Abstract
Pvlea-18 is a novel stress gene whose transcript is present in the dry embryo and the endosperm from bean (Phaseolus vulgaris) seeds. It accumulates in vegetative tissues in response to water deficit and abscisic acid application (J.M. Colmenero-Flores, F. Campos, A. Garciarrubio, A.A. Covarrubias [1997] Plant Mol Biol 35: 393–405). We show that the Pvlea-18 gene encodes a 14-kD protein that accumulates during late embryogenesis. Related proteins have been detected in both monocots and dicots, indicating that PvLEA-18 is a member of a new family of LEA (Late Embryogenesis Abundant) proteins. We also show that the PvLEA-18 transcript and protein accumulate not only in different organs of the bean seedlings during water stress but also in well-irrigated seedlings. This accumulation occurs in seedling regions with more negative values of water and osmotic potentials, such as the growing region of the hypocotyl. This phenomenon has not previously been described for LEA proteins. Immunohistochemical localization showed that the PvLEA-18 protein is present in the nucleus and cytoplasm of all cell types, with a higher accumulation in the epidermis and vascular cylinder tissues, particularly in protoxylem cells and root meristematic tissues. We found a similar localization but a higher abundance in water-stressed seedlings.
LEA (Late Embryogenesis Abundant) proteins are a broad family of plant proteins that are stored in the dry seed. Their presence in vegetative tissues is restricted to osmotic stress situations. Characteristically, they are very hydrophilic and seem to be structured in an extended form instead of being folded in a stable globular manner, as can be deduced from the randomly coiled moieties of some LEA proteins and from their boiling resistance (heat stability). These properties (which are consistent with a role in binding water), together with their high intracellular concentration and their expression patterns, have led to the suggestion that LEA proteins can protect specific cellular structures or ameliorate the effects of drought stress by maintaining a minimum cellular water requirement (for review, see Dure, 1993a; Ingram and Bartles, 1996). Other hypothetical roles assigned to the LEA proteins are sequestration of ions (Dure, 1993b), molecular chaperone activity (Close, 1996), and transport of nuclear-targeted proteins during stress (Goday et al., 1994).
Based on regions of significant similarity between LEA proteins of different species (Dure, 1993a), five classes of LEA proteins have been established (Ingram and Bartels, 1996). Two of these have a functional role in stress tolerance: HVA1, a group 3 LEA protein from barley, and LE25, a group 4 LEA protein from tomato. Overexpression of HVA1 improves drought and salinity resistance in transgenic rice plants (Xu et al., 1996). The LE25 protein was expressed in yeast and found to confer improved resistance to high salinity and freezing (Imai et al., 1996). During normal seed development, most of the characterized LEA proteins have been located in all embryo cell types (Close et al., 1993; Roberts et al., 1993; Goday et al., 1994). However, tissue-specific and temporal-dependent expression has also been observed in Rab28 (which is restricted to the meristem and vascular elements of the plumule) and the root and scutellum of the developing maize embryo (Niogret et al., 1996). Little information is available concerning the localization of LEA proteins in the vegetative tissues of plants subjected to water deficit. When localization was investigated, LEA proteins were found predominantly in the cytosol and nucleus of meristematic, vascular, and provascular cells (Mundy and Chua, 1988; Close et al., 1993; Godoy et al., 1994; Houde et al., 1995; Niogret et al., 1996).
We previously described the water-stress and ABA inducibility of Pvlea-18, a novel lea-like gene whose transcript is stored in dry seeds (Colmenero-Flores et al., 1997). Here we report evidence that the Pvlea-18 gene encodes a 14-kD protein that establishes a new group of LEA proteins in plants. Additionally, we show that the PvLEA-18 transcript and protein accumulate not only in response to stress conditions but also during normal development, particularly in the growing regions of bean seedlings. Immunolocalization experiments indicate that this protein is present in the cytosols and nuclei of different cell types along the bean hypocotyl. The high accumulation of PvLEA-18 in the embryo radicle during the early stages of germination suggests that it may have a protective role during this process.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Bean (Phaseolus vulgaris L. cv Negro Jamapa; National Institute for Forest and Agricultural Research, México) seeds were surface-sterilized in 10% (v/v) sodium hypochlorite for 10 min, rinsed in running tap water for 2 h, sown on water-saturated paper towels, and germinated in the dark at 27°C ± 1°C and 100% RH. After 5 d, we selected seedlings for uniform size and transplanted them to vermiculite containing different amounts of water. The control growth condition was 5 mL of water per gram of vermiculite (Ψw = −0.074 MPa). The water-deficit conditions corresponded to Ψw = −0.35 MPa, one-twelfth the amount of water relative to the control. We maintained the vermiculite at a constant Ψw throughout the experiment. Seedlings grew in the dark at 27°C ± 1°C and 75% RH. The duration of the water deficit in the different treatments appears in the text and in the corresponding figure legends. Plant material was frozen immediately in liquid nitrogen after harvesting and stored at −80°C until extraction.
Ψw and Ψs Measurements
We determined Ψw and Ψs using the dew-point method with the C-52 sample chamber, as described in the instruction manual from the manufacturer of the dew-point microvoltimeter (model HR-33T, Wescor, Logan, UT). We calculated the Ψp values from the experimentally obtained Ψw and Ψs values according to the equation Ψp = Ψs − Ψw.
Protein Purification and Antibody Production
The open reading frame encoded by the gene Pvlea-18 (Colmenero-Flores et al., 1997) was expressed in Escherichia coli using the pGEX3 expression vector. The PvLEA-18 protein fused to GST was purified by affinity chromatography using glutathione-agarose beads and a standard method (Ausubel et al., 1994). Antibodies were raised in rabbits by conventional procedures using purified preparations of the GST-PvLEA-18 fusion protein (Harlow and Lane, 1988a). Antibodies that specifically recognized PvLEA-18 were purified by immunoaffinity using the antigen GST-PvLEA-18 covalently coupled to bromine cyanide-activated Sepharose-4B (Sigma). The coupled resin was packed in a column and used for the immunoaffinity purification as described by Harlow and Lane (1988b).
Protein Extraction and Analysis in SDS-PAGE
Plant tissues were homogenized in a mortar in the presence of liquid nitrogen and extracted in a solution containing 10% (w/v) TCA and 0.3% (v/v) 2-mercaptoethanol prepared in 100% (v/v) acetone. After the sample was centrifuged, the pellet was washed three times with cold 100% acetone (−20°C). The resulting pellet was vacuum-dried and resuspended in PBS buffer for concentration measurement before 2 volumes of 2× Laemmli buffer was added (Laemmli, 1970). We analyzed total proteins by electrophoresis in SDS-PAGE gels after boiling the extracts for 3 min (Laemmli, 1970).
