Abstract
AIM:
In this study, we evaluated the effect of silver nanoparticles (AgNPs) on the production of aflatoxin B1 (AFB1) through assessment the transcription activity of aflatoxin biosynthesis pathway genes in Aspergillus flavus ATCC28542.
MATERIAL AND METHODS:
The mRNAs were quantitative by Real Time-polymerase chain reaction (qRT-PCR) of A. flavus grown in yeast extract sucrose (YES) medium containing AgNPs. Specific primers that are involved in the AFB1 biosynthesis which highly specific to A. flavus, O-methyltransferase gene (omt-A), were designed and used to detect the fungus activity by quantitative PCR assay. The AFB1 production (from A. flavus growth) which effected by AgNPs were measured in YES medium by high-pressure liquid chromatography (HPLC).
RESULTS:
The AFB1 produced by A. flavus have the highest reduction with 1.5 mg -100 ml of AgNPs were added in media those records 88.2%, 67.7% and 83.5% reduction by using AgNP HA1N, AgNP HA2N and AgNP EH, respectively. While on mycelial growth give significantly inhibitory effect. These results have been confirmed by qRT-PCR which showed that culture of A. flavus with the presence of AgNPs reduced the expression levels of omt-A gene
CONCLUSION:
Based on the results of the present study, AgNPs inhibit growth and AFB1 produced by Aspergillus flavus ATCC28542. This was confirmed through RT-PCR approach showing the effect of AgNPs on omt-A gene involved in aflatoxin biosynthesis.
Keywords: Aflatoxin B1, Silver nanoparticles, qRT-PCR, HPLC, Omt-A gene
Introduction
Aflatoxins are secondary polyketide metabolites mainly produced by aflatoxigenic fungi of Aspergillus flavus and Aspergillus parasiticus. Among the aflatoxins that have been identified, aflatoxin B1 (AFB1) is most prevalent form, presents the highest potential toxic [1]. AFB1 is classified as group 1 carcinogenic to mammals by the International Agency for Cancer Research (IARC) [2]. AFB1 has potential hepatic [3], carcinogenicity [4], cytotoxicity, genotoxicity [5] [6] [7] and immunotoxicity [8].
Swallowing of AFB1 has a toxic to the immune system works [9] and mainly reduces the function of cell-mediated immunity [8]. Biochemical pathways and gene regulation it is vital to aflatoxin (AF) also characterized. Many studied already were published in genetics about the AF biosynthesis [10] [11]. Approximately Twenty Seven enzymatic and at least 25 genes involved in the AF biosynthesis in A. flavus, this genes are cluster within 75 kilobytes (kb) in the fungal genome [12] [13]. Most of that seems to be co-organized by the DNA binding protein AflR, encoded by the genes aflR [14]. Yabe and Nakajima [15] demonstrated that the three genes pksA, ver-1 and omt-A encode enzyme proteins were involved in the AF biosynthetic pathway. On the other hand, gene aflR is a regulatory gene whose product regulates transcription of some genes; pksA, ver-1 and omt-A [16]. Many strategies have been developed to reduce aflatoxins contamination, either by preventing the growth of aflatoxigenic fungi or by blocking the production of the toxin after infection [17]. Silver nanoparticles (AgNPs) is recent advancements in the field of nanotechnology were used as a novel therapeutic agent as antibacterial, antifungal, antiviral, anti-inflammatory and anti-cancer agents [18] [19]. The effect of Silver nanoparticles on growth and production of other types mycotoxins by toxigenic fungi were studied but; the effect Silver nanoparticles AgNPs on gene expression of AFs biosynthesis pathway genes still need more studies. This study aims to evaluate the effects of three types AgNPs on the production of AFB1 through assessment the transcription activity (gene expression) of AFB1 biosynthesis pathway genes in Aspergillus flavus ATCC28542.
Material and Methods
Fungal strain
Fungal Strain: a Toxigenic strain of Aspergillus flavus (ATCC 28542) was obtained from Microbial Research Center, Faculty of Agriculture, Ain Shams University Cairo, Egypt (MIRCEN).
