ABSTRACT
Co-stimulatory and inhibitory receptors expressed by immune cells in the tumor microenvironment modulate the immune response and cancer progression. Their expression and regulation are still not fully characterized and a better understanding of these mechanisms is needed to improve current immunotherapies. Our previous work has identified a novel ligand/receptor pair, LLT1/CD161, that modulates immune responses. Here, we extensively characterize its expression in non-small cell lung cancer (NSCLC). We show that LLT1 expression is restricted to germinal center (GC) B cells within tertiary lymphoid structures (TLS), representing a new hallmark of the presence of active TLS in the tumor microenvironment. CD161-expressing immune cells are found at the vicinity of these structures, with a global enrichment of NSCLC tumors in CD161+ CD4+ and CD8+ T cells as compared to normal distant lung and peripheral blood. CD161+ CD4+ T cells are more activated and produce Th1-cytokines at a higher frequency than their matched CD161-negative counterparts. Interestingly, CD161+ CD4+ T cells highly express OX40 co-stimulatory receptor, less frequently 4-1BB, and display an activated but not completely exhausted PD-1-positive Tim-3-negative phenotype. Finally, a meta-analysis revealed a positive association of CLEC2D (coding for LLT1) and KLRB1 (coding for CD161) gene expression with favorable outcome in NSCLC, independently of the size of T and B cell infiltrates. These data are consistent with a positive impact of LLT1/CD161 on NSCLC patient survival, and make CD161-expressing CD4+ T cells ideal candidates for efficient anti-tumor recall responses.
KEYWORDS: LLT1, CD161, non-small cell lung cancer, tertiary lymphoid structures, germinal center, tumor-infiltrating lymphocytes;, co-stimulatory receptor, th1 response, immune checkpoints
Introduction
Cancer progression is associated with defective immunosurveillance by innate and adaptive immune cells. 1 However, numerous studies have demonstrated over the past decades that high densities of immune cells within the tumor microenvironment, especially T cells and B cells, correlate with increased patient survival.2-4 Besides their density, the organization of these tumor-infiltrating immune cells has also been shown to strongly determine clinical outcome.5 Based on a better understanding of the molecular mechanisms underlying T-cell activation and inhibition,6 recent immunotherapies reprogramming the anti-tumoral immune responses with immune check-point (ICP) blockers have yielded durable positive results.7 New strategies combining ICP blockers with agonistic antibodies for co-stimulatory receptors are also under investigation.8 The study of the immune contexture thus provides an informative way to predict prognosis and responses to treatments.9
Our team and others previously identified LLT1 as the ligand of CD161,10,11 a C-type lectin-like receptor expressed on the majority of Natural Killer (NK) cells and on T cell subsets.12 We found that LLT1/CD161 interaction inhibits NK cell functions and co-stimulates T cell proliferation, IFN-γ- and Il-17-secretion.10,13 Using specific anti-LLT1 monoclonal antibodies (mAb),14 we characterized LLT1 expression profile which is restricted to hematopoietic cells.13 LLT1 is not expressed at the cell surface of resting peripheral blood mononuclear cells (PBMCs) but can be transiently expressed on activated B cells, dendritic cells (DCs), T cells and NK cells with a synergic effect of IFN-γ.13 This indicates that LLT1 up-regulation is preferentially triggered by a T helper 1 (Th1)-polarizing environment, and suggests that LLT1/CD161 interaction may modulate Th1-adaptive immune responses. Importantly, we reported high expression of LLT1 on germinal center (GC) B cells in tonsils and lymph nodes (LN) and a sustained expression in GC-derived B cell lymphomas.15
In the tumor microenvironment of Non-Small Cell Lung Cancer (NSCLC), we previously identified the presence of tertiary lymphoid structures (TLS) that display the same organization as Secondary Lymphoid Organs (SLOs).16,17 TLS are composed of a T cell zone with mature DCs, and of a B cell zone characterized by a mantle of naive B cells surrounding a germinal center (GC) with proliferating B cells, follicular dendritic cells (FDCs) and follicular T helper cells (Tfh).18 We hypothesized that like in SLOs, LLT1 may be expressed within tumor-associated TLS, on GC-B cells. Because a high density of TLS is associated with improved patient survival5,16,17 and has been shown to signal a Th1 immune contexture,19 we hypothesized that the ligand/receptor pair LLT1/CD161 could represent an additional important immune regulation mechanism. A recent cancer wide analysis of prognostic gene signatures, identifying KLRB1 coding for CD161 receptor as the gene most frequently associated with favorable outcomes, further supports that hypothesis.20
We thus undertook a thorough analysis of LLT1 and CD161 expression in NSCLC. We report that like in SLOs, LLT1 is prominently expressed on the cell surface of GC-B cells within TLS. No expression was detected on tumor cells, neither on adjacent non-tumoral lung tissue. We also found that lung tumors are highly infiltrated by CD161-expressing CD4+ and CD8+ T cells displaying an effector-memory (EM) phenotype. The CD161+ CD4+ tumor infiltrating lymphocytes (TILs) express less FoxP3, are more prone to produce Th1 cytokines, and are more activated and less exhausted than their matched CD161-negative counterparts. CD161 expression on CD4+ TILs parallels OX40 co-stimulatory receptor expression, which suggests that CD161 could similarly play a role in favoring rapid antigen recall responses. Lastly, we found CLEC2D (LLT1) and KLRB1 (CD161) gene expression associated with a favorable outcome in NSCLC. All together, these findings document that LLT1/CD161 interaction may actively participate to the anti-tumoral immune response.