Western Analysis
Proteins separated by SDS-PAGE were transferred to nitrocellulose according to the method of Towbin et al. (1979). We used a 1:1,500 dilution of the anti-PvLEA-18 antibody and a 1:10,000 dilution of goat anti-rabbit IgG conjugated to peroxidase. An ECL chemiluminescence kit (Amersham) was used to detect the cross-reacting polypeptides.
Quantitation of PvLEA-18 in Plant Tissues
We extracted the proteins from fresh plant tissues by the TCA/acetone method, as described above. Different dilutions of the total protein extract were separated in 12% SDS-PAGE and transferred to nitrocellulose. We included different known amounts of purified GST-PvLEA fusion protein as an internal standard. The protein bands reacting with the anti-PvLEA-18 antibody were analyzed by western blot and developed using the ECL kit. We quantified the chemiluminescent signal in the cross-reacting bands with a one-dimensional analysis program (Bio Image Products, Millipore). We constructed a standard curve from the values obtained for the different amounts of the purified GST-PvLEA-18 fusion protein, which was used as the internal standard. This allowed an accurate estimation of the amount of the PvLEA-18 protein in the plant extracts. The molar concentration of the PvLEA-18 protein was determined according to the water content in the sample tissues.
RNA Isolation and Northern Analysis
We followed the procedure described by de Vries et al. (1991) to prepare total RNA. Northern blots were carried out by electrophoresis of 5 μg of total RNA on 1.5% (w/v) agarose gels containing 1.1% formaldehyde, according to the method of Sambrook et al. (1989), and transferred onto nylon membranes (Hybond N+, Amersham). The hybridization and washes took place at high stringency as described by Church and Gilbert (1984). Filters were exposed on Kodak XAR film at −80°C using an intensifying screen. We quantified the mRNA levels using the one-dimensional analysis program.
Labeling of Probes
The complete Pvlea-18 cDNA and a 3′-noncoding fragment of the PvleaIV-25 cDNA clone (Colmenero-Flores et al., 1997) were labeled with a commercial random primer kit (DuPont-NEN) using [α-32P]dCTP (3000 Ci mmol−1) (Amersham).
Immunocytochemistry
We dissected 6-d-old etiolated seedlings (either well watered or subjected to water deficit for 72 h) and 36-h-soaked embryos into 4- to 8-mm sections. The sections were fixed overnight at room temperature in a solution containing 3.7% (v/v) formaldehyde, 50% (v/v) ethanol, and 5% (v/v) acetic acid. The 1st h of the fixation procedure took place under a vacuum. We dehydrated the fixed tissues and embedded them in paraffin using the method described by Van de Wiel et al. (1990). Seven-micrometer-thick sections were rehydrated and preincubated overnight at 4°C in blocking buffer (5% BSA in PBS/0.1% Tween 20). We subsequently incubated the sections at room temperature for 3 h in a solution containing the anti-PvLEA-18 affinity-purified antibody, diluted 1:50 in blocking buffer. After the sections were washed three times for 10 min each time with blocking buffer, we incubated them with the secondary antibody coupled to alkaline phosphatase for 1 h at room temperature. The complex developed in 100 mm Tris, pH 9.5, 100 mm NaCl, 50 mm MgCl2, 0.2 mg mL−1 nitroblue tetrazolium, and 0.2 mg mL−1 5-bromo-chloro-3-indolyl phosphate. After the sections were dehydrated through an ethanol series and two changes of xylene, we mounted them in Permount.
RESULTS
Identification of the PvLEA-18 Protein
The identification of the PvLEA-18 protein and the study of its expression during development and in response to environmental stress required the production of polyclonal antibodies. With this aim, we purified the PvLEA-18 protein from E. coli as a GST-fusion protein. The Pvlea-18 cDNA (Colmenero-Flores et al., 1997) was subcloned into the pGEX3 expression vector, giving rise to a translational fusion of the GST gene with the bean Pvlea-18 cDNA. After we verified the correct reading frame of the chimera by sequence analysis, the gene was overexpressed in E. coli and we used glutathione-agarose beads to purify the GST-PvLEA-18 fusion protein by affinity chromatography. Purified fusion protein elicited antibodies in rabbits, as described in Methods. We carried out all of the experiments in this work with polyclonal antibodies purified by affinity chromatography, coupling the GST-PvLEA-18 fusion protein to Sepharose-4B.
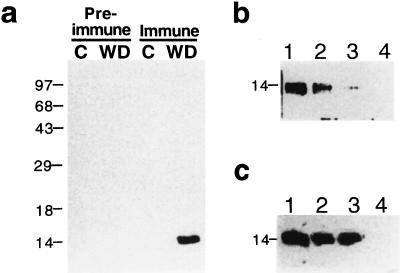
To identify the product of the Pvlea-18 gene, we subjected bean seedlings to water deficit and obtained total protein extracts from roots, the organs that present the highest transcript level (Colmenero-Flores et al., 1997). The western blot in Figure 1a shows that the antibodies cross-react with a 14-kD polypeptide, which, like the Pvlea-18 transcript, accumulated in response to water-stress conditions. To test the specificity of the detection reaction, the purified GST-PvLEA-18 fusion protein was added as a competitor when the immunoblot was incubated with the purified anti-GST-PvLEA-18 antibody. The results shown in Figure 1b indicate that an incubation with 5 μg of GST-PvLEA-18 protein was sufficient to completely block the cross-reaction with the 14-kD polypeptide. To discard the interference of antibodies raised against the GST domain of the fusion protein during the immunodetection, we performed a similar experiment using the purified GST protein as the competitor.
Figure 1.
Characterization of the anti-PvLEA-18 antibody. Bean seedlings grown in the dark for 5 d were transferred to well-irrigated or water-stressed vermiculite and harvested after 24 h of treatment. Total protein extracts were purified from roots, separated by SDS-PAGE, transferred to nitrocellulose, and incubated with antisera as follows: a, Immunodetection of PvLEA-18 in protein extracts obtained from control (C) or water-deficient (WD) tissues using immune, anti-PvLEA-18, or preimmune antisera. b, Competition of the antiserum by preincubation with different concentrations of purified PvLEA-18-GST fusion protein: 0 ng (lane 1), 50 ng (lane 2), 500 ng (lane 3), and 5 μg (lane 4). c, Competition analysis of the antiserum by addition of purified GST protein in different amounts: 0 ng (lane 1), 5 μg (lane 2), and 50 μg (lane 3). As a control, 5 μg of purified PvLEA-18-GST fusion protein was added in lane 4. Numbers at the left indicate the corresponding molecular masses in kD.
The results in Figure 1c show that even 50 μg of GST was unable to block the detection of the 14-kD protein, indicating that the detected protein was not related to GST. Therefore, we can conclude that the antibodies specifically recognized the PvLEA-18 protein. As indicated above, the immunodetected signal corresponding to the PvLEA-18 protein showed an apparent molecular mass of 14 kD, which did not correspond to the 9 kD inferred from the protein deduced from the cDNA sequence. This inconsistency could be explained by an anomalous migration in SDS-PAGE given the PvLEA-18 amino acid composition. This possibility is supported by the fact that the Pvlea-18 cDNA expressed in Saccharomyces cerevisiae and E. coli also produced a 14-kD protein that was immunodetected with the anti-PvLEA-18 antibodies (data not shown).
Accumulation of the Pvlea-18 Transcript and Protein during Embryogenesis and Germination
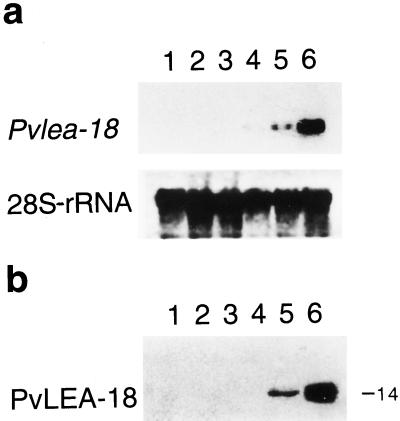
We reported that the Pvlea-18 transcript accumulated in response to water-deficit conditions and ABA treatment and that it was stored in the dry bean seed. These data, together with the amino acid composition of the deduced polypeptide, suggested that the Pvlea-18 gene encoded a LEA-like protein (Colmenero-Flores et al., 1997). To define whether the PvLEA-18 protein was a bona fide LEA protein, we analyzed the pattern of Pvlea-18 transcript and protein accumulation during embryogenesis. Figure 2 shows that the PvLEA-18 protein and its transcript accumulated during late embryogenesis (after 20 d postanthesis; Fig. 2). Embryos 20 d postanthesis initiated the desiccation process and at 26 d postanthesis had reached maturity and their lowest water content. By quantitation of the immunoreactive proteins, we estimated that the concentration of the PvLEA-18 protein was approximately 10 μm at this developmental stage. These accumulation patterns allowed us to unambiguously classify this protein as a bean LEA protein.
Figure 2.
Analysis of the accumulation of the Pvlea-18 transcript and protein during embryogenesis. a, Pvlea-18 transcript accumulation as determined by northern analysis of RNA extracts obtained from bean embryos at different days postanthesis (dpa): lane 1, 5 dpa; lane 2, 15 dpa; lane 3, 20 dpa; lane 4, 22 dpa; lane 5, 24 dpa; and lane 6, 26 dpa. Five micrograms of total RNA was electrophoresed, blotted on nylon membranes, and hybridized against the indicated probes. Hybridization against a 28S-rRNA probe was used as an RNA-loading control. b, Accumulation of the PvLEA-18 protein was analyzed by western blot of total protein extracts obtained from the same samples described in a. The molecular mass of PvLEA-18 is indicated on the right in kilodaltons. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes before incubation with the immunopurified anti-PvLEA-18 antiserum.
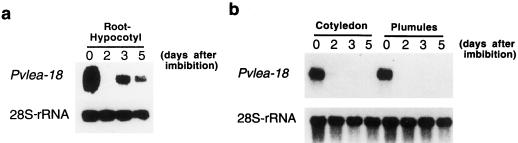
We previously determined that the Pvlea-18 transcript decreased gradually during the germination process: the transcript levels remained the same as those in the dry seed during the first 12 h of imbibition and then decreased after 24 h, and no transcript was observed after 48 h of imbibition (Fig. 3a; Colmenero-Flores et al., 1997). When the analysis was extended during seedling establishment in the dark and under optimal irrigation, a reinduction of the transcript occurred after 72 h in the hypocotyl/radicle system (Fig. 3a) but not in the cotyledons or plumules (Fig. 3b).
Figure 3.
Analysis of the accumulation of the Pvlea-18 transcript during germination. Bean seeds were germinated in the dark and seedlings were harvested at different times after imbibition (0, 2, 3, and 5 d postimbibition). Five micrograms of total RNA was purified from different seed or seedling organs, blotted onto nylon membranes, and hybridized as indicated. Hybridization against a 28S-rRNA probe was used as an RNA-loading control. a, Northern-blot analysis of RNA extracts obtained from the hypocotyl-root system. b, Northern-blot analysis of total RNA extracts obtained from cotyledons and plumules.
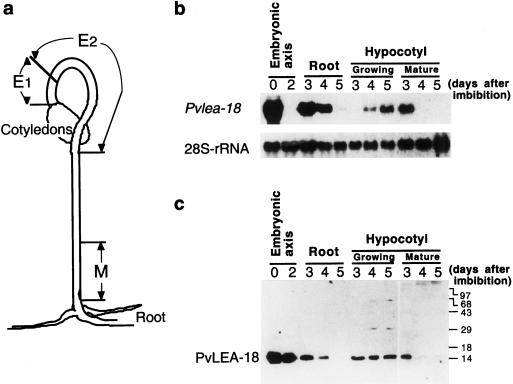
Analysis of the accumulation of the Pvlea-18 transcript in the growing and mature regions of the hypocotyls and roots showed that a transient reinduction took place at d 3 of imbibition in the hypocotyl basal region (nongrowing) and in roots, whereas we detected a reinduction only 1 d later in the hypocotyl-growing regions (Fig. 4, a and b). In the latter case, we observed that the Pvlea-18 transcript levels progressively increased, even 8 d after imbibition (data not shown). To correlate the transcript levels with those of the protein, we performed western blots using anti-PvLEA-18 and protein extracts obtained from the same samples described above (Fig. 4). In contrast to the transcript accumulation pattern, the PvLEA-18 protein levels remained without a major change even after 2 d of imbibition (Fig. 4c), when the transcript was no longer detectable (Fig. 4b). We observed a similar phenomenon in the hypocotyl-growing regions after d 3 of imbibition and again in cotyledons in which the PvLEA-18 protein was still present even though its transcript was not detectable (Figs. 4, b and c, and 5), which suggested that there was a low turnover of the protein. We did not observe this characteristic in the hypocotyl mature region or in the root at these germination stages when the accumulation patterns for the transcript and the protein are similar (Fig. 4, b and c).
Figure 4.
Analysis of the accumulation of Pvlea-18 transcript and protein in roots and in different regions of the hypocotyl during seedling establishment. a, Schematic description of the hypocotyl regions used. E1 and E2, Hypocotyl-growing regions (E2 shows the highest elongation rate); M, nongrowing or mature zone. b, Northern-blot analysis of the Pvlea-18 transcript. Bean seeds were germinated in the dark and seedlings were harvested at different times (0, 2, 3, 4, and 5 d after imbibition). Five micrograms of total RNA was purified from different seed or seedling organs and from the hypocotyl regions indicated above, blotted on nylon membranes, and hybridized. Hybridization against a 28S-rRNA probe was used as an RNA-loading control. c, Western-blot analysis of the PvLEA-18 protein accumulation from total protein extracts obtained from the same samples as described in b. Numbers at the right indicate the corresponding molecular masses in kilodaltons. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes before incubation with the immunopurified anti-PvLEA-18 antiserum.