Chemicals and solvents
Potato dextrose agar (PDA) and yeast extract sucrose (YES) liquid growth medium and Sodium sulphate anhydrous were obtained from Sigma-Aldrich, France. Aflatoxin B1 (AFB1) standard was purchased from Sigma, Chemical Co. (St. Louis, MO, U.S.A). Stock solutions and standard were prepared and assayed according to Association of Official Analytical Chemists (AOAC) [20] Method 990.33A. Methanol and acetonitrile HPLC grade were produced by BDH, England. The water was double distilled with Millipore water purification system (Bedford, M A, USA).
Synthesis and characterised of AgNPs
AgNPs in this study were synthesised by Aspergillus terreusHA1N (KR364880) and Penicillium expansum HA2N (KR269857) which isolated and identified according to Ammar and El-Desouky [21]. Also, the characterisation of AgNPs has been done by UV-Visible Spectrophotometer, Dynamic Light Scattering (DLS), Fourier Transform Infrared Spectroscopy (FTIR), and Transmission Electron Microscope (TEM) in the previous study [21]. On the other hand, in the same study, we synthesised the AgNPs by Egyptian honey (EH) preparation and production according to El-Desouky and Ammar [22].
Inhibition of A. flavus growth and aflatoxin B1 production in the presence of silver nanoparticles
A hundred ml of YES medium were put in a 500 ml flasks and then autoclaved at 120°C for 15 min. Inoculation was carried out by adding 1 ml of a suspension of spores (105 spores) of a toxigenic A. flavus ATCC28542 strains without AgNPs (control) or with 0.5, 1.0 and 1.5 mg/100ml YES medium of one of the tested AgNPs (6a, 6b, 6c AgNps HA1N were synthesized by Aspergillus terreus (KR364880),3a, 3b, 3C AgNps EH were synthesized by Egyptian honey, and 1a, 1b, 1C AgNps HA2N were synthesized by Penicillium expansum (KR269857). The flasks were incubated in the dark for 14 days at 28°C. After the incubation period, extraction of AFB1 from in the YES culture according to the method of Munimbazi and Bullerman [23]. Where, the mycelium of each flask contained YES medium was harvested by filtration through Whatman paper (No. 4), then extracted with 100 ml chloroform. The chloroform extract was dried by addition of anhydrous sodium sulfate. The residue was transferred to a vial and evaporated off using a stream of nitrogen at a temperature below 60oC. The dry film was used for the detection and determination of AFB1 by (HPLC) according to (Deabes et al., [24,6] the retention time of AFB1 standard separation is 4.061. The percentage of inhibition of AFB1 is calculated using equation: % inhibition = (control- treatment /control) X100
Effect of AgNPs on biosynthesis pathway genes of AFB1 in Aspergillus flavus ATCC28542
DNA extraction
DNA was extracted from 25 mg of the harvested mycelia, which was frozen in liquid N2 and ground in a mortar, according to the protocol recommended for the DNA Tissue purification mini kit (Qiagen). The genomic DNA was checked by agarose gel electrophoresis, and the concentrations of the purified total genomic DNA were determined with a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and stored at -20°C for further use.
Primer design
Primers presented in Table 1 were selected according to the sequence of the omt-A gene of A. flavus from GenBank database (http://www.ncbi.nlm.nih.gov/).
Table 1.
Primer used in this study for amplification of AFB1 gene
| Primer | 5’-3’ nucleotide sequence |
|---|---|
| omt-F | GACCAATACGCCCACACAG |
| omt-R | CTTTGGTAGCTGTTTCTCGC |
Polymerase Chain Reaction amplification
PCR reactions were carried out in a total reaction volume of 20 μl, containing (10 μl of 2 X Go Taq master mix (Promega Corporation, Madison, WI) and 10 pmol of each primer of 50 ng template DNA. Amplification was performed in a T100-Bio-Rad Gradient Thermal cycler. The following programmer was used to amplify the DNA: 5 min at 94°C (1 cycle); 1 min at 94°C, 1 min at 59°C, and 1 min at 72°C (35 cycles); and 10 min at 72°C. A 10 μl aliquot of PCR products were separated on a 1.5% agarose gel stained with ethidium bromide (0.1 mg/l) and photographed under Gel Doc™ XR+ Gel Documentation System. Thermo Scientific GeneRuler 100 bp DNA Ladder was used as a size standard.
Gene Expression Analysis
Extraction of mRNA and cDNA synthesis
mRNA was extracted from selected A. flavus isolates treated with nanoparticles using mRNA Isolation Kit (Roche Applied Science).
Quantitative Real Time-PCR (qRT-PCR)
A Step One Real-Time PCR System (Applied Biosystem, USA) was used to assess the copy of the cDNA of A.flavus strain treated with AgNps HA1N, AgNps HA2N and AgNps EH, to detect the expression values of the tested genes according to El-Baz et al. [25].
Statistical analysis
All data were statistically analysed using the General Linear Model procedure of the SPSS ver. 18 (IBM Corp, NY).The significance of the differences among treatment groups was determined by Waller–Duncan k-ratio [26]. All statements of significance were based on the probability of P < 0.05.
Results
The present study was undertaken to investigate the antifungal activity of silver nanoparticles on A. flavus in vitro study. The addition of AgNPs including (AgNps HA1N, AgNps HA2 N and AgNps EH) individually to the (YES) growth medium at a level of 0.5, 1.0 and 1.5 (mg/100ml). The percentages of AFB1 reduction with AgNPs HA1N were 22.8, 50.7 and 88.2% after treating by 0.5, 1 and 1.5 mg AgNPs /100mL medium, respectively. On the other hand, in case of AgNps HA2N AFB1 reduced to 13.3, 37.3 and 67.7%, while AgNps EH reduced AFB1to 22.1, 42.9 and 83.5% as (Fig. 1).
Figure 1.

The percentages of inhibition of AFB1 production by Aspergillus flavus ATCC 28542 in YES medium
The results have also indicated that the AgNps HA1N at concentration 1.5 mg/100 mL medium gave the highest reduction of AFB1 were determined by HPLC (Fig. 2).
Figure 2.
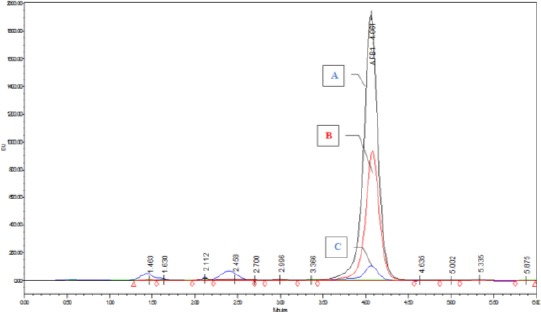
HPLC chromatogram of AFB1 with different concentration of AgNPs HA1N. A = AF; B1 treated by 0.5 mg AgNp HA1N/100 ml media; B = AFB1 treated by1 mg media AgNp HA1N/100 ml media; C = AFB1 treated by 1.5 mg AgNp HA1N/100 ml media
The obtained data in (Table 2) showed the AFB1 concentrations in YES medium treated with three types of AgNPs, which showed the significant differences between the different concentrations as well as between the types of AgNPs (Table 3). On the other hand, data in (Table 4) display the effect of AgNPs on mycelial production from A. flavus ATCC 28542. The reduction on fungal growth and AFB1 creation was dependent on the concentration of AgNPs. The antifungal activity of AgNPs was demonstrated by the diminution of AFB1 production by A. flavus. This reduction is due to the interfering with growth and fungal proliferation that AgNPs are altering protein activity and leads to cell death.
Table 2.
