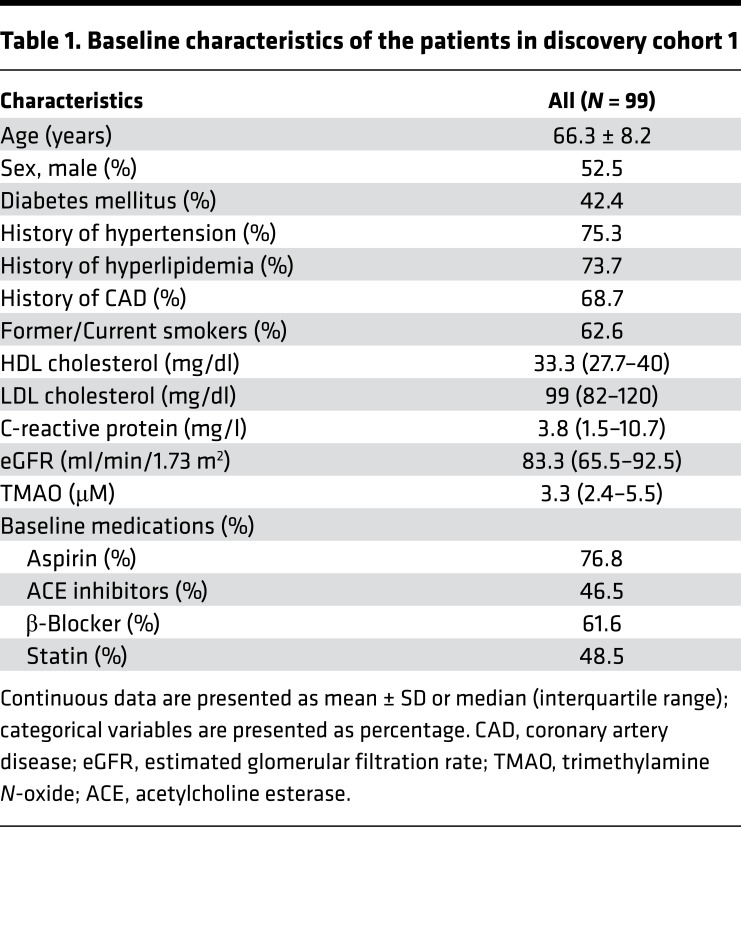
Table 1. Baseline characteristics of the patients in discovery cohort 1.
Figure 1. Untargeted metabolomics studies discover candidate compound with m/z of 189.1598 is associated with CVD and is trimethyllysine.
(A) Forest plot indicating plasma metabolite of unknown structure with m/z of 189.1 is associated with risk for cardiovascular disease (CVD) according to relative peak area intensity ranked by tertiles among subjects (N = 99) in discovery cohort 1. (B) Forest plot indicating plasma metabolite of unknown structure with high-resolution m/z of 189.1598 is associated with the risk for incident CVD and mortality risks according to relative peak intensity from untargeted mass spectrometry analyses of subjects (N = 1,162; discovery cohort 2). MACE, major adverse cardiac events, including myocardial infarction, stroke, or death. The analyses were performed using R 3.4.1. (C) Collision-induced dissociation (CID) spectrum in positive-ion mode of the metabolite m/z of 189.1598 in plasma. (D) CID spectra in positive mode of synthetic trimethyllysine (TML) standard. (E) Demonstration of cochromatography of multiple unique parent→daughter ion transitions for plasma analyte m/z 189.1598 and synthetic d9-TML.
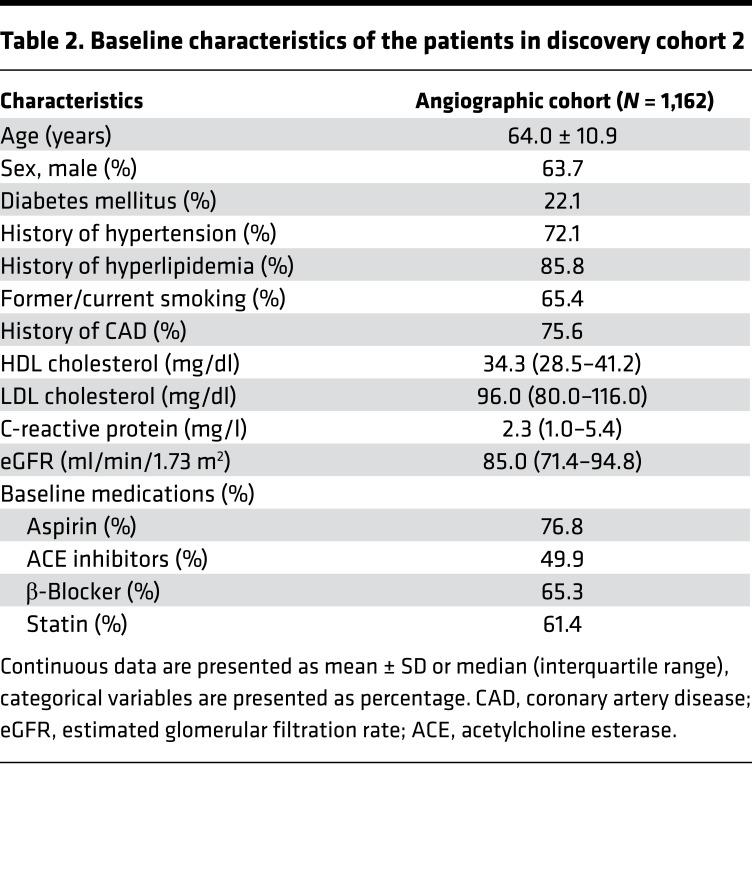
Table 2. Baseline characteristics of the patients in discovery cohort 2.
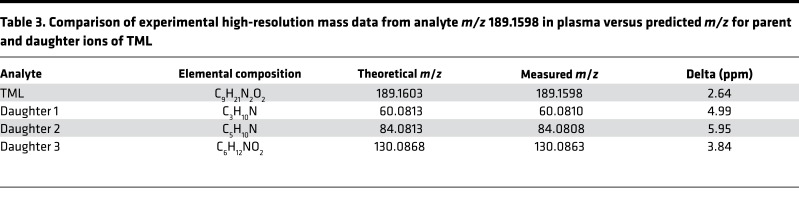
The collision-induced-dissociation mass spectrum of the plasma metabolite with m/z 189.1598 revealed a molecular cation (M+) with a predicted elemental composition of C9H21N2O2 (Figure 1C). This information, coupled with the observed high-resolution fragmentation pattern (daughter ion m/z and intensities), strongly suggested the identity of the metabolite as TML (Figure 1D and Table 3). Thus, in subsequent studies we compared analyte m/z 189.1598 with an authentic synthetic TML standard, and observed identical MSn and retention time characteristics using multiple different column matrices and chromatography conditions, as described under Methods. These data unambiguously confirmed the identity of analyte m/z 189.1598 as TML. We therefore synthesized d9(trimethyl)TML for use as an internal standard (Supplemental Methods; Supplemental Figure 1 and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.99096DS1), and developed a stable-isotope-dilution liquid chromatography with on-line tandem MS (LC-MS/MS) based method for quantitative analyses of TML that was used in all subsequent investigations. The method employs 3 unique and characteristic parent→daughter ion transitions for monitoring TML and its heavy isotope–labeled internal standard, and thus both quantitatively measured and further confirmed the identity of the analyte as TML (Figure 1E and Supplemental Figure 2). It should also be noted that MS/MS spectra later deposited in MassBank of North America from GNPS (13) and SPLASH (i.e., splash10-001i-5900000000-3e873bca7eaaf8953194) (14), identified as TML, also matched the unknown.
Table 3. Comparison of experimental high-resolution mass data from analyte m/z 189.1598 in plasma versus predicted m/z for parent and daughter ions of TML.
Clinical validation studies confirm that systemic TML levels predict incident MACE risks independent of CVD risk factors.