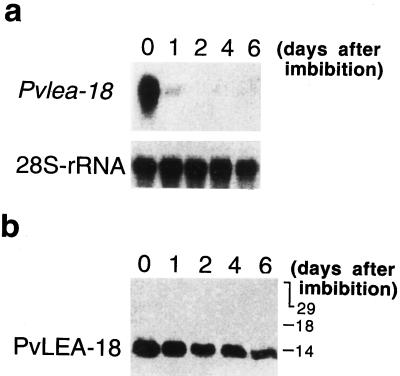
Figure 5.
Analysis of the Pvlea-18 transcript and protein accumulation in cotyledons during seedling establishment. a, Northern-blot analysis of the Pvlea-18 transcript. Bean seeds were germinated in the dark and seedlings were harvested after different times (0, 1, 2, 4, and 6 d postimbibition). Five micrograms of total RNA was purified from the seed or seedling cotyledons, blotted onto nylon membranes, and hybridized as indicated. Hybridization against a 28S-rRNA probe was used as an RNA-loading control. b, Western-blot analysis of the PvLEA-18 protein accumulation from total protein extracts obtained from the same samples described in a. Numbers at the right indicate the corresponding molecular masses in kilodaltons. Proteins were separated by SDS-PAGE and transferred onto nitrocellulose membranes before incubation with the immunopurified anti-PvLEA-18 antiserum.
Distribution of the Pvlea-18 Transcript and Protein in the Different Hypocotyl-Growing Regions from Well-Irrigated and Water-Stressed Bean Seedlings
The Pvlea-18 gene not only responded to water-stress situations but was also reinduced in the hypocotyl during seedling establishment. Because it is known that water status changes along the stem region (Nonami and Boyer, 1993), we investigated in more detail the water status of the different bean hypocotyl regions from well-irrigated and water-stressed seedlings. For this, we divided the bean hypocotyls into discrete regions: E1 and E2 corresponding to the elongating regions (E2 was the section with the highest elongation rate) and M, the most basal or mature region (Fig. 4a). The water status of these regions was determined as described in Methods. The results shown in Table I indicate that the most negative values of Ψp and Ψs were found in the most apical regions of well-irrigated and water-stressed seedlings. However, as expected, the values for both potentials were lower in water-stressed than in well-irrigated seedlings. Furthermore, the most apical region showed the highest turgor pressure, which did not change after the water-deficit treatment. In contrast, turgor in the mature region showed an 8-fold decrease in water-stressed plants (Table I).
Table I.
Water status of the hypocotyl regions
| Region | Ψw
|
Ψs
|
Ψpa
|
|||
|---|---|---|---|---|---|---|
| Cb | WDc | C | WD | C | WD | |
| bars | ||||||
| E1 | −7.03 (±1.02) | −9.83 (±0.70) | −10.96 (±1.35) | −13.05 (±1.55) | 3.85 | 3.22 |
| E2 | −6.50 (±0.74) | −8.94 (±1.04) | −9.20 (±0.96) | −11.93 (±1.78) | 2.70 | 2.80 |
| Mature | −5.88 (±1.14) | −8.51 (±1.31) | −8.24 (±0.85) | −8.82 (±1.16) | 2.36 | 0.31 |
The data in this table are expressed as the means ± sd (shown in parentheses) from 10 measurements of three independent experiments, according to Student's t distribution analysis with 95% confidence intervals.
Ψp was not experimentally determined; it was calculated from the difference between the Ψw and the Ψs values.
C, Control.
WD, Water deficient.
We investigated the accumulation pattern of the Pvlea-18 transcript in the expanding hypocotyl regions described above. In agreement with the results in the previous section, Figure 6 shows that the Pvlea-18 mRNA was present in the hypocotyl-elongating regions (E1 and E2) but not in the nongrowing region (M) of nonstressed bean seedlings (Fig. 6a). These results also showed that the highest accumulation of the transcript occurred in the region with the highest elongation rate (E2). In contrast, the Pvlea4–25 mRNA, corresponding to a member of a typical LEA family, was not detected in any region from the nonstressed seedlings (Fig. 6c). When seedlings grew under water-stress conditions, both Pvlea-18 and PvleaIV-25 transcripts accumulated in all regions and had the highest levels in the mature zone, the area with the lowest turgor values.
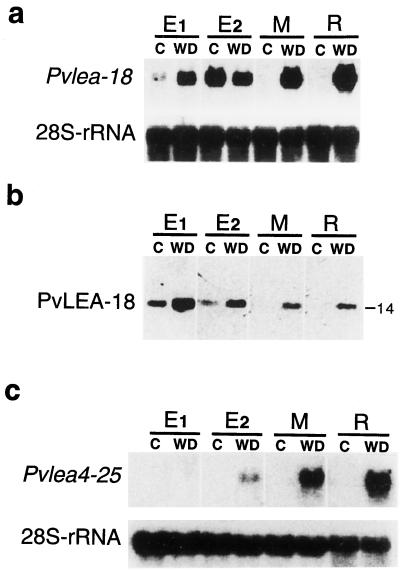
Figure 6.
Pvlea-18 transcript and protein accumulation patterns in different hypocotyl regions from seedlings grown under well-irrigated and water-stress conditions. E1 and E2, Hypocotyl-growing regions; M, nongrowing or mature zone; R, root. a, Northern-blot analysis of the Pvlea-18 transcript. Five-day-old bean seedlings grown in the dark were transferred to well-irrigated (C) or water-deficient (WD) vermiculite and harvested after 24 h of treatment. Five micrograms of total RNA was purified from the indicated seedling regions, blotted onto nylon membranes, and hybridized against the indicated probes. Hybridization against a 28S-rRNA probe was used as an RNA-loading control. b, PvLEA-18 protein accumulation pattern obtained by western analysis of total protein extracts purified from the same samples described in a. The molecular mass of PvLEA-18 is indicated on the right in kilodaltons. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes before incubation with the immunopurified anti-PvLEA-18 antiserum. c, Northern-blot analysis of the PvleaIV-25 transcript by blotting total RNA obtained from the samples described in a. Five micrograms of total RNA was blotted onto nylon membranes and hybridized against the indicated probes. Hybridization against an 28S-rRNA probe was used as an RNA-loading control.
The analysis of the PvLEA-18 protein levels along the different hypocotyl regions showed a similar accumulation pattern, i.e. the protein accumulated in the growing but not in the mature regions of well-irrigated plants. However, in contrast to its transcript, which showed a higher accumulation in the E2 than in the E1 region, the accumulation of the protein was similar in both sections (Fig. 6b). Additionally, in water-stressed seedlings, the protein showed different levels along the hypocotyl, presenting the highest accumulation in the most apical region and the lowest in the basal zone (Fig. 6b), where we had detected the highest accumulation of transcript (Fig. 6a). Roots from water-stressed seedlings and the mature hypocotyl region had the lowest protein-to-transcript ratio (Fig. 6, a and b).