Effect of AgNPs on AFB1 produced by Aspergillus flavus ATCC 28542
| Concentration of AgNPs (mg)/100ml media | AFB1 (µg/100 ml media)* | Mean for concentration | ||
|---|---|---|---|---|
| AgNP HA1N | AgNP HA2N | AgNP EH | ||
| 0.5 | 0.826 ± 0.051 | 0.928 ± 0.041 | 0.833 ± 0.044 | 0.863 ± 0.063c |
| 1.0 | 0.602 ± 0.010 | 0.746 ± 0.029 | 0.685 ± 0.018 | 0.677 ± 0.065b |
| 1.5 | 0.126 ± 0.054 | 0.346 ± 0.036 | 0.176 ± 0.058 | 0.216 ± 0.109a |
| Mean for type AgNPs | 0.518 ± 0.312a | 0.673 ± 0.259c | 0.564 ± 0.301b | |
Control = 1.07 ± 0.11;
Mean ± SD.
Table 3.
Analysis of variance of the effect of type and different concentration of AgNPs on AFB1 produced by Aspergillus flavus ATCC 28542
| Source | SS | df | MS | F | P |
|---|---|---|---|---|---|
| Intercept | 9.258 | 1 | 9.258 | 5423.688 | 0.000 |
| Type Nano | 0.114 | 2 | 0.057 | 33.377 | 0.000 |
| Con. Nan | 1.996 | 2 | 0.998 | 584.762 | 0.000 |
| Type Nano x Con. Nan | 0.016 | 4 | 0.004 | 2.376 | 0.091 |
| Error | 0.031 | 18 | 0.002 | ||
| Total | 11.415 | 27 |
SS - the sum of squares; df - degree of freedom; MS - mean square; P - probability at confidence 0.95.
Table 4.
Effect of AgNPs on mycelial production from Aspergillus flavus ATCC 28542
| Concentration of AgNPs (mg)/100ml media | The weight of mycelial production (mg/ml)* | ||
|---|---|---|---|
| AgNP HA1N | AgNP HA2N | AgNP EH | |
| 0.5 | 11.7±1.25 | 17.2±0.76 | 14.5±0.5 |
| 1.0 | 8.5±1.32 | 13.2±0.77 | 10.6±1.27 |
| 1.5 | 4.4±1.15 | 8.6±0.87 | 6.5±0.51 |
Control=25mg/ml; Mean ±SD
This action may be attributed to the associated with the cation of silver, and it’s the soluble complexes [27] [28] [29] [30]. With the result of Ionic silver (Ag+) binding to the thiol groups in NADPH enzyme, and hamper the bacterial respiratory chain creating interactive oxygen species that cause oxidative stress and cell damage [30].
Some previous research suggests that the metabolic activity of fungi is also overlapped with AgNPs by decreasing the mycotoxins production, cytotoxicity and organic acid production. Moreover, a change in the extracellular enzyme profile is observed [31]. The antimicrobial effects of AgNPs depend on their size and silver rate release ion, and also the decrease of the size of AgNPs increased the antimicrobial activity [32] [33] [34].
The obtained data recorded in Fig. 3 showed that the bands of the omt_A gene fragments which could be visualised at 300 bp. The patterns showed in all treatments isolates indicating that the presence of this structure gene, omt-A, enclosed in aflatoxin biosynthetic pathway which regulates the production of aflatoxin aflR gene [16].The results of the gene expression analysis using quantitative real-time RT-PCR are summarised in (Fig. 4). The gene encoding AFB1 was determined in all isolates of A. flavus (ATCC 28542) after treatment with three types of AgNPs at 1.5 mg/100 ml media 6a AgNps HA1N, 1C AgNps EH, and 3c AgNps HA2N.
Figure 3.
