ABSTRACT
The clinical utility of circulating tumor cells (CTCs) has been investigated in numerous publications, but CTCs that express very typical immune cell markers have not been reported. Here we report a novel class of CTCs—CSV-positive macrophage-like CTCs (ML-CTCs). This nomenclature was based on the fact that this class of CTCs can be captured from blood samples of gastrointestinal stromal tumors (GISTs) patients using either the macrophage marker CD68 or our proprietary tumor-specific cell-surface vimentin (CSV) antibody 84–1; likewise, the captured ML-CTCs can be co-stained with both typical macrophage markers (CD14, CD68) and tumor cell markers (DOG-1, C-kit) but not CD45. Patients with metastatic GIST had significantly greater numbers of ML-CTCs than patients with localized GIST or cancer-free blood donors (P<0.0001). Unexpectedly, the classic CSV positive CTCs was abundant in metastatic disease but failed to predict GIST metastasis. Only CSV-positive ML-CTCs was able to serve as a solid and novel biomarker for prediction of metastatic risk in GIST patients.
KEYWORDS: circulating tumor cells, cell-surface vimentin, gastrointestinal stromal tumor, sarcoma, tumor-associated macrophage
Introduction
Gastrointestinal stromal tumor (GIST) is the most common sarcoma in the gastrointestinal tract.1 The majority of GISTs are driven by activating mutations of either the KIT or PDGFRA gene, which result in high sensitivity to treatment with inhibitors of the KIT and PDGFRA proteins (imatinib and sunitinib).2,3 These drugs are not curative, however, as GISTs become resistant to treatment, leading to disease recurrence or distant metastasis within 2 years of treatment.4,5 Reliable biomarkers to predict therapeutic resistance and metastasis of GISTs are lacking.
Tumor-associated macrophages (TAMs) are specialized and differentiated macrophages which are found within many tumors and have an important role in prediction of tumor invasiveness and immune suppression.6–8 The immune system is active in the microenvironment of GISTs and potentiates the antitumor effects of imatinib.9 Notably, TAMs are present in the greatest abundance in GISTs among all solid tumors, and the presence of polarized M2-like TAMs has been correlated with GIST progression.10 However, detection of TAMs in tumors for prognosis of GIST's progression and metastasis requires invasive tumor biopsies.11 A none invasive sampling and analysis method for TAM is needed.
Circulating tumor cells (CTCs) in blood, which have emerged as a clinical tool during recent years, are shed from primary tumors and enter the circulation, potentially forming distant metastasis.12–14 CTC enumeration via liquid biopsy is highly correlated with disease status in patients with epithelial cancers.15,16 The CTCs for epithelial tumors were enumerated by utilizing the epithelial cell adhesion molecule (EpCAM) biomarker and the CellSearch-based CTC capturing system.17 However, the EpCAM biomarker cannot be used to capture CTCs from mesenchymal tumor-bearing patients.18 We recently discovered that cell-surface vimentin (CSV) may serve as a universal biomarker for capturing CTCs across tumor types including mesenchymal tumors.19,20 We have developed monoclonal antibody 84–1, which is specific to CSV, and have used it successfully to capture and enumerate CTCs from different types of human sarcomas with high sensitivity and specificity.21,22 However, studies of the clinical utility of 84–1 in GIST patients are lacking. Additionally, CTCs were able to extravasate capillaries and set up secondary metastatic lesions under the protection of CTC-educated TAMs.23 Nevertheless, to the best of our knowledge, there are no reports on the clinical utility of circulating TAMs in predicting disease status in GIST.
Using a modified immunomagnetic method (described in our previous publication)19,20 and CSV antibody 84–1 antibody as circulating cell capturing tool, we identified a novel class of CSV-positive (CSV+) CTCs with macrophage-like features. We defined this novel class of CTCs as macrophage-like because these cells were not only positive for macrophage markers CD14 and CD68 but also for tumor markers C-kit, DOG-1, and CSV. However, these CTCs were not bona fide macrophages but derived from the microenvironment of metastatic GIST. While these novel ML-CTCs were found rarely in patients with localized GIST or cancer-free blood donors, they were found in much greater numbers and much more frequently in patients with metastatic GIST. Therefore, this novel class of CTCs may serve as a novel and robust biomarker of metastatic GIST.
Results
CSV+ macrophage-like CTCs (ML-CTC) found in peripheral blood samples from patients with metastatic GIST
We previously established a method of enumerating CTCs in different types of sarcoma using the universal specific CTC marker CSV.19 CSV was thoroughly validated as a tumor-specific CTC marker in our previous studies19–22. In the present study, we used the same method to isolate CTCs from samples of peripheral blood from GIST patients using the anti-CSV monoclonal antibody 84–1 (Fig. 1A). Unlike CTCs from patients with other types of sarcomas, more than half of the CTCs captured from the GIST patients were not only CSV+ but also CD14+ and CD68+ (Fig. 1B). CD14 and CD68 are typical markers of monocytes and macrophages, respectively. Despite expressing CD68, these CTCs were not typical macrophages, as they were CD45 negative (CD45–). This points toward a novel, specialized class of CTC cells. Further analysis showed that these novel CTCs were negative for CD3 (T cell marker), CD31 (endothelial marker), and CD133/SOX2 (stem cell markers), suggesting that the lineage of this cell population is irrelevant to other types of cells and related only to macrophages and CTCs. Therefore, this novel class of CTCs, which expressed CSV, CD14, and CD68 but not CD45, was defined as CSV+ macrophage-like CTCs (ML-CTCs).
Figure 1.
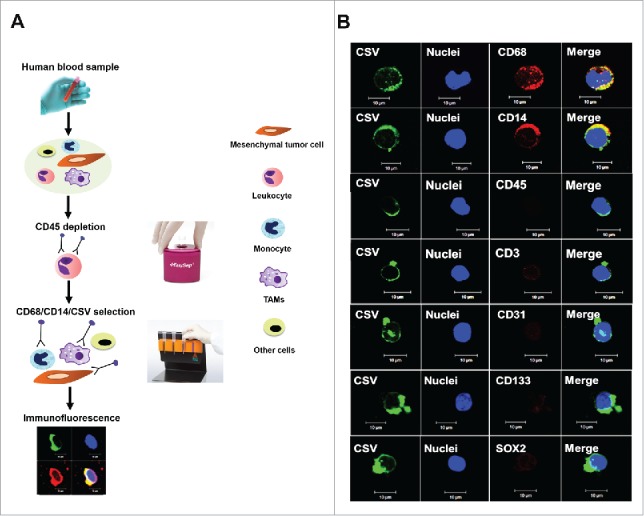
Isolation of CSV+ ML-CTCs in peripheral blood samples from patients with metastatic GIST. (A) Schematic representation of CSV+ CTC selection, enumeration, and analysis. (B) Circulating CSV+ cells were isolated from 104 blood samples of sarcoma patients. Immunofluorescent staining results for CSV (84–1), nuclei, and the markers CD14, CD68, CD45, CD3, CD31, CD133, and SOX2 from one representative metastatic GIST patient are shown. Scale indicates 10 µm.
ML-CTCs occur primarily in metastatic GIST tumors
To ascertain whether the ML-CTCs detected in the blood were linked to metastatic tumors, single-cell suspensions was prepared from freshly resected GIST tumors and analyzed with the same set of markers used for the ML-CTC analysis. As we expected, the single tumor cells from all four metastatic GIST tumors expressed the same set of molecular markers as for ML-CTCs (CD14+ CD68+ CSV+ CD45–) (Fig. 2B). However, single tumor cells dissociated from 2 primary GIST tumors expressed neither CD14 nor CD68, though they did express the tumor-specific marker CSV, DOG-1 or C-kit (Fig. 2A). Furthermore, 2 GIST cell lines tested did not express these macrophage markers (Figs. 2C-D), suggesting that this novel class of CTC is specific to metastatic GIST.
Figure 2.
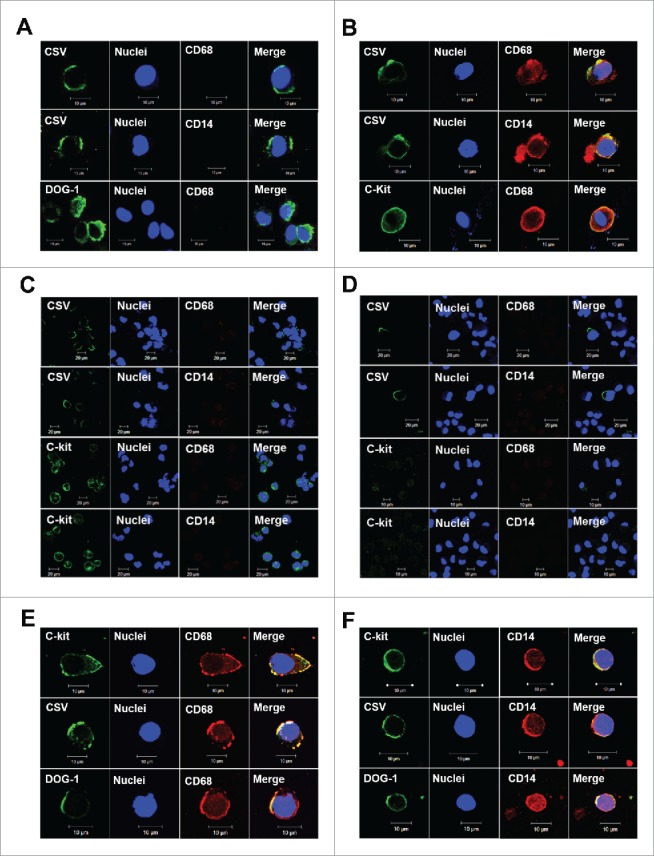
GIST microenvironment derivation. (A, B) After single-cell dissociation from primary GIST tumors (A) or metastatic GIST tumors (B), cells were subjected to CD45 depletion and CSV+ cell selection. Results of immunofluorescent staining of representative cells for CSV, nuclei, CD68, CD14, C-kit and DOG-1 are shown. (C, D) GIST cell lines T1 (C) and 882 (D) were subjected to immunofluorescent staining for CSV (84–1), nuclei, C-kit, CD68, CD14. Results for representative cells are shown. (E, F) CD68+ macrophages (E) and CD14+ monocytes (F) were selected from blood samples of patients with metastatic GIST after CD45 depletion. Immunofluorescent staining results for representative cells are shown. Scale indicates 10 µm.