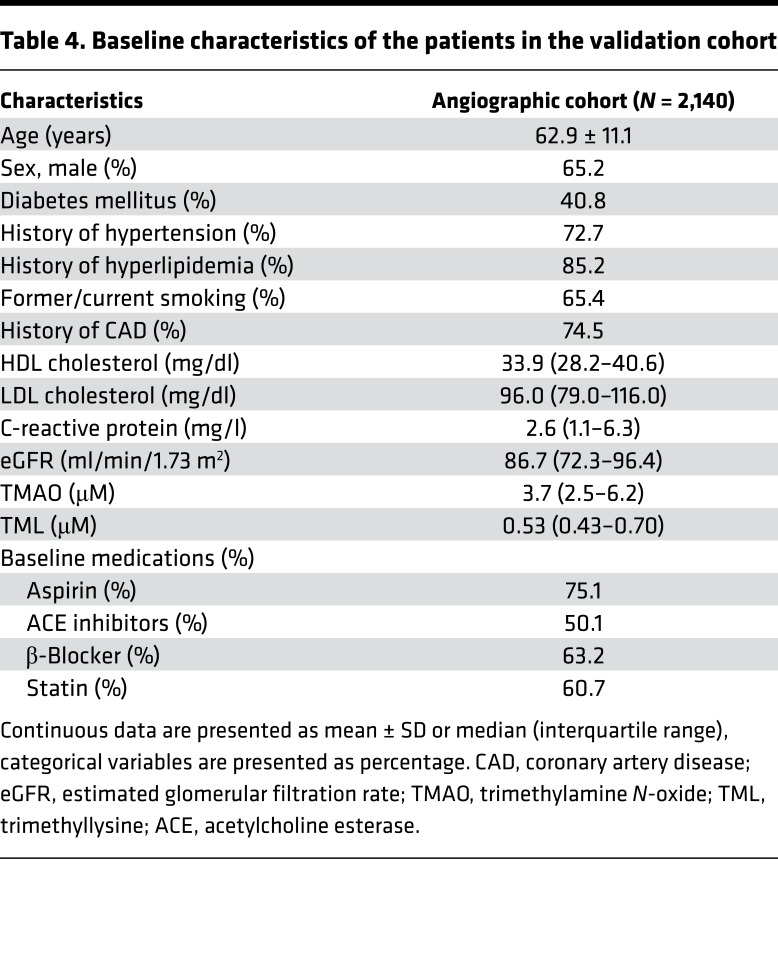
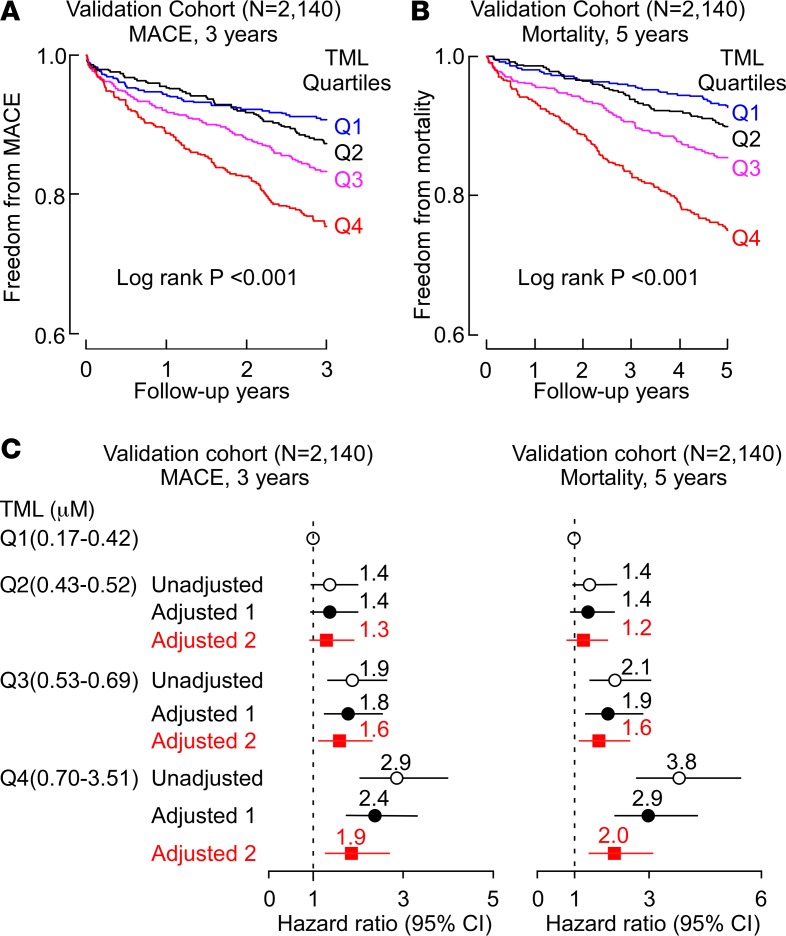
We next examined whether the association between plasma levels of TML and CVD risk could be confirmed using the quantitative stable-isotope-dilution LC-MS/MS assay in an independent group of subjects (validation cohort). Samples from sequential stable consented patients (N = 2,140) undergoing elective diagnostic coronary angiography and for whom adjudicated long-term adverse outcomes data were available were examined. Baseline demographic, clinical, and laboratory characteristics of the subjects in the validation cohort are shown in Table 4. In general, the subjects were middle-aged (62.9 ± 11.1 years), 65% male, had relatively preserved renal function, and in majority had either a high burden of CVD risk factors or CAD. Initial examination of the distribution of TML levels within the cohort (range 0.17–3.51 μM) showed a significantly higher TML level (P < 0.001) among those who experienced a MACE over the ensuing 3-year period following enrollment (Supplemental Figure 3). Similarly, higher TML levels were also observed among those who died over a 5-year follow-up period (P < 0.001) (Supplemental Figure 3). Kaplan-Meier survival analyses revealed a dose-dependent relationship between higher TML levels and incident risk for MACE (3 years) and mortality (5 years) (log-rank P < 0.001 for each) (Figure 2, A and B). Moreover, compared with subjects in the lowest quartile (Q1) of TML levels, subjects in the highest quartile (Q4) demonstrated a significant increased risk of incident MACE (HR, 2.9; 95% CI, 2.0–4.0; P < 0.001), or death (HR, 3.8; 95% CI, 2.7–5.5; P < 0.001) (Figure 2C). Elevated TML (Q4) levels remained an independent predictor of incident MACE (3 years) and mortality (5 years) risks even after adjustments for traditional cardiac risk factors and C-reactive protein levels (adjusted model 1, Figure 2C).
Table 4. Baseline characteristics of the patients in the validation cohort.
Figure 2. Stable-isotope-dilution LC-MS/MS analyses verify systemic levels of TML are associated with incident cardiovascular disease risks independent of TMAO.
Kaplan-Meier estimates and 3-year risks (HR [95% CI]) for (A) major adverse cardiac events (MACE, including myocardial infarction, stroke or death); and (B) all-cause mortality (5 years) ranked by trimethyllysine (TML) quartiles in the validation cohort (N = 2,140). (C) Forest plots indicate the HR (95% CI) for incident (3-year) risks for MACE and all-cause mortality (5-year) according to TML quartiles. HR (unadjusted, open circles) and multivariable Cox model 1 adjusted (filled black circles; adjusted for age, sex, high-density lipoprotein [HDL], low-density lipoprotein [LDL], smoking, diabetes mellitus, hypertension, C-reactive protein level), or model 2 adjusted (filled red squares, adjusted for model 1 plus trimethylamine N-oxide [TMAO]). The 5%–95% confidence interval is indicated by line length. The analyses were performed using R 3.4.1. HR, hazard ratio.
Examination of potential processes impacting TML and CVD risks.
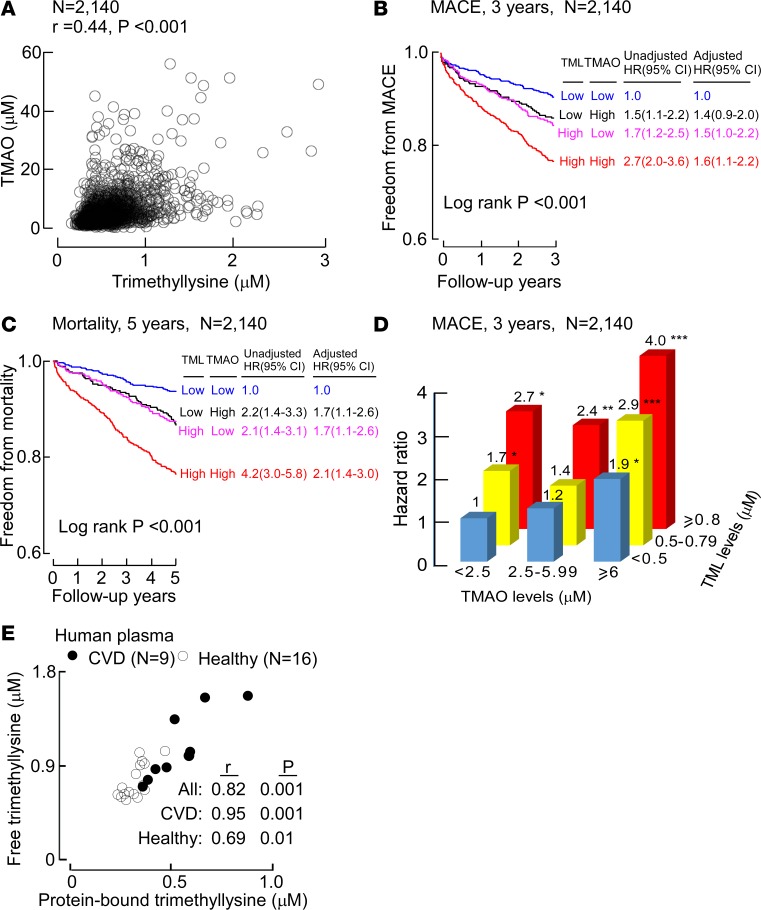
Because the structure of TML contains a TMA moiety, we next sought to investigate whether there was a relationship between TML and TMAO levels, as well as whether the prognostic association between TML and CVD risks was dependent on TMAO. TMAO was therefore quantified in the same subjects using established stable-isotope-dilution LC-MS/MS methods in the entire cohort (15). Of note, Spearman’s correlation analyses revealed a highly significant association between plasma TML and TMAO levels (r = 0.44, P < 0.001; N = 2,140) (Figure 3A). Moreover, as previously observed in multiple cohorts (16–18), TMAO levels within the present cohort were associated with all monitored incident adverse CVD events including MACE (3 years) (HR, 2.5; 95% CI, 1.8–3.5; P < 0.001) and mortality (5 years) risks (HR, 4.0; 95% CI, 2.7–5.8; P < 0.001). When TMAO levels were included in the Cox regression models, elevated levels of both TMAO and TML remained significant predictors of incident (3-year) MACE and incident (5-year) mortality risks after multivariate adjustments including CAD status, traditional CVD risk factors, indices of renal function, and medications (Figure 3, B and C). The strength of the prognostic value of TML observed in the presence of TMAO is different from that observed with other TMA-containing compounds like choline, carnitine, or betaine, since in past studies, the prognostic value of these TMA and TMAO precursors (choline, carnitine, or betaine) are completely abolished following inclusion of TMAO into statistical models (8, 19). The data observed (Figure 2C and Figure 3, B and C) thus suggest that some of the prognostic signal associated with TML and incident CVD event risks appears to be independent of TMAO levels.
Figure 3. Relationship between plasma TML and both CVD risks and TMAO.
