Abstract
Rotavirus A species (RVA) is the leading cause of severe diarrhea among children in both developed and developing countries. Among different RVA G types, humans are most commonly infected with G1, G2, G3, G4 and G9. During 2003–2004, G3 rotavirus termed as “new variant G3” emerged in Japan that later disseminated to multiple countries across the world. Although G3 rotaviruses are now commonly detected globally, they have been rarely reported from Pakistan. We investigated the genetic diversity of G3 strains responsible RVA gastroenteritis in children hospitalized in Rawalpindi, Pakistan during 2014. G3P[8] (18.3%; n = 24) was detected as the most common genotype causing majority of infections in children less than 06 months. Phylogenetic analysis of Pakistani G3 strains showed high amino acid similarity to “new variant G3” and G3 strains reported from China, Russia, USA, Japan, Belgium and Hungary during 2007–2012. Pakistani G3 strains belonged to lineage 3 within sub-lineage 3d, containing an extra N-linked glycosylation site compared to the G3 strain of RotaTeqTM. To our knowledge, this is the first report on the molecular epidemiology of G3 rotavirus strains from Pakistan and calls for immediate response measures to introduce RV vaccine in the routine immunization program of the country on priority.
Introduction
Rotavirus is the most common etiologic agent of severe diarrhea in infants and children worldwide causing approximately 0.2 million deaths annually [1]. Being a member of Reoviridae family, it has a unique genome comprising 11 segments of double stranded RNA which encode six structural (VP1-VP4, VP6, VP7) and six non-structural proteins (NSP1-NSP6) [2]. Among the nine recognized and one proposed rotavirus species (RVA—RVI and RVJ respectively) [3,4] species A, B, C and H are known to infect humans [5] while RVA cause majority of infections [6]. RVA is classified into G and P genotypes based on their outer capsid proteins VP7 and VP4 respectively and 35 G-types and 50 P-types have been identified so far [3,7,8]. Epidemiological studies demonstrated that humans are mostly infected with G1, G2, G3, G4 and G9 in combination with P[4], P[6] and P[8] [9,10] whereas G12 has recently emerged in most parts of the world including Pakistan [11,12].
G3 rotavirus is one of the most common RVA strain reported worldwide [9]. G3 was detected at low rates during 1990’s however during the last decade, it has re-emerged in different countries of the world [13–23]. Moreover, a variant of G3 referred to as "new variant G3" was reported from Japan during 2003–2004 [18]. Later, these variants were also detected from different parts of the world [19,20,23–30]. To date, limited data is available on circulation of RVA genotypes from Pakistan indicating G1, G2 and G9 among the most common genotypes infecting Pakistani children [12,31–34] whereas G3 strains are rarely detected [31]. Therefore, the objective of this study was to investigate the genetic diversity of G3 rotaviruses and to compare them with the two licensed rotavirus vaccines.
Materials and methods
Stool specimen collection
Stool samples were collected from children hospitalized with acute dehydrating gastroenteritis at Benazir Bhutto Hospital, Rawalpindi (BBH) as per WHO standard case definition [35] during January to December 2014. Benazir Bhutto Hospital, previously known as Rawalpindi General Hospital is a tertiary care teaching hospital in district Rawalpindi with its pediatric department providing health facilities to population of Rawalpindi (the fourth largest city by population in the country) and its adjacent regions. The inclusion criteria of enrolled cases followed was: any child under five years of age admitted to the hospital for the treatment of acute gastroenteritis as primary illness and onset of symptoms in ≤ 7 days. The cases aged above 5 years and those presented with bloody diarrhea and non-infectious gastric disorders were excluded from the study. Informed written consent was obtained from patient’s parent/guardian before sample collection. Demographic and clinical data including age, gender, area of residence, hospital admission date, date of stool sample collection, vomiting (duration and episodes per 24 hours), diarrhea (duration and episodes per 24 hours), body temperature and patient’s recovery status at the time of discharge from hospital was recorded on a standard questionnaire. Stool samples and the relevant clinical records of patients were transported to Department of Virology, National Institute of Health (NIH), Islamabad for laboratory testing. The study design was approved by Internal review Board of the National Institute of Health, Islamabad.
Enzyme immunoassay
The presence of Group A rotavirus in stool was confirmed by use of a commercial immunoassay (ProSpecT, Oxoid Ltd.) following manufacturer instructions.
Viral RNA extraction and G & P genotyping
RNA was extracted from 10% fecal suspension using QIAamp Viral RNA Mini Kit (Qiagen Hilden, Germany) according to manufacturer’s instructions and stored at -20°C until further analysis. Reverse-transcription polymerase chain reaction (RT-PCR) assay and G and P typing was carried out as described previously [36,37].
Sequence analysis of VP7 and VP4 genes
Partial sequencing of VP7 and VP4 genes was directly done with primers Beg9 & End9 [36] and Con2 & Con3 [37] respectively using BigDye Terminator Cycle Sequencing kit v3.0 (Applied Biosystems) on an automated sequencer ABI 3100 (Applied Biosystems). Analysis of nucleotide sequences was done by using Sequencher version 4.1 (Gene Codes Corp., USA). Closely matched nucleotide sequences of VP7 and VP4 genes were retrieved from GenBank using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic tree was constructed using neighbor joining method (with 1000 bootstrap replicates) and distances were calculated with Kimura 2-parameter method using MEGA 5 [38]. Lineage designation for phylogenetic trees of G3 and P[8] were based on criteria reported earlier [19,39]. Structural analysis of VP7 (PBD 3FMG) and VP8* (PDB 1KQR) were performed using the UCSF Chimera-Molecular Modeling System [40]. Nucleotide sequences of rotavirus VP4 and VP7 gene segments sequenced in this study are submitted to the GenBank under the accession numbers KX681820-KX681833.
Results
Clinical features of RVA G3 patients
From a total of 502 patients, G3P[8] (18.3%; n = 24/131) was detected as the most common genotype. In the current study the highest rate of infection due to rotavirus G3 was found in children aged 1–6 months (56.6%, 17 out of 30) followed by those between 7–12 months of age (40%, 12 out of 30). Infection with G3 rotavirus was observed in a single child of 24 months age. Frequency of G3 infection in male was higher (63.3%) than female (36.7%) patients. Infections with G3 were found throughout the year and the most common clinical signs and symptoms of children infected with G3 included dehydration (100%), vomiting (mean duration = 2.3±4.1 days, range = 1–6 days and mean episodes per 24 hours = 3.6±6.21, range = 1–10 days) and diarrhea (mean duration = 2.3±3.1 days, range = 1–6 days and mean episodes per 24 hours = 12.5±15.3, range = 10–16 days).
Phylogenetic analysis
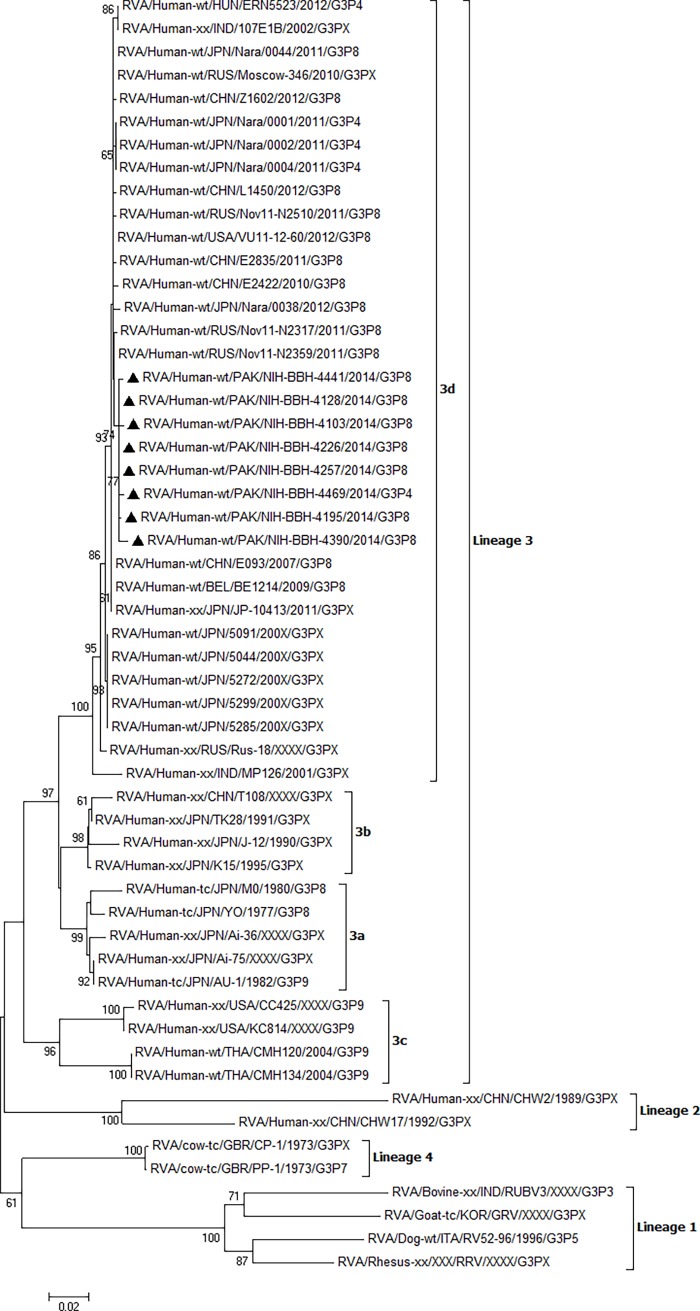
In order to explore the genetic diversity of G3, eight strains were randomly selected and their VP7 gene was partially sequenced (942bp). Phylogenetic analysis showed that all Pakistani G3 strains shared high nucleotide and amino acid similarity (99–100%) with each other and 98.9–99.8% similarity was observed between Pakistani and recent G3 strains reported from China, Russia, USA, Japan, Belgium and Hungary within lineage 3 and sub-lineage 3d (Fig 1). These strains showed 98.7–99.1% similarity at nucleotide and 99–99.7% at amino acid level respectively with new variant G3 isolated in Japan during 2003–2004 [18]. Deduced amino acid sequence of VP7 gene between Pakistani and new variant G3 was highly conserved except for position 221 with Adenine substituted to Aspartic acid (A to D).
Fig 1. Phylogenetic analysis of VP7 gene segment of G3 rotavirus strains.
Phylogenetic tree was reconstructed using neighbor-joining method. Bootstrap values were calculated using 1000 replicates. Bootstrap values less than 60 are not shown. Filled triangles represent G3 strains detected in this study.
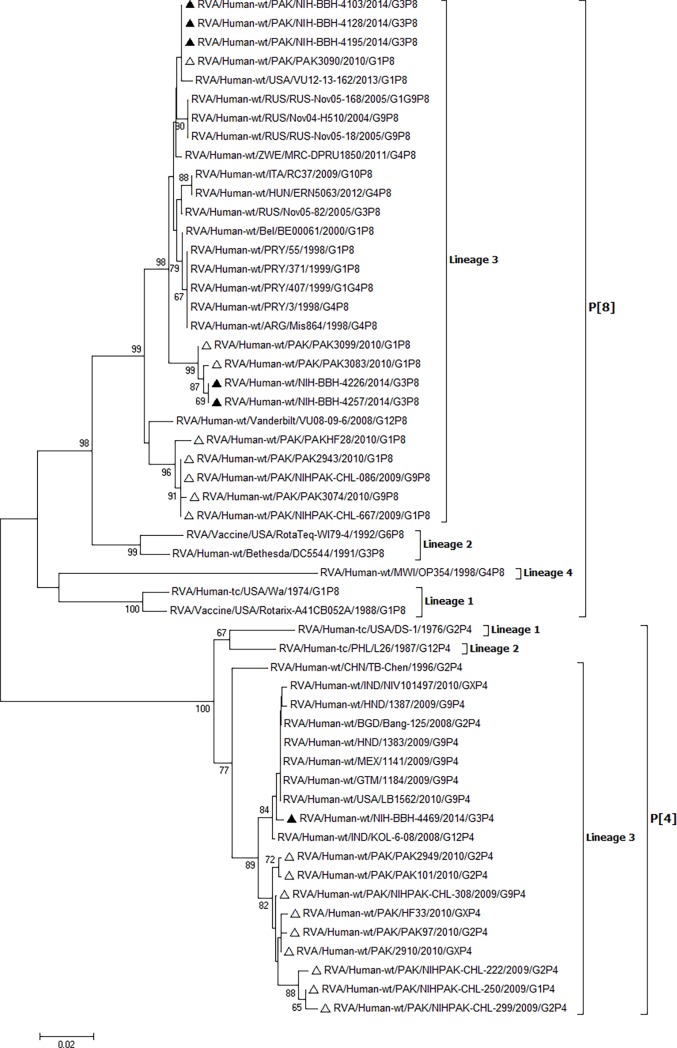
Partial sequencing of VP4 gene of five G3P[8] and one G3P[4] strain was also carried out. All five Pakistani G3P[8] strains shared high identities (98.2–100% at nucleotide and 98.9–100% at amino acid level) with each other and were closely related to P[8] strains isolated from USA, Russia, Belgium, Italy, Argentina, Hungary, Zimbabwe and Paraguay (Fig 2). The Pakistani G3P[4] clustered in lineage 3 with P[4] strains from USA, Mexico, Honduras, Guatemala, Bangladesh and India (Fig 2).
Fig 2. Phylogenetic analysis of rotavirus VP4 gene segment.
Phylogenetic tree was reconstructed using neighbor-joining method. Bootstrap values were calculated using 1000 replicates. Bootstrap values less than 60 are not shown. Filled triangle represent P[8] and P[4] strains detected in this study. Empty triangle represent P[8] and P[4] strains reported previously from Pakistan.
Comparison of Pakistani G3 strains with RVA vaccine strains
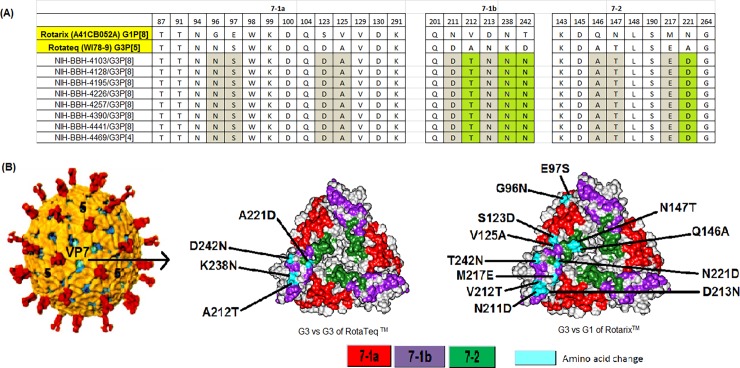
In the present study, we compared the VP7 and VP4 (VP8*) antigenic epitopes of Pakistani G3 with those of RVA vaccine strains. Of the 29 amino acid residues comprising VP7 antigenic epitopes, four differences were found between RotaTeqTM (WI78-9) and Pakistani G3 strains, three of which occurred in 7-1b region (A212T, K238N, D242N) and one difference was found in the antigenic epitope 7–2 (A221D) (Fig 3). Interestingly, all Pakistani G3 strains showed a K238N substitution, which created a potential N-linked glycosylation site in contrast to G3 strain of RotaTeqTM. When compared with RotarixTM, twelve amino acid changes were observed within the VP7 antigenic epitopes that were equally distributed among 7-1a, 7-1b and 7–2 (Fig 3).
Fig 3. Comparison of VP7 antigenic epitope sites between Pakistani G3 strains and rotavirus vaccine RotarixTM and RotaTeqTM.
(A) Antigenic residues are divided into three antigenic epitopes 7-1a, 7-1b and 7–2. Amino acid highlighted in green are those that differ from G3 strain of RotaTeqTM while those in gray are different from RotarixTM. (B) Surface representation of VP7 trimer (PDB 3FMG). Antigenic epitopes are colored in red 7-1a, purple 7-1b and green 7–2. Surface exposed residues that differ between Pakistani G3 and vaccine strains of RotarixTM and RotaTeqTM are shown in cyan.
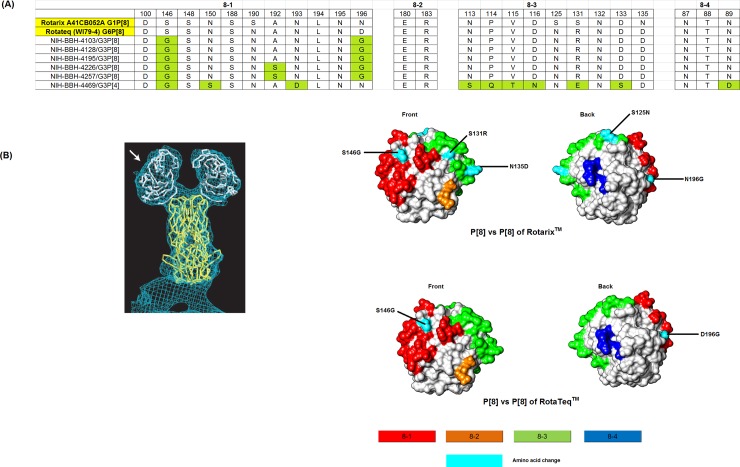
VP8* antigenic epitopes (8–1, 8–2, 8–3, 8–4) of RotarixTM and RotaTeqTM were compared with those observed in P[8] and P[4] strains detected in this study. Out of total 25 amino acids in the VP8*, ten were found conserved among vaccine and Pakistani strains (Fig 4A). Pakistani G3P[8] strains showed 6 amino acid differences when compared with RotarixTM in 8–1 and 8–3 epitopes, while two variations were found when compared with RotaTeqTM (Fig 4). Thirteen amino acid differences were observed between Pakistani G3P[4] (strain NIH-BBH-4469) and RotarixTM while 11 variations were observed when compared to RotaTeqTM (Fig 4A).
Fig 4. Alignment of antigenic residues in VP4 between the strains contained in RotarixTM and RotaTeqTM and Pakistani P[8] and P[4].
(A) Antigenic residues are divided in three antigenic epitopes in VP8* (8–1, 8–2, 8–3 and 8–4). Amino acids in green are different from both RotarixTM and G3 strain of RotaTeqTM. (B) Surface representation of the VP8* core (PDB 1KQR). Antigenic epitopes are colored red 8–1, orange 8–2, green 8–3 and blue 8–4. Surface exposed residues that differ between Pakistani G3P[8] and vaccine RotarixTM and RotaTeqTM are shown in cyan.
Discussion
Studies on clinical epidemiology of G3 rotavirus strains reported globally are very scarce. In a 2009 study conducted by Day et al [41] in Bangladesh, 2.6% of the total 917 enrolled patients were detected positive for G3 rotavirus. In their report, the majority (78%) of G3 positive cases was below 12 months of age and all patients showed dehydration. Vomiting was reported in 55% patients with 5–8 diarrheal episodes in a day. In our study, 100% of the patients were presented with vomiting, dehydration and watery diarrhea. The mean diarrheal episodes in our patients were remarkably high with mean number of 12.5±15.3 episodes per 24 hours. In another study conducted in Japan, Phan et al [18] tested 402 fecal specimen from patients with acute gastroenteritis reported during 2003–2004, out of which 21% (n = 83/402) were found positive for rotavirus. Genotyping results indicated that 81 out of 83 positive patients were infected with G3 rotavirus with the highest positivity rate among children above 12 months of age. This is in contrast to our findings where we found 56% infants positive for rotavirus below 6 months of age.
RVA G3 is a ubiquitous genotype but it was only reported from Faisalabad region of Pakistan in 2010 at very low levels [31]. Notably, our results highlight a greater degree of G3 detection compared to previous studies in the region [31]. The high detection rate (22.9%) of G3 in our study is consistent with reports from Japan [18,19,24], China [20,42], Vietnam [43] and Hong Kong [25].
G3 rotaviruses detected in Japan during 2003–2004 contained multiple amino acid changes in the VP7 gene consequently named as “new variant G3” represented by 5091 strain [18]. These viruses continued to circulate in Japan from 2007 to 2011 [19,24]. Pakistani G3 strains detected in this study were highly identical to the new variant G3 found in Japan as well as to the contemporary G3 strains circulating worldwide [44–46]. Similarly, high degree of amino acid similarities (99–99.7%) were observed between Pakistani G3 and Chinese new variant G3 isolated during 2010–2012 [46]. China has a long border with Pakistan and there is frequent movement of people across the border for trade reflecting a possible source of introduction from China into Pakistan or vice versa.
Comparison of antigenic epitopes of RVA vaccine RotaTeqTM and Pakistani strains revealed that all Pakistani G3 strains contained K238N change in VP7 gene which creates a potential N-linked glycosylation site. The presence of this glycosylation site might change the antigenicity of G3 strains and calls for further investigations. Previous reports have shown that glycosylation of 238 residue can reduce neutralization of animal RVA strains by hyper-immune sera and monoclonal antibodies [47,48]. Furthermore, the role of glycosylation of viral proteins in altering the immunogenicity of different viruses like influenza A virus, human immunodeficiency virus, and human respiratory syncytial virus has previously been reported [49–51].
Pakistan has an annual birth rate of approximately 5 million and is among the five countries where rotavirus related deaths are the highest [52] hence a suitable rotavirus vaccine is the need of time. Two vaccines (monovalent RotarixTM and pentavalent RotaTeqTM) are currently licensed for immunization against rotavirus worldwide [53]. WHO recommends the inclusion of these vaccines into national immunization programs of all countries especially those where diarrheal deaths account for ≥10% of under-five mortality [54]. The effectiveness of these vaccines is higher in developed countries compared to low and middle income countries [55–59]. Pakistan is expected to introduce RVA vaccine in its Expanded Program on Immunization (EPI) in 2017 with the support of GAVI (Global Alliance for Vaccines and Immunization). The success of the rotavirus vaccines in Pakistan will depend on their ability to provide protection against the RVA genotypes circulating in the country. Considering the variations observed in the VP7 and VP4 epitopes of Pakistani G3 strains and those of vaccine strains, further investigation of the antigenic variability and their impact would be required to understand the significance of these differences and their influence on vaccine efficacy.
To our knowledge, this is the first report on the molecular epidemiology of G3 and detection of new variant G3 rotavirus from Pakistan. We could not compare our G3 strains with those previously reported from Faisalabad, Pakistan [31] due to the unavailability of VP7 sequences in the GenBank. In addition, the small sample size was a limitation of the current study. However, our present findings conclude that rotavirus G3P[8] genotype is a major cause of acute gastroenteritis in pediatric population admitted in a tertiary care hospital of Rawalpindi. Comparative analysis of Pakistani G3 with RVA vaccine RotateqTM showed a K238N change in the VP7 gene which creates a potential N-linked glycosylation site. The genotypic diversity and high prevalence of G3 strains detected in the current study highlights the need for robust large scale surveillance to formulate future vaccine strategies.
Acknowledgments
We would like to acknowledge the Pediatrics Department of Benazir Bhutto Hospital, Rawalpindi, Pakistan, especially Dr. Rai Asghar and Saif Aqeel for their support and assistance in sample collection.
Data Availability
All relevant data are within the paper.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Tate JE, Burton AH, Boschi-Pinto C, Parashar UD, Network WHOCGRS, Agocs M, et al. Global, regional, and national estimates of rotavirus mortality in children< 5 years of age, 2000–2013. Clinical Infectious Diseases. 2016;62(suppl_2):S96–S105. [DOI] [PubMed] [Google Scholar]
- 2.Estes M, Greenberg H. Rotaviruses In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2013. [Google Scholar]
- 3.Rotavirus Classification Working Group (RCWG), 2017. Newly assigned genoypes. (https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg).
- 4.Bányai K, Kemenesi G, Budinski I, Földes F, Zana B, Marton S, et al. (2017) Candidate new rotavirus species in Schreiber's bats, Serbia. Infection, Genetics and Evolution 48: 19–26. doi: 10.1016/j.meegid.2016.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthijnssens J, Otto PH, Ciarlet M, Desselberger U, Van Ranst M, Johne R. VP6-sequence-based cutoff values as a criterion for rotavirus species demarcation. Archives of virology. 2012;157(6):1177–82. doi: 10.1007/s00705-012-1273-3 [DOI] [PubMed] [Google Scholar]
- 6.Diggle L. Rotavirus diarrhoea and future prospects for prevention. British Journal of Nursing. 2007;16(16). [DOI] [PubMed] [Google Scholar]
- 7.Rotavirus Classification Working Group (RCWG), 2016. Newly assigned genoypes. (https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification/rcwg).
- 8.Li K, Lin X-D, Huang K-Y, Zhang B, Shi M, Guo W-P, et al. Identification of novel and diverse rotaviruses in rodents and insectivores, and evidence of cross-species transmission into humans. Virology. 2016;494:168–77. doi: 10.1016/j.virol.2016.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos N, Hoshino Y. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Reviews in medical virology. 2005;15(1):29–56. doi: 10.1002/rmv.448 [DOI] [PubMed] [Google Scholar]
- 10.Bányai K, László B, Duque J, Steele AD, Nelson EAS, Gentsch JR, et al. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30:A122–A30. doi: 10.1016/j.vaccine.2011.09.111 [DOI] [PubMed] [Google Scholar]
- 11.Rahman M, Matthijnssens J, Yang X, Delbeke T, Arijs I, Taniguchi K, et al. Evolutionary history and global spread of the emerging G12 human rotaviruses. Journal of virology. 2007;81(5):2382–90. doi: 10.1128/JVI.01622-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamim S, Hasan F, Matthijnssens J, Sharif S, Shaukat S, Alam M, et al. Epidemiology and phylogenetic analysis of VP7 and VP4 genes of rotaviruses circulating in Rawalpindi, Pakistan during 2010. Infection, Genetics and Evolution. 2013;14:161–8. doi: 10.1016/j.meegid.2012.10.009 [DOI] [PubMed] [Google Scholar]
- 13.Kawai K, O’Brien MA, Goveia MG, Mast TC, El Khoury AC. Burden of rotavirus gastroenteritis and distribution of rotavirus strains in Asia: a systematic review. Vaccine. 2012;30(7):1244–54. doi: 10.1016/j.vaccine.2011.12.092 [DOI] [PubMed] [Google Scholar]
- 14.Fang Z-Y, Yang H, Qi J, Zhang J, Sun L-W, Tang J-Y, et al. Diversity of rotavirus strains among children with acute diarrhea in China: 1998–2000 surveillance study. Journal of clinical microbiology. 2002;40(5):1875–8. doi: 10.1128/JCM.40.5.1875-1878.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orenstein EW, Fang ZY, Xu J, Liu C, Shen K, Qian Y, et al. The epidemiology and burden of rotavirus in China: a review of the literature from 1983 to 2005. Vaccine. 2007;25(3):406–13. doi: 10.1016/j.vaccine.2006.07.054 [DOI] [PubMed] [Google Scholar]
- 16.Duan Z-j, Liu N, Yang S-h, Zhang J, Sun L-W, Tang J-Y, et al. Hospital-based surveillance of rotavirus diarrhea in the People’s Republic of China, August 2003–July 2007. The Journal of infectious diseases. 2009;200(Supplement_1):S167–S73. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Li Z, Han D, Cui D, Chen X, Zheng S, et al. Viral agents associated with acute diarrhea among outpatient children in southeastern China. The Pediatric infectious disease journal. 2013;32(7):e285–e90. doi: 10.1097/INF.0b013e31828c3de4 [DOI] [PubMed] [Google Scholar]
- 18.Phan TG, Trinh QD, Khamrin P, Kaneshi K, Shigekazu Nakaya YU, Nishimura S, et al. Emergence of new variant rotavirus G3 among infants and children with acute gastroenteritis in Japan during 2003–2004. Clinical laboratory. 2007;53(1–2):41–8. [PubMed] [Google Scholar]
- 19.Thongprachum A, Chan-it W, Khamrin P, Okitsu S, Nishimura S, Kikuta H, et al. Reemergence of new variant G3 rotavirus in Japanese pediatric patients, 2009–2011. Infection, Genetics and Evolution. 2013;13:168–74. doi: 10.1016/j.meegid.2012.09.010 [DOI] [PubMed] [Google Scholar]
- 20.Wang YH, Kobayashi N, Zhou X, Nagashima S, Zhu ZR, Peng JS, et al. Phylogenetic analysis of rotaviruses with predominant G3 and emerging G9 genotypes from adults and children in Wuhan, China. Journal of medical virology. 2009;81(2):382–9. doi: 10.1002/jmv.21387 [DOI] [PubMed] [Google Scholar]
- 21.Cashman O, Collins P, Lennon G, Cryan B, Martella V, Fanning S, et al. Molecular characterization of group A rotaviruses detected in children with gastroenteritis in Ireland in 2006–2009. Epidemiology & Infection. 2012;140(2):247–59. [DOI] [PubMed] [Google Scholar]
- 22.Iturriza-Gómara M, Dallman T, Bányai K, Böttiger B, Buesa J, Diedrich S, et al. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiology & Infection. 2011;139(6):895–909. [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Laso J, Roman A, Rodriguez M, Cervera I, Head J, Rodríguez-Avial I, et al. Diversity of the G3 genes of human rotaviruses in isolates from Spain from 2004 to 2006: cross-species transmission and inter-genotype recombination generates alleles. Journal of General Virology. 2009;90(4):935–43. [DOI] [PubMed] [Google Scholar]
- 24.Chan-it W, Thongprachum A, Dey SK, Phan TG, Khamrin P, Okitsu S, et al. Detection and genetic characterization of rotavirus infections in non-hospitalized children with acute gastroenteritis in Japan, 2007–2009. Infection, Genetics and Evolution. 2011;11(2):415–22. doi: 10.1016/j.meegid.2010.11.018 [DOI] [PubMed] [Google Scholar]
- 25.Mitui MT, Chan PK, Nelson EAS, Leung TF, Nishizono A, Ahmed K. Co-dominance of G1 and emerging G3 rotaviruses in Hong Kong: A three-year surveillance in three major hospitals. Journal of Clinical Virology. 2011;50(4):325–33. doi: 10.1016/j.jcv.2011.01.008 [DOI] [PubMed] [Google Scholar]
- 26.Stupka JA, Carvalho P, Amarilla AA, Massana M, Parra GI, Diarrheas ANSNf. National Rotavirus Surveillance in Argentina: high incidence of G9P [8] strains and detection of G4P [6] strains with porcine characteristics. Infection, Genetics and Evolution. 2009;9(6):1225–31. doi: 10.1016/j.meegid.2009.07.002 [DOI] [PubMed] [Google Scholar]
- 27.Lennon G, Reidy N, Cryan B, Fanning S, O'shea H. Changing profile of rotavirus in Ireland: predominance of P [8] and emergence of P [6] and P [9] in mixed infections. Journal of medical virology. 2008;80(3):524–30. doi: 10.1002/jmv.21084 [DOI] [PubMed] [Google Scholar]
- 28.Sánchez-Fauquier A, Montero V, Moreno S, Solé M, Colomina J, Iturriza-Gomara M, et al. Human rotavirus G9 and G3 as major cause of diarrhea in hospitalized children, Spain. Emerging infectious diseases. 2006;12(10):1536 doi: 10.3201/eid1210.060384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seheri L, Page N, Dewar J, Geyer A, Nemarude A, Bos P, et al. Characterization and molecular epidemiology of rotavirus strains recovered in Northern Pretoria, South Africa during 2003–2006. Journal of Infectious Diseases. 2010;202(Supplement_1):S139–S47. [DOI] [PubMed] [Google Scholar]
- 30.Pietsch C, Schuster V, Liebert U. A hospital based study on inter-and intragenotypic diversity of human rotavirus A VP4 and VP7 gene segments, Germany. Journal of Clinical Virology. 2011;50(2):136–41. doi: 10.1016/j.jcv.2010.10.013 [DOI] [PubMed] [Google Scholar]
- 31.Iftikhar T, Butt A, Nawaz K, Sarwar Y, Ali A, Mustafa T, et al. Genotyping of rotaviruses detected in children admitted to hospital from Faisalabad Region, Pakistan. Journal of medical virology. 2012;84(12):2003–7. doi: 10.1002/jmv.23402 [DOI] [PubMed] [Google Scholar]
- 32.Qazi R, Sultana S, Sundar S, Warraich H, Rais A, Zaidi AK. Population-based surveillance for severe rotavirus gastroenteritis in children in Karachi, Pakistan. Vaccine. 2009;27:F25–F30. doi: 10.1016/j.vaccine.2009.08.064 [DOI] [PubMed] [Google Scholar]
- 33.Kazi AM, Warraich GJ, Qureshi S, Qureshi H, Khan MMA, Zaidi AKM, et al. Sentinel hospital-based surveillance for assessment of burden of rotavirus gastroenteritis in children in Pakistan. PloS one. 2014;9(10):e108221 doi: 10.1371/journal.pone.0108221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alam MM, Khurshid A, Shaukat S, Suleman RM, Sharif S, Angez M, et al. Epidemiology and genetic diversity of rotavirus strains in children with acute gastroenteritis in Lahore, Pakistan. PLoS One. 2013;8(6):e67998 doi: 10.1371/journal.pone.0067998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.World Health Organization (2002). Generic portocols for (i) hospital-based surveillance to estimate the burden of rotavirus gastroenteritis in children and (ii) a community-based survey on utilization of health care services for gastroenteritis in children: field test version.