To further validate the origin of the ML-CTCs, we isolated monocytes and macrophages from GIST patients’ peripheral blood samples using the same isolation procedure used for CSV (Fig. 1A) with CD14 and CD68 selection microbeads. As shown in Fig. 2E and 2 F, the isolated ML-CTCs were positive not only for CSV but also for C-kit and DOG-1, which agreed with our findings in tumor cells isolated from metastatic GIST fresh tumors. Taken together, these data provide further support for our working hypothesis that ML-CTCs are derived from the microenvironment of metastatic GIST tumors.
ML-CTCs are associated with metastatic GIST
To ascertain the clinical relevance of ML-CTC enumeration in different types of sarcomas, we analyzed freshly obtained blood samples from 104 sarcoma patients (77 GIST and 27 other sarcomas). Table 1 showed the baseline characteristics of these patients. Immunofluorescent staining showed co-expression of CSV and CD14 or CD68 in the ML-CTCs captured from patients with metastatic GIST but not in those captured from patients with localized GIST or other types of sarcomas (Fig. 3A). Of note, ML-CTCs were found in a majority of the tested blood samples from GIST patients (32 of 44 patients, 73%) but in a much lower percentage of the samples from patients with other types of sarcomas (7 of 27; 26%). Likewise, the number of ML-CTCs was significantly greater in the GIST patients (as high as 15/mL) than in the patients with other types of sarcomas (maximum 1/mL), except for one leiomyosarcoma patient (6/mL) (Fig. 3B). Notably, numbers of ML-CTCs were significantly greater in the patients with metastatic GIST than in the patients with localized GIST (Fig. 3C). The numbers of ML-CTCs were significantly lower in GIST patients who were receiving adjuvant therapy. There was no association between number of ML-CTCs and patient sex, age, lesion site, mitotic rate, or genomic information (Table 2). However, the 27 blood samples obtained from patients with other types of sarcomas contained lower numbers of ML-CTCs, and the difference in ML-CTC count between the localized and metastatic groups of these 27 non-GIST sarcoma patients was not significant (P = 0.715) (Fig. 3D).
Table 1.
Clinical characteristics of sarcoma patients in study (N = 104).
| Characteristic | Number(%) |
|---|---|
| Disease | |
| GIST | 77 (74) |
| Other sarcoma | 27 (26) |
| Sex | |
| Male | 55 (53) |
| Female | 49 (47) |
| Age, years | |
| ≤56 | 54 (52) |
| >56 | 50 (48) |
| Disease status | |
| Localized | 42 (44) |
| Metastasis | 62 (56) |
Figure 3.
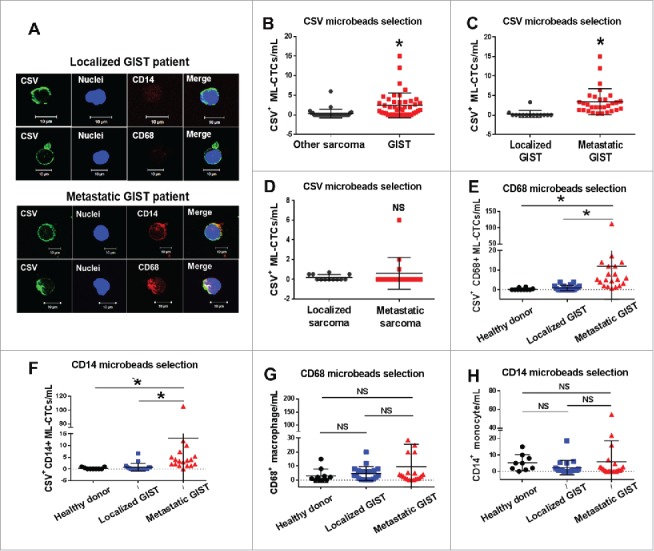
Association of CSV+ ML-CTCs with metastatic GIST. (A) Results of immunofluorescent staining for CSV and other markers showing CSV+ ML-CTCs isolated from blood samples of representative patients with localized (upper) or metastatic (lower) GIST. (B-D) Results of CSV+ microbeads selection from blood tumor samples obtained from 14 patients with localized GIST, 30 patients with metastatic GIST, and 27 patients with another type of sarcoma. (B) Comparison of CSV+ ML-CTC counts in samples from 44 patients with GIST and 27 patients with another type of sarcoma. (C) Comparison of CSV+ ML-CTC counts in samples from 14 patients with localized GIST and 30 with metastatic GIST. (D) Comparison of CSV+ ML-CTC counts in samples from 12 patients with localized non-GIST sarcoma and 15 with metastatic non-GIST sarcoma. (E, F) Comparison of CSV+ ML-CTC counts in blood samples from 10 cancer-free donors, 20 patients with localized GIST, and 20 with metastatic GIST using CD68 (E) and CD14 (F) microbeads selection. (G, H) Comparison of total macrophage (G) and monocyte (H) counts in blood samples from cancer-free donors, patients with localized GIST, and patients with metastatic GIST, using CD68 (G) and CD14 (H) microbeads selection. NS, not significant; *P<0.001.
Table 2.
Association of CSV+ ML-CTC count with clinical features in 77 GIST patients.
| Characteristic | Number (%) | CSV+ ML-CTCs (mean ± SD) | P | CSV+classic CTCs (mean ± SD) | P |
|---|---|---|---|---|---|
| Sex | 0.08 | 0.20 | |||
| Male | 43 (56) | 2.1 ± 3.3 | 6.4 ± 10.0 | ||
| Female | 34 (44) | 8.0 ± 21.6 | 3.2 ± 3.8 | ||
| Age, years | 0.54 | 0.90 | |||
| ≤60 | 39 (51) | 3.7 ± 11.6 | 4.9 ± 9.9 | ||
| >60 | 38 (49) | 5.7 ± 17.3 | 5.2 ± 5.5 | ||
| Metastasis | <0.001 | 0.41 | |||
| No | 30 (40) | 0.4 ± 0.9 | 2.8 ± 3.6 | ||
| Yes | 47 (60) | 7.5 ± 18.3 | 6.0 ± 9.3 | ||
| Primary lesion site | 0.65 | 0.93 | |||
| Small intestine | 41 (53) | 6.2 ± 19.5 | 5.9 ± 10.0 | ||
| Other | 36 (47) | 2.9 ± 4.0 | 4.0 ± 5.2 | ||
| Metastatic lesion site | 0.97 | 0.91 | |||
| Liver | 30 (64) | 6.3 ± 13.3 | 4.3 ± 4.6 | ||
| Other | 17 (36) | 9.5 ± 24.9 | 9.1 ± 14.0 | ||
| Mitotic rate | 0.62 | 0.25 | |||
| ≤5 mitoses/50 HPFs | 20(26) | 7.2 ± 23.3 | 8.6 ± 12.8 | ||
| >5 mitoses/50 HPFs | 25(32) | 2.1 ± 2.4 | 2.4 ± 2.8 | ||
| NAa | 32(42) | 5.1 ± 13.1 | 4.0 ± 4.4 | ||
| Lines of therapies | <0.001 | 0.35 | |||
| Adjuvant | 29 (38) | 0.3 ± 0.9 | 2.8 ± 3.6 | ||
| First line | 27 (35) | 7.7 ± 19.8 | 7.0 ± 11.3 | ||
| Second line | 15 (19) | 7.3 ± 18.3 | 4.0 ± 6.1 | ||
| Other | 6 (8) | 4.7 ± 5.5 | 8.5 ± 8.4 | ||
| Last therapy | 0.76 | 0.85 | |||
| Imatinib | 42 (55) | 4.8 ± 16.2 | 5.0 ± 9.7 | ||
| Sunitinib | 9 (12) | 2.8 ± 2.9 | 5.4 ± 7.6 | ||
| Other | 5 (6) | 2.1 ± 1.7 | 6.3 ± 6.6 | ||
| No treatment | 21 (27) | 5.6 ± 15.8 | 4.6 ± 5.6 | ||
| Mutation | 0.55 | 0.78 | |||
| Wild type | 4 (5) | 1.6 ± 1.4 | 2.3 ± 1.9 | ||
| KIT exon 11 | 48 (62) | 2.3 ± 3.3 | 5.2 ± 8.6 | ||
| KIT exon 9 | 7 (9) | 1.6 ± 1.2 | 3.1 ± 1.6 | ||
| Otherb | 4 (6) | 2.0 ± 2.8 | 4.8 ± 2.5 | ||
| NAa | 14 (18) | 15.5 ± 32.1 | 8.0 ± 10.0 |
NA: not available
Other: KIT exon 13 (N = 3) and exon 17 (N = 1)
To validate this ML-CTC cell population further, monocyte- and macrophage-capturing microbeads were used to enumerate CD45– circulating cells in blood samples from both cancer-free donors and GIST patients. As shown in Figs. 3E and F, the tested 20 patients with metastatic GIST had significantly greater numbers of ML-CTCs than the tested 20 patients with localized GIST or the 10 cancer-free blood donors (P < 0.001). In contrast, there were no significant differences in the total number of normal macrophages among these 3 groups in the same set of samples (P > 0.05) (Figs. 3G-H). These findings clearly showed that this novel class of CTCs, the ML-CTCs, may serve as a diagnostic biomarker for metastatic GIST, regardless of which marker (CSV, CD14, or CD68) is used to capture this novel class of CTCs.
ML-CTCs, but not classic CTCs, may predict risk of GIST metastasis
Given that CSV is a universal and tumor-specific marker for enumerating classic CTCs across different types of sarcoma, as shown by our previous studies19,22, we compared counts of classic CSV+ CTCs in patients with GIST and in patients with other types of sarcomas. Classic CTCs were defined as both CD68– and CD14– but CSV+ CTCs with a diameter of approximately 10 µm or larger. The classic CTC count did not differ significantly in these 2 groups (P > 0.05) (Fig. 4A) or between patients with metastatic GIST and those with localized disease (P > 0.05) (Fig. 4B). Additionally, we observed no differences in classic CTC count in terms of patient sex, age, lesion site, mitotic rate, therapeutic strategy, or genomic information (Table 2). The only significant difference was found between localized and metastatic non-GIST sarcoma (1.7 ± 2.2 cells/mL vs. 5.7 ± 5.3 cells/mL; P = 0.03, Fig. 4C). Together, these data suggest that the presence of ML-CTCs but not classic CTCs is an effective diagnostic marker for GIST metastasis.
Figure 4.

The novel class of CTCs, CSV+ ML-CTCs, but not classic CTCs, may predict the risk of GIST metastasis. (A) Comparison of CSV+ classic CTC counts in patients with GIST and patients with another type of sarcoma using CSV+ microbeads selection. (B) Comparison of CSV+ classic CTC counts in patients with localized GIST and patients with metastatic GIST using CSV+ microbeads selection. (C) Comparison of CSV+ classic CTC counts in patients with localized non-GIST sarcoma and patients with metastatic non-GIST sarcoma using CSV+ microbeads selection. NS, not significant; *P<0.05.
Discussion
As tumor dissemination occurs primarily through the circulating blood, CTCs that are shed into the blood circulation and seed potential metastatic sites are highly clinically relevant.12–14 Associations of CTCs with metastasis have been reported in multiple cancer types,24 but such studies on mesenchymal tumors have been limited by the lack of tumor-specific CTC capture tools for this class of tumors. Our previous findings that CSV+ tumor cells have high metastatic potential in mice and can be detected in more than 90% of metastatic colon tumors suggest that CSV+ CTCs may serve as a biomarker for metastasis across tumor types.19 However, studies of the clinical relevance of CTCs in GIST have been absent because of the lack of a specific marker. Furthermore, the GIST microenvironment is heavily infiltrated with immune cells, especially TAMs, which are differentiated from circulating monocytes recruited by tumors.10 CTCs and circulating TAMs originating from the primary tumor microenvironment was shown in one study to bind together in the blood, and distinguishing between them proved difficult.23
Because CSV is a marker for CTCs in progressive and metastatic disease across different tumor types, including mesenchymal tumors,19–21 we expected to capture/detect CSV+ CTCs from patients with GIST, a mesenchymal type of tumor. As expected, we are able to capture this classic CTC using the CSV capturing antibody from GIST patients; unexpectedly, this classic CSV+ CTCs are not specifically associated with metastatic GIST. Instead, a novel class of CTCs, which we call CSV+ ML-CTC or simply ML-CTC cells were found in almost all samples from patients with metastatic GIST. Unlike TAMs, these ML-CTCs were CD45–, were as large as tumor cells (10 μm in diameter), and expressed GIST-specific marker DOG-1. The numbers of these rare cells were much lower in samples from patients with localized GIST or other types of sarcomas than in patients with metastatic GIST. This novel class of CTCs was neither monocyte nor macrophage because the total counts of circulating monocyte and macrophage in blood samples between cancer-free blood donors and GIST patients (localized or metastatic) are the same by either the CD14 or CD68 capturing assay. Thus, ML-CTCs is a novel class of CTC that reflects disease status in GIST cases. We propose that this population of cells could be utilized to predict GIST relapse/metastasis in future studies.
Several types of unique circulating cells have been reported by other groups to associate with tumor metastasis and prognosis. A recently published study showed a type of circulating megakaryocyte that was negative for CD45, cytokeratin, and intracellular vimentin expression but positive for expression of the megakaryocytic lineage–specific marker CD41 in patients with advanced prostate cancer.25 These circulating megakaryocytes were potentially associated with good prognosis in patients with metastatic castration-resistant prostate cancer.25 Furthermore, a population of large hyperploid cells that were negative for epithelial CTC markers was reported in liver cancer.26 As the authors reporting that study did not include any specific immune cell markers, these hyperploid cells could be categorized only as epithelial-mesenchymal transition–CTCs without analyzing their clinical relevance. Another circulating cell, cancer-associated macrophage-like cells (CAMLs), was found in blood samples from patients with breast, pancreatic, or prostate cancer.27 These giant cells expressed CD14 and vacuoles of material that had undergone phagocytosis. The authors found that CAMLs could bind to CTCs expressing epithelial, monocyte, and endothelial protein markers in the circulation of patients with a solid tumor. Nevertheless, no data were provided that allowed evaluation of the potential utility of CAMLs in cancer diagnosis or prognosis. Our data plus the published data showed that a mixture of classic CTCs, ML-CTCs, and other types of circulating cells may be present in the peripheral blood circulation of cancer patients, which could be highly valuable biomarkers for different diagnostic purposes. This mixed population of circulating cells in peripheral blood might not stand alone. CAMLs or ML-CTCs might interact with CTCs or other surrounding immune cells in the blood to promote CTC-dependent metastasis. This speculation is supported by published data indicating that CTCs could increase tumor invasiveness through recruiting and “educating” TAMs in small cell lung cancer.
In conclusion, we have identified a novel class of CTCs, which we have called CSV+ ML-CTCs or ML-CTCs. The findings presented here suggest that the metastatic risk of GIST is linked to these ML-CTCs and thus that they are a novel biomarker of metastatic GIST. Establishing rigorous criteria for monitoring ML-CTCs will provide an important approach to liquid biopsy analysis for GIST metastasis. With further validation and optimization in large sample cohorts, ML-CTCs have the potential to be translated into clinical use for pro-diagnosis of GIST metastasis and relapse.
Patients and methods
Patient eligibility and recruitment
This study was approved by The University of Texas MD Anderson Cancer Center Institutional Review Board. Informed written consent was obtained from all patients enrolled in this study. Peripheral blood samples from 104 patients diagnosed with localized or metastatic sarcoma were collected in the Sarcoma Center and the Department of Laboratory Medicine at MD Anderson. Fresh tumor tissues were obtained from 2 patients with localized GIST and 4 with metastatic GIST who underwent surgical resection in the Department of Surgical Oncology at MD Anderson. Key inclusion criteria for patients with localized tumors were that the tumor be completely resectable and locally advanced without evidence of distant metastasis. The main inclusion criterion for patients with metastatic tumors was distant metastasis. Patients with infections or secondary malignant disease were excluded. Clinicopathological information for all patients was recorded before blood collection. Blood samples from cancer-free donors were obtained from the MD Anderson Blood Bank. The donors had no infections or known malignant disease at the time of the blood draw.
Blood collection and peripheral blood mononuclear cell preparation
Blood was drawn from the study participants before or at least 7 days after intravenous chemotherapy. Blood collection and PBMC preparation were carried out as described previously. Briefly, blood samples from patients and cancer-free donors were collected in 10-mL BD Vacutainer tubes with K2 ethylenediaminetetraacetic acid (EDTA, BD Diagnostics). All samples were processed within 48 hours of blood draw to ensure the best results. Collected blood was mixed with phosphate-buffered saline solution (PBS) containing 2% fetal bovine serum (FBS) at a dilution of 1:1. Later, the diluted blood was layered carefully over 15 mL of Ficoll-Paque PLUS density gradient medium (Ficoll-Paque PREMIUM) in a 50-mL SepMate tube (StemCell Technologies) and then subjected to centrifugation at 1200 × g for 10 minutes at room temperature with the brake on. PBMCs were harvested by pipetting the top layer, transferred to a new tube, and washed twice with PBS containing 2% FBS at room temperature. The cell pellet was suspended in fresh medium for further analysis.
Selection of CSV+ macrophages and other cells
The selection method used to enrich CSV+ cells from the human blood samples was described previously. Briefly, PBMCs were depleted of CD45+ cells using an EasySep Human CD45 Depletion Kit (StemCell Technologies) according to the manufacturer's directions. The CD45– cell population was subjected to CSV+ selection using the 84–1 antibody followed by mouse IgG-microbeads (Miltenyi Biotec) binding. The cells labeled with 84–1 antibody were then extracted using a magnetic column according to the manufacturer's protocol (Miltenyi Biotec). Macrophages were selected from the PBMCs using the same method.
Immunofluorescent imaging and analysis
For immunofluorescent imaging, the selected cell pellet was mixed with MACS buffer (Miltenyi Biotec) and stained with the 84–1 antibody in a tube at room temperature for 1 hour. Later, cells were cytospun onto Polylysine-coated microscope adhesion slides (Thermo Fisher Scientific) using a CytoFuge (Iris). Next, the cells were fixed in 4% paraformaldehyde (Fisher Scientific) for 10 minutes and blocked in a blocking buffer (1% FBS in PBS) with 0.25% NP-40 for 1 hour.
For detection of markers, including CD68 (Bioss Antibodies), CD14 (BioVision), CD133 (Novus Biologicals), CD117 (Cell Signaling Technology), CD31 (Bioss Antibodies), CD3, SOX2, and CD45 (all three, Abcam Inc.), the cells were incubated with the primary antibodies overnight at 4°C. The next day, the slides were stained with secondary antibodies—Alexa Fluor-647 for 84–1, SYTOX Green for nuclei, and Alexa Fluor-555 for the other markers (Invitrogen) —for 1 hour at room temperature. After mounting using SlowFade antifade reagent (Invitrogen), all slides were visualized under an LSM 510 confocal microscope using the LSM 5 3.2 image capture and analysis software program (Carl Zeiss).
Cell culture
The human GIST cell line T1 and 882 were kindly provided by Dr. Ronald P. DeMatteo (Memorial Sloan-Kettering Cancer Center). The cells were cultured in complete medium containing RPMI-1640 with 10% FBS and 10 U/mL penicillin and streptomycin (Life Technologies) in a 5% CO2 atmosphere at 37°C. All cells were passaged every 3 days and harvested in the logarithmic phase of growth. Cell viability was evaluated using a trypan blue assay (Life Technologies). Live cells with greater than 98% viability were used in the experiments.
Tumor dissociation
Immediately after surgical resection, the GIST specimens were dissociated into single-cell suspensions using RPMI medium (without supplements; Invitrogen) containing collagenase IV (1000 U/mL; Worthington Biochemical Corporation) and DNase I (0.1 mg/mL; Roche). The cell suspensions were incubated at 37°C for 1 hour with slow shaking. The resulting cell suspensions were filtered through a 40-μm nylon cell strainer (BD Biosciences), and single cells were harvested into a 50-mL conical tube. Total cells were counted after red blood cell lysis and two washes in PBS containing 2% FBS at room temperature.
Statistical analysis
The data presented herein are expressed as means ± standard deviation and are representative of at least three independent experiments. Differences in baseline characteristics and CTC counts between patients with primary GIST and those with metastatic GIST were analyzed using the Fisher exact test and t-tests. Statistical analysis was performed using the Prism software program (GraphPad Software). P values less than 0.05 were considered significant.
Funding Statement
This work was supported by grants from the National Institutes of Health [NIH R01CA120895] and PCRF to Dr. Shulin Li; by an MD Anderson Institutional Research Grant to Dr. Qing H. Meng; and by a grant from the National Natural Science Foundation of China to Dr. Heming Li [81502124].
Abbreviations
- CTC
circulating tumor cell
- CSV
cell-surface vimentin
- GIST
gastrointestinal stromal tumor
- ML-CTC
CSV+ macrophage-like CTC
- TAM
tumor-associated macrophage
Disclosure of potential conflicts of interest
The authors have declared no conflicts of interest.
Acknowledgment
The authors would like to thank all the patients who took their time to participate in this research study. The authors would like to acknowledge the Department of Scientific Publications of MD Anderson Cancer Center for language editing.
References
- 1.Sorour MA, Kassem MI, Ghazal Ael-H, El-Riwini MT, Abu Nasr A. Gastrointestinal stromal tumors (GIST) related emergencies. Int J Surg. 2014;12(4):269–80. doi: 10.1016/j.ijsu.2014.02.004. PMID:24530605. [DOI] [PubMed] [Google Scholar]
- 2.Heinrich MC, Rubin BP, Longley BJ, Fletcher JA. Biology and genetic aspects of gastrointestinal stromal tumors: KIT activation and cytogenetic alterations. Human Pathol. 2002;33(5):484–95. doi: 10.1053/hupa.2002.124124. PMID:12094373. [DOI] [PubMed] [Google Scholar]
- 3.Heinrich MC, Corless CL, Duensing A, McGreevey L, Chen C-J, Joseph N, Singer S, Griffith DJ, Haley A, Town A. PDGFRA activating mutations in gastrointestinal stromal tumors. Science. 2003;299(5607):708–10. https://doi.org/ 10.1126/science.1079666. PMID:12522257. [DOI] [PubMed] [Google Scholar]
- 4.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiguro S, Kawano K, Hanada M, Kurata A, Takeda M, et al.. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279(5350):577–80. doi: 10.1126/science.279.5350.577. PMID:9438854. [DOI] [PubMed] [Google Scholar]
- 5.Verweij J, Casali PG, Zalcberg J, LeCesne A, Reichardt P, Blay J-Y, Issels R, van Oosterom A, Hogendoorn PC, Van Glabbeke M. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: randomised trial. The Lancet. 2004;364(9440):1127–34. doi: 10.1016/S0140-6736(04)17098-0. PMID:15451219. [DOI] [PubMed] [Google Scholar]
- 6.Heusinkveld M, van der Burg SH. Identification and manipulation of tumor associated macrophages in human cancers. J Trans Med. 2011;9:216. doi: 10.1186/1479-5876-9-216. PMID:22176642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ries CH, Cannarile MA, Hoves S, Benz J, Wartha K, Runza V, Rey-Giraud F, Pradel LP, Feuerhake F, Klaman I, et al.. Targeting tumor-associated macrophages with anti-CSF-1R antibody reveals a strategy for cancer therapy. Cancer Cell. 2014;25(6):846–59. doi: 10.1016/j.ccr.2014.05.016. PMID:24898549. [DOI] [PubMed] [Google Scholar]
- 8.Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers. 2014;6(3):1670–90. doi: 10.3390/cancers6031670. PMID:25125485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan Y, Garcia-Buitrago MT, Trent JC, Rosenberg AE.The immune system and gastrointestinal stromal tumor: a wealth of opportunities. Curr Opin Oncol. 2015;27(4):338–42. doi: 10.1097/CCO.0000000000000201. PMID:26049274. [DOI] [PubMed] [Google Scholar]
- 10.van Dongen M, Savage ND, Jordanova ES, Briaire-de Bruijn IH, Walburg KV, Ottenhoff TH, Hogendoorn PC, van der Burg SH, Gelderblom H, van Hall T. Anti-inflammatory M2 type macrophages characterize metastasized and tyrosine kinase inhibitor-treated gastrointestinal stromal tumors. Int J Cancer. 2010;127(4):899–909. doi: 10.1002/ijc.25113. PMID:20013807. [DOI] [PubMed] [Google Scholar]
- 11.Cavnar MJ, Zeng S, Kim TS, Sorenson EC, Ocuin LM, Balachandran VP, Seifert AM, Greer JB, Popow R, Crawley MH, et al.. KIT oncogene inhibition drives intratumoral macrophage M2 polarization. J Exp Med. 2013;210(13):2873–86. doi: 10.1084/jem.20130875. PMID:24323358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plaks V, Koopman CD, Werb Z: Circulating tumor cells. Science. 2013;341(6151):1186–88. doi: 10.1126/science.1235226. PMID:24031008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mitra A, Mishra L, Li S. EMT, CTCs and CSCs in tumor relapse and drug-resistance. Oncotarget. 2015;6(13):10697–711. doi: 10.18632/oncotarget.4037. PMID:25986923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Batth I, Mitra A, Manier S, Ghobrial I, Menter D, Kopetz S, Li S. Circulating tumor markers: harmonizing the yin and yang of CTCs and ctDNA for precision medicine. Ann Oncol. 2016;28(3):468–77. doi: 10.1093/annonc/mdw619. PMID:27998963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Riethdorf S, Fritsche H, Müller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F, et al.. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13(3):920–28. doi: 10.1158/1078-0432.CCR-06-1695. PMID:17289886. [DOI] [PubMed] [Google Scholar]
- 16.De Bono JS Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008;14(19):6302–09. doi: 10.1158/1078-0432.CCR-08-0872. PMID:18829513. [DOI] [PubMed] [Google Scholar]
- 17.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, et al.. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004;351(8):781–91. doi: 10.1056/NEJMoa040766. PMID:15317891. [DOI] [PubMed] [Google Scholar]
- 18.Andrew J, Armstrong Matthew S, Marengo Sebastian Oltean, Kemeny Gabor, Rhonda L, Bitting James Turnbull, Christina I, et al.. Circulating Tumor Cells from Patients with Advanced Prostate and Breast Cancer Display Both Epithelial and Mesenchymal Markers. Mol Cancer Res. 2011;9(8):997–1007. doi: 10.1158/1541-7786.MCR-10-0490. PMID:3157566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Satelli A, Mitra A, Cutrera JJ, Devarie M, Xia X, Ingram DR, Dibra D, Somaiah N, Torres KE, Ravi V, et al.. Universal marker and detection tool for human sarcoma circulating tumor cells. Cancer Res. 2014;74(6):1645–50. doi: 10.1158/0008-5472.CAN-13-1739. PMID:24448245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Satelli A, Brownlee Z, Mitra A, Meng QH, Li S. Circulating Tumor Cell Enumeration with a Combination of Epithelial Cell Adhesion Molecule–and Cell-Surface Vimentin–Based Methods for Monitoring Breast Cancer Therapeutic Response. Clin Chem. 2015;61(1):259–66. doi: 10.1373/clinchem.2014.228122. PMID:25336717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Satelli A, Mitra A, Brownlee Z, Xia X, Bellister S, Overman MJ, Kopetz S, Ellis LM, Meng QH, Li S. Epithelial–mesenchymal transitioned circulating tumor cells capture for detecting tumor progression. Clin Can Res. 2015;21(4):899–906. doi: 10.1158/1078-0432.CCR-14-0894. PMID:25516888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Meng QH, Noh H, Batth IS, Somaiah N, Torres KE, Xia X, Wang R, Li S. Detection of circulating tumor cells from cryopreserved human sarcoma peripheral blood mononuclear cells. Cancer Lett. 2017;403:216–23. doi: 10.1016/j.canlet.2017.05.032. PMID:28652021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamilton G, Rath B, Klameth L, Hochmair MJ. Small cell lung cancer: recruitment of macrophages by circulating tumor cells. Oncoimmunology. 2016;5(3):e1093277. doi: 10.1080/2162402X.2015.1093277. PMID:27141354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massagué J, Obenauf AC. Metastatic colonization by circulating tumour cells. Nature. 2016;529(7586):298–306. doi: 10.1038/nature17038. PMID:26791720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu L, Mao X, Guo T, Chan PY, Shaw G, Hines J, Stankiewicz E, Wang Y, Oliver RTD, Ahmad AS, et al.. The novel association of circulating tumor cells and circulating megakaryocytes with prostate cancer prognosis. Clin Cancer Res. 2017;23(17):5112–22. doi: 10.1158/1078-0432.CCR-16-3081. PMID:28615267. [DOI] [PubMed] [Google Scholar]
- 26.Ogle LF, Orr JG, Willoughby CE, Hutton C, McPherson S, Plummer R, Boddy AV, Curtin NJ, Jamieson D, Reeves HL. Imagestream detection and characterisation of circulating tumour cells–A liquid biopsy for hepatocellular carcinoma? J Hepatol. 2016;65(2):305–13. doi: 10.1016/j.jhep.2016.04.014. PMID:27132171. [DOI] [PubMed] [Google Scholar]
- 27.Adams DL, Martin SS, Alpaugh RK, Charpentier M, Tsai S, Bergan RC, Ogden IM, Catalona W, Chumsri S, Tang C-M, et al.. Circulating giant macrophages as a potential biomarker of solid tumors. Proc Natl Acad Sci. 2014;111(9):3514–19. doi: 10.1073/pnas.1320198111. PMID:24550495. [DOI] [PMC free article] [PubMed] [Google Scholar]