(A) Correlation between plasma levels of trimethyllysine (TML) and trimethylamine N-oxide (TMAO) in the validation cohort (N = 2,140). (B) Kaplan–Meier plot illustrating the relationship between plasma TML and risk of incident (3-year) major adverse cardiac events (MACE); and (C) incident (5-year) mortality risk according to TML and TMAO levels where each marker is categorized above versus below the median level in the validation cohort (N = 2,140). Also shown are hazard ratio (HR [95% CI]) for the indicated TML and TMAO grouping using either an unadjusted model, or following adjustments for traditional cardiovascular disease (CVD) risk factors (age, sex, HDL, LDL, smoking, diabetes mellitus, hypertension), high-sensitivity C-reactive protein level, estimated glomerular filtration rate (eGFR ), history of CAD and medications. Median plasma concentration of TML (0.53 μM) and TMAO (3.69 μM) within the cohort was used to stratify subjects as high (≥median) or low (<median) values. (D) Plot of HR for incident 3-year MACE risk stratified by indicated low, intermediate, and high levels of TMAO (cutoff values of 2.5 and 6 μM) and TML (cutoff values of 0.5 and 0.8 μM). *P < 0.05, **P < 0.01, ***P < 0.001 relative to low/low TMAO/TML group. (E) Comparison of plasma levels of TML in free form versus protein-bound TML levels in random samples from both healthy subjects and subjects with CVD, as indicated. The analyses were performed using R 3.4.1.
To further examine and illustrate both the independence of TML and TMAO and their potential additive prognostic value, we stratified the entire cohort into low, intermediate, and high groups of both TML (cutoffs 0.5, 0.8 μM) and TMAO (cutoffs 2.5, 6.0 μM) levels (Figure 3D and Supplemental Table 2). Notably, compared with subjects with low levels of both TML and TMAO (reference group), who experienced an absolute annual MACE rate of 2.8%/year, subjects with both high TML and TMAO levels experienced a remarkable 10%/year annual event rate, which represents a 4.0-fold increased risk of incident MACE (HR, 4.0; 95% CI, 2.6–6.2; P < 0.001) (Figure 3D and Supplemental Table 2). Moreover, the independence of TML and TMAO clinical prognostic value is further illustrated by noting that even among subjects with low TMAO levels, increasing levels of TML are associated with significant increase in MACE risk (2.7-fold risk when TML is high; P = 0.04); and similarly, among subjects with a low TML level, increasing TMAO levels dose dependently is associated with increased MACE risk (e.g., 1.9-fold risk when TMAO is concurrently high; P = 0.03) (Figure 3D and Supplemental Table 2). Thus, unlike other TMAO nutrient precursors like choline, carnitine, or betaine (8, 19), TML provides independent and additive prognostic value in combination with TMAO for prediction of incident MACE risks.
Given the above results, we next proceeded to explore the potential origins of the association between TML and incident CVD risks. TML can be generated in both free and protein-bound form via lysine methylation. Methylation of free lysine to form TML is an initial step in the biosynthetic pathway for carnitine (i.e., lysine→TML→γ-butyrobetaine→carnitine) (20, 21). Within the same subjects examined for TML levels (N = 2,140) we therefore also performed stable-isotope-dilution LC-MS/MS analyses of each of the metabolites in the carnitine biosynthetic pathway using established methods (1, 8, 9, 15). Of note, each metabolite demonstrated a significant correlation with plasma TML levels (Spearman’s correlation coefficients of TML with lysine [r = 0.16, P < 0.001]; γ-butyrobetaine [r = 0.43, P < 0.001]; and carnitine [r = 0.28, P < 0.001]). Protein-bound TML is also generated as a common posttranslational modification of protein lysine residues, particularly histones during chromatin remodeling and transcriptional regulation (22, 23); however, the presence and distribution of protein-bound TML in secreted proteins within plasma has been largely unexplored. We therefore examined the relationship between plasma free TML levels versus protein-bound TML in plasma proteins recovered (see Methods) from a random set of subjects (N = 25). Remarkably, the content of total protein-bound TML in plasma proteins was highly correlated with free TML among all subjects (Spearman’s correlation r = 0.82, P < 0.001), as well as among both healthy controls (r = 0.69, P < 0.01; N = 16), and subjects with CVD (r = 0.95, P < 0.001; N = 9) (Figure 3E).
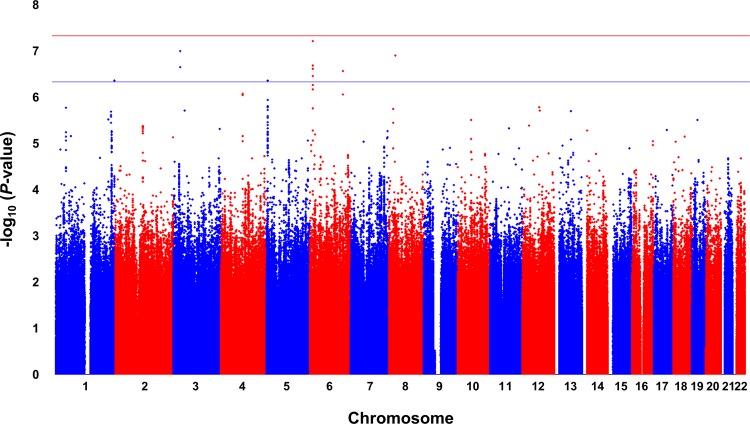
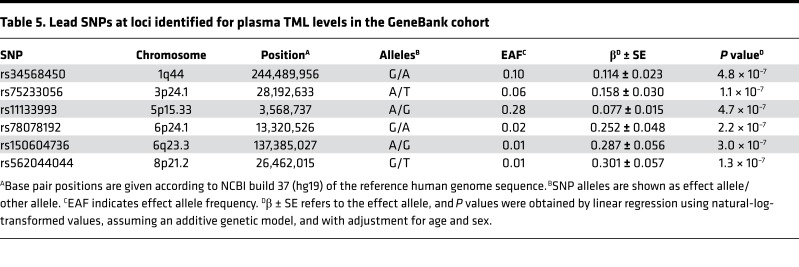
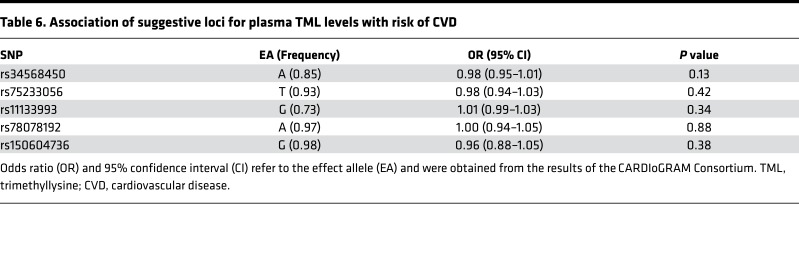
The factors that contribute to variations in plasma TML levels among subjects are largely unexplored. We therefore sought to test whether we could identify genetic determinants of plasma TML levels using an unbiased genome-wide association study (GWAS) approach. Of the 2,140 subjects with TML levels, 1,297 also had data for 9,012,028 imputed autosomal single-nucleotide polymorphisms (SNPs). As shown by the Manhattan plot (Figure 4), no locus exceeded the genome-wide significant P value of 5.0 × 10–8, but several SNPs (mostly with minor allele frequencies [MAFs] ≤ 5%) on chromosomes 1, 3, 5, 6, and 8 exhibited suggestive evidence of association (P < 5.0 × 10–7) (Table 5). We also specifically looked at 400-kb regions surrounding the genes encoding 27 known lysine methyltransferases (24), but none of these candidate loci yielded evidence of association with TML levels either. We next used the results from the CARDIoGRAM Consortium (25) to determine whether any of the 6 suggestively associated regions/loci for plasma TML levels were associated with risk of CVD. Data were available for 5 of these loci, but none of these were associated with risk of CVD in CARDIoGRAM (Table 6).
Figure 4. Results of a GWAS for plasma TML levels in the GeneBank cohort.
The Manhattan plot for plasma TML levels (N = 1,297 subjects) shows 6 suggestively associated loci on chromosomes 1q44, 3p24.1, 5p15.33, 6p24.1, 6q23.3, and 8p21.2. The genome-wide thresholds for significant (P = 5.0 × 10–8) and suggestive (P = 5.0 × 10–7) association are indicated by the horizontal red and blue lines, respectively. P values were obtained using linear regression with natural-log-transformed values and adjustment for age and sex.
Table 5. Lead SNPs at loci identified for plasma TML levels in the GeneBank cohort.
Table 6. Association of suggestive loci for plasma TML levels with risk of CVD.
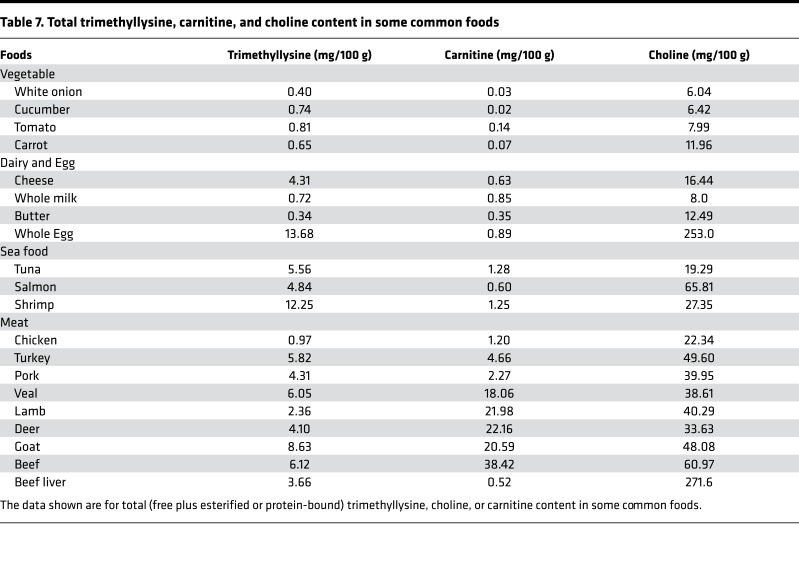
Since both plant and animal cells alike use histone protein methylation to regulate gene expression, we wondered whether TML (free plus protein bound) could be a potentially unrecognized yet abundant nutrient precursor for TMAO generation from plant- and animal-derived foods. To examine the relative levels and distribution of TML versus other major TMAO nutrient precursors in common foods, we quantified total (free + esterified or protein-bound) levels of TML, choline, and carnitine in a variety of animal- and plant-derived foods (Table 7). Surprisingly, TML was found in virtually all food sources, though in general, at a greater level in animal- versus plant-derived products. Thus, when compared with total choline or total carnitine content in foods, TML was present at surprisingly abundant levels (Table 7).