Distribution of the PvLEA-18 Protein in Different Tissues of Bean Seedlings
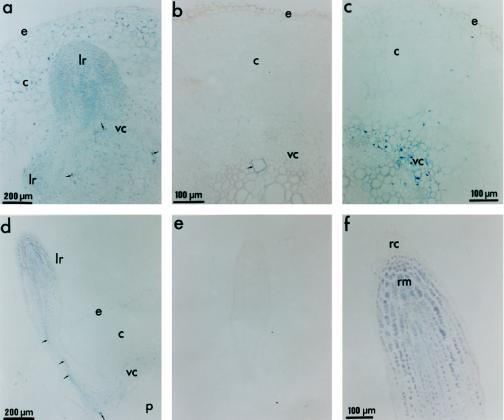
Given the differential accumulation of the PvLEA-18 protein during normal development and stress conditions, we decided to investigate its tissue distribution. The immunolocalization in Figure 7 shows that, in agreement with the results obtained from the immunoblot-detection experiments, the PvLEA-18 protein responded to water deficit and preferentially accumulated in the most apical regions of hypocotyls (Fig. 7, a–h). Additionally, the PvLEA-18 protein was present in all hypocotyl cell types, although we did detect a preferential accumulation in the vascular cylinder and epidermal tissues. This distribution was more remarkable in the mature or nongrowing regions from stressed seedlings because we detected less accumulation of the protein in the parenchymal tissues, pith, and cortex (Fig. 7, g, h, and i, respectively). The immunolocalization experiments also showed that the PvLEA-18 protein was present in a high concentration in the nuclei of most cell types (Fig. 8). Electron microscopic analysis of mature embryo tissues supported these results by showing the presence of the protein in cytoplasm and nuclei (data not shown). Another characteristic of PvLEA-18 protein distribution was its accumulation in protoxylem cells present in young, growing tissues (Fig. 8, a and d) and in mature tissues (Fig. 8, e and f).
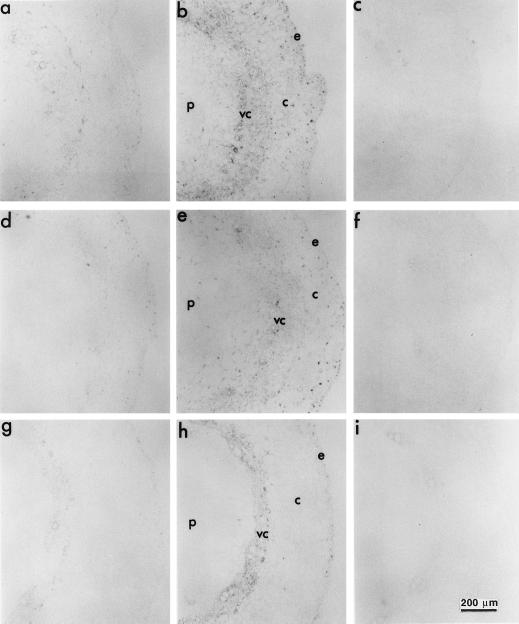
Figure 7.
Immunohistochemical detection of the PvLEA-18 protein in transverse sections of different hypocotyl regions from well-watered and water-stressed seedlings using immunopurified anti-PvLEA-18 antibody. Bean seedlings grown in the dark for 5 d were transferred to well-irrigated or water-stressed vermiculite and harvested after 36 h of treatment. Five-millimeter fragments corresponding to the E1 (a–c), E2 (d–f), and mature (g–i) hypocotyl regions were embedded in paraffin for subsequent immunohistochemical analysis of microtome sections. Sections from well-irrigated seedlings are shown in a, d, and g; water-stressed sections are shown in b, e, and h; and water-stressed sections incubated with preimmune antiserum are shown in c, f, and i. c, Cortex; e, epidermis; p, pith; vc, vascular cylinder.
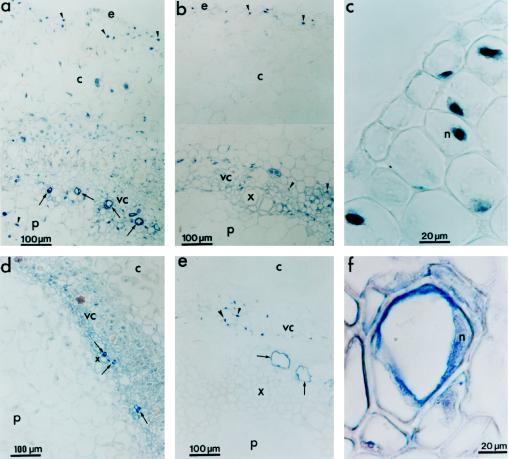
Figure 8.
Immunohistochemical detection of PvLEA-18 in protoxylem cells and nuclei from root and hypocotyl tissues using immunopurified anti-PvLEA-18 antibody. Tissues were dissected and soaked in paraffin as described in Figure 7. a and c, Sections from the hypocotyl-elongating region E1 obtained from water-stressed seedlings corresponding to ×2 and ×5 magnifications of Figure 7b, respectively. b, Section from the hypocotyl mature region obtained from water-stressed seedlings corresponding to a ×2 magnification of Figure 7h. d, Section from the emerging hypocotyl obtained from well-irrigated seedlings after 36 h of imbibition. e, Section from a root obtained from well-irrigated seedlings grown as described in Figure 7. f, Detail of a protoxylem cell from the mature region of the hypocotyl of a 6-d-old well-irrigated seedling. Arrows indicate protoxylem cells and arrowheads indicate nuclei. c, Cortex; e, epidermis; p, pith; vc, vascular cylinder; x, xylem.
The immunolocalization results in Figure 9 show that in nonstressed seedlings the PvLEA-18 protein was abundantly and homogeneously distributed in the radicle during the early stages of germination (36 h of imbibition) (Fig. 9a). In contrast, the PvLEA-18 protein was barely detectable in roots from the older, well-irrigated seedlings (6 d postimbibition; Fig. 9b), in agreement with the results obtained from western-blot analysis (Fig. 4c). As in the mature region of water-stressed hypocotyls, the PvLEA-18 protein in roots responded to water deficit, showing a higher accumulation in the epidermis and vascular cylinder (Fig. 9c). Emerging secondary roots had a high accumulation and a uniform distribution of PvLEA-18 (Fig. 8a). The same characteristics can be seen at the tip of water-stressed secondary roots (Fig. 8d) as can be seen at their basal region: as in the hypocotyl, the protein accumulated preferentially at the epidermal and vascular tissues. The magnification in Figure 8f shows that, although the PvLEA-18 protein was present in the root tip, it could not be detected in the root cap cells.
Figure 9.
Immunohistochemical detection of PvLEA-18 protein in root tissues from well-watered and water-stressed seedlings. Tissues were dissected and soaked in paraffin as described in Figure 7. a, Section from a radicle obtained from germinating seedlings grown under well-irrigated conditions. Seeds were germinated in the dark and harvested 36 h after imbibition. b, Section from a root obtained from well-irrigated 6-d-old seedlings. Root sections were obtained from the same seedlings transplanted to well-irrigated vermiculite described in Figure 7. c, Section from a root obtained from water-stressed 6-d-old seedlings. Root sections were obtained from the same seedlings transplanted to water-limited vermiculite described in Figure 7. d, Longitudinal section of a lateral root from a water-stressed seedling. e, Longitudinal section of a lateral root from a water-stressed seedling corresponding to the section described in d incubated with preimmune antiserum. f, Detail of the secondary root tip corresponding to a ×2 magnification of d. Arrows indicate protoxylem cells and arrowheads indicate nuclei. c, Cortex; e, epidermis; lr, lateral root; n, nucleus; p, pith; rc, root cap; rm, root meristem; vc, vascular cylinder.
The PvLEA-18 Protein Is a Member of a Novel Family of LEA Proteins
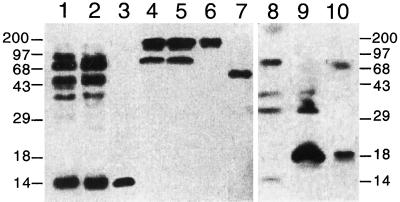
Previous analysis of the deduced amino acid sequence of the PvLEA-18 protein did not show significant homology with known LEA proteins (Colmenero-Flores et al., 1997). We investigated the possibility that the PvLEA-18 protein is a member of a new family of LEA proteins by looking for homologous proteins in other plant species, using the anti-PvLEA-18 antibodies for detection. As shown in Figure 10, related proteins were identified in total protein extracts obtained from seeds of tobacco, tomato, soybean, pea, maize, and Arabidopsis. The cross-reacting proteins from the different plant species varied in their molecular masses, from higher than 97 kD to close to 14 kD (Fig. 10, lanes 4–10). Characteristically, for several LEA protein families described, polypeptide sizes were variable among members of the same family (Close et al., 1993; Houde et al., 1995; Ingram and Bartels, 1996). Other bean cultivars also had high-molecular-mass PvLEA-18-related proteins (Fig. 10).
Figure 10.
Detection of PvLEA-18-related proteins in different plant species by western analysis. Protein extracts obtained from the seeds of the different species described below were separated by SDS-PAGE and transferred to nitrocellulose before incubation with the immunopurified anti-PvLEA-18 antiserum. PvLEA-18 cross-reacting proteins in dry seeds from: lane 1, cv Cacahuate of bean; lane 2, cv Flor de Mayo of bean; lane 3, cv Negro jamapa of bean; lane 4, tobacco; lane 5, maize; lane 6, Arabidopsis; lane 8, tomato; lane 9, soybean; and lane 10, pea. For S. lepidophylla, the protein extract was obtained from vegetative tissues of desiccated whole plants (lane 7). Numbers at both sides indicate the corresponding molecular masses in kilodaltons.
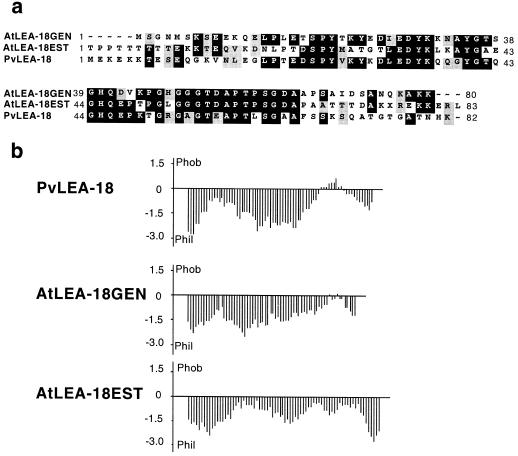
We obtained similar results from the immunoblot analysis of total protein extracts from desiccated tissue of a representative of an ancestral vascular plant, Selaginella lepidophylla (Lycopodiophyta) (Fig. 10, lane 7). In some cases, such as in maize and soybean, we detected an 18-kD protein when seedlings were subjected to water stress (data not shown). The existence of similar proteins is supported by the presence of a homologous open reading frame (40% identity and 46% similarity) in the Arabidopsis genome (accession no. U78721/ATU78721), as well as a similarly expressed sequence tag gene (accession no. AA395541) from Arabidopsis (34% identity and 44% similarity) (Fig. 11a). This homology was significant in the central Gly-rich domain. Furthermore, these proteins presented very similar hydropathic patterns (Fig. 11b). Taken together, these data suggested that the PvLEA-18 protein is a representative member of a novel family of LEA proteins.
Figure 11.
Putative PvLEA-18 homologous proteins from Arabidopsis. a, Alignment of the amino acid sequence predicted from the bean Pvlea-18 cDNA with the amino acid sequence deduced from Arabidopsis genomic (Atlea-18GEN) and partial cDNA sequences (Atlea-18EST). b, Comparison of the corresponding hydropathy profiles.
DISCUSSION
In a previous report we described a novel stress gene from common bean whose transcript accumulated in dry seeds and in different organs of plants subjected to water-deficit conditions or ABA treatment. Although this gene and its deduced protein did not show significant similarity with any known genes or proteins in the database, analysis of the protein sequence and amino acid composition revealed structural characteristics similar to those of the LEA proteins, such as high hydrophilicity, high content of Gly and charged amino acids, and high percentages of randomly coiled moieties. Given the characteristics of the putative protein and the accumulation pattern of its transcript, we named the corresponding gene “Pvlea-18” (Colmenero-Flores et al., 1997). In this work we show that the PvLEA-18 protein is a bona fide LEA protein, because, in addition to its response to dehydration and ABA treatment, it also accumulated in the seed during the final stage of embryogenesis. Also, as was the case for other lea genes (for review, see Dure, 1993b; Ingram and Bartels, 1996), the Pvlea-18 transcript accumulated to high levels in dry seed and decreased to undetectable levels during the germination process (Figs. 3 and 4).
Analysis of the protein-accumulation pattern revealed that, although the transcript was not detected after 2 d of imbibition, the protein maintained levels comparable to those present in the dry seed. A similar phenomenon occurred in the emerging hypocotyls 3 d postimbibition (Fig. 4) and in cotyledons, in which the transcript was undetectable after 2 d of seed imbibition, although the protein was present even after 6 d (Fig. 5). These observations suggest the existence of posttranscriptional mechanisms favoring differential turnover rates of the protein and/or the transcript in these regions during seedling development (see below).