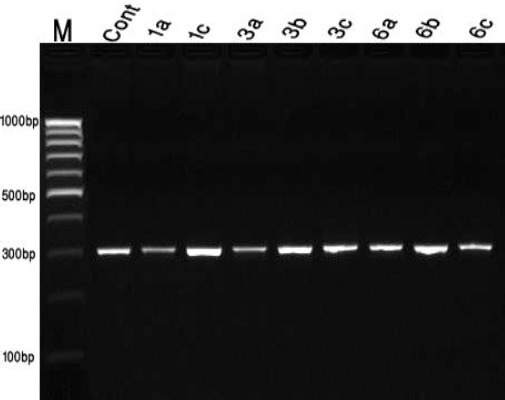
Sensitivity of PCR assay using an omt-A primer of A. flavus. DNA as template Lane1 M, Thermo Scientific GeneRuler 100bp DNA Ladder; lane 2 to lane8 isolates treated with nanoparticles
Figure 4.
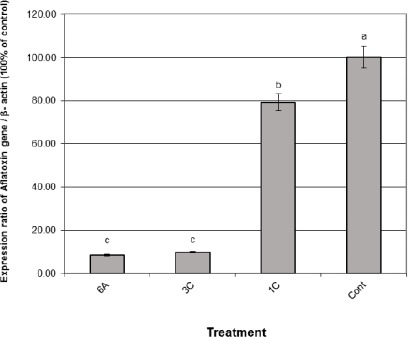
Expression levels of AFB1 gene in A. flavus isolates treated with 1.5 mg /100ml medium AgNPs of 6a AgNps HA1N, 3C AgNps EH and 1C AgNps HA2N. Data are presented as mean ± SEM a,b,c followed by different superscripts are significantly different (P ≤ 0.05)
Discussion
The results found that samples of A. flavus treated by AgNps HA2N (1C) were very lower expression levels of AFB1 gene than those collected from AgNps HA1N (6A) treatment and AgNps HA2N (3C). In control sample (without treated) isolate increased significantly (P < 0.01) the expression levels of AFB1 gene with highest up-regulation action of the gene compared with the other treatments.
Moreover, samples 6a (AgNPs HA1N) aflatoxin gene compared with 1C (AgNps HA2N) and 3C (AgNps EH) samples. The RT-PCR experiment elucidated that the AgNpS consolidated expression of aflatoxins pathway omt-A gene [35]. On the other hand, Jing et al. [36] found that the treatment of AgNps significantly decreased the secretion of AF from A. flavus. Also, they explained the mechanisms of AgNps could cause the depression of AF production by A. flavus.
In conclusion, based on the results of the present study, AgNPs inhibit growth and AFB1 produced by Aspergillus flavus ATCC28542. This was confirmed through RT-PCR approach showing the effect of AgNPs on omt-A gene involved in aflatoxin biosynthesis.
Footnotes
Funding: This research did not receive any financial support
Competing Interests: The authors have declared that no competing interests exist
References
- 1.Diaz GJ, Murcia HW, Cepeda SM. Cytochrome P450 enzymes involved in the metabolism of aflatoxin B1 in chickens and quail. Poult Sci. 2010;89:2461–2469. doi: 10.3382/ps.2010-00864. https://doi.org/10.3382/ps.2010-00864. PMid:20952710. [DOI] [PubMed] [Google Scholar]
- 2.IARC. Some traditional herbal medicines, some mycotoxins, naphthalene and styrene. IARC Monographs Evaluation of Carcinogenic Risks to Humans. 2002;82:1–556. PMid:12687954. PMCid: PMC4781602. [PMC free article] [PubMed] [Google Scholar]
- 3.Lu X, Hu B, Shao L, Tian Y, Jin T, Jin Y, Ji S, Fan X. Integrated analysis of transcriptomics and metabonomics profiles in aflatoxin B1-induced hepatotoxicity in rat. Food Chem Toxicol. 2013;55:444–455. doi: 10.1016/j.fct.2013.01.020. https://doi.org/10.1016/j.fct.2013.01.020. PMid:23385219. [DOI] [PubMed] [Google Scholar]
- 4.Groopman JD, Kensler TW. Role of metabolism and viruses in aflatoxin-induced liver cancer. Toxicol Appl Pharmacol. 2005;206:131–137. doi: 10.1016/j.taap.2004.09.020. https://doi.org/10.1016/j.taap.2004.09.020. PMid:15967201. [DOI] [PubMed] [Google Scholar]
- 5.Golli-Bennour EE, Kouidhi B, Bouslimi A, Abid-Essefi S, Hassen W, Bacha H. Cytotoxicity and genotoxicity induced by aflatoxin B1, ochratoxin A, and their combination in cultured Vero cells. J Biochem Mol Toxicol. 2010;24:42–50. doi: 10.1002/jbt.20310. https://doi.org/10.1002/jbt.20310. PMid:20175139. [DOI] [PubMed] [Google Scholar]
- 6.Deabes MM, Darwish HR, Abdel-Aziz KB, Farag IM, Nada SA, Tewfik NS. Protective effects of Lactobacillus rhamnosus GG on Aflatoxins-induced Toxicities in male Albino Mice. J Environmental & Analytical Toxicology. 2012;2:2–9. [Google Scholar]
- 7.Corcuera L, Vettorazzi A, Arbillaga L, Pérez N, Gil A G, Azqueta A, et al. Genotoxicity of Aflatoxin B1 and Ochratoxin A after simultaneous application of the in vivo micronucleus and comet assay. Food and Chemical Toxicology. 2015;76:116–12. doi: 10.1016/j.fct.2014.12.003. https://doi.org/10.1016/j.fct.2014.12.003. PMid:25530104. [DOI] [PubMed] [Google Scholar]
- 8.Meissonnier GM, Pinton P, Laffitte J, Cossalter AM, Gong YY, Wild CP, et al. Immunotoxicity of aflatoxin B1: impairment of the cell-mediated response to vaccine antigen and modulation of cytokine expression. Toxicol Appl Pharmacol. 2008;231:142–149. doi: 10.1016/j.taap.2008.04.004. https://doi.org/10.1016/j.taap.2008.04.004. PMid:18501398. [DOI] [PubMed] [Google Scholar]
- 9.Quist CF, Bounous DI, Kilburn JV, Nettles VF, Wyatt RD. The effect of dietary aflatoxin on wild turkey poults. J Wildl Dis. 2000;36:436–444. doi: 10.7589/0090-3558-36.3.436. https://doi.org/10.7589/0090-3558-36.3.436. PMid:10941727. [DOI] [PubMed] [Google Scholar]
- 10.Brown RL, Chen Z-Y, Cleveland TE, Russin JS. Advances in the development of host resistance in corn to aflatoxin contamination by Aspergillus flavus (A mini-review) Phytopathology. 1999;89:113–117. doi: 10.1094/PHYTO.1999.89.2.113. https://doi.org/10.1094/PHYTO.1999.89.2.113. PMid:18944783. [DOI] [PubMed] [Google Scholar]
- 11.Woloshuk CP, Prieto R. Genetic organization and function of the aflatoxin B1 biosynthetic genes. FEMS Microbiol Lett. 1998;160:169–176. doi: 10.1111/j.1574-6968.1998.tb12907.x. https://doi.org/10.1111/j.1574-6968.1998.tb12907.x. [DOI] [PubMed] [Google Scholar]
- 12.Yu J, Chang PK, Ehrlich KC, Cary JW, Bhatnagar D, Cleveland TE, Payne GA, Linz JE, Woloshuk CP, Bennett JW. Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol. 2004;70(3):1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. https://doi.org/10.1128/AEM.70.3.1253-1262.2004. PMid:15006741. PMCid: PMC368384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ehrlich KC, Yu J, Cotty PJ. Aflatoxin biosynthesis gene clusters and flanking regions. J Appl Microbiol. 2005;99(3):518–527. doi: 10.1111/j.1365-2672.2005.02637.x. https://doi.org/10.1111/j.1365-2672.2005.02637.x. PMid:16108793. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich KC, Montalbano BG, Cotty PJ. Sequence comparison of aflR from different Aspergillus species provides evidence for variability in regulation of aflatoxin production. Fungal Genetics and Biology. 2003;38:63–74. doi: 10.1016/s1087-1845(02)00509-1. https://doi.org/10.1016/S1087-1845(02)00509-1. [DOI] [PubMed] [Google Scholar]