Results
Expression of LLT1 and its receptor CD161 in NSCLC primary tumors
We first investigated the expression of LLT1 and CD161 in tumor samples from untreated NSCLC patients by immunohistochemistry (IHC) (Fig. 1 and Sup Fig. 1). We observed the presence of LLT1+ cells organized in follicles at the invasive margin (Fig. 1A, 1B) and LLT1+ cells within NSCLC tumor stroma (Fig. 1A, 1C). No LLT1 staining could be observed in tumor cells (Fig. 1C), nor in the adjacent non-tumoral lung tissue (Fig. 1D), underlying that LLT1 is specifically expressed in immune cells within the tumor microenvironment. We also looked at LLT1 expression in lung tissue sections from hyper pulmonary arterial pressure disease, another highly inflammatory lung pathology, and we did not detect any LLT1-expressing cells (Sup Fig. 1C, 1D).
Figure 1.

Expression of LLT1 and CD161 in NSCLC tumors. (A-H) Hematoxylin (HE) counterstained IHC stainings of (A-D) FFPE and (E-H) frozen sections of two representative tumors from untreated NSCLC patients using (A-D) anti-human LLT1 clone 2F1 and (E-H) anti-human CD161 clone DX12. (B-D) represent higher magnifications (x100) of areas (black rectangles) in (A) (magnification x10). (F-H) represent higher magnifications (x100) of areas (black rectangles) in (E) (magnification x10). (A-D) Solid adenocarcinoma (ADC) subtype. (E-H) Lepidic ADC subtype. Str, Stroma; Tu, Tumor Nests.
Similarly, we observed the presence of CD161+ cells within NSCLC tumor stroma (Fig. 1E, 1G), and at the vicinity of lymphoid aggregates (Fig. 1E, 1F). But by contrast to LLT1, CD161 expression was also detected within adjacent non-tumoral lung tissue (Fig. 1H).
These results highlight the presence of CD161-expressing cells within the lung and identify LLT1 expression as being restricted to the tumor microenvironment.
LLT1 is predominantly expressed on GC-B cells within NSCLC-associated TLS
We next characterized LLT1 expression within the tumor microenvironment. As depicted in Fig. 1A and 1B, a strong labeling was detected in cells organized in follicles. On serial sections of tumors from untreated NSCLC patients, we showed that LLT1+ cells (Fig. 2A) are part of a CD20+ B-cell follicle, characterized by a GC of proliferating Ki67+ B cells (Fig. 2B) and a network of CD21+ FDCs (Fig. 2C), hallmarks of TLS. A moderate positive correlation could be observed between the number of LLT1+ follicles and the number of CD21+ follicles in a cohort of 32 tumors from untreated NSCLC patients (Fig. 2D), indicating that LLT1 can be used as a marker of active TLS-derived GC-B cells, in addition to CD21 marker. We also stained tumor sections from 20 NSCLC patients treated with neo-adjuvant chemotherapy and found similar expression in active TLS (Sup Fig. 1E–1I). This pattern of expression resembles what can be observed in SLOs, with LLT1 (Sup Fig. 2A, 2B) co-localizing with CD21+ (Sup Fig. 2C) and CD20+ Ki67+ (Sup Fig. 2D) cells within GC.
Figure 2.