Although LEA proteins have been associated with dehydration, seed development, and adverse environmental conditions, we show in this work that the PvLEA-18 protein and its transcript also accumulated during the growth of etiolated bean seedlings under conditions of optimal irrigation. In etiolated seedlings with no expanded leaves, evapotranspiration in the apical zone was not a major driving force for water transport. However, a Ψw gradient must be generated along the hypocotyl to ensure water flux (Meyer and Boyer, 1972). It has been proposed that this Ψw gradient was generated by the accumulation of osmolytes in the apical zones, which is also important for the maintenance of cell growth in growing tissues (Meyer and Boyer, 1972, 1981; Creelman et al., 1990; Nonami and Boyer, 1993).
As indicated above, the Pvlea-18 transcript decreased to undetectable levels during the germination process; however, it was transiently reinduced in the radicle, the earliest emerging organ, and hours later in the emerging hypocotyls, where we detected a progressive increase in the transcript levels (Fig. 4). The possibility that this transcript reinduction was related to the elongation process under optimal growth conditions was investigated by looking at the transcript-accumulation pattern along the different hypocotyl-growing regions. The data described in Results and shown in Figures 4 and 6 indicate that the Pvlea-18 transcript accumulated in those hypocotyl regions that were in active elongation (E1 and E2). Furthermore, the fastest-elongating zone, E2, coincided with the highest Pvlea-18 transcript accumulation. A similar accumulation pattern was observed for the protein, although the protein-to-transcript ratio in the most apical region (E1), which exhibited the lowest Ψs and Ψw, was higher than that in the region below (E2). As indicated above, a similar situation (a high protein-to-transcript ratio) was observed in emerging hypocotyls (Fig. 4) and in cotyledons (Fig. 5) during seedling establishment. It is interesting that the lowest osmotic potentials were detected in cotyledons (data not shown). These results led us to the hypothesis that the low turnover of the PvLEA-18 protein is associated with a stabilization mechanism that is favored in tissues with a high concentration of osmolytes.
We observed a different transcript accumulation pattern in seedlings grown under water-deficit conditions (Fig. 6). The Pvlea-18 transcript accumulated in all hypocotyl regions, presenting its highest levels in the most basal or mature region, the one with less turgor under water stress, and where no transcript was detected in well-irrigated seedlings. The high transcript accumulation in the low-turgor regions has also been observed for other genes whose expression is induced under drought conditions (J.M. Colmenero-Flores, unpublished results). Even though the PvLEA-18 protein also accumulated in all hypocotyl regions under water-deficit conditions, it showed higher levels in the most apical regions, again suggesting a correlation with the osmotic status. The fact that transcript accumulation occurred in the growing regions of well-irrigated and water-stressed seedlings suggests that the Pvlea-18 gene is modulated by at least two different control mechanisms, one that is mediated by ABA during water stress (Colmenero-Flores et al., 1997) and another possibly mediated by hormones involved in growth induction. In both cases the stimuli could be the low Ψs. The Pvlea-18 expression pattern resembles that of some plant aquaporin genes that are preferentially expressed in zones of cell division and elongation, as well as in the epidermis and youngest portions of the xylem (Yamada et al., 1995; Barrieu et al., 1998; Chaumont et al., 1998).
The PvLEA-18 protein-immunolocalization experiments clearly showed that many cell types are able to accumulate the protein in their cytoplasm, even in well-watered seedlings. The highest accumulation of the protein occurred in cells from the epidermis and the vascular cylinder (Figs. 7 and 8), tissues that may have exhibited more negative Ψw because they were more exposed to the changing environment (Nonami and Boyer, 1983; Davies, 1986). The accumulation of the PvLEA-18 protein in the apical regions of elongating roots (radicle and lateral roots) from either stressed or nonstressed seedlings could also be related to the low Ψs present in the emerging roots (Creelman et al., 1990). The absence of the protein in the root cap cells indicates that its accumulation was in response to specific regulatory factors. We also observed a high accumulation of the PvLEA-18 protein in immature xylem cells in hypocotyls and roots (Fig. 8). Although we do not know the role of this protein during xylogenesis, its accumulation may be related to the osmotic status of this cell type. It has been proposed that high concentrations of sugars, auxins, and cytokinins are needed to induce xylem development (Aloni, 1987). In addition, a high concentration of potassium ions has been detected during xylogenesis (McCully, 1994), suggesting low Ψs during the development of this cell type.
The presence in Arabidopsis of genes encoding putative PvLEA-18-homologous proteins, together with the detection of PvLEA-18-immunorelated polypeptides in protein extracts from different plant species, such as Arabidopsis, maize, tomato, tobacco, pea, soybean, and the primitive vascular plant S. lepidophylla, indicates that PvLEA-18 is a member of a novel LEA protein family. Even though the putative proteins deduced from the expressed sequence tag and genomic sequences in Arabidopsis have low molecular masses, polypeptides of this size were not detected in the western-blot experiments. Because the protein extracts analyzed were obtained only from seeds, we cannot discard the presence of the expected low-molecular-mass proteins in tissues from different developmental stages or growth conditions. More experiments are needed to clarify this observation.
The fact that the PvLEA-18 protein belongs to a different LEA protein group than the ones described indicates that it may carry out a different function. Although the Pvlea-18 expression pattern and protein localization during water-deficit conditions were very similar to other lea genes reported (Mundy and Chua, 1988; Close et al., 1993; Godoy et al., 1994; Houde et al., 1995; Ingram and Bartles, 1996; Niogret et al., 1996), no other LEA protein has been found during seedling establishment and growth under optimal irrigation conditions. These characteristics seem to be particular for the Pvlea-18 gene and protein. The possibility that other proteins from the PvLEA-18 group present a similar behavior awaits testing.
Finally, the presence of PvLEA-18 in cells with high osmolyte accumulation (embryos, germinating embryonic axes, hypocotyl growing tissues, root tips, cotyledons, and water-stressed tissues) may be related to the capability of this loosely structured hydrophilic protein to capture water and/or to a protective function in conditions of low Ψw. The ubiquitous presence of PvLEA-18 among many cell types and in different cell compartments, such as the cytoplasm and the nucleus, suggests a nonspecific protective role. Experiments are being carried out to elucidate the function of PvLEA-18 and the factors involved in the regulation of its expression.
ACKNOWLEDGMENTS
We thank G. Cassab and S. Gilmore for critical reading of the manuscript. We also thank P.C. Zambryski for her support during the establishment of the microscopic techniques, R.M. Solórzano for technical assistance, P. Gaytán and E. López for the synthesis of oligonucleotides, and E. Mata for animal care during the antibody production. We are also grateful to N. Capote for her constant support.
Abbreviations:
- GST
glutathione S-transferase
- Ψp
pressure potential
- Ψs
osmotic potential
- Ψw
water potential
Footnotes
This work was partially supported by grants from the Consejo Nacional de Ciencia y Tecnología, México (no. 0131P-N), and from the Dirección General de Asuntos del Personal Académico, Universidad Nacional Autónoma de México (no. IN204496 to A.A.C.).