- 36.Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, et al. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. Journal of clinical microbiology. 1990;28(2):276–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gentsch J, Glass R, Woods P, Gouvea V, Gorziglia M, Flores J, et al. Identification of group A rotavirus gene 4 types by polymerase chain reaction. Journal of clinical microbiology. 1992;30(6):1365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution. 2011;28(10):2731–9. doi: 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Degiuseppe JI, Parra GI, Stupka JA. Genetic diversity of G3 rotavirus strains circulating in Argentina during 1998–2012 assessed by full genome analyses. PloS one. 2014;9(10):e110341 doi: 10.1371/journal.pone.0110341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25(13):1605–12. doi: 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 41.Dey SK, Thongprachum A, Islam AR, Phan GT, Rahman M, Mizuguchi M, et al. Molecular analysis of G3 rotavirus among infants and children in Dhaka City, Bangladesh after 1993. Infection, Genetics and Evolution. 2009;9(5):983–6. doi: 10.1016/j.meegid.2009.06.016 [DOI] [PubMed] [Google Scholar]
- 42.Jin Y, Ye X-H, Fang Z-Y, Li Y-N, Yang X-M, Dong Q-L, et al. Molecular epidemic features and variation of rotavirus among children with diarrhea in Lanzhou, China, 2001–2006. World Journal of Pediatrics. 2008;4(3):197–201. doi: 10.1007/s12519-008-0036-4 [DOI] [PubMed] [Google Scholar]
- 43.Cuong NT, Minh NB, Anh DD, Thu NH, Tu NT, Van Nam T, et al. Molecular epidemiology of rotavirus diarrhoea among children in Haiphong, Vietnam: the emergence of G3 rotavirus. Vaccine. 2009;27:F75–F80. doi: 10.1016/j.vaccine.2009.08.074 [DOI] [PubMed] [Google Scholar]
- 44.Dóró R, Mihalov-Kovács E, Marton S, László B, Deák J, Jakab F, et al. Large-scale whole genome sequencing identifies country-wide spread of an emerging G9P [8] rotavirus strain in Hungary, 2012. Infection, Genetics and Evolution. 2014;28:495–512. doi: 10.1016/j.meegid.2014.09.016 [DOI] [PubMed] [Google Scholar]
- 45.Zeller M, Patton JT, Heylen E, De Coster S, Ciarlet M, Van Ranst M, et al. Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in belgium and rotaviruses in rotarix™ and RotaTeq™. Journal of clinical microbiology. 2011:JCM. 05590–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y-H, Pang B-B, Ghosh S, Zhou X, Shintani T, Urushibara N, et al. Molecular epidemiology and genetic evolution of the whole genome of G3P [8] human rotavirus in Wuhan, China, from 2000 through 2013. PloS one. 2014;9(3):e88850 doi: 10.1371/journal.pone.0088850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ciarlet M, Hoshino Y, Liprandi F. Single point mutations may affect the serotype reactivity of serotype G11 porcine rotavirus strains: a widening spectrum? Journal of virology. 1997;71(11):8213–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ciarlet M, Reggeti F, Piña CI, Liprandi F. Equine rotaviruses with G14 serotype specificity circulate among venezuelan horses. Journal of clinical microbiology. 1994;32(10):2609–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skehel J, Stevens D, Daniels R, Douglas A, Knossow M, Wilson I, et al. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proceedings of the National Academy of Sciences. 1984;81(6):1779–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palomo C, Cane PA, Melero JA. Evaluation of the antibody specificities of human convalescent‐phase sera against the attachment (G) protein of human respiratory syncytial virus: Influence of strain variation and carbohydrate side chains. Journal of medical virology. 2000;60(4):468–74. [PubMed] [Google Scholar]
- 51.Utachee P, Nakamura S, Tokunaga K, Sawanpanyalert P, Ikuta K, Auwanit W, et al. Two N-linked glycosylation sites in the V2 and C2 regions of human immunodeficiency virus type 1 CRF01_AE envelope glycoprotein gp120 regulate viral neutralization susceptibility to the human monoclonal antibody specific for the CD4 binding domain. Journal of virology. 2010;84(9):4311–20. doi: 10.1128/JVI.02619-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. The Lancet infectious diseases. 2012;12(2):136–41. doi: 10.1016/S1473-3099(11)70253-5 [DOI] [PubMed] [Google Scholar]
- 53.Dennehy PH. Rotavirus vaccines: an overview. Clinical microbiology reviews. 2008;21(1):198–208. doi: 10.1128/CMR.00029-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.World Health Organization (2009) Meeting of the immunization Strategic Advisory Group of Experts, April 2009-conclusions and recommendations. Weekly Epidemiological Record. 84(23):220–36. [PubMed] [Google Scholar]
- 55.Vesikari T, Matson DO, Dennehy P, Van Damme P, Santosham M, Rodriguez Z, et al. Safety and efficacy of a pentavalent human–bovine (WC3) reassortant rotavirus vaccine. New England Journal of Medicine. 2006;354(1):23–33. doi: 10.1056/NEJMoa052664 [DOI] [PubMed] [Google Scholar]
- 56.Vesikari T, Karvonen A, Prymula R, Schuster V, Tejedor J, Cohen R, et al. Efficacy of human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in European infants: randomised, double-blind controlled study. The Lancet. 2007;370(9601):1757–63. [DOI] [PubMed] [Google Scholar]
- 57.Linhares AC, Velázquez FR, Pérez-Schael I, Sáez-Llorens X, Abate H, Espinoza F, et al. Efficacy and safety of an oral live attenuated human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in Latin American infants: a randomised, double-blind, placebo-controlled phase III study. The Lancet. 2008;371(9619):1181–9. [DOI] [PubMed] [Google Scholar]
- 58.Armah GE, Sow SO, Breiman RF, Dallas MJ, Tapia MD, Feikin DR, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in sub-Saharan Africa: a randomised, double-blind, placebo-controlled trial. The Lancet. 2010;376(9741):606–14. [DOI] [PubMed] [Google Scholar]
- 59.Zaman K, Anh DD, Victor JC, Shin S, Yunus M, Dallas MJ, et al. Efficacy of pentavalent rotavirus vaccine against severe rotavirus gastroenteritis in infants in developing countries in Asia: a randomised, double-blind, placebo-controlled trial. The Lancet. 2010;376(9741):615–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.