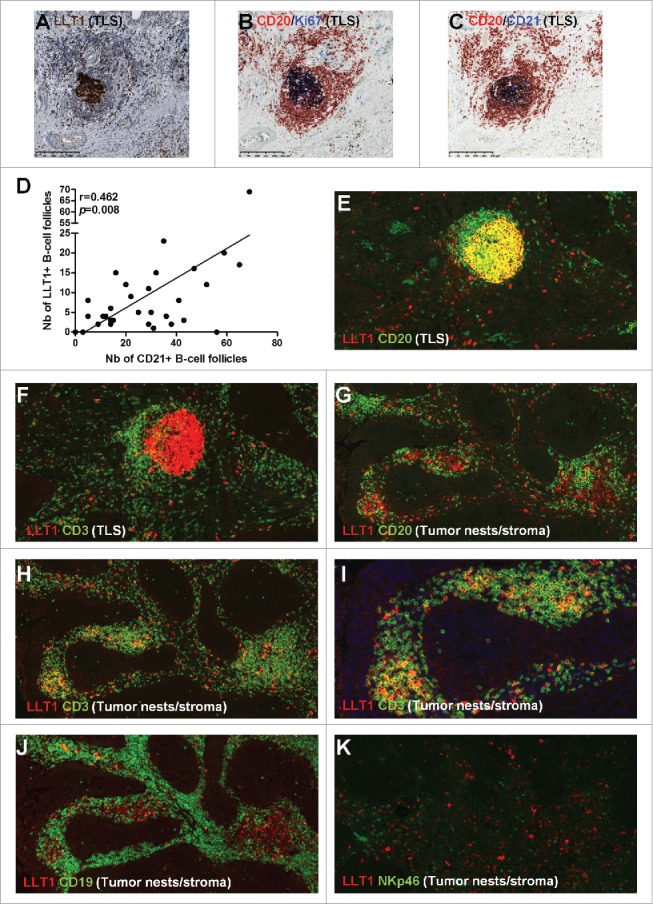
LLT1co-localizes with GC-B cells within NSCLC-associated TLS and with CD3+ (NKp46−) T cells within tumor stroma. (A) HE counterstained IHC staining of a FFPE section of tumor from untreated NSCLC patient using anti-human LLT1 clone 2F1, with a focus on a TLS at high magnification (x100). (B,C) represent the same TLS on serial sections stained with anti-human CD20 and anti-human Ki67 (B) or anti-human CD20 and anti-human CD21 (C). (D) Correlation between the number of LLT1+ follicles and the number of CD21+ follicles in 32 tumors from untreated NSCLC patients. Linear regression curve, spearman r value and p value (95% confidence interval) are shown. (E-K) Double IF stainings of FFPE sections from NSCLC tumors (magnification x200) using anti-human LLT1 clone 2F1 associated with anti-human CD20 (E, G), anti-human CD3 (F, H), anti-human CD19 (J), or anti-human NKp46 (K). (E, F) represent the same TLS on serial sections. (G-J) represent the same area of tumor stroma on serial sections. K represents another area of NSCLC tumor stroma.
By immunofluorescence (IF) on FFPE NSCLC sections (Fig. 2 and Sup Fig. 3), LLT1 mainly co-localized with CD20+ cells within GC of TLS (Fig. 2E), similarly to what was observed in tonsil (Sup Fig. 2E and in a previous report15). LLT1 was also found expressed on scattered cells around GC (Fig. 2E, 2F and Sup Fig. 2F) and in non-TLS areas, within tumor stroma (Fig. 2G–K), mostly in CD3+ T cells (Fig. 2F, 2H, 2I and Sup Fig. 2F), and to a lesser extend in CD20+ and/or CD19+ B cells (Fig. 2G, 2J). No colocalization of LLT1 could be observed with NKp46+ NK cells (Fig. 2K), neither with DC-LAMP+ mature DCs (Sup Fig. 3E) nor with CD68+ macrophages (Sup Fig. 3F).
By multiparametric flow cytometry (Sup Fig. 4), we selectively detected cell surface expression of LLT1 on NSCLC tumor-infiltrating CD19+ cells, more precisely on a subset of CD38+ GC/pre-GC B cells (Fig. 3A).21 This was in agreement with the staining of LN cells (Sup Fig. 2G), tonsils15 and with IHC stainings. Importantly, in 6 out of 10 NSCLC patients, the frequency of LLT1-expressing GC/pre-GC B cells was significantly increased compared to matched distant lung (Fig. 3B). These data demonstrate a specific enrichment of LLT1+ CD38+ GC/pre-GC B cells within the tumor microenvironment. By contrast, no expression of LLT1 was detected on the cell surface of other B cell subsets, T cells, myeloid cells or plasmacytoid dendritic cells (pDCs) (Fig. 3A, 3C) or in blood (data not shown).
Figure 3.

LLT1 is expressed on GC-B cells in NSCLC tumors. (A) Representative dot plots of LLT1 expression by flow cytometry on all B cells and B cell subsets in two different NSCLC tumors (upper and lower panels), using anti-human LLT1 mAb clone 4F68 and isotype control. Percentages of cells expressing LLT1 among each B cell subset in NSCLC tumors are indicated, n = 10. (B) Representative dot plots of LLT1 expