Summary
Innate immune memory is the phenomenon whereby innate immune cells such as monocytes or macrophages undergo functional reprogramming after exposure to microbial components such as lipopolysaccharide (LPS). We apply an integrated epigenomic approach to characterize the molecular events involved in LPS-induced tolerance in a time-dependent manner. Mechanistically, LPS-treated monocytes fail to accumulate active histone marks at promoter and enhancers of genes in the lipid metabolism and phagocytic pathways. Transcriptional inactivity in response to a second LPS exposure in tolerized macrophages is accompanied by failure to deposit active histone marks at promoters of tolerized genes. In contrast, β-glucan partially reverses the LPS-induced tolerance in vitro. Importantly, ex vivo β-glucan treatment of monocytes from volunteers with experimental endotoxemia re-instates their capacity for cytokine production. Tolerance is reversed at the level of distal element histone modification and transcriptional reactivation of otherwise unresponsive genes.
Introduction
Accumulating evidence suggests that monocytes can be reprogrammed by exposure to microbe-associated molecular patterns (MAMPs) during their time in the circulation (Quintin et al., 2014). In this model, immune tolerance in myeloid cells, be they monocytes in the circulation or macrophages in the tissues (lipopolysaccharide macrophages [LPS-Mfs]), represents one extreme in the spectrum of innate immune memory and can be induced by high bacterial burden in vivo or lipopolysaccharide (LPS) exposure in vitro (Netea et al., 2016). On the other hand, trained immunity can be induced by exposure to certain vaccines, microbial components, or metabolites, and is a state characterized by increased pro-inflammatory response to secondary unrelated infections (Netea et al., 2016). We recently showed that tolerance (induced by LPS) and trained immunity (induced by Candida albicans β-glucan [BG]) are both associated with specific epigenomic states (Cheng et al., 2014; Saeed et al., 2014). Most notably, the identity of these macrophage subtypes was specified by differences in primed and active distal element repertoires (Saeed et al., 2014).
Monocytes and macrophages play an important role in the pathophysiology of sepsis and inflammation, along with other innate and adaptive immune cells (Biswas and Lopez-Collazo, 2009). Transcriptome analysis of tolerant monocytes from sepsis patients (Shalova et al., 2015) and a mouse sepsis model (Foster et al., 2007) reveals that the tolerized phenotype cannot be explained purely through failure of specific signaling pathways induced by pattern recognition receptors to activate downstream genes. This implicates a role for local chromatin architecture and specific transcriptional regulators in controlling the expression of tolerized genes (Glass and Natoli, 2016). Further, studies in human cancers have revealed commonalities between inflammation and cancer associated tolerance, including the role for IDO1 in both (Bessede et al., 2014). Accordingly, several anti-cancer drugs, such as bromodomain and extraterminal domain family (BET) inhibitors and a topoisomerase inhibitor, have proven efficacious in blocking inflammation-associated death in mice (Nicodeme et al., 2010; Rialdi et al., 2016). The specific epigenetic and transcriptional remodeling induced by the initial LPS exposure and the extent to which it specifies tolerance to future LPS exposure are unknown.
Here, as part of the BLUEPRINT epigenome consortium (http://www.blueprint-epigenome.eu), we report the time-resolved, comprehensive epigenomes of human monocyte-to-macrophage differentiation and induction of tolerance with LPS and training with BG. Our epigenomic analysis revealed that tolerance and trained immunity involve opposing regulation of common pathways during early exposure to MAMPs, leading to distinct epigenomic states in the two macrophage subtypes. We therefore hypothesized that BG may be capable of reversing LPS-induced tolerance. We show that ex vivo BG exposure can reinstate a responsive phenotype in both monocytes tolerized by ex vivo LPS exposure and monocytes tolerized by in vivo experimental endotoxemia in healthy volunteers. This reversal of tolerance involves epigenomic reprogramming of macrophages.
Results
Distinct Temporal Epigenetic Remodeling in Response to Microbial Components
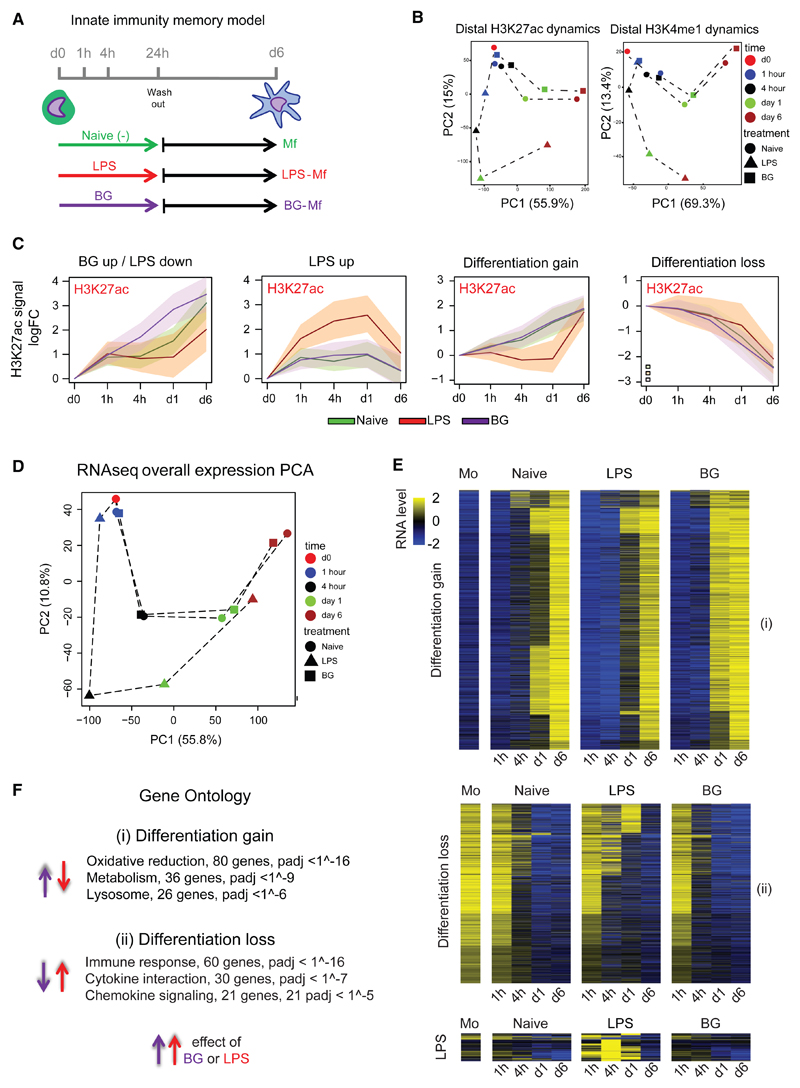
Two innate immune memory states can be induced in culture through an initial exposure of primary human monocytes to either LPS or BG for 24 hr, followed by removal of stimulus and differentiation to macrophages for an additional 5 days (Figures 1A and S1; Quintin et al., 2012; Saeed et al., 2014). The three subtypes of macrophages generated in this study were untreated naive macrophages (naive-Mfs), LPS-exposed tolerized macrophages (LPS-Mfs), and BG-exposed trained macrophages (BG-Mfs). To gain insight into the mechanisms and order of events that ultimately lead to these three subtypes, we generated epigenomic data at several time points during this process (two donors; summarized in Table S1 and Figure S1; GEO: GSE85246). Depending on the modification, 2%–31% of marked regions showed dynamics during differentiation or LPS or BG exposure, with H3K27ac at promoters and enhancers being the most dynamic mark in number and range (Figure S1; Table S1). Interestingly, epigenetic changes were observable as early as 1 hr in response to LPS and 4 hr to BG (Figure 1B). The overall H3K27ac pattern at dynamic promoters and enhancers indicates that the most pronounced changes are associated with differentiation (principal component 1 [PC1]), with BG- and RPMI-treated monocytes partially establishing macrophage-specific active regions already by day 1 (Figure 1B). Conversely, LPS treatment results in establishment of pro-inflammatory associated active elements (PC2) and stunted differentiation, followed by partial “catch up” establishment of differentiation marks following removal of stimulus (Figures 1B and S1). Contrary to this catch up of H3K27ac marked enhancers, the H3K4me1 marked enhancer repertoire of LPS-Mfs is significantly different from those of naive-Mfs and BG-Mfs (Figure 1B). Repressive marks, H3K9me3 and H3K27me3, showed no dynamics during the first 24 hr, indicating little role in the early, priming phase of innate immune memory (Figure S1C; Table S1).
Figure 1. Epigenomic and Transcriptomic Remodeling of Monocytes Induced by Exposure to LPS or BG.
(A) Experimental setup for epigenomic interrogation of monocyte-to-macrophage differentiation and induction of tolerance (with LPS) or trained immunity (with BG).
(B) PCA plots of H3K27ac and H3K4me1 dynamic enhancers (monocytes, red circle; naive, circle; LPS, triangle; BG, square; 1 hr, blue; 4 hr, black; day 1, green; and day 6, brown). Dynamic H3K27ac patterns show a clear deviation from the differentiation pathway (PC1) in LPS-treated cells. On the other hand, BG-treated cells at day 1 are well on their way toward a full macrophage epigenetic profile.
(C) A total of 17,500 H3K27ac dynamic gene-distal regions were identified and can be clearly separated into four clusters: BG up/LPS down, LPS up, differentiation gain, and differentiation loss. Solid lines are median log-FC relative to day 0, and shaded areas represent the 25th and 75th quartile. Naive cells are shown as a green line, LPS as a red line, and BG as a purple line. H3K4me1 at these regions can be seen in Figure S1B; LPS induces early H3K27ac accumulation, followed by long-term H3K4me1 marking, while BG induces concurrent accumulation of H3K27ac and H3K4me1.
(D) PCA plots showing the relationships among all samples based on dynamic gene expression. PC1 explains most of the variation and is associated with differentiation. PC2 is LPS related, with LPS 4 hr and LPS day 1 samples separating from the corresponding naive and BG samples.
(E) Heatmap of differentiation associated genes, as well as those induced by LPS or BG exposure. The general trend in expression is that BG exposed cells start to express differentiation associated genes faster (at day 1) than naive cells, while LPS exposed cells lag behind.
(F) Top pathways associated with differentiation and showing opposing directions in response to BG and LPS.
See also Figures S1, S2, and S3 and Tables S1, S2, S3, and S4.
In total, 17,500 enhancers with dynamic H3K27ac were identified (Figure S1B). The two largest clusters show gain (n = 4,028) or loss (n = 6,462) of H3K27ac during differentiation in all three macrophage subtypes (Figure 1C and S1B). The closest genes associated with differentiation gain or loss clusters are associated with leukocyte differentiation, activation, metabolism, and phagocytosis (Table S2). Upon monocyte exposure to LPS, H3K27ac induction precedes a temporally delayed H3K4me1 (Figures 1C and S1B). The closest genes associated with these enhancers are involved in cytokine response and nuclear factor κB (NF-κB) signaling, among other well-known LPS-response pathways (Table S2). The “BG up/LPS down” enhancer cluster shows accelerated H3K27ac deposition in BG-exposed monocytes and little to no H3K27ac accumulation in LPS-exposed monocytes relative to naive-Mfs (Figure 1C). This cluster is composed of >3,200 enhancers and shows concordant increase in H3K4me1 to day 6 (Figure S1B). Chromatin segmentation analysis using EpicSeq (Mammana and Chung, 2015) revealed that these regions gain H3K4me1 at the expense of repressive H3K27me3 markings in naive-Mfs and BG-Mfs (Figure S1D). Conversely, LPS-Mfs maintain a chromatin state more similar to monocytes, primarily low H3K4me1 with the presence of H3K27me3 (Figure S1D). The closest genes to these enhancers are involved in lipid biosynthesis and lysosome and leukocyte differentiation (Table S2), indicating that BG exposure leads to the accumulation of membrane components necessary for phagocytosis and cytokine release, whereas LPS exposure prevents their activation (Figure 1C).
Transcriptome Changes Modulated by LPS and β-Glucan
RNA sequencing (RNA-seq) was performed on the same time points as epigenetic marks (n = 2 donors; Figure 1A). General kinetics similar to those unveiled for epigenetic remodeling was observable, with monocytes clustering after a short exposure to BG and LPS (Figure 1D). Over the time course, the major changes in gene expression patterns were associated with differentiation (PC1, 55.8% of the variance) and LPS exposure (PC2, 10.8% of the variance), which is most pronounced at 4 hr and day 1 (Figure 1D). Over 5,700 protein-coding genes showed dynamic expression (fold change [FC] > 2, adjusted p value [padj] < 0.05) in our model between either treatments or time points (Table S3; Figures 1E and S2A). LPS-induced genes were involved in immune response, whereas LPS-delayed genes were generally differentiation associated (Figure 1E; Table S4). The major ontologies of BG-induced genes (662 genes at day 1) were lipid biosynthesis, metabolism, and the lysosome pathway (Figure 1E; Table S4). Intersection between exposure-dependent gene expression and promoter acetylation patterns showed a strong overlap between H3K27ac and gene expression temporal profiles (Figures S2B and S2C).
LPS-Specific DNA (De)methylation Signatures
Recent studies have revealed extensive DNA methylation remodeling during B cell (Kulis et al., 2015) and osteoclast differentiation (de la Rica et al., 2013; Nishikawa et al., 2015). Considering that our ex vivo differentiation model occurs in the absence of cell division, we were interested to see the extent to which (de)methylation plays a role during monocyte-to-macrophage differentiation and innate immune memory. Unlike the comprehensive histone modification remodeling, consistent DNA methylation change (at least 30% change and four or more significant differentially methylated CpGs per differentially methylated region [DMR]) was limited to a few hundred genomic regions (Figure S3). The vast majority of DMRs showed loss of methylation during monocyte-to-macrophage differentiation irrespective of MAMP exposure (Figure S3B), consistent with recent findings in macrophages and dendritic cells (Vento-Tormo et al., 2016). We did not observe a role for DNA methylation in “training” the macrophages for future transcriptional response to infection. More than 90% of DMRs occurred at distal elements marked by H3K4me1, and only 6% occurred at promoters (Figure S3C). Cumulatively, our data indicate that LPS-specific DNA methylation changes occur and, due to the more stable nature of this mark, may represent a useful biomarker for LPS-induced macrophage tolerance (Figures S3D and S3E).
LPS- and β-Glucan-Specific Transcriptional Networks
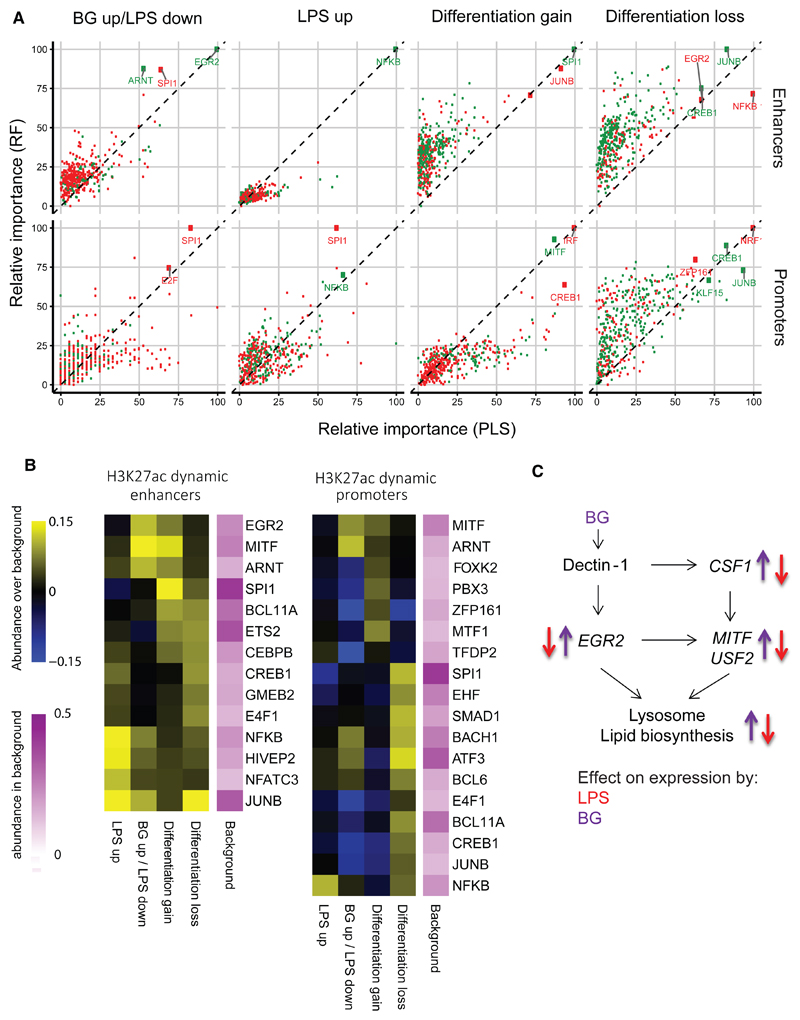
Motif analysis was used to gain insight into which pathways and transcription factors (TFs) regulate the epigenetic changes associated with differentiation and LPS or BG exposure. Four clusters of enhancers and promoters were designated based on H3K27ac dynamics over time: BG up/LPS down, “LPS up,” “differentiation gain,” and “differentiation loss” (Figures 1C and S1B). Two classifiers (random forest [RF] and a partial least-squares [PLS]) were trained, using the TF motifs found by GIMME (van Heeringen and Veenstra, 2011). Both score features (TF motifs) were based on their ability to separate the clusters—the so-called feature importance score (between 0 and 100)—which is a measure of how characteristic the presence or absence of the TF motif is for the considered cluster. Both classifiers were trained with the caret R-package using 10-fold cross-validation, repeated five times. We define positive (green dots) and negative predictors (red dots) as TF motifs that are more or less abundant of the considered cluster compared to the other clusters, respectively.
Enhancers that show differentiation gain in H3K27ac were enriched for the SPI1 (PU.1) motif, while LPS-induced active enhancers were enriched for NF-κBmotif (Figures 2A and 2B). The top positive predictor motif for the BG up/LPS down cluster was EGR2, with a score of 100, followed by ARNT (Figure 2A). EGR2 is downstream of dectin-1 (Goodridge et al., 2007) and shows prominent, transient induction in BG-exposed monocytes (Figure S4A). Enhancers with EGR2 motifs are mainly associated with genes involved in lipid metabolism and biosynthesis and lysosome function (Figures 1E and 1F). The early activation of these pathways in BG may account for the higher expression of LAMP1, the major component of the mature lysosome, in BG-Mfs (Figure S4A).
Figure 2. Motif Enrichment at Epigenetically Dynamic Promoters and Enhancers and Associated Transcription Factor Networks.
Motif enrichment analysis was performed on ATAC-sequencing (nucleosome-free) peaks that overlap H3K27ac dynamic enhancers and H3K27ac promoters.
(A) Random forest (RF) and a partial least-squares (PLS) classifiers were trained using the TF motifs found by GIMME to determine features (TF motifs) based on their ability to separate the 4 H3K27ac clusters shown in Figure 1C. Both classifiers produce a feature importance score (between 0 and 100), which is a measure of how “characteristic” the presence or absence of the TF motif is for the considered cluster. Green dots represent positive features (motif over-represented in cluster), and red dots represent negative features (motif under-represented in cluster). The EGR2 motif was the strongest positive feature for the BG up/LPS down enhancer cluster, NF-κB for the LPS up cluster, SPI1 (PU.1) for the differentiation gain cluster, and JUNB for the differentiation loss cluster. At the promoter regions NF-κB was a positive feature for LPS up cluster, MITF for the differentiation gain cluster, and CREB1 and JUNB for the differentiation loss cluster.
(B) Motif enrichment is plotted as absolute difference in abundance compared to background (yellow, higher abundance than background; blue, lower abundance than background) for the top enriched motifs. Consistently identified transcription factor motifs include SPI1 at differentiation associated enhancers, NF-κB at LPS enhancers, and EGR2 and MITF at BG enhancers. Abundance increase over background supports the level of importance score.
(C) A diagram of the transcription factor network based on EGR2 and MITF motif occurrence at BG-induced lysosome and lipid metabolism genes. Purple arrows indicate the direction of expression induced by BG exposure, and red arrows indicate the direction of expression induced by for LPS exposure. BG exposure induces transient expression of the genes, while LPS exposure inhibits activation. The full network based on promoter abundance is shown in Figure S4B.
See also Figure S4.
Interestingly, LPS-exposed monocytes do not transiently activate EGR2 (Figure S4A). The discordant effect of BG and LPS on EGR2 expression, the differential H3K27ac deposition at associated enhancers, and expression of downstream lipid metabolism genes suggests that this pathway plays a role in inducing trained immunity as opposed to tolerance. In order to further confirm the relationship between EGR2 and downstream lipid pathways, the DNA-binding motif of EGR2 was scanned at the promoters of known transcription factors, as well as lipid metabolism and lysosome genes that are induced in BG-Mfs compared to monocytes (Figure S4B). The EGR2 motif was found at the promoter of several highly expressed TFs, including MITF, which is a positive identifier for the differentiation gain promoter cluster (Figure 2A), and is also not activated in LPS-exposed monocytes (Figure S4A). Cumulatively, EGR2, MITF, and downstream TF motifs were found at the promoters of 79% of induced lipid metabolism and lysosome genes (Figure 2B). This analysis suggests that BG/dectin-1-induced EGR2 activation leads to higher expression of downstream TFs (e.g., MITF) and the establishment of promoters and enhancers that drive the expression of lysosomal and lipid metabolism genes (Figure 2C). Given the importance of lipid pathways in macrophage function, the opposing effect of LPS and BG on these genes suggests that this pathway may play a critical role in the low cytokine release in LPS-Mfs and elevated release in BG-Mfs.