Table 7. Total trimethyllysine, carnitine, and choline content in some common foods.
Dietary TML can generate TMA and TMAO via gut microbiota.
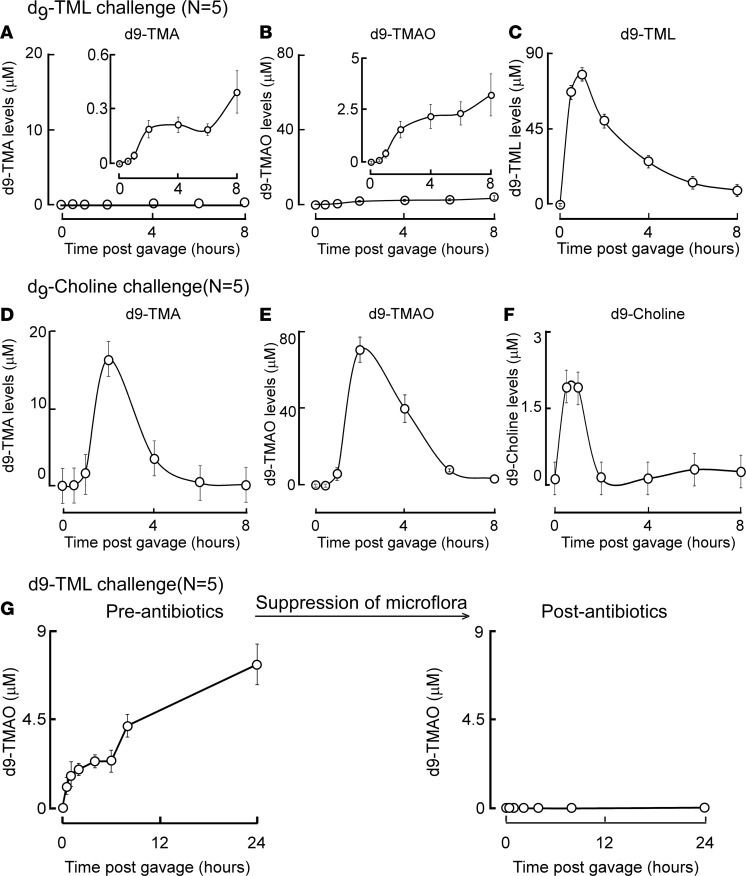
Given the presence of TML in so many food sources, we next examined whether dietary TML could serve as a substrate for microbiota-dependent generation of TMA and TMAO. We initially performed oral (gastric gavage) challenges in mice using either synthetic d9(trimethyl)-TML (see Supplemental Methods for synthesis and characterization) or d9(trimethyl)-choline, and monitored serial plasma samples over time for quantification of isotope-labeled parent compound and d9-TMA and d9-TMAO levels. Following oral d9-TML gavage of conventionally reared C57BL/6J mice, plasma levels of both d9-TMA and d9-TMAO rose only minimally following a lag phase, despite a rapid (within 1 hour) increase in d9-TML level following ingestion (Figure 5, A–C). By comparison, identically challenged C57BL/6J mice with the same (molar) amount of oral d9-choline demonstrated markedly (30- to 50-fold) higher peak levels of d9-TMA and d9-TMAO, and substantially lower plasma levels of isotope-labeled precursor (d9-choline) (Figure 5, D–F). These results directly demonstrate that oral TML can serve as a precursor for TMA and TMAO generation. Of note, however, compared with comparable oral challenge of choline, TML was a much poorer precursor for TMA and TMAO formation in vivo.
Figure 5. d9-TML and d9-choline oral isotope tracer studies.
Synthetic d9-TML or d9-choline was administered by gastric gavage to the indicated numbers of C57BL/6J mice and serial plasma levels of the indicated isotope-labeled compounds were quantified by LC-MS/MS, as described under Methods. (A–C) Plasma levels of d9-trimethylamine (d9-TMA), d9-trimethylamine N-oxide (d9-TMAO), and d9-trimethyllysine (d9-TML) are shown at the indicated times following oral challenge with time of d9-TML gavage designated as T = 0. (D–F) Plasma d9-TMA, d9-TMAO, or d9-choline at the indicated times following oral d9-choline challenge. (G) Plasma concentrations of d9-TMAO were also quantified by LC-MS/MS in C57BL/6J female mice at the indicated times following challenge with d9-TML via gastric gavage, either before or following 3-week administration of a cocktail of broad-spectrum poorly absorbed antibiotics, as described under Methods. All data are presented as mean ± standard error.
To determine whether gut microbes are involved in dietary TML-dependent generation of TMAO, parallel studies were performed in mice before versus after a 3-week exposure to a cocktail of oral poorly absorbed broad-spectrum antibiotics previously shown to markedly suppress gut microbiota (1, 26). Notably, despite initial generation of TMAO before antibiotics (Figure 5G), no detectable plasma d9-TMAO was observed following d9-TML oral gavage in mice treated with the oral antibiotics (Figure 5G). Further, the pretreatment with oral antibiotics had no impact on plasma d9-TML levels following oral gavage (P = 0.87). Collectively, these results are consistent with gut microbial involvement in TMA and TMAO generation from dietary TML, albeit at a level substantially lower than observed with dietary choline.
Characterization of mouse and human gut bacteria–dependent generation of TMA from TML.
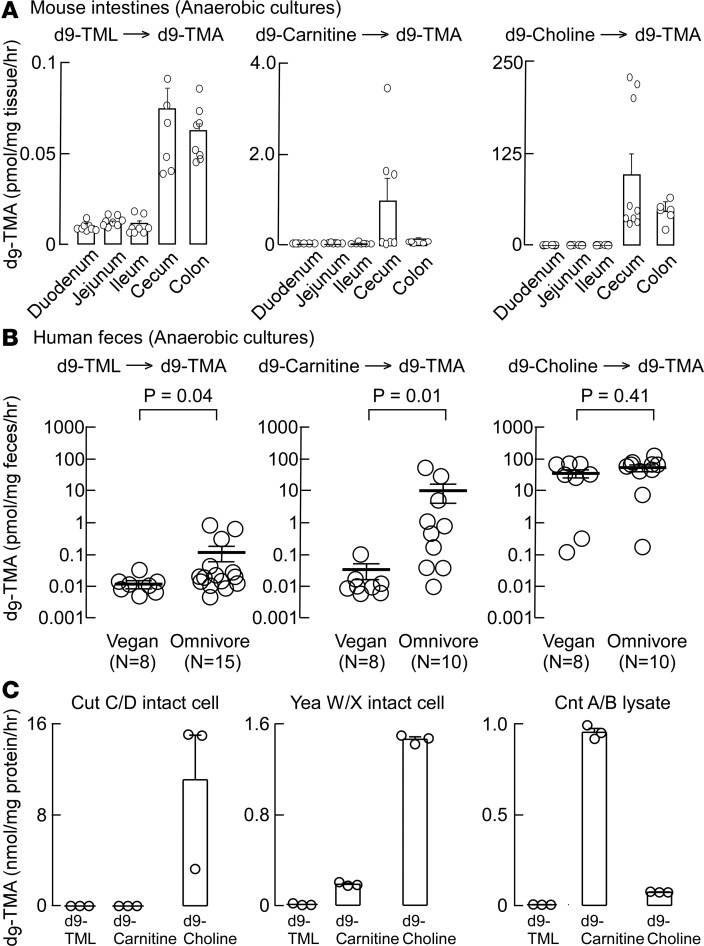
Initial studies examining cecal contents recovered from conventionally reared mice revealed d9-TMA generation from d9-TML under anaerobic (but not aerobic) culture conditions, as described under Methods. We therefore examined the anatomical distribution of gut microbiota capable of producing d9-TMA from d9-TML in intestinal contents recovered from segments harvested from conventionally raised mice and cultured in an anaerobic chamber. To permit direct comparisons, we also performed side-by-side anaerobic cultures using comparable amounts of 2 alternative TMA-producing nutrient precursors, d9-carnitine and d9-choline (Figure 6A). Interestingly, microbes recovered from both cecal and colonic segments generated d9-TMA from d9-TML under anaerobic culture, and significantly lower but detectable levels of d9-TMA were also formed from d9-TML incubated with small-bowel microbes. By comparison, only cecal and colonic microbiota converted d9-carnitine or d9-choline into d9-TMA. Notably, the synthetic capacity of cecal microbiota to generate d9-TMA from d9-carnitine was 10-fold higher than that observed with d9-TML, and 1,000-fold higher from a comparable exposure to d9-choline. These data thus reveal a nutrient TMA-generating potential from murine gut commensals of TML < carnitine << choline (Figure 6A).
Figure 6. Characterization of microbial TMA formation from TML versus alternative TMA-generating nutrients in mouse intestines, human fecal cultures, and cloned microbial TMA lyases.
(A) Intestines from conventionally reared C57BL/6J mice (N = 7) were sectioned as indicated, incubated with either d9-trimethyllysine (d9-TML), d9-carnitine, or d9-choline anaerobically, and then production of d9-trimethylamine (d9-TMA) quantified as described in Methods. (B) Human feces from vegans (N = 8) and omnivores (N = 10–15 as indicated) were incubated with either d9-TML, d9-carnitine, or d9-choline anaerobically, and then production of d9-TMA quantified as described in Methods. Student’s t test (2 tailed) was used to examine the difference between groups. All data are presented as mean ± standard error. (C) Recombinant microbial TMA lyases were cloned and expressed, and then activity (production of d9-TMA) with the indicated d9-labeled substrates determined as described under Methods.
We next sought to examine known culturable human commensals to identify specific microbial species capable of using TML as substrate to generate TMA. We began with studies employing a panel of 68 culturable human commensals, representing a large fraction of the diversity observed in the human gut, including the 7 most abundant bacterial phyla and 28 different genera. This library was previously used to identify species capable of converting choline into TMA (i.e., through choline TMA lyase activity) (27). Eighteen individual members of the human commensal library were initially screened under anaerobic conditions as previously described for detecting choline TMA lyase activity using d9-choline as substrate, but this time adding d9-TML to media, as described under Methods. Despite the ability of several strains to convert d9-choline→d9-TMA under these conditions, none of these species significantly promoted conversion of d9-TML→d9-TMA (data not shown). We next examined whether a pool containing all 68 species (grown individually and mixed in similar proportions prior to the assay) could promote the conversion of d9-TML→d9-TMA, speculating that perhaps the observed activity in cultured cecal/colonic contents, and in vivo (mouse isotope tracer studies), might occur via sequential metabolic transformations promoted by more than one microbe. Unfortunately, however, virtually no detectable d9-TML→d9-TMA transformation was observed with pooled microbes in prolonged (4 days) incubation (assay limit of detection, ~1 nM). These results suggest that conversion of TML to TMA is a relatively rare trait, possibly restricted to a specific taxonomic group not represented in our screen, or possibly inactive under the screening conditions.
Given that d9-TML is clearly converted into d9-TMA in a gut microbiota–dependent fashion in vivo in mice (Figure 5), and in anaerobic cultures of either mouse cecal or colonic contents (Figure 6A), we next decided to examine human polymicrobial fecal anaerobic cultures for their capacity to convert d9-TML→d9-TMA. Human feces from both healthy volunteer omnivores (N = 10 or more) and vegans (N = 8) were examined for d9-TMA generation from equivalent amounts of d9-TML versus the alternative TMA precursor nutrients, d9-carnitine and d9-choline, under anaerobic culture conditions. Notably, feces from omnivores and vegans alike promoted the transformation of d9-TML→d9-TMA, with omnivores having significantly greater activity than vegans (P < 0.04; Figure 6B). However, the overall level of TMA generation from TML was relatively low, with d9-carnitine→d9-TMA transformation showing a comparable degree of d9-TMA formed in vegans. Remarkably, over a 100-fold greater production of d9-TMA was produced from d9-carnitine incubated with polymicrobial fecal cultures from omnivores (Figure 6B). Even more notable was the observation that d9-choline→d9-TMA conversion within anaerobic cultures from human feces recovered from vegan and omnivore alike were both approximately 3 orders of magnitude greater than the levels of d9-TMA produced from d9-TML (Figure 6B). These studies thus revealed a nutrient TMA-generating potential from human fecal commensals of TML < carnitine << choline, similar to results observed with mouse cecal and colonic microbiota.
In a final series of studies aimed at examining gut microbial participants in TMA/TMAO generation, we examined whether recombinant forms of known microbial enzymes capable of generating TMA with alternative substrates could instead use TML as substrate to produce TMA. We thus examined transformed E. coli expressing the genes cutC/D, yeaX/W, or cntA/B, microbial TMA-generating enzyme systems previously reported for their capacity to use choline-related, or carnitine-related substrates (28–30). With E. coli transformed individually with each recombinant enzyme system, and despite observing enzymatic activity (TMA generation) with previously reported substrates for generating TMA from the given microbial TMA lyases, none of the known recombinant microbial TMA generating enzyme systems examined supported d9-TML conversion into d9-TMA (Figure 6C).
Impact of chronic dietary TML versus choline supplementation on systemic TMAO levels and in vivo thrombosis potential.
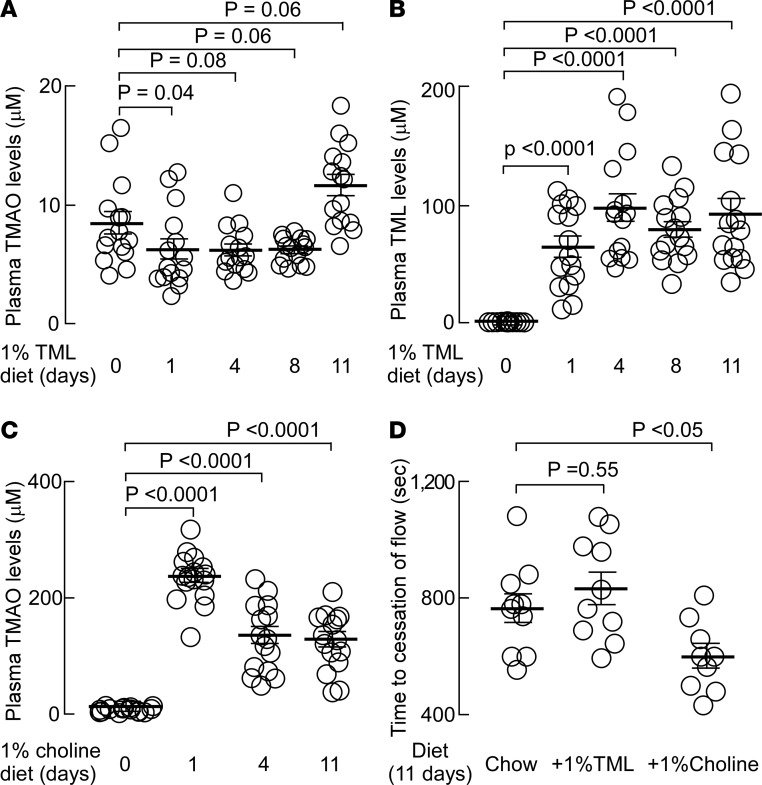
In previous studies we have shown that chronic provision of a dietary nutrient can lead to enhanced gut microbial community capacity to form TMA and raise systemic TMAO levels (8). Since TML→TMA conversion in human fecal polymicrobial cultures derived from omnivores was significantly greater than that observed from vegans (Figure 6B), we hypothesized that chronic dietary exposure may enhance gut microbiota–dependent conversion of TML→TMA. We therefore sought to investigate the impact of chronic dietary TML supplementation on plasma TMAO levels and in vivo thrombosis potential in mice. Since a large amount of TML is not readily commercially available, before proceeding, we synthesized and chemically characterized sufficient quantities of TML for the proposed in vivo studies (Supplemental Figure 4 and Supplemental Table 3). In addition, for direct comparison, parallel mouse studies were performed using chronic supplementary (1%) dietary choline. Provision of supplemental dietary TML (1%) to mice for up to 11 days resulted in a highly significant, nearly 100-fold increase in plasma TML levels (P < 0.0001) but no significant increase in systemic TMAO levels (Figure 7, A and B). By comparison, provision of the comparable amount of supplemental dietary choline (1%) significantly (~20-fold) raised plasma TMAO levels (P < 0.0001, Figure 7C). We further tested if elevated plasma TML in mice fostered enhanced thrombosis potential in vivo using a FeCl3-induced carotid artery injury model, as described in Methods. The impact of a comparable amount of choline supplementation on in vivo thrombosis potential was similarly examined for comparison. As shown in Figure 7D, chronic dietary TML failed to significantly change thrombus formation rate, or time to cessation of blood flow, in the carotid artery injury in vivo thrombosis model (Figure 7D). In contrast, however, as previously observed with choline supplementation (11), significant TMAO elevations were accompanied by enhanced in vivo thrombosis potential, as monitored via reduction in time to cessation of blood flow of the carotid artery following FeCl3-induced injury (Figure 7D).
Figure 7. Impact of TML- versus choline-supplemented diet on plasma levels of TML and TMAO, and in vivo thrombosis potential.
Groups of mice (N = 15) were placed on chemically defined diets supplemented with either 1% trimethyllysine (TML) (A and B) or 1% choline (C) for 11 days as described under Methods. At the indicated times, plasma levels of trimethylamine N-oxide (TMAO) (A and C) or TML (B) were quantified by stable-isotope-dilution LC-MS/MS. (D) In addition, after 11 days of the indicated diet (N = 10), the impact of supplemental dietary TML versus choline on both TMAO levels and in vivo thrombosis potential, as monitored using the FeCl3 carotid artery injury model, was determined as described under Methods. Plasma levels of metabolites after 11 days of the indicated diets were as follows: for 1% TML group, TMAO =4.5 ± 0.7 μM, TML = 37.0 ± 4.9 μM. For 1% choline group, TMAO = 90.6 ± 19.5 μM, TML = 0.8 ± 0.1 μM. For chemically defined chow group, TMAO= 2.6 ± 0.4 μM; TML = 0.9 ± 0.2 μM. Student’s t test (2 tailed) was used to examine the difference between groups. All data are presented as mean ± standard error.
Discussion
Herein we utilized untargeted metabolomics to discover that systemic levels of the unusual free amino acid, TML, are strikingly associated with incidence of CVD risks independent of multiple cardiometabolic risk factors, prevalent CVD, and medication use. Our studies also reveal that TML, a TMA-containing compound, is surprisingly abundant in both plant- and animal-derived foods alike, and that gut microbiota have evolved enzymatic machinery to metabolize TML. Nonetheless, our studies show that in gut commensals from both mice and humans, the utilization of TML as nutrient precursor for TMA and TMAO formation is a relatively modest microbial activity compared with microbiota-dependent conversion of choline, and to lesser extent, carnitine (especially in omnivores). Further, gut microbial transformation of TML into TMA occurs via enzyme(s) distinct from known microbial choline or carnitine TMA lyases, such as cutC/D, cntA/B, or yeaW/X.
Despite the observed association between TML and incident CVD risks, the underlying mechanism(s) accounting for the strong association remain unclear. One obvious mechanism that was extensively explored was the potential capacity of TML to form TMA and the atherogenic and prothrombotic metabolite TMAO. However, multiple lines of evidence suggest that mechanisms alternative to TMAO account for the observed association between TML and incident CVD risks. First, in clinical studies, adjustment for TMAO in Cox models failed to significantly attenuate the clinical prognostic value of TML and incident CVD risks. This behavior contrasts with that observed with other TMA nutrient precursors like choline, betaine, or carnitine, which lose their clinical prognostic value for incident CVD risks when TMAO levels are included in multilogistic regression models (8, 19). Further, despite observing gut microbiota–dependent generation of TMA and TMAO from TML, plasma levels of TMAO were not affected by dietary TML. Thus, in contrast to choline supplementation, which raises TMAO levels and increases in vivo thrombosis potential (11), chronic TML supplementation both failed to raise TMAO levels and did not enhance in vivo thrombosis potential as monitored using a carotid artery injury model. Lastly, to try and identify factors that may be linked to systemic TML levels and CVD risk, we performed a GWAS study in approximately 1,300 subjects. While several suggestive loci were identified, none reached genome-wide significance or yielded evidence of association with risk of CVD.