- 15.Yabe K, Nakajima H. Enzyme reactions and genes in aflatoxin biosynthesis. Appl Microbiol Biotechnol. 2004;64:745–755. doi: 10.1007/s00253-004-1566-x. https://doi.org/10.1007/s00253-004-1566-x. PMid:15022028. [DOI] [PubMed] [Google Scholar]
- 16.Woloshuk CP, Foutz KR, Brewer J, Bhatnagar D, Cleveland TE, Payne GA. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl Environ Microbiol. 1994;60:2408–2414. doi: 10.1128/aem.60.7.2408-2414.1994. PMid:8074521. PMCid: PMC201664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmes RA, Boston RS, Payne GA. Diverse inhibitors of aflatoxin biosynthesis. Appl Microbiol Biotechnol. 2008;78:559–572. doi: 10.1007/s00253-008-1362-0. https://doi.org/10.1007/s00253-008-1362-0. PMid:18246345. [DOI] [PubMed] [Google Scholar]
- 18.Otari SV, Patil RM, Ghosh SJ, Thorat ND, Pawar SH. Intracellular synthesis of silver nanoparticle by actinobacteria and its antimicrobial activity. Spectrochim Acta A Mol Biomol Spectrosc. 2015;136:1175–1180. doi: 10.1016/j.saa.2014.10.003. https://doi.org/10.1016/j.saa.2014.10.003. PMid:25456659. [DOI] [PubMed] [Google Scholar]
- 19.Yassin MA, El-Samawaty AMA, Dawoud TM, Abd-Elkader OA, Al Maary KS, Hatamleh AA, Elgorban AM. Characterization and anti-Aspergillus flavus impact of nanoparticles synthesized by Penicillium citrinum. Journal of Biological Sciences. 2017;24:1243–1248. doi: 10.1016/j.sjbs.2016.10.004. https://doi.org/10.1016/j.sjbs.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.AOAC Official methods of analysis of AOAC international. Published by AOAC International suite 500 481 North Fredrick Avenue Gaithersburg, MARYLAND 20877-2417, USA. Chapter 49. 19th ed. Washington: Association of Official Analytical Chemists; 2012. pp. 1–125. [Google Scholar]
- 21.Ammar HAM, El-Desouky TA. Green synthesis of nanosilver particles by Aspergillus terreus HA1N and Penicillium expansum HA2N and its antifungal activity against mycotoxigenic fungi. J Appl Microbiol. 2016;121:89–100. doi: 10.1111/jam.13140. https://doi.org/10.1111/jam.13140. PMid:27002915. [DOI] [PubMed] [Google Scholar]
- 22.El-Desouky TA, Ammar HAM. Honey mediated silver nanoparticles and inhibitory effect on aflatoxins and ochratoxin A. Journal of Applied Pharmaceutical Science. 2016;6(6):83–90. https://doi.org/10.7324/JAPS.2016.60615. [Google Scholar]
- 23.Munimbazi C, Bullerman LB. Isolation and partial characterization of antifungal metabolites of Bacillus pumilus. J Appl Micro. 1998;84:959–969. doi: 10.1046/j.1365-2672.1998.00431.x. https://doi.org/10.1046/j.1365-2672.1998.00431.x. [DOI] [PubMed] [Google Scholar]
- 24.Deabes MM, Aboelsoud NH, Taha L. In vitro Inhibition of growth and aflatoxin B1 production of Aspergillus flavus strain (ATCC 16872) by various medicinal plant essential oils. Macedonian J Medical Sciences. 2011;4(4):345–350. https://doi.org/10.3889/MJMS.1857-5773.2011.0190. [Google Scholar]
- 25.El-Baz FK, Wagdy KB, Hanan FA, Hoda FB. Berry extracts improved inflammatory cytokines, antioxidant enzyme and suppressed the gene expression alterations in diabetic rats. Int J Pharm Sci Rev Res. 2016;38(2):219–226. https://doi.org/10.22159/ijpps.2016v8i11.14480. [Google Scholar]
- 26.Walter A, Duncan DB. Multiple range and multiple test. Biometries. 1969;11:1–24. [Google Scholar]
- 27.Chen M, Yan L, He H, Chang Q, Yu Y, Qu J. Catalytic Sterilization of Escherichia coli K12 on Ag/Al2O3 Surface. J Inorg Biochem. 2007;101:817–823. doi: 10.1016/j.jinorgbio.2007.01.008. https://doi.org/10.1016/j.jinorgbio.2007.01.008. PMid:17350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carlson C, Hussain SM, Schrand AM, Braydich-Stolle LK, Hess KL, Jones RL, Schlafer JJ. Unique Cellular Interaction of Silver Nanoparticles: Size-Dependent Generation of Reactive Oxygen Species. J Phys Chem. B. 2008;112:13608–13619. doi: 10.1021/jp712087m. https://doi.org/10.1021/jp712087m. PMid:18831567. [DOI] [PubMed] [Google Scholar]
- 29.Lubick N. Nanosilver Toxicity: Ions, Nanoparticles- or Both? Environ Sci Technol. 2008;42:8617. doi: 10.1021/es8026314. https://doi.org/10.1021/es8026314. PMid:19192768. [DOI] [PubMed] [Google Scholar]
- 30.Asharani PV, Mun GLK, Hande MP, Valiyaveettil S. Cytotoxicity and Genotoxicity of Silver Nanoparticles in Human Cells. ACS Nano. 2009;3:279–290. doi: 10.1021/nn800596w. https://doi.org/10.1021/nn800596w. PMid:19236062. [DOI] [PubMed] [Google Scholar]
- 31.Logeswari P, Silambarasan S, Abraham J. Synthesis of silver nanoparticles using plant extracts and analysis of their antimicrobial activity. J Saudi Chem Soc. 2015;19:311–317. https://doi.org/10.1016/j.jscs.2012.04.007. [Google Scholar]
- 32.Morones JR, Elechiguerra JL, Camacho A, Holt K, Kouri JB, Yacaman MJ. The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16:2346–2353. doi: 10.1088/0957-4484/16/10/059. https://doi.org/10.1088/0957-4484/16/10/059. PMid:20818017. [DOI] [PubMed] [Google Scholar]
- 33.Martınez-Castanon GA, Nino-Martınez N, Martınez- Gutierrez F, Martınez-Mendoza JR, Ruiz F. Synthesis and antibacterial activity of silver nanoparticles with different sizes. J Nanopart Res. 2008;10:1343–1348. https://doi.org/10.1007/s11051-008-9428-6. [Google Scholar]
- 34.Martinez-Gutierrez F, Olive PL, et al. Synthesis, characterization, and evaluation of antimicrobial and cytotoxic effect of silver and titanium nanoparticles. Nanomedicine. 2010;6:681–688. doi: 10.1016/j.nano.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 35.Cuero R, Ouellet T, Yu J, Mogongwa N. Metal ion enhancement of fungal growth, gene expression and aflatoxin synthesis in Aspergillus flavus: RT-PCR characterization. J Appl Microbiol. 2003;94:953–961. doi: 10.1046/j.1365-2672.2003.01870.x. https://doi.org/10.1046/j.1365-2672.2003.01870.x. PMid:12752802. [DOI] [PubMed] [Google Scholar]
- 36.Jing Z, Ling W, Dan X, Zhisong L. Involvement of ROS in nanosilver-caused suppression of aflatoxin production from Aspergillus flavus. RSC Adv. 2017;7:23021–23026. https://doi.org/10.1039/C7RA02312J. [Google Scholar]