Transcriptional Response of Tolerized Macrophages to LPS Re-exposure
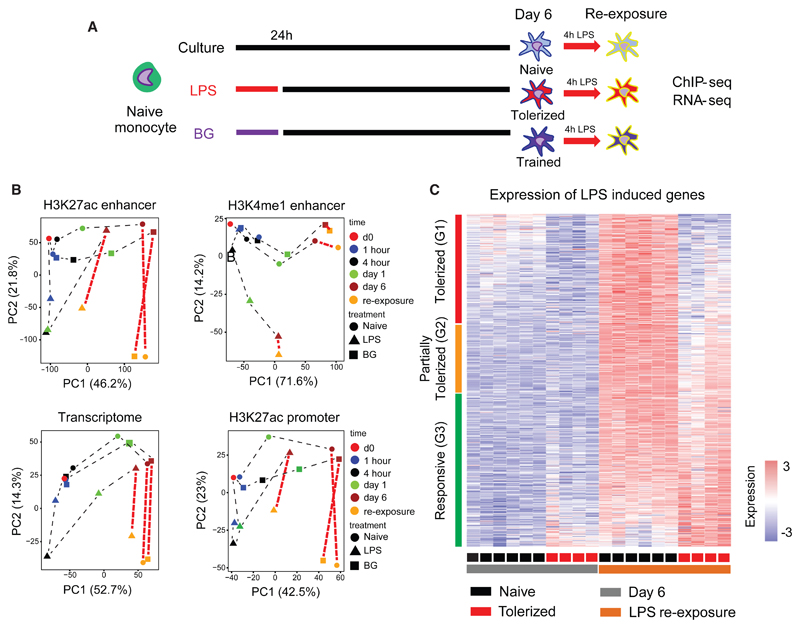
Previous analysis in an ex vivo mouse model showed that tolerant LPS-Mfs are impaired in their ability to produce pro-inflammatory cytokines, but maintain their ability to express other genes, such as those required for tissue repair (Foster et al., 2007). Given the wide-ranging epigenetic alterations in LPS-Mfs (Figure 1C; Table S1), we sought to investigate the epigenetic basis for endotoxin tolerance by exposing differentiated naive-Mfs, LPS-Mfs, and BG-Mfs to LPS for 4 hr (LPS re-exposure) (Figure 3A). The overall transcriptional and histone modification changes induced in macrophages by LPS re-exposure are shown in Figure 3B, and few differences were observed between naive-Mfs and BG-Mfs. LPS-Mfs show an avid response to LPS re-exposure both transcriptionally and with H3K27ac deposition at promoters and distal enhancers (observable as large shift in PC2; Figure 3B). This indicates that tolerized macrophages can and do respond to LPS at the epigenetic and transcriptional level. However, from H3K27ac and H3K4me1 principal-component analysis (PCA), it is clear that the epigenetic profile of LPS-Mfs is markedly different from that of naive-Mfs and BG-Mfs (observable as an LPS-Mfs lag on PC1; Figure 3B).
Figure 3. Macrophage Endotoxin Tolerance Defined at the Transcriptional Level following LPS Re-exposure.
(A) The innate immune memory model, including data collection at LPS re-exposure at day 6.
(B) PCA plots of dynamic RNA-seq, H3K27ac at promoters and enhancers, and H3K4me1 peaks, including LPS re-exposure samples. After re-exposure to LPS, significant enhancer H3K27ac changes occur in LPS-Mfs, indicating that they are capable of activating their enhancers. However, the level of their response is lower compared to monocytes, naive-Mfs, and BG-Mfs, which can be seen on the second principal component. Unlike RNA and H3K27ac, H3K4me1 does not show significant changes following LPS re-exposure in any of the three macrophage subtypes.
(C) The total macrophage transcriptional response (750 genes) to LPS was separated into three groups based on the induction of genes in LPS-Mfs, relative to naive-Mfs and BG-Mfs, revealing a gradient in LPS-Mf response to LPS re-exposure. The groups are (G1) tolerized genes, (G2) partially tolerized genes, and (G3) responsive genes.
See also Figure S5.
Polytomous modeling was used to separate genes based on their transcriptional response to LPS re-exposure (4 hr) in macrophages at day 6. In total, 780 genes showed higher expression (FC > 2, posterior probability > 0.3) in naive-Mfs following 4-hr LPS exposure (Figure 3C; Table S5). Transcriptional responsiveness to LPS re-exposure in LPS-Mfs is a gradient, with genes showing complete tolerance (unresponsiveness) (cluster G1), a partial response (G2), or a full response comparable to naive-Mfs (G3) (Figures 3C and S5A). Cytokine genes were the most enriched group and were spread across the LPS re-exposure response gradient, with CXCL9 (G1) and TNF (G2) showing complete or partial tolerance and IL6 and IL8 showing comparable responsiveness to naive-Mfs (G3) (Figures S5B and S5C). The normal induction of interleukin 6 (IL-6) mRNA expression and the absence of response in ELISA assays therefore suggests that tolerance is a complex phenotype that involves both dampened transcriptional responses to LPS re-exposure and an inability to release some cytokines (Figure S5C). The top tolerance-specific biological process was “cytokine production,” while the top pathway was “RIG-I-like signaling” and “p53 signaling” (Figure S5D).
Epigenetic Profile of Tolerized Genes
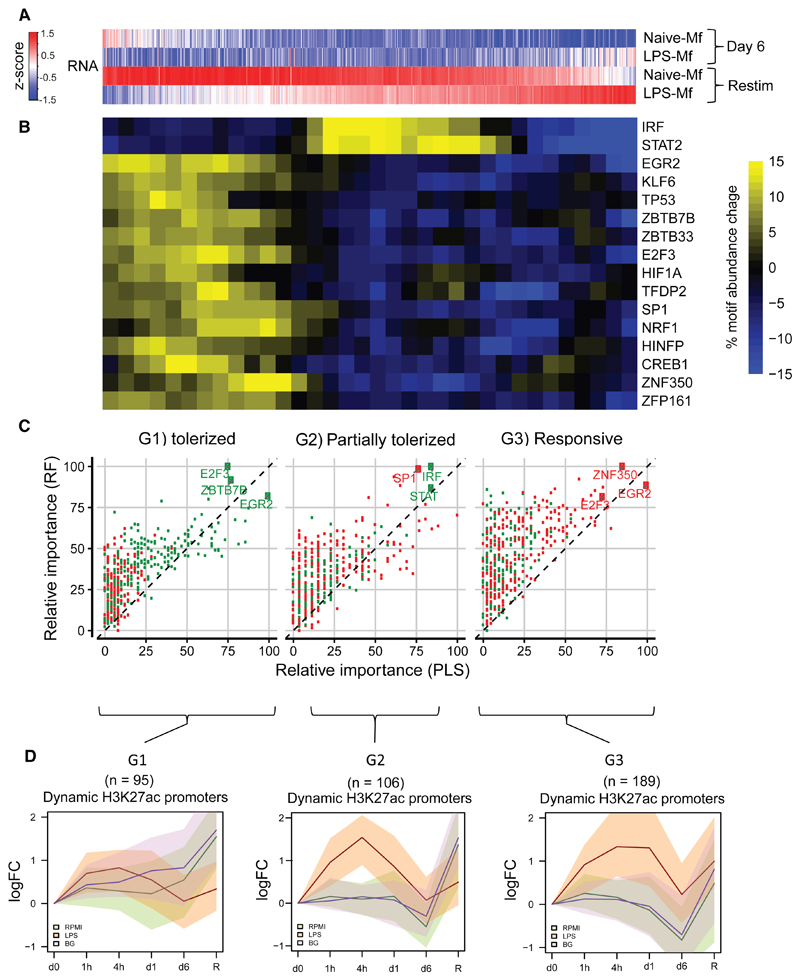
To understand the molecular mechanisms involved in the altered gene induction by LPS re-exposure in LPS-Mfs, we investigated promoter motif enrichment at overlapping assay for transposase accessible chromatin (ATAC) peaks. Because transcriptional responsiveness to LPS re-exposure in tolerized macrophages occurs on a gradient (Figure 4A), motif enrichment at promoters was scanned in a sliding window of 100 promoters throughout the response gradient from most tolerized (G1) to responsive (G3) genes (Figure 4B). This analysis identified discrete motif signatures in the G1 and G2 tolerized groups. The G1 gene promoters were enriched for several TF motifs, including EGR2, HIF1A, and p53. The latter TF was also identified as a top tolerized pathway (Figure S5D). The partially tolerized genes are enriched for IRF and STAT motifs (Figure 4B). Random Forest analysis also indicated that EGR2 was the top identifier for the G1 group, while IRF and STAT motifs are top identifiers for the G2 group. Interestingly, the G3 group does not contain positive identifiers (Figure 4C). IRF and STAT genes show a tolerized pattern (Figure S6A), indicating that their unresponsiveness to LPS effects downstream partially tolerized genes. On the other hand, NFKB1 and RELA showed normal induction in LPS-Mfs (Figure S6B).
Figure 4. Histone Modification Dynamics and Open Chromatin Analysis at Tolerized Gene Promoters.
(A) Heatmap showing average expression of 777 LPS-responsive genes in naive-Mfs. Genes are ranked based on their induction in LPS-Mfs, first by tolerance group (G1, G2, and G3) and then by relative induction compared to naive-Mfs within each group. Response to LPS re-exposure is a gradient in LPS-Mfs, with the most tolerized genes on the left and the most responsive genes on the right.
(B) Heatmap showing abundance of significant motifs in the promoter regions of the three macrophage LPS-responsive gene groups. The tolerized gene promoters are enriched for several transcriptional repressors, such as EGR2 and TP53, while the partially tolerized gene promoters are enriched for IRF and STAT motifs.
(C) Random forest (RF) and a partial least-squares (PLS) classifiers importance score (between 0 and 100) for each tolerized gene cluster (G1, tolerized; G2, partially tolerized; and G3, responsive). Green dots represent over-represented motifs, and red dots under-represented motifs. The top features of G1 gene promoters are E2F3, EGR2, and ZBTB7B motifs. The top features for G2 gene promoters are IRF and STAT, while G3 promoters do not have over-represented features but are depleted of EGR2, E2F3, and ZNF350.
(D) Median H3K27ac at dynamic promoters of G1, G2, and G3 group genes, shaded areas represent the 25th and 75th quartile. This shows that LPS-Mfs do not accumulate H3K27ac at tolerized genes but do so at the promoters of responsive genes. See also Figure S6 for H3K4me3.
See also Figure S6.
Dynamic H3K27ac change during differentiation and LPS or BG exposure was plotted over the promoter regions of G1, G2, and G3 genes (Figure 4D). Dynamic promoter H3K27ac was observed for roughly half of all genes, with the rest showing consistent high acetylation during all time-points, including LPS re-exposure (not shown). Tolerized genes (G1) and partially tolerized genes (G2) showed no or impaired accumulation of H3K27ac, respectively, after LPS re-exposure in LPS-Mfs compared to naive-Mfs and BG-Mfs (Figure 4D), while responsive genes (G3) were equally acetylated after LPS re-exposure in all subtypes (Figure 4D). H3K4me3 patterns at these promoters closely matched those of H3K27ac (Figure S5B). This finding suggests that LPS-Mfs fail to accumulate H3K27ac at tolerized genes either through absence of pro-inflammatory activators, such as IRF and STATs in the case of G2 genes, or through presence of tolerance inducing TFs, such as HIF1A in the case of G1 genes.
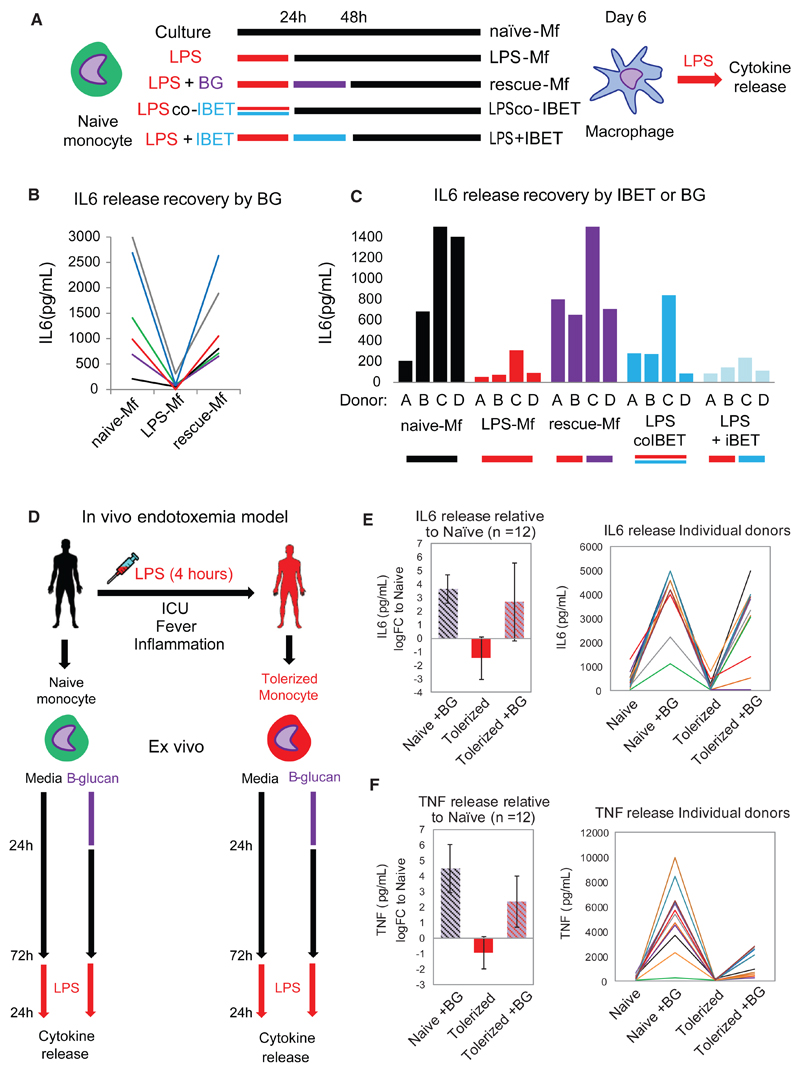
β-Glucan Exposure Can Reverse Tolerance in Both In Vitro and In Vivo LPS-Exposed Monocytes
As indicated before, BG and LPS have an opposing effect on EGR2 and MITF expression (Figure S4A), accumulation of H3K27ac at target enhancers and promoters (Figure 1C), and expression of genes involved in macrophage function, such as lipid metabolism and lysosome and cytokine production (Figure 1E). These findings point to a potential for reversal of LPS-induced tolerance by using BG to stimulate the dectin-1 pathway. To test this hypothesis, monocytes were exposed to LPS for 24 hr and then to BG for 24 hr, followed by a rest period before LPS re-exposure (Figure 5A). We refer to these macrophages as “rescue-Mfs”. Additionally, we used the clinically relevant small molecular histone mimic bromodomain and extra-terminal domain family (BET) inhibitor (IBET)151 in a co-treatment with LPS (“preventative”) or following LPS exposure (“reversal”) setting (Figure 5A). ELISAs showed that BG exposure was able to reverse LPS-induced tolerance and reinstate normal levels of cytokine release in rescue-Mfs (Figure 5B). On the other hand, IBET151 was only effective in preventing tolerance when used to block the LPS-induced response, but it did not reverse tolerance when administered after LPS (Figure 5C). This is in line with the finding that IBET151 is effective in blocking inflammation-associated death in mice (Nicodeme et al., 2010) but suggests that IBET151 is not an effective treatment in monocytes that have already experienced an inflammatory response. Therefore, BG represents a possible treatment option for restoring proper macrophage cytokine release during the post-inflammation tolerance phase.
Figure 5. BG Can Reverse Both In Vitro and In Vivo LPS-Induced Tolerance and Reinstate Proper Cytokine Production in Macrophages.
(A) The in vitro monocyte tolerance reversal model, with BG added therapeutically after 24 hr of LPS exposure (rescue-Mfs). The histone-mimic and inflammation blocker IBET was used in a preventative (co-culture with LPS for 24 hr LPS-co-IBET-Mfs) and a therapeutic (added after 24 hr of LPS exposure [LPS + IBET-Mfs]) manner. Following several days of rest, macrophages were re-exposed to LPS and cytokine release measured after 24 hr.
(B) BG re-instates IL-6 release in tolerized macrophages. Data from six donors are shown for naive-Mfs, LPS-Mfs, and rescue-Mfs.
(C) Preventative use of IBET blocks the first LPS response in monocytes, resulting in differentiation of macrophages that can release cytokines at the second LPS exposure. Therapeutic use of IBET does not re-instate cytokine release in macrophages.
(D) Experimental human endotoxemia model, with ex vivo BG administration. Monocytes were isolated from 12 healthy volunteers before (naive) and 4 hr after LPS injection (tolerized). Naive or tolerized monocytes were exposed to BG for 24 hr, followed by culture media, or culture media alone. After 3 days ex vivo, monocytes were re-exposed to LPS, and cytokines were measured 24 hr later.
(E and F) BG recovered IL-6 release in 9 out of 12 tolerized monocytes (E) and TNF release in 8 out of 12 monocytes (F). Data are presented as mean ± SD.
The suitability of the in vitro tolerance model to mimic the in vivo situation is a major question. Chiefly, does LPS exposure in vivo induce the same transcriptional responses in monocytes, and can in vivo LPS-induced tolerance be reversed by BG? To answer these questions, we used an in vivo experimental human endotoxemia model (Draisma et al., 2009) (Figure 5D). In this model, healthy volunteers are injected with 2 ng/kg US Standard Reference Endotoxin Escherichia coli O:113 LPS (Pharmaceutical Development Section of the National Institute of Health, Bethesda, MD, USA), which leads to a sepsis-like state (reviewed in Bahador and Cross, 2007). Study protocols were approved by the local ethics committee of the Radboud University Nijmegen Medical Centre. The volunteers experience transient fever and cold chills as well as pro- and anti-inflammatory cytokine signatures. The in vivo LPS-exposed monocytes show elevated mRNA expression of key cytokines at 4 hr (not shown) and fail to release cytokines in response to a second ex vivo LPS exposure. In this regard, they behave much like in vitro LPS-tolerized monocytes. Monocytes were isolated from peripheral blood taken before and after LPS administration, and then exposed ex vivo to either culture medium alone, or with BG. Cytokine release was measured following LPS re-exposure in culture (Figure 5D). Ex vivo BG exposure increased the release of tumor necrosis factor (TNF) and IL-6 in tolerized monocytes at LPS reexposure (Figures 5B and 5C). This finding indicates that BG can restore cytokine production of in-vivo-tolerized monocytes. Cumulatively, this confirms that the mechanisms involved in the establishment of tolerance by LPS in vivo and in vitro are similar, validating the use of the in vitro model to study reversal of tolerance by BG. More importantly, it suggests that the BG effect on monocyte tolerance may be transferred to the clinic in the future.
β-Glucan Recovers the Transcriptional Response to LPS at Tolerized Genes
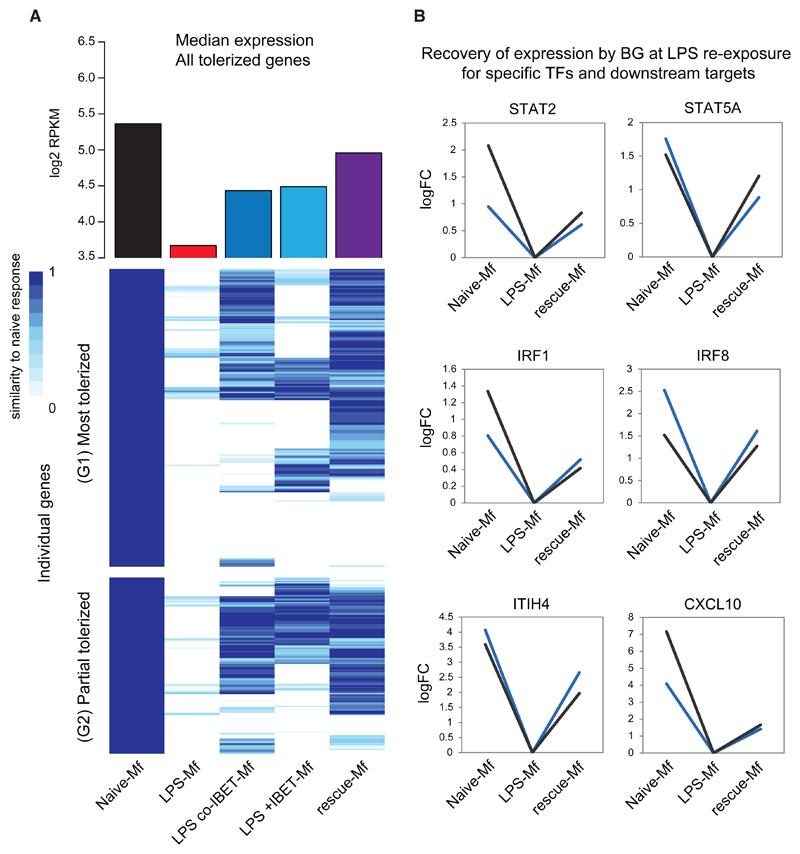
Next, we assessed whether BG reverses tolerance at the transcriptional level. In this experimental setup, monocytes were exposed to LPS followed by BG and then left to rest 24 hr or 4 days before LPS re-exposure for 4 hr (Figure 6A). Additionally, monocytes were treated with a combination of LPS and IBET151 (preventative) and LPS followed by IBET151 (reversal). BG was able to recover the induction of 60% of tolerized genes at day 6 (Figure 6A), including several pro-inflammatory TFs (Figure 6B). Similar effects were observed when IBET151 was used in a preventative model, indicating that BG reversal of LPS-induced tolerance leads to an outcome similar to that produced by blocking LPS-induced tolerance altogether. Overall BG reversal led to a higher median expression of tolerized genes compared to both preventative and reversal use of IBET151 (Figure 6A).
Figure 6. Reversal of Tolerance by BG at the Transcriptional Level.