LITERATURE CITED
- Aloni R. Differentiation of vascular tissues. Annu Rev Plant Physiol. 1987;38:179–204. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1994) Current Protocols in Molecular Biology, Vol 2. John Wiley & Sons, New York, pp 16.7.1–16.7.7
- Barrieu F, Chaumont F, Chrispeels MJ. High expression of the tonoplast aquaporin ZmTIP1 in epidermal and conducting tissues of maize. Plant Physiol. 1998;117:1153–1163. doi: 10.1104/pp.117.4.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F, Barrieu F, Herman EM, Chrispeels MJ. Characterization of a maize tonoplast aquaporin expressed in zones of cell division and elongation. Plant Physiol. 1998;117:1143–1152. doi: 10.1104/pp.117.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close TJ. Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant. 1996;97:795–803. [Google Scholar]
- Close TJ, Fenton RD, Yang A, Asghar R, DeMason DA (1993) Dehydrin: the protein. In TJ Close, EA Bray, eds, Plant Responses to Cellular Dehydration during Environmental Stress. American Society of Plant Physiologists, Rockville, MD, pp 104–118
- Colmenero-Flores JM, Campos F, Garciarrubio A, Covarrubias AA. Characterization of Phaseolus vulgaris cDNA clones responsive to water deficit: identification of a novel late embryogenesis abundant-like protein. Plant Mol Biol. 1997;35:393–405. doi: 10.1023/a:1005802505731. [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mason HS, Bensen RJ, Boyer JS, Mullet JE. Water deficit and abscisic acid cause differential inhibition of shoot versus root growth in soybean seedlings. Plant Physiol. 1990;92:205–214. doi: 10.1104/pp.92.1.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies WJ (1986) Transpiration and the water balance of plants. In FC Steward, ed; JF Sutcliffe, JE Dale, co-eds, Plant Physiology: A Treatise, Vol IX: Water and Solutes in Plants. Academic Press, New York, pp 49–154
- de Vries S, Hoge H, Bisseling T. Isolation of total and polysomal RNA from plant tissues. In: Gelvin SB, Schilperoot RA, Verma DPS, editors. Plant Molecular Biology Manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1991. p. B6. : 1–13. [Google Scholar]
- Dure L III (1993a) Structural motifs in LEA proteins. In TJ Close, EA Bray, eds, Plant Responses to Cellular Dehydration during Environmental Stress. American Society of Plant Physiologists, Rockville, MD, pp 91–103
- Dure L., III A repeating 11-mer amino acid motif and plant desiccation. Plant J. 1993b;3:363–369. doi: 10.1046/j.1365-313x.1993.t01-19-00999.x. [DOI] [PubMed] [Google Scholar]
- Goday A, Jensen A, Culiáñez-Macià FA, Albà MM, Figueras M, Serratosa J, Torrent M, Pagés M. The maize abscisic acid responsive protein rab17 is located in the nucleus and cytoplasm and interacts with nuclear localization signals. Plant Cell. 1994;6:351–360. doi: 10.1105/tpc.6.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godoy JA, Luna R, Torres-Schumann S, Moreno J, Rodrigo RM, Pintor-Toro JA. Expression, tissue distribution and subcellular localization of dehydrin TASI4 in salt-stressed tomato plants. Plant Mol Biol. 1994;26:1921–1934. doi: 10.1007/BF00019503. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D (1988a) Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 53–138
- Harlow E, Lane D (1988b) Antibodies. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 283–318
- Houde M, Daniel C, Lachapelle M, Allard F, Laliberté S, Sarhan F. Immunolocalization of freezing-tolerance-associated proteins in the cytoplasm and nucleoplasm of wheat crown tissues. Plant J. 1995;8:583–593. doi: 10.1046/j.1365-313x.1995.8040583.x. [DOI] [PubMed] [Google Scholar]
- Imai R, Chang L, Ohta A, Bray EA, Takagi M. A lea-class gene of tomato confers salt and freezing tolerance when overexpressed in Saccharomyces cerevisiae. Gene. 1996;170:243–248. doi: 10.1016/0378-1119(95)00868-3. [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:377–403. doi: 10.1146/annurev.arplant.47.1.377. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- McCully ME. Accumulation of high levels of potassium in the developing xylem elements in roots of soybean and some other dicotyledons. Protoplasma. 1994;183:116–125. [Google Scholar]
- Meyer RF, Boyer JS. Sensitivity of cell division and cell elongation to low water potentials in soybean hypocotyls. Planta. 1972;108:77–87. doi: 10.1007/BF00386508. [DOI] [PubMed] [Google Scholar]
- Meyer RF, Boyer JS. Osmoregulation in soybean seedling having low water potentials. Planta. 1981;151:482–489. doi: 10.1007/BF00386543. [DOI] [PubMed] [Google Scholar]
- Mundy J, Chua N. Abscisic acid and water-stress induce the expression of a novel rice gene. EMBO J. 1988;7:2279–2286. doi: 10.1002/j.1460-2075.1988.tb03070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niogret MF, Culiáñez-Macià FA, Goday A, Albà MM, Pagès M. Expression and cellular localization of rab28 mRNA and Rab28 protein during maize embryogenesis. Plant J. 1996;9:549–557. doi: 10.1046/j.1365-313x.1996.09040549.x. [DOI] [PubMed] [Google Scholar]
- Nonami H, Boyer JS. Turgor and growth at low water potentials. Plant Physiol. 1983;89:798–804. doi: 10.1104/pp.89.3.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonami H, Boyer JS. Direct demonstration of a growth-induced water potential gradient. Plant Physiol. 1993;102:13–19. doi: 10.1104/pp.102.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts JK, DeSimone NA, Lingle WL, Dure L., III Cellular concentrations and uniformity of cell-type accumulation of two lea proteins in cotton embryos. Plant Cell. 1993;5:769–780. doi: 10.1105/tpc.5.7.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning. A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp 7.37–7.52
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Wiel C, Scheres B, Franssen H, Van Lierop MJ, Van Lammeren A, Van Kammen A, Bisseling T. The early nodulin transcript ENOD2 is located in the nodule parenchyma (inner cortex) of pea and soybean root nodules. EMBO J. 1990;9:1–7. doi: 10.1002/j.1460-2075.1990.tb08073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Duang X, Wang B, Hong B, Ho TDH, Wu R. Expression of a late embryogenesis abundant protein gene, HVA1, from barley confers tolerance to water deficit and salt stress in transgenic rice. Plant Physiol. 1996;110:249–257. doi: 10.1104/pp.110.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Katsuhara M, Kelly WB, Michalowski CB, Bohnert HJ. A family of transcripts encoding water channel proteins: tissue-specific expression in the common ice plant. Plant Cell. 1995;7:1129–1142. doi: 10.1105/tpc.7.8.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]