An intriguing finding in the present study is the remarkably tight correlation observed between protein-bound TML and free TML levels. These results raise the prospects that free TML levels may reflect the level of protein-bound TML in the vascular compartment. They thus suggest that further investigation into defining which proteins in plasma are modified through lysine methylation is of interest. Even more so, it will be of interest to explore whether posttranslational modification of plasma or vascular wall proteins alters their function. In this respect, it is intriguing to note that lysine methylation plays a critical role in the histone code and the overall regulation of chromatin remodeling and transcriptional gene control. While lysine methylation was originally proposed to be an irreversible posttranslational modification on proteins, it has recently been recognized that protein lysine residue methylation (at least for histones) can be reversible, with multiple demethylases discovered (31–33). With discovery of protein-bound TML present in plasma and its remarkably high correlation with free TML, the role of protein lysine methylation in both the secreted proteome and proteins within the vascular wall and other organs is of interest. Moreover, whether the protein-bound TML detected in the vascular compartment is a reversible posttranslational modification, like in histones, is similarly of interest.
It is remarkable that the present studies identify yet another TMA-containing metabolite, TML, with links to both TMA and TMAO generation and significant clinical prognostic value for incident adverse cardiovascular event risks. Of note, in contrast to choline or carnitine, chronic dietary TML fails to significantly change plasma TMAO levels, and in this respect, TML behaves somewhat like betaine, which similarly was identified through untargeted metabolomics as having plasma levels associated with incident MACE risks (1), and which can be a TMA nutrient precursor, albeit also at a rate about 100-fold less than choline (19). The present studies reaffirm the promise of untargeted metabolomics as a platform for discovery of candidate metabolites and pathways with links to disease processes. They substantially expand our understanding of TML and its association with CVD risks, and suggest further exploration of the pathobiology of protein lysine methylation, and the biological processes that modulate free TML levels are of interest.
Methods
Clinical study population.
Plasma samples, feces, and associated clinical data were collected as part of 2 studies at a tertiary care center. Metabolomics studies were performed on GeneBank samples, a large (N > 10,000) well-characterized and longitudinal tissue repository with associated clinical database. It comprised sequential participants enrolled in the study GeneBank, which consists of sequential stable subjects without evidence of acute coronary syndrome (cardiac troponin I < 0.03 ng/ml) who underwent elective diagnostic coronary angiography (cardiac catheterization or coronary computed tomography) for evaluation of CAD (1, 16, 34). All subjects had extensive clinical and longitudinal outcome data monitored, including adjudicated outcomes over the ensuing 3 to 5 years for all participants after enrollment. MACE was defined as death, nonfatal myocardial infarction, or nonfatal cerebrovascular accident (stroke) following enrollment. Initial discovery metabolomics analyses began with an unbiased search for plasma analytes linked to CVD risk using a case/control design (49 cases and 50 controls, discovery cohort 1). Cases were randomly selected from GeneBank subjects who experienced a myocardial infarction, stroke, or death over the ensuing 3-year period following enrollment (1). For controls, an age- and gender-matched group was randomly selected from GeneBank subjects that did not experience a CVD event. An independent nonoverlapping cohort (N = 1,162, discovery cohort 2) for untargeted metabolomics was also derived from sequential consenting subjects enrolled in GeneBank. A third large independent nonoverlapping validation cohort (N = 2,140) was also from sequential consenting subjects enrolled in GeneBank. Fasting blood glucose, high-sensitivity C-reactive protein, and lipid profiles were measured on the Roche Cobas platform (Roche Diagnostics).
In additional studies, healthy volunteers (N = 15 omnivores and N= 8 vegetarians or vegans) were consented and subjected to extensive dietary questioning and stool collection as part of a study examining gut microbiota–dependent metabolism of dietary carnitine and CVD (8). Only feces at baseline were used. Male and female volunteers were at least 18 years of age. Volunteers participating were excluded if they were pregnant, had chronic illness (including a known history of heart failure, renal failure, pulmonary disease, gastrointestinal disorders, or hematologic diseases).
Initial metabolomics analyses.
In general, the initial untargeted metabolomics approach used was similar to that described previously (1, 8). Plasma proteins were precipitated with 4 volumes of ice-cold methanol and small-molecule analytes within supernatants were analyzed after injection onto a phenyl column (4.6 × 250 mm, 5 μm Rexchrom Phenyl; Regis) at a flow rate of 0.8 ml/min using a Cohesive HPLC interfaced with a PE Sciex API 365 triple-quadrupole mass spectrometer (Applied Biosystems) with Ionics HSID1, EP101, XT1 redesigned source and collision cell as upgrades in positive MS1 mode. LC gradient (LC1) starting from 10 mM ammonium formate over 0.5 minutes, then to 5 mM ammonium formate, 25% methanol and 0.1% formic acid over 3 minutes, held for 8 minutes, followed by 100% methanol and water washing for 3 minutes at a flow rate of 0.8 ml/min was used to resolve analytes. Spectra were continuously acquired after the initial 4 minutes. Peaks within reconstructed ion chromatograms at 1–atomic mass unit increments were integrated and both retention times and m/z of analytes were used for statistical analyses.
To chemically define the structures of the plasma analyte selected for further investigation (that is, candidate compound with m/z 189.1 in positive MS1 mode). TML in plasma was identified by HPLC/high-resolution mass spectrometer with the same retention time, high-resolution mass, and fragmented ions as standard. Plasma supernatant after methanol precipitation of protein was analyzed by injection onto a silica column (2.0 × 150 mm, 5 μm Luna silica; catalog 00G-4274-B0, Phenomenex) using a 2 LC-20AD Shimadazu pump system, SIL-HTC autosampler interfaced with a TripleTOF 5600 mass spectrometer (AB SCIEX). Separation was performed employing a gradient starting at 100% A (10 mM ammonium acetate aqueous solution) for 2 minutes, then increasing linearly to 20% B (acetonitrile/methanol/acetic acid, 50:50:0.1, v/v/v) over 8 minutes followed by 100% B washing for 5 minutes and 100% A balancing for 5 minutes. The parameters for the ion monitoring were as follows: spray voltage, 5.5 kV; curtain gas, 20; GS1, 20; GS2, 15; DP, 30; CE, 30 volts, CES, 15 volts. Nitrogen (99.95% purity) was used as the only gas.
To confirm the identity of plasma analyte 189.1 as TML, the plasma analyte and synthetic d9-TML standard were examined on multiple different column matrices and chromatography conditions:
System 1: column (2.0 × 150 mm, 5 μm Luna silica; catalog 00G-4274-B0, Phenomenex), separated on discontinuous gradient composed of mobile phase A (0.1% propionic acid in water ) versus mobile phase B (0.1% acetic acid in methanol) starting at 100% A for 2 minutes, then increasing linearly to 15% B over 8 minutes followed by 100% B washing for 5 minutes and balancing for 5 minutes; the flow rate was 0.2 ml/min. System 2: column (2.0 × 50 mm, 1.9 μm Titan silica; catalog 581530-U, Supelco), separated on discontinuous gradient composed of mobile phase A (10 mM ammonium formate and 0.1% formic acid in water ) versus mobile phase B (10 mM ammonium formate and 0.1% formic acid in acetonitrile) starting at 100% B linearly decreasing to 100% A over 4 minutes followed by washing with 100% A for 0.5 minutes and balancing with 100% B for 0.5 minutes; the flow rate was 0.4 ml/min. System 3: column (2.0 × 150 mm, 3 μm Scherzo SM-C18; catalog SM025, Imtakt) separated on discontinuous gradient composed of mobile phase A (0.1% formic acid in water ) versus mobile phase B (0.1% formic acid in methanol) starting at 100% A for 2 minutes, then increasing linearly to 15% B over 7 minutes followed by 100% B washing for 3 minutes and balancing for 3 minutes; the flow rate was 0.3 ml/min.
Untargeted analysis of plasma samples using HILIC-MS.
Untargeted analysis of plasma samples was similar to that described previously (35, 36). In brief, extraction of plasma metabolites was carried out using an acetonitrile/isopropanol/water (3:3:2, v/v/v) mixture. Aliquots were evaporated, resuspended using an acetonitrile/water (4:1, v/v) mixture, vortexed, centrifuged, and the upper layer transferred to a glass vial with a microinsert. Hydrophilic interaction chromatography (HILIC) analysis was performed on a system including an Agilent 1290 Infinity LC system (Agilent Technologies) with a pump (G4220A), a column oven (G1316C), an autosampler (G4226A), and a TripleTOF 5600+( SCIEX). Extracts were separated on an Acquity UPLC BEH Amide column (150 × 2.1 mm; 1.7 μm, Waters) coupled to a Waters Acquity UPLC BEH Amide VanGuard precolumn (5 × 2.1 mm; 1.7 μm). The column was maintained at 45°C at a flow rate of 0.4 ml/min. The mobile phases consisted of (A) water with ammonium formate (10 mM) and formic acid (0.125%) and (B) 95:5 (v/v) acetonitrile/water with ammonium formate (10 mM) and formic acid (0.125%). The separation was conducted under the following gradient: 0 min 100% B; 0–2 min 100% B; 2–7.7 min 70% B; 7.7–9.5 min 40% B; 9.5–10.25 min 30% B; 10.25–12.75 min 100% B; 12.75–17.75 min 100% B. A sample volume of 1 μl was used for the injection. Sample temperature was maintained at 4°C. The QTOFMS instrument was operated in electrospray ionization in positive mode with the following parameters: curtain gas, 35 psi; ion source gas 1, 50 psi; ion source gas 2, 50 psi; temperature, 300°C; ion spray voltage floating, 4.5 kV; declustering potential, 100 V; acquisition speed, 2 spectra/s. For data processing MZmine 2 (37) and MultiQuant (SCIEX) software programs were used.
Quantification of TML, TMAO, TMA, lysine, butyrobetaine, and carnitine.
Stable-isotope-dilution LC-MS/MS was used for quantification of TML, TMAO, TMA, lysine, γ-butyrobetaine, and carnitine similar to that described previously (1, 8, 9, 15). For TML standard curves, varying levels of TML were spiked into 20 μl control plasma followed by precipitation with 80 μl methanol containing 0.8 nmol d9-TML. Supernatant was loaded for LC-MS/MS analysis. Analyses were performed using electrospray ionization in positive-ion mode with multiple reaction monitoring of parent and characteristic daughter ions and retention times specific for components monitored. The transitions monitored for TML were m/z 189.1→84.1, 189.1→60.1, and 189.1→130.1. The internal standards (i.e., d9-trimethyl-TML, d9-trimethyl-TMAO) were added to plasma before protein precipitation. Various concentrations of analytes and a fixed amount of internal standards were spiked into control plasma to prepare the calibration curves for quantification of plasma analytes. Their concentrations in human and mouse plasma samples were determined by HPLC with a Shimadzu 8050 triple-quadrupole mass spectrometer interfaced to a Shimadzu Nexera ultra high performance liquid chromatography (UHPLC) system.
Genotyping and imputation.
Genome-wide genotyping was carried out on 3031 GeneBank subjects of European ancestry using the Affymetrix Genome-Wide Human Array 6.0 SNP chip. After conversion of genomic coordinates to GRCh37/hg19, exclusion of SNPs with duplicates, call rates < 97%, MAFs < 1%, and without chromosome and base pair position, and exclusion of 44 subjects with genotype call rates < 90%, 642,766 were available for imputation in 2,972 participants. Imputation was carried out on the forward (+) strand using the University of Michigan Imputation Server (https://imputationserver.sph.umich.edu) and data from the 1000 Genomes Project (Phase 3, Version 5). Application of the same quality control filters described above to the 46,180,700 imputed SNPs, with the addition of excluding SNPs with Hardy-Weinberg equilibrium P values < 0.0001 and imputation Rsq scores < 0.3, resulted in 9,012,028 autosomal SNPs that were available for analysis in 1,297 GeneBank subjects for whom plasma TML levels were also available.
Total TML, carnitine, and choline quantification in food.
Plant- and animal-derived products were purchased in various local supermarkets. A portion of samples was accurately weighed into a 2.0-ml polypropylene Eppendorf tube in a total of 1.5 ml water (if homogenization was needed). Tissues were homogenized using a TissueLyser II (Qiagen) with a 5-mm stainless steel ball and with the adapters (pre-chilled at –20°C) operated at 30 Hz for 2 minutes, 5 to 10 times (until samples appeared homogeneous). After thorough mixing, a 20-μl aliquot of homogenate was transferred to a baked glass test tube with mininert valve screw cap, TML, carnitine, and choline isotope-labeled internal standards were added, and 0.5 ml 6N HCl was added. Samples were hydrolyzed by incubation under argon atmosphere at 110°C for 20 hours. Samples and standards were cooled down, 2 ml water added to the hydrolysate, and then the mixture was processed using solid-phase extraction (Discovery DSC-SCX 3 ml Tube, 500 mg, Supelco). Activation and conditioning of the solid-phase column before sample addition consisted of two 2.5-ml aliquots of methanol followed by two 2.5-ml aliquots of 0.2N formic acid. The sample was then loaded and subsequently washed with two 2.5-ml aliquots of formic acid. Analytes were eluted using 70% methanol, 5% ammonium hydroxide, and 0.1 M ammonium formate. The eluent was evaporated using a heated nitrogen evaporation system, and continued drying of recovered material using speed-vacuum, and then the sample was reconstituted to 100 μl with 50% aqueous methanol for stable-isotope-dilution LC-MS/MS analysis. Total plasma TML content of samples followed a similar overall solid protocol except for the use of d9(trimethyl)-TML as internal standard.
Metabolic challenges in mice.
Whole blood (50 μl) was collected via the saphenous vein from C57BL/6J (Jackson Laboratory, 0664) female mice prior to gavage for the measurement of baseline analyte levels. Mice were administered an oral dose of stable-isotope-labeled TML or choline. The gastric gavage consisted of 150 μl of either 150 mM synthetic d9-TML or 150 mM choline-N,N,N-trimethyl-d9 (d9-choline, Cambridge Isotope Lab), followed by postgavage blood collection at the indicated times. Where indicated, gut microbiota were suppressed in mice by placement of a cocktail of poorly absorbed antibiotics in drinking water for 3 weeks prior to the d9-TML challenge, as previously described (1, 19). Quantification of the abundance of native and isotope-labeled forms of TML, TMA, and TMAO in mouse plasma was performed using stable-isotope-dilution LC-MS/MS using established methods for TMA and TML (19), and using the parent daughter ion transitions for TML and its internal standard as described above. In additional studies, the role of gut microbiota in TMAO formation from dietary TML was determined in C57BL/6J mice by performance of oral (gavage) d9-TML challenge. Mice were housed in conventional cages in the absence or presence of a cocktail of broad-spectrum antibiotics added to the drinking water previously shown to suppress gut microbiota (1, 19).
Gut microbial TMA lyase activity assay.
TMA lyase activity was monitored by quantifying d9-TMA production from d9(trimethyl)-labeled potential nutrient precursors by stable-isotope-dilution LC-MS/MS analysis. Briefly, TMA lyase activity was typically quantified by incubating the indicated enzyme source (C57BL/6J female mouse intestine segments or human feces from healthy vegans/vegetarians and omnivores). Reactions were initiated by the addition of d9-labeled substrates (d9-TML, d9-carnitine, or d9-choline) incubated under anaerobic conditions. Following 20 hours incubation at 37°C, reactions were halted, [13C3,15N]-TMA (Sigma-Aldrich) in 0.2N formic acid was added as an internal standard, followed by immediate extraction of TMA isotopologues into the organic layer produced following mixing with successive additions of 2 ml hexane, 1 ml butanol, and 0.2 ml 1N NaOH (to deprotonate the TMA isotopologues). Mixtures were then vortexed, centrifuged, and the upper hexane layer was transferred to a 12 × 75 mm PTFE capped threaded glass tube, acidified with the addition of 200 μl of 0.2N formic acid, vortexed, and the organic and aqueous layers were separated by centrifugation. The now protonated TMA isotopologues were recovered in the aqueous phase for quantification by stable-isotope-dilution LC-MS/MS analysis. Sample aliquots were injected onto a reverse phase C18 HPLC column (2.0 × 150 mm, 5 μm, Phenomenex Prodigy) eluted at a flow rate of 0.2 ml/min and resolved using a linear gradient between 0.2% formic acid in water and 0.2% formic acid in acetonitrile/methanol (95:5, v/v). HPLC column effluent was introduced into an AB Sciex 5500 QTRAP mass spectrometer using electrospray ionization in the positive-ion mode. Both d9-TMA and [13C3,15N]-TMA were monitored using multiple reaction monitoring of parent and characteristic daughter ions: m/z 69→49 for d9-TMA; and m/z 64→47 for [13C3,15N]-TMA. Reactions without microbial enzyme source (tissues, cells, cell lysates, or isolated proteins) were used as blank. Control studies for each d9-TMA–containing substrate showed negligible (~0.25 nM) background d9-TMA production under the conditions employed.
In vitro screening for the conversion of d9-TML by human commensals.
Human feces and all bacteria including individual strain and fresh combined 68 strains were grown with 100 μM d9-TML or d9-choline (as a positive control) in complex reduced medium in a Coy anaerobic chamber (5% H2, 20% CO2, and 75% N2, 37°C) for 0–4 days as previously described (27). The reaction was stopped by adding 6N HCl to samples (1:100 dilution to 60 mM final), and freezing at –80°C until d9-TMA extraction and analysis by LC-MS/MS, as described above in Methods.
In vivo carotid artery thrombosis model.
Conventional mice (female, 10 weeks old, C57BL/6J, 10 per treatment group) were placed on chemically defined diets for 11 days. Mice were assessed for in vivo thrombosis potential with the common carotid injury model by application of 10% FeCl3 for 1 minute (11, 38). To test this, platelets were labeled with rhodamine 6G (100 μl; 1 mg/ml) and injected directly into the right jugular vein. The left carotid artery was exposed and injured by placing a FeCl3-soaked filter paper for 1 minute. Thrombus formation was observed in real time and captured with a video camera linked to an intravital fluorescence microscope. Time to cessation of blood flow was determined by visual inspection of computer images of the thrombotic process from 30 seconds to 30 minutes by investigators blinded to mouse treatment group.
Statistics.
Student’s t test (2 tailed) or Wilcoxon’s rank-sum test for continuous variables and χ2 test for categorical variables were used to examine the differences between groups. Numerical data are presented as mean ± SD or median (25th–75th percentile = Q1–Q3). Categorical data are presented as n (%). HR for MACE and death at 5-year follow-up and corresponding 95% CI were estimated using both univariable (unadjusted) and multivariable (adjusted) Cox models. Kaplan–Meier analysis with Cox proportional hazards regression was used for time-to-event analysis to determine HR and 95% 95% CI for MACE, and all-cause mortality. Adjustments were made for individual traditional cardiac risk factors including but not limited to age, sex, HDL, LDL, smoking, diabetes mellitus, history of hypertension, and high-sensitivity C-reactive protein level. All analyses were performed using R 3.4.1 (Vienna, Austria, 2017). P values < 0.05 were considered statistically significant.