(A) Heatmap of the transcriptional response of naive-Mfs, LPS-Mfs, and rescue-Mfs (BG reversed LPS-Mfs) to LPS re-exposure at day 6. The scale represents relative expression between LPS exposed naive-Mfs (1) and LPS-Mfs (0). Rescue-Mfs exposed to LPS show the most similar profile to naive-Mfs. On the top of the heatmap is median expression (log2 RPKM) of tolerized genes at day 6 and LPS re-exposure in naive-Mfs (black), LPS-Mfs (red), LPS-co-IBET-Mfs (blue), LPS + IBET-Mfs (light blue), and rescue-Mfs (purple).
(B) BG reverses the tolerization of key LPS-induced transcription factors, such as STAT2, STAT5A, IRF1, and IRF8. Log2 fold change increase in mRNA expression is shown.
Epigenomic Analysis of β-Glucan Recovery of Tolerized Macrophages
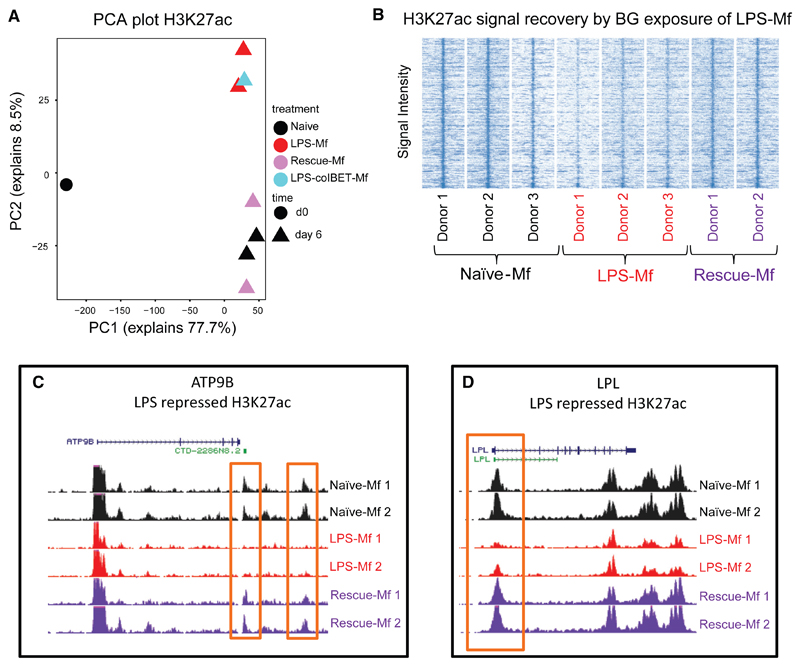
BG exposure following LPS exposure recovers the expression of genes involved in lipid biosynthesis, phagocytosis, and cytokine transport (Figures S7A and S7B). Recovery of expression was observed as early as day 3 and maintained at a higher level in macrophages at day 6 (Figure S7B). Interestingly, addition of BG to naive and tolerized monocytes at day 1, elicited the expression of EGR2 and MITF within 4 hr, with a lower induction in tolerized monocytes (Figure S7C). These findings indicate that BG-induced receptor pathways remain at least partially inducible after the LPS-induced cytokine response and that these pathways can partially recover the naive macrophage epigenetic and transcriptional programs. Analysis of dynamic H3K27ac promoters and enhancers in naive-Mfs, LPS-Mfs, rescue-Mfs, and LPS-co-IBET151-Mfs revealed that BG exposure restores H3K27ac deposition at regions where H3K27ac increase was not obtained following LPS exposure (Figure 7). Interestingly, while IBET151 blocks ∼75% of the transcriptional response to LPS in monocytes at 4 hr (data not shown), LPS-co-IBET151-Mfs look more like LPS-Mfs at day 6, indicating no effect of IBET151 on the overall epigenomic profile of LPS-Mfs (Figure 7A, blue square). The effect of BG exposure on H3K27ac deposition in LPS-Mfs was observable at both promoters and distal enhancers of genes involved in metabolism and lipid biosynthesis (Figures 7C and 7D).
Figure 7. Reversal of Tolerance by BG at the Chromatin Level.
(A) PCA plot of H3K27ac dynamics among monocytes, naive-Mfs, LPS-Mfs, rescue-Mfs, and LPS-co-IBET-Mfs. BG exposure of tolerized monocytes results in a H3K27ac profile more similar to naive macrophages, while co-incubation of monocytes with LPS and IBET does not lead to activation of these regions.
(B) Heatmap showing re-establishment of the naive-Mf H3K27ac signal by BG exposure in tolerized macrophages.
(C) PCA plot of H3K4me1 dynamics among monocytes, naive-Mfs, LPS-Mfs, Rescue Mfs, and LPS-co-IBET-Mfs, the effect is similar to H3K27ac, but to a lesser extent. (C) H3K27ac tracks at ATP9B (glucose transport) gene enhancer and (D) LPL (lipid metabolism) gene promoter.
See also Figure S7.
Discussion
Perturbation of normal monocyte-to-macrophage differentiation by exogenous signals, such as high bacterial burden in sepsis, can lead to a changed chromatin state and an associated deviation from steady-state function (Amit et al., 2016). This phenomenon is known as innate immune memory, with the best-characterized outcomes being endotoxin tolerance or trained innate immunity (Netea et al., 2016). Trained immunity can have beneficial effects through priming of macrophages for stronger responses to subsequent infection and can be induced by a variety of MAMPs, such as C. albicans (Quintin et al., 2012), Bacille Calmette-Gueérin (BCG) vaccine (Kleinnijenhuis et al., 2012), and BG (Saeed et al., 2014). Conversely, exposure to high levels of LPS can induce a tolerized macrophage phenotype, which is a major cause of sepsis-associated mortality (SepsisReport, 2012). Previously, we showed that tolerized macrophages (LPS-Mfs) and trained macrophages (BG-Mfs) have distinct epigenetic (Saeed et al., 2014), and metabolic states (Cheng et al., 2014). Mouse studies have shown that such distal element markings are important for appropriate responses to infection (Ghisletti et al., 2010; Ostuni et al., 2013) and identity of tissue-resident macrophages (Amit et al., 2016; Lavin et al., 2014). Nevertheless, until now, the epigenetic basis for endotoxin tolerance in humans has not been explored.
In the current study, our aim was to unveil the early epigenetic and transcriptional events following monocyte exposure to LPS or BG and how the resulting epigenetic landscapes determine the function of tolerized and trained macrophages. LPS- and BG-induced active histone dynamics were observed as early as 1 hr and 4 hr after exposure, respectively (Figures 1B and 1C). Generally, H3K27ac accumulation was accompanied by H3K4me1 accumulation, most obviously at BG-induced enhancers (Figure S1B). Contrary to this general pattern, LPS-induced active enhancers, associated with an inflammation response, showed discordance in time with accumulation of H3K4me1 (Figure S1B), which remained at higher levels in LPS-Mfs compared to naive-Mfs and BG-Mfs. This persistence of H3K4me1 in LPS-Mfs contributes to the overall epigenetic signature of this macrophage subtype and may account for some of the tolerized phenotype (Figure 1B). More pointedly, we discovered a set of more than 3,000 de novo macrophage established distal enhancers that were modulated in the opposite direction by BG or LPS exposure (Figure 1D). Deposition of H3K27ac and H3K4me1 at these regions was accelerated by BG exposure and delayed or completely blocked by LPS exposure. Accordingly, expression of genes near these elements was induced by BG, peaking at 24 hr post-exposure, while they remained lowly expressed in LPS-exposed monocytes (Figure 1F). These genes were involved in lipid metabolism and biosynthesis, phagocytosis, and lysosome maturation (Figures 1G and S2) and have clear TF motif signatures for EGR2, MITF, and ARNT (Figure 2A). Interestingly, EGR2, a TF downstream of the BG receptor dectin-1, showed clear transient upregulation by BG but remained inactive in LPS-exposed monocytes, suggesting a possible role in modulating these pathways (Figure S4). TFs and pathways linking lipid biosynthesis and inflammation have been described (Spann et al., 2012). Further, macrophage response to infection requires a substantial amount of energy, and shifts in metabolism and energy production are a whole mark of macrophage polarization to M1 or M2 subtypes (Ghesquière et al., 2014), as well as for establishment of trained immunity (Cheng et al., 2014).
Reversal of tolerance after the initial inflammation phase has garnered interest because of the limited success of inflammation-blocking treatments to reduce overall sepsis mortality (Angus and van der Poll, 2013) and because the majority of sepsis deaths occur due to secondary hospital infection during the tolerized phase (Gilroy and Yona, 2015). Our hypothesis was that BG can reverse LPS-induced tolerance because it discordantly regulated pathways that LPS also affected. Specifically, LPS fails to activate key regulators of lipid, lysosome, and metabolism genes, EGR2 and MITF, while BG induces their expression (Figures 1 and 2). Recently, IFNG was shown to partially recover metabolic function in tolerized monocytes from sepsis patients, indicating that reversal of tolerance using innate immune “trainers” is a viable therapeutic strategy (Cheng et al., 2016). We show that BG exposure can indeed reverse the tolerance in macrophages induced by LPS exposure, with rescue-Mfs showing higher release of cytokines in response to a second LPS stimulus (Figures 5A–5C). This was in contrast to the inflammation blocker IBET151, which only prevented tolerance when used to block the initial LPS response but could not reverse it when given to cells after LPS-induced inflammation (Figure 5). In order to further relate our findings to the in vivo situation, we used an experimental human endotoxemia model to induce tolerance in vivo (Draisma et al., 2009; Kox et al., 2014). In terms of cytokine production, in-vivo-tolerized monocytes behave similarly to their in-vitro-tolerized counterparts. The tolerized state of in vivo LPS-exposed monocytes is similar to that of ex-vivo-exposed monocytes and, most importantly, can also be rescued by ex vivo BG exposure (Figures 5D–5F), indicating that the mechanisms controlling monocyte tolerance in vivo can also be reverted to a more responsive phenotype.
In order to determine the ability of BG to reverse tolerance at the molecular level, we first characterized the transcriptional and epigenetic response to a second LPS exposure in tolerized macrophages (Figures 3 and 4). Studies in mouse sepsis models and human sepsis patients have shown that rather than being inert in response to a second LPS exposure, tolerized macrophages show a shift in the specific pathways that they activate (Foster et al., 2007; Shalova et al., 2015). In line with these studies, we show that LPS-Mfs remodel both H3K27ac and gene expression in response to LPS (Figure 3B). However, the starting point of LPS-Mfs is significantly different from that of naive and BG-Mfs, most clearly for H3K4me1 marked enhancers, suggesting that while activation is occurring, the available enhancer repertoire of these cells is limited or not suited. Our analysis identified a gradient in the LPS-Mf response to LPS, with some genes showing a tolerized pattern (no induction) and others showing a responsive pattern (Figure 3C). The most tolerized gene promoters were enriched for EGR2, HIF1A, and p53 motifs, among many others. A potential role for HIF1A is in agreement with a recent transcriptional analysis in monocytes from sepsis patients (Shalova et al., 2015), while the p53 pathway was a top-ranked tolerized identified by Gene Ontology (GO) analysis (Figure S5D). The strongest enrichment at partially tolerized genes was for the IRF and STAT TF motifs that show strong tolerized expression patterns themselves, ie, IRF1, IRF8, STAT2, and STAT5A (Figure S6A). IRF8 and its downstream target, KLF4, both of which are important regulators of monocyte differentiation (Kurotaki et al., 2013), show a tolerized profile and enrichment at tolerized gene promoters (Figure 4B). Other, non-TF regulators of tolerance, such as IRAK3, HIF1A, SOCS3, and IDO1, are all more highly expressed in LPS-Mfs compared to naive-Mfs and BG-Mfs and have previously been associated with endotoxin-induced tolerance (Bessede et al., 2014; Saeed et al., 2014; Shalova et al., 2015).