A GWAS for plasma TML levels was carried out using linear regression analyses with natural-log-transformed values and adjustment for age and sex. All analyses were performed using PLINK 1.07 (39) assuming additive genetic models. The results of the Coronary Artery Disease Genome-wide Replication and Meta-Analysis (CARDIoGRAM) Consortium were used to determine whether variants identified for plasma TML levels were associated with CVD. CARDIoGRAM represents a GWAS meta-analysis of CVD comprising 60,801 cases and 123,504 controls from 48 studies, in which logistic regression was first used in each cohort to test for association with CVD using a log-additive model with adjustment for age and sex and taking into account the uncertainty of possibly imputed genotypes. Subsequently, a meta-analysis was performed separately for every SNP from each study that passed the quality-control criteria using a fixed-effects model with inverse-variance weighting or a random-effects model, depending on the presence of heterogeneity between studies (25). The results of these meta-analyses were used to determine whether loci for TML were associated with CVD.
Study approval.
All clinical study protocols and informed consent for human subjects were approved by the Cleveland Clinic Institutional Review Board. Written informed consent was obtained from all subjects. All animal model studies were approved by the Institutional Animal Care and Use Committee at the Cleveland Clinic.
Author contributions
XSL participated in the design, performance, and analysis of most studies presented, and participated in the drafting of the manuscript. AGH and XG aided in chemical synthesis and characterization of all compounds. ZW, TC, IN, CJS, and OF participated in the performance of mass spectrometry analyses. JAB, MAW, and SMS assisted in the animal studies. ABR and JAD cloned, expressed, and performed studies using the various recombinant choline TMA lyases. YW and LL provided statistical support. JAH, YH, HA, and WHWT assisted with GWAS studies. RLK, KAR, and FER assisted in the design and performance of studies employing the human commensal library. SO, TFL, and WHWT provided advice on clinical studies. SLH conceived, designed, and supervised all studies, and participated in the drafting of the manuscript. All authors contributed to the critical review of the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the NIH and the Office of Dietary Supplements (HL113452, HL103866, DK106000). GeneBank was partially supported by NIH grants R01HL126827, P01 HL076491, and R01HL130819. H. Allayee and his team were supported by NIH grants R01ES021801, R01ES025786, and R01HL133169. Some of the mass spectrometry studies were performed on instruments housed in a facility supported in part by a Center of Excellence Award by Shimadzu Scientific Instruments. T.F. Lüscher was supported by grants from the Swiss National Science Foundation (SPUM 33CM30-124112 and 32473B_163271). S.L. Hazen was partially supported by a gift from the Leonard Krieger endowment. This work was also supported in part by a grant from a Transatlantic Networks of Excellence Award from the Leducq Foundation.
Version 1. 03/22/2018
Electronic publication
Footnotes
AGH’s present address is: Department of Biostatistics, University of Pittsburgh, Pittsburgh, Pennsylvania, USA. KAR’s present address is: Department of Cellular and Molecular Medicine, Cleveland Clinic, Cleveland, Ohio, USA.
Conflict of interest: SLH and ZW are named as coinventors on patents held by the Cleveland Clinic relating to cardiovascular diagnostics and therapeutics. SLH and ZW report being eligible to receive royalty payments for inventions or discoveries related to cardiovascular diagnostics or therapeutics from Cleveland Heart Lab and Procter & Gamble (P&G). SLH is a paid consultant for P&G, and has received research funds from P&G, Pfizer Inc., Roche Diagnostics, and Takeda.
Reference information: JCI Insight. 2018;3(6):e99096. https://doi.org/10.1172/jci.insight.99096.
Contributor Information
Zeneng Wang, Email: WANGZ2@ccf.org.
Tomas Cajka, Email: tomas.cajka@gmail.com.
Jennifer A. Buffa, Email: buffaj@ccf.org.
Ina Nemet, Email: nemeti@ccf.org.
Xiaodong Gu, Email: gux2@ccf.org.
Yuping Wu, Email: wuy2@ccf.org.
Lin Li, Email: lil@ccf.org.
Jaana A. Hartiala, Email: hartiala@usc.edu.
Yi Han, Email: han003@usc.edu.
Slayman Obeid, Email: Slayman.Obeid@usz.ch.
Hooman Allayee, Email: hallayee@usc.edu.
Joseph A. DiDonato, Email: didonaj@ccf.org.
Oliver Fiehn, Email: ofiehn@gmail.com.
W.H. Wilson Tang, Email: tangw@ccf.org.
Stanley L. Hazen, Email: hazens@ccf.org.
References
- 1.Wang Z, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472(7341):57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albert CJ, Anbukumar DS, Monda JK, Eckelkamp JT, Ford DA. Myocardial lipidomics. Developments in myocardial nuclear lipidomics. Front Biosci. 2007;12:2750–2760. doi: 10.2741/2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beger RD, et al. Metabolomics enables precision medicine: “A White Paper, Community Perspective”. Metabolomics. 2016;12(10):149. doi: 10.1007/s11306-016-1094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang K, Han X. Lipidomics: techniques, applications, and outcomes related to biomedical sciences. Trends Biochem Sci. 2016;41(11):954–969. doi: 10.1016/j.tibs.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ussher JR, Elmariah S, Gerszten RE, Dyck JR. The emerging role of metabolomics in the diagnosis and prognosis of cardiovascular disease. J Am Coll Cardiol. 2016;68(25):2850–2870. doi: 10.1016/j.jacc.2016.09.972. [DOI] [PubMed] [Google Scholar]
- 6.Gross RW. The evolution of lipidomics through space and time. Biochim Biophys Acta. 2017;1862(8):731–739. doi: 10.1016/j.bbalip.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Afshinnia F, et al. Impaired β-oxidation and altered complex lipid fatty acid partitioning with advancing CKD. J Am Soc Nephrol. 2018;29(1):295–306. doi: 10.1681/ASN.2017030350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koeth RA, et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19(5):576–585. doi: 10.1038/nm.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koeth RA, et al. γ-Butyrobetaine is a proatherogenic intermediate in gut microbial metabolism of L-carnitine to TMAO. Cell Metab. 2014;20(5):799–812. doi: 10.1016/j.cmet.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregory JC, et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem. 2015;290(9):5647–5660. doi: 10.1074/jbc.M114.618249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu W, et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165(1):111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kind T, et al. Identification of small molecules using accurate mass MS/MS search. Mass Spectrom Rev. 2017;9999:1–20. doi: 10.1002/mas.21535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34(8):828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wohlgemuth G, et al. SPLASH, a hashed identifier for mass spectra. Nat Biotechnol. 2016;34(11):1099–1101. doi: 10.1038/nbt.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Levison BS, Hazen JE, Donahue L, Li XM, Hazen SL. Measurement of trimethylamine-N-oxide by stable isotope dilution liquid chromatography tandem mass spectrometry. Anal Biochem. 2014;455:35–40. doi: 10.1016/j.ab.2014.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang WH, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med. 2013;368(17):1575–1584. doi: 10.1056/NEJMoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senthong V, Wang Z, Fan Y, Wu Y, Hazen SL, Tang WH. Trimethylamine N-oxide and mortality risk in patients with peripheral artery disease. J Am Heart Assoc. 2016;5(10):e004237. doi: 10.1161/JAHA.116.004237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang WH, et al. Increased trimethylamine N-oxide portends high mortality risk independent of glycemic control in patients with type 2 diabetes mellitus. Clin Chem. 2017;63(1):297–306. doi: 10.1373/clinchem.2016.263640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Z, et al. Prognostic value of choline and betaine depends on intestinal microbiota-generated metabolite trimethylamine-N-oxide. Eur Heart J. 2014;35(14):904–910. doi: 10.1093/eurheartj/ehu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vaz FM, Wanders RJ. Carnitine biosynthesis in mammals. Biochem J. 2002;361(Pt 3):417–429. doi: 10.1042/0264-6021:3610417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Servillo L, Giovane A, Cautela D, Castaldo D, Balestrieri ML. Where does N(ε)-trimethyllysine for the carnitine biosynthesis in mammals come from? PLoS One. 2014;9(1):e84589. doi: 10.1371/journal.pone.0084589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Stallcup MR. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene. 2001;20(24):3014–3020. doi: 10.1038/sj.onc.1204325. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X, Wen H, Shi X. Lysine methylation: beyond histones. Acta Biochim Biophys Sin (Shanghai) 2012;44(1):14–27. doi: 10.1093/abbs/gmr100. [DOI] [PubMed] [Google Scholar]
- 25.Nikpay M, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47(10):1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Romano KA, Vivas EI, Amador-Noguez D, Rey FE. Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. MBio. 2015;6(2):e02481. doi: 10.1128/mBio.02481-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Z, et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell. 2015;163(7):1585–1595. doi: 10.1016/j.cell.2015.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Craciun S, Balskus EP. Microbial conversion of choline to trimethylamine requires a glycyl radical enzyme. Proc Natl Acad Sci USA. 2012;109(52):21307–21312. doi: 10.1073/pnas.1215689109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y, et al. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc Natl Acad Sci USA. 2014;111(11):4268–4273. doi: 10.1073/pnas.1316569111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scoumanne A, Chen X. Protein methylation: a new mechanism of p53 tumor suppressor regulation. Histol Histopathol. 2008;23(9):1143–1149. doi: 10.14670/hh-23.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 33.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, et al. Protein carbamylation links inflammation, smoking, uremia and atherogenesis. Nat Med. 2007;13(10):1176–1184. doi: 10.1038/nm1637. [DOI] [PubMed] [Google Scholar]
- 35.Tsugawa H, et al. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat Methods. 2015;12(6):523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsugawa H, et al. Hydrogen rearrangement rules: computational MS/MS fragmentation and structure elucidation using MS-FINDER software. Anal Chem. 2016;88(16):7946–7958. doi: 10.1021/acs.analchem.6b00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics. 2010;11:395. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen K, Febbraio M, Li W, Silverstein RL. A specific CD36-dependent signaling pathway is required for platelet activation by oxidized low-density lipoprotein. Circ Res. 2008;102(12):1512–1519. doi: 10.1161/CIRCRESAHA.108.172064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.