Rescue macrophages (BG exposed following LPS exposure) were able to induce ∼60% of tolerized genes at LPS re-exposure (Figure 6). This indicates that BG reversal of tolerance at the transcriptional level is not complete (Figure 6A). Fascinatingly, BG recovered the expression of tolerized genes to a level greater than that observed in macrophages treated with IBET and LPS together (Figure 6A). This indicates that BG can reinstate a responsive state at a higher level than that obtained by actually blocking the initial LPS transcriptional response. This important observation suggests that BG-associated pathways remain intact even after large-scale epigenetic and transcriptional programs are induced by LPS. At the level of histone modifications, BG recovers H3K27ac at regions that are silent in LPS-Mfs, further supporting the notion that the molecular mechanisms required for BG-induced chromatin remodeling remain after the initial LPS response (Figures 7 and S7).
In conclusion, the hypothesis-free epigenomic and transcriptomic analysis of monocyte-to-macrophage differentiation and innate immune memory generated a number of testable hypotheses. Our findings show that the innate immune “training stimulus” β-glucan can reverse macrophage tolerance ex vivo. This is an important step toward understanding how the tolerized phenotype can be reversed in sepsis patients and ultimately provides the framework for future therapeutic developments in innate immune diseases.
Star⋆Methods
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-H3K27ac | Diagenode | pAb-196-050 |
| Rabbit polyclonal anti-H3K4me1 | Diagenode | pAb-037-050 |
| Rabbit polyclonal anti-H3K4me3 | Diagenode | pAb-003-050 |
| Rabbit polyclonal anti-H3K27me3 | Diagenode | pAb-195-050 |
| Rabbit polyclonal anti-H3K9me3 | Diagenode | pAb-193-050 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| IBET-151 | GSK Epinova and Cellzome | GSK1210151A |
| Human Serum | Sigma-Aldrich | H4522-100ML |
| RPMI 1640 Medium, GlutaMAX | Thermo Fisher Scientific | 61870036 |
| Gentamycin | Thermo Fisher Scientific | 15750060 |
| L-glutamine | Thermo Fisher Scientific | 25030081 |
| Sodium Pyruvate | Thermo Fisher Scientific | 11360070 |
| Percoll | Sigma-Aldrich | P1644-1L |
| Ficoll Paque Plus | Sigma-Aldrich | GE17-1440-03 |
| Lipopolysaccharides from Escherichia coli 055:B5 | Sigma-Aldrich | L2880-10MG |
| β1,3(D)glucan (β-glucan) | (Saeed et al., 2014) | N/A |
| 2-Mercaptoethanol | Thermo Fisher Scientific | 21985023 |
| Actinomycin D | Thermo Fisher Scientific | 11805017 |
| IGEPAL CA-630 | Sigma-Aldrich | I8896-50ML |
| Critical Commercial Assays | ||
| KAPA library preparation kit | Kapa Biosystems | KK8400 |
| riboZero gold rRNA removal kit | Illumina | MRZG12324 |
| Nextera DNA Library Prep Kit | Illumina | FC-121-1031 |
| TruSeq SBS KIT v3 - HS (50 cycles) | Illumina | FC-401-3002 |
| NextSeq 500/550 High Output v2 kit (75 cycles) | Illumina | FC-404-2005 |
| NEBNext High-Fidelity 2 × PCR Master Mix | New England Biolabs | M0541 |
| iQ SYBR Green Supermix | Bio-Rad | 1708880 |
| 100 × SYBR Green I Nucleic Acid Gel Stain | Thermo Fisher Scientific | S7563 |
| Human IL-6 elisa | Sanquin | M9316 |
| Human TNFα elisa | R&D | DY210 |
| SPRIselect reagent kit | Beckman Coulter | B23218 |
| E-Gel SizeSelect Agarose Gels, 2% | Thermo Fisher Scientific | G661002 |
| CD3 MicroBeads, human | Miltenyi Biotec | 130-050-101 |
| CD19 MicroBeads, human | Miltenyi Biotec | 130-050-301 |
| CD56 MicroBeads, human | Miltenyi Biotec | 130-050-401 |
| dNTP set 100 mM | Life Technologies | 10297-018 |
| dUTP 100 mM | Promega | U119A |
| Glycogen (20 mg/ml) | Life Technologies | 10814-010 |
| Random Hexamer primers | Sigma-Aldrich | 11034731001 |
| Second Strand Buffer | Life Technologies | 10812-014 |
| Superscript III Reverse Transcriptase | Life Technologies | 18080-044 |
| DNA polymerase I, E. coli | New England Biolabs | M0209S |
| USER enzyme | New England Biolabs | M5505L |
| E.Coli Ligase | New England Biolabs | M0205L |
| Rnasin Plus Rnase Inhibitor | Promega | N2615 |
| Ribonuclease H | Life Technologies | AM2293 |
| T4 DNA polymerase | New England Biolabs | M0203L |
| Sodium Acetate (3M) | Life Technologies | AM9740 |
| DNase I | QIAGEN | 79254 |
| Qubit RNA HS assay kit | Life Technologies | Q32852 |
| Ribozero Gold Kit | Illumina | MRZG12324 |
| Rneasy Mini Kit | QIAGEN | 74106 |
| Deposited Data | ||
| Raw data files for RNA sequencing | This paper | GEO: GSE85243 |
| Raw data files for ChIP sequencing | This paper | GEO: GSE85245 |
| Raw data files for ATAC sequencing | This paper | GEO: GSE87218 |
| Raw data files for WGBS sequencing | This paper | EGA: EGAD00001002693 |
| Experimental Models: Organisms/Strains | ||
| Human: primary monocytes from healthy volunteers | Sanquin Blood Bank | N/A |
| Sequence-Based Reagents | ||
| NEXTflex DNA Barcodes - 48 | Bioo Scientific | 514104 |
| Primer EGR2: F 5′ TTGACCAGATGAACGGAGTG 3′R 5′ GTTGAAGCTGGGGAAGTGAC 3′ | This paper | N/A |
| Primer MITF: F 5′ AACTCATGCGTGAGCAGATG 3′R 5′ TACTTGGTGGGGTTTTCGAG 3′ | This paper | N/A |
| Primer CSF1: F 5′ CAGATGGAGACCTCGTGCC 3′R 5′ GCATTGGGGGTGTTATCTCTG 3′ | This paper | N/A |
| Primer LAMP1: F 5′ TGAACAAGACAGGCCTTCCC 3′R 5′ TGTGCAGCTCCAGAGTCACC 3′ | This paper | N/A |
| Software and Algorithms | ||
| Bedtools | (Quinlan and Hall, 2010) | http://bedtools.readthedocs.io/en/latest/ |
| Bamtools | (Barnett et al., 2011) | https://github.com/pezmaster31/bamtools |
| Samtools | (Li and Durbin, 2009) | http://samtools.sourceforge.net/ |
| GSNAP | (Wu and Nacu, 2010) | http://research-pub.gene.com/gmap/ |
| GimmeMotifs | (van Heeringen and Veenstra, 2011) | https://github.com/simonvh/gimmemotifs |
| Caret | (Kuhn, 2008) | http://cran.r-project.org/web/packages/caret/index.html |
| HOMER | (Heinz et al., 2010) | http://homer.salk.edu/homer/motif/ |
| DAVID | (Huang da et al., 2009) | https://david.ncifcrf.gov/ |
| bwa | (Li and Durbin, 2009) | http://bio-bwa.sourceforge.net/ |
| bowtie | (Langmead et al., 2009) | http://bowtie-bio.sourceforge.net/index.shtml |
| MACS2 | (Zhang et al., 2008) | https://github.com/taoliu/MACS |
Contact for Reagents and Resource Sharing
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Hendrik G. Stunnenberg (h.stunnenberg@ncmls.ru.nl).
Experimental Model and Subject Details
Monocytes from Healthy Donors
All primary cells were isolated from healthy volunteers who gave written informed consent (Sanquin Blood bank, Nijmegen, the Netherlands). Volunteers are of Northern European descent. Peripheral blood mononuclear cells were isolated by centrifugation in Ficoll-Paque (GE Healthcare), followed by removal of T cells using an additional Percoll gradient. Monocytes were purified from PBMCs using negative selection in an LD column magnet separator, with beads for CD3+ (T cells), CD19+ (B cells) and CD56+ (NK cells) positive cells (Miltenyi Biotech), yielding > 95% pure monocytes. Successful isolation of monocytes was confirmed with FACS, as previously described (Saeed et al., 2014).
In Vitro Monocyte-to-Macrophage Differentiation and Induction of Innate Immune Memory
Monocytes were differentiated into resting macrophages by ex vivo culture in RPMI 1640 medium (Sigma Aldrich) with 10% Human Serum. Media was supplemented with 10 μg/mL gentamycin, 10 mM L-glutamine and 10 mM pyruvate (Life Technologies). Tolerization was induced by treatment of monocytes with 10-100ng/mL LPS for 24 hr, followed by washout and five days culture in RPMI + 10% human serum, while trained innate immunity was induced by treatment with 5 μg/mL BG for 24 hr, followed by washout and 5 days in culture. Establishment of tolerance or training in the resulting macrophages at day 6 was determined by TNF and IL6 release at 24 hr following LPS stimulation using ELISA. For ChIP-seq, 10x106 monocytes were seeded in 10cm dishes, for RNaseq and ATAC-seq 1.5 x 106 monocytes were seeded in 6 well plates. IBET151 (GSK) was diluted to 50 mM stock using DMSO. Following dosage titration 5 μM was determined as the appropriate final concentrations to prevent tolerization, without causing cell death. IBET-151 was added to monocytes at the same time as LPS for 24 hr, followed by washout and five days culture in RPMI + 10% human serum to macrophage differentiation.
Experimental Human Endotoxemia Model
In vivo endotoxin tolerance was examined in 12 healthy nonsmoking volunteers who participated in an experimental human endotoxemia study. The study is registered at Clinicaltrials.gov (NCT02602977) and study protocols were approved by the local ethics committee of the Radboud University Nijmegen Medical Centre (NL53584.091.15/CMO 2015-1796). Written informed consent was obtained from all study participants. Subjects were screened before the start of the experiment and had a normal physical examination, electrocardiography, and routine laboratory values. Throughout the study period, subjects were not allowed to take any drugs, including acetaminophen, and were asked to refrain from alcohol and caffeine 24 hr and from food 12 hr before the start of the endotoxemia experiment. All study procedures were conducted in accordance with the declaration of Helsinki including current revisions and Good Clinical Practice guidelines. Experimental human endotoxemia was conducted as described previously (Kox et al., 2014). Briefly, all subjects received an intravenous bolus injection of LPS (lipopolysaccharide derived from Escherichia coli O:113, Clinical Center Reference Endotoxin, National Institutes of Health (NIH), Bethesda, MD) at a dose of 2 ng/kg. Blood was obtained before LPS administration and 4 hr afterward, and monocytes were isolated. Monocytes were exposed to culture or BG ex vivo, and cytokine production in the supernatants was measured following ex vivo LPS (10ng/ml) exposure. Cytokine production was determined by ELISA following the protocol of the manufactures (IL-6, sanquin and TNFα, R&D systems).
Method Details
Cytokine Assays
TNFα and IL-6 were measured using ELISA according to the manufacturer protocol (IL6: Sanquin; and TNFα: R&D). For cytokines production assays the differences between groups were analyzed using the Wilcoxon signed-rank test. The level of significance was defined as a p value < 0.05.
RNA Extraction and cDNA Synthesis
Total RNA was extracted from cells using the QIAGEN RNeasy RNA extraction kit (QIAGEN, Netherlands), using on-column DNaseI treatment. Ribosomal RNA was removed using the riboZero rRNA removal kit (Illumina). RNA was then fragmented into 200bp fragments by incubation for 7.5 min at 95°C in fragmentation buffer (200 mM Tris-acetate, 500 mM Potassium Acetate, 150 mM Magnesium Acetate [pH 8.2]). First strand cDNA synthesis was performed using SuperScript III (Life Technologies), followed by synthesis of the second cDNA strand. Library preparation was performed using the KAPA hyperprep kit (KAPA Biosystems). Quality of cDNA and the efficiency of ribosomal RNA removal was confirmed using quantitative RT-PCR using the IQ Sybr Supermix, with primers for GAPDH, 18S and 28S rRNA.
Chromatin Immunoprecipitation
Purified cells were fixed with 1% formaldehyde (Sigma) at a concentration of approximately 10 million cells/ml. Fixed cell preparations were sonicated using a Diagenode Bioruptor UCD-300 for 3x 10 min (30 s on; 30 s off). 67 μl of chromatin (1 million cells) was incubated with 229 μl dilution buffer, 3 μl protease inhibitor cocktail and 0.5-1 μg of H3K27ac, H3K4me3, H3K4me1, H3K27me3, H3K9me3 or H3K36me3 antibodies (Diagenode) and incubated overnight at 4°C with rotation. Protein A/G magnetic beads were washed in dilution buffer with 0.15% SDS and 0.1% BSA, added to the chromatin/antibody mix and rotated for 60 min at 4°C. Beads were washed with 400μl buffer for 5 min at 4°C with five rounds of washes. After washing chromatin was eluted using elution buffer for 20 min. Supernatant was collected, 8 μl 5M NaCl, 3μl proteinase K were added and samples were incubated for 4 hr at 65°C.Finally samples were purified using QIAGEN; Qiaquick MinElute PCR purification Kit and eluted in 20 μl EB. Detailed protocols can be found on the Blueprint website (http://www.blueprint-epigenome.eu/UserFiles/file/Protocols/Histone_ChIP_May2013.pdf).
Library Preparation for Sequencing
Illumina library preparation was done using the Kapa Hyper Prep Kit. For end repair and A-tailing double stranded DNA was incubated with end repair and A-tailing buffer and enzyme and incubated first for 30 min at 20°C and then for 30 min at 65°C.Subsequently adapters were ligated by adding 30μl ligation buffer, 10 Kapa l DNA ligase, 5 μl diluted adaptor in a total volume of 110μl and incubated for 15 min at 15°C. Post-ligation cleanup was performed using Agencourt AMPure XP reagent and products were eluted in 20 μl elution buffer. Libraries were amplified by adding 25 μl 2x KAPA HiFi Hotstart ReadyMix and 5μl 10x Library Amplification Primer Mix and PCR, 10 cycles. Samples were purified using the QIAquick MinElute PCR purification kit and 300bp fragments selected using E-gel. Correct size selection was confirmed by BioAnalyzer analysis. Sequencing was performed using Illumina HiSeq 2000 machines and generated 43bp single end reads. Samples for RNA-seq were treated to the above protocol exactly, except for a single additional step: After post-ligation cleanup, and before library amplification, samples were incubated with 3 uL USER enzyme for 15 min at 37°C to digest the 2nd cDNA strand.
Assay for Transposase Accessible Chromatin
Monocytes or macrophages (100,000 cells) were scrapped in a well of a 6-well plate with cold PBS and then spun down at 800 x g for 5 min at 4°C. Cells were washed with 50 μl of cold 1x PBS buffer, incubated in 50 μl of cold lysis buffer (10 mM Tris-HCL (pH 7.4), 10 mM NaCl, 3 mM MgCl2 0,1% IGEPAL) and spun down at 800 x g for 10 min at4°C.The nuclei were immediately resuspended in the transposition reaction mix (22.5 μl TD buffer, 2.5 μl Tn5 Transposase, 25 μl NF H2O) and incubated for 30 min at 37°C. Following transposition, 100 μl AMPure beads were added to the reaction (sample-to-bead ratio of 1:2), mixed thoroughly by pipetting, and incubated for 15 min at RT. Samples and beads were washed on the magnetic rack with 80% ethanol, dried for 5 min, and resuspended in 15 μl EB buffer. DNA was amplified with 10 - 15 PCR cycles using the mix (15 μl transposed DNA, 0.3 μl 100x SYBR Green I, 25 μl NEBNext High-Fidelity master mix, 2.5 μl Nextera Primer index N7.. (25 μM), 2.5 μl Nextera Primer index S5.. (25 μM), 4.7 μl NF H2O). In order to reduce GC and size bias in PCR, the PCR reaction is monitored using qPCR to stop amplification prior to saturation. Following amplification, samples were incubated purified twice using SPRI beads, first using negative selection with a sample-to-bead ratio of 1-0.65 and then positive selection with a sample-to-bead ratio of 1-1.8. After 80% Ethanol wash and drying, the sample was eluted in 20 μl EB buffer, and quality checked before sequencing. Detailed protocol can be found on the Blueprint website (http://www.blueprint-epigenome.eu/UserFiles/file/Protocols/ATAC_Seq_Protocol.pdf).
Whole Genome Bisulfite Sequencing
Genomic DNA (1-2 μg) was spiked with unmethylated λ DNA (5ng of λ DNA per μg of genomic DNA) (Promega). The DNA was sheared by sonication to 50-500bp using a Covaris E220 and fragments of size 150-300 bp were selected using AMPure XP beads (Agencourt Bioscience). Genomic DNA libraries were constructed using the Illumina TruSeq Sample Preparation kit (Illumina) following the lllumina standard protocol: end repair was performed on the DNA fragments, an adenine was added to the 3′ extremities of the fragments and Illumina TruSeq adapters were ligated at each extremity. Adter adaptor ligation, the DNA was treated with sodium bisulfite using the EpiTexy Bisulfite kit (QIAGEN) following the manufacturer's instructions for formalin-fixed and paraffin-embedded (FFPE) tissue samples. Two rounds of bisulfite conversion were performed to assure a high conversion rate. An enrichment for adaptor-ligated DNA was carried out through 7 PCR cycles using the PfuTurboCx Hotstart DNA polymerase (Stratagene). Library quality was monitored using the Agilent 2100 BioAnalyzer (Agilent), and the concentration of viable sequencing fragments (molecules carrying adaptors at both extremities) estimated using quantitative PCR with the library quantification kit from KAPA Biosystem. Paired-end DNA sequencing (2x100 nucleotides) was then performed using the Illumina Hi-Seq 2000. WGBS data are available upon request from the BLUEPRINT consortium.
RNA-Seq Data Analysis
For quality control and visualization, RNA-seq reads were aligned to the hg19 reference genome using GSNAP (Wu and Nacu, 2010) with non-default parameters -m 1 -N 1 -n 1 -Q -s Ensembl_splice_68. Each RNA-seq sample was subjected to a quality control step, where, based on read distribution over the annotated genome, libraries that are outliers were identified and discarded from further analysis. To infer gene expression levels, RNA-seq reads were aligned to the Ensembl v68 human transcriptome using Bowtie. Quantification of gene expression was performed using MMSEQ. Differential expression was determined using MMDIFF. A two model comparison was used to identify differentially expressed genes that confer cellular identity Mo/Mf. The null-model is that the mean expression levels are the same in both cell types, and the alternative model is that the mean expression levels are allowed to differ between the two cell types. Genes with a larger posterior probability for the second model, an RPKM value greater than 2 in any of Mo or Mf and minimally a 2-fold expression change were considered as differentially expressed. Expression changes related to differentiation of each treatment were studied using a 52-model comparison, a.k.a. polytomous comparison, under the null-model that assumes the mean expression levels are the same across each time-point. Expression differences related to the treatments at each time-point were studied using a 5-model comparison, under the null-model that assumes the mean expression levels are the same across each treatment.
ChIP-Seq Data Analysis
Sequencing reads were aligned to human genome assembly hg19 (NCBI version 37) using bwa. Duplicate reads were removed after the alignment with the Picard tools. For peak calling the BAM files were first filtered to remove the reads with mapping quality less than 15, followed by fragment size modeling (https://code.google.com/archive/p/phantompeakqualtools/). MACS2 (https://github.com/taoliu/MACS/) was used to call the peaks. H3K4me1,H3K9me3 and H3K27me3 peaks were called using the broad setting of MACS2 while H3K27ac and H3K4me3 were called using the default (narrow) setting. For each histone mark dataset, the data were normalized using the R package DESeq2 and then pair-wise comparisons were performed (fold change 3, adjusted p-adjvalue < 0.05 and RPKM ≥ 2 in at least in any condition) to determine the differentially expressed genes per condition. The results from all possible pairwise comparisons (within each condition and similar time points across all conditions per mark) were pooled and merged to define the dynamic set of enriched regions. Promoters were defined as regions between ± 2kb from TSSs for each ensemble gene and enhancers were determined as enriched H3K27ac/H3K4me1 regions more than ± 2kb away from the TSS. To find different patterns over dynamic promoters or enhancers, we applied a K-means clustering procedure (with optimal number of clusters per each dataset) to the dynamic datasets as described above.
ATAC-Seq Data Analysis
The full ATAC-seq protocol is available at the BLUEPRINT website (http://www.blueprint-epigenome.eu/UserFiles/file/Protocols/ATAC_Seq_Protocol.pdf). ATAC-seq reads were mapped to the hg19 reference genome using BWA (Li and Durbin, 2009) with default parameters. Non-uniquely mapped reads and PCR duplicates were removed. MACS2 (Zhang et al., 2008) was used to identify regions of open chromatin (peaks) with parameters “-nomodel -p 1e-9.” Overlap peaks from different samples were merged.
DNA-Binding Motif Scanning
All the DNA-binding motifs used in this study are based on the cis-bp database described in (Weirauch et al., 2014). Only motifs with direct evidence of binding in the species of vertebrate were selected. Within each motif family, as annotated by cis-bp, all motifs were clustered using 'gimme cluster' from the GimmeMotifs package (van Heeringen and Veenstra, 2011) with a threshold of 0.9999. The annotation of motifs is based on the annotation of human in the cis-bp database. Motifs were used for scanning if the assigned TF is expressed (> 1 RPKM) in at least one time-point during the differentiation. Total ATAC-seq peaks were scanned for the presence of motifs. We used Gimme motifs for scanning with dynamic motif scoring cut-offs targeting a false discovery rate (FDR) of both 0.01 and 0.05. To look at the motif enrichments in each set of regions (epigenetic cluster or gene cluster), ATAC-seq peaks were assigned to the epigenomic cluster or the gene promoters by intersection. Motif occurrences were acquired by intersection of the assigned ATAC-seq peaks with the motif scanning results on total ATAC-seq peaks. Total ATAC-seq peaks were divided into promoter set and non-promoter set as the background for the calculation of motif enrichment. Enrichment of motifs in each set of regions was defined by applying a hypergeometric test using the motif frequency in the corresponding background. This results in TFs that putatively regulate the activities of the regulatory regions. Motifs in each heat map satisfy an arbitrary cutoff of > 5% motif presence and a fixed minimal presence difference from background in at least one cluster. Hierarchical clustering (Pearson correlation) was performed in each heat map using the motif occurrence frequencies in the clusters. Based on the gene activity and dynamics, only one TF was selected to represent a motif if multiple genes are assigned to the same motif. Scanning results from FDR of 0.01 and 0.05 were compared and do not affect the result of enrichment analysis.
Gene Ontology Analysis
Gene ontology analysis on dynamic lists of genes was performed using DAVID (Huang da et al., 2009). Gene ontology on dynamic enhancer clusters was performed using GREAT (McLean et al., 2010). KEGG pathways and Biological Processes were ranked by p value and the top terms were plotted.
Quantification and Statistical Analysis
Statistical parameters including the exact value of n, the definition of center, dispersion, and precision measures (mean ± SEM) and statistical significance are reported in the Figures and the Figure Legends. Data are judged to be statistically significant when p < 0.05 by two-tailed Student's T-Test or 2-way ANOVA, where appropriate.
Data and Software Availability
Data Resources
Raw data files for the RNA, ATAC, and ChIP sequencing and analysis have been deposited in the NCBI Gene Expression Omnibus under accession number: GSE85246.
Links to GEO SubSeries linked to GSE85246:
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE85243
Supplemental Information
Acknowledgments
The research leading to the results described in this paper has received funding from the European Union’s Seventh Framework Programme (FP7/2007-2013) under grant agreement 282510-BLUEPRINT. B.N. is supported by an NHMRC (Australia) CJ Martin Early Career Fellowship. M.G.N. was supported by an ERC Consolidator Grant (310372). The authors would like to thank GSK Epinova and Cellzome for providing the IBET reagent and Prof. David L. Williams (University of Tennessee) for β1,3(D)glucan (β-glucan).
Footnotes
Author Contributions
Conceptualization, H.G.S., C.L., J.H.A.M., and M.G.N.; Methodology, B.N., R.D., B.K., R.J.W.A., T.K., F.M., C.W., E.M.J.-M., J.Z., M.K., P.P., and N.S.; Investigation, B.N., E.H., S.-Y.W., and W.M.; Writing – Original Draft, B.N., H.G.S., and C.L.; Writing – Review & Editing, E.H., S.-Y.W., M.G.N., J.H.A.M., R.J.W.A., and T.K.; Funding Acquisition, H.G.S.; Resources, P.F., L.C., S.J.v.H., P.P., and I.G.; Supervision, H.G.S., C.L., J.H.A.M., and M.G.N.
References
- Amit I, Winter DR, Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol. 2016;17:18–25. doi: 10.1038/ni.3325. [DOI] [PubMed] [Google Scholar]
- Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369:840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- Bahador M, Cross AS. From therapy to experimental model: a hundred years of endotoxin administration to human subjects. J Endotoxin Res. 2007;13:251–279. doi: 10.1177/0968051907085986. [DOI] [PubMed] [Google Scholar]
- Barnett DW, Garrison EK, Quinlan AR, Strömberg MP, Marth GT. BamTools: a C++ API and toolkit for analyzing and managing BAM files. Bioinformatics. 2011;27:1691–1692. doi: 10.1093/bioinformatics/btr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessede A, Gargaro M, Pallotta MT, Matino D, Servillo G, Brunacci C, Bicciato S, Mazza EM, Macchiarulo A, Vacca C, et al. Aryl hydrocarbon receptor control of a disease tolerance defence pathway. Nature. 2014;511:184–190. doi: 10.1038/nature13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, Giamarellos-Bourboulis EJ, Martens JH, Rao NA, Aghajanirefah A, et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345 doi: 10.1126/science.1250684. 1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SC, Scicluna BP, Arts RJ, Gresnigt MS, Lachmandas E, Giamarellos-Bourboulis EJ, Kox M, Manjeri GR, Wagenaars JA, Cremer OL, et al. Broad defects in the energy metabolism of leukocytes underlie immunoparalysis in sepsis. Nat Immunol. 2016;17:406–413. doi: 10.1038/ni.3398. [DOI] [PubMed] [Google Scholar]
- de la Rica L, Rodríguez-Ubreva J, García M, Islam AB, Urquiza JM, Hernando H, Christensen J, Helin K, Gómez-Vaquero C, Ballestar E. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 2013;14:R99. doi: 10.1186/gb-2013-14-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draisma A, Pickkers P, Bouw MP, van der Hoeven JG. Development of endotoxin tolerance in humans in vivo. Crit Care Med. 2009;37:1261–1267. doi: 10.1097/CCM.0b013e31819c3c67. [DOI] [PubMed] [Google Scholar]
- Foster SL, Hargreaves DC, Medzhitov R. Gene-specific control of inflammation by TLR-induced chromatin modifications. Nature. 2007;447:972–978. doi: 10.1038/nature05836. [DOI] [PubMed] [Google Scholar]
- Ghesquière B, Wong BW, Kuchnio A, Carmeliet P. Metabolism of stromal and immune cells in health and disease. Nature. 2014;511:167–176. doi: 10.1038/nature13312. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Barozzi I, Mietton F, Polletti S, De Santa F, Venturini E, Gregory L, Lonie L, Chew A, Wei CL, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- Gilroy DW, Yona S. HIF1α allows monocytes to take a breather during sepsis. Immunity. 2015;42:397–399. doi: 10.1016/j.immuni.2015.02.016. [DOI] [PubMed] [Google Scholar]
- Glass CK, Natoli G. Molecular control of activation and priming in macrophages. Nat Immunol. 2016;17:26–33. doi: 10.1038/ni.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge HS, Simmons RM, Underhill DM. Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J Immunol. 2007;178:3107–3115. doi: 10.4049/jimmunol.178.5.3107. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Ifrim DC, Saeed S, Jacobs C, van Loenhout J, de Jong D, Stunnenberg HG, et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci USA. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kox M, van Eijk LT, Zwaag J, van den Wildenberg J, Sweep FC, van der Hoeven JG, Pickkers P. Voluntary activation of the sympathetic nervous system and attenuation of the innate immune response in humans. Proc Natl Acad Sci USA. 2014;111:7379–7384. doi: 10.1073/pnas.1322174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn M. Building predictive models in R using the caret package. J Stat Softw. 2008;28:1–26. [Google Scholar]
- Kulis M, Merkel A, Heath S, Queirós AC, Schuyler RP, Castellano G, Beekman R, Raineri E, Esteve A, Clot G, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47:746–756. doi: 10.1038/ng.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurotaki D, Osato N, Nishiyama A, Yamamoto M, Ban T, Sato H, Nakabayashi J, Umehara M, Miyake N, Matsumoto N, et al. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood. 2013;121:1839–1849. doi: 10.1182/blood-2012-06-437863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammana A, Chung HR. Chromatin segmentation based on a probabilistic model for read counts explains a large portion of the epigenome. Genome Biol. 2015;16:151. doi: 10.1186/s13059-015-0708-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Joosten LA, Latz E, Mills KH, Natoli G, Stunnenberg HG, O’Neill LA, Xavier RJ. Trained immunity: A program of innate immune memory in health and disease. Science. 2016;352 doi: 10.1126/science.aaf1098. aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodeme E, Jeffrey KL, Schaefer U, Beinke S, Dewell S, Chung CW, Chandwani R, Marazzi I, Wilson P, Coste H, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010;468:1119–1123. doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K, Iwamoto Y, Kobayashi Y, Katsuoka F, Kawaguchi S, Tsujita T, Nakamura T, Kato S, Yamamoto M, Takayanagi H, Ishii M. DNA methyltransferase 3a regulates osteoclast differentiation by coupling to an S-adenosylmethionine-producing metabolic pathway. Nat Med. 2015;21:281–287. doi: 10.1038/nm.3774. [DOI] [PubMed] [Google Scholar]
- Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin J, Saeed S, Martens JH, Giamarellos-Bourboulis EJ, Ifrim DC, Logie C, Jacobs L, Jansen T, Kullberg BJ, Wijmenga C, et al. Candida albicans infection affords protection against reinfection via functional reprogramming of monocytes. Cell Host Microbe. 2012;12:223–232. doi: 10.1016/j.chom.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin J, Cheng SC, van der Meer JW, Netea MG. Innate immune memory: towards a better understanding of host defense mechanisms. Curr Opin Immunol. 2014;29:1–7. doi: 10.1016/j.coi.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Rialdi A, Campisi L, Zhao N, Lagda AC, Pietzsch C, Ho JS, Martinez-Gil L, Fenouil R, Chen X, Edwards M, et al. Topoisomerase 1 inhibition suppresses inflammatory genes and protects from death by inflammation. Science. 2016;352 doi: 10.1126/science.aad7993. aad7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, Cheng SC, Ratter J, Berentsen K, van der Ent MA, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345 doi: 10.1126/science.1251086. 1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SepsisReport. Focus on sepsis. Nat Med. 2012;18:997. doi: 10.1038/nm0712-997. [DOI] [PubMed] [Google Scholar]
- Shalova IN, Lim JY, Chittezhath M, Zinkernagel AS, Beasley F, Hernández-Jiménez E, Toledano V, Cubillos-Zapata C, Rapisarda A, Chen J, et al. Human monocytes undergo functional re-programming during sepsis mediated by hypoxia-inducible factor-1α. Immunity. 2015;42:484–498. doi: 10.1016/j.immuni.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, Reichart D, Fox JN, Shaked I, Heudobler D, et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heeringen SJ, Veenstra GJ. GimmeMotifs: a de novo motif prediction pipeline for ChIP-sequencing experiments. Bioinformatics. 2011;27:270–271. doi: 10.1093/bioinformatics/btq636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vento-Tormo R, Company C, Rodríguez-Ubreva J, de la Rica L, Urquiza JM, Javierre BM, Sabarinathan R, Luque A, Esteller M, Aran JM, et al. IL-4 orchestrates STAT6-mediated DNA demethylation leading to dendritic cell differentiation. Genome Biol. 2016;17:4. doi: 10.1186/s13059-015-0863-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS. Modelbased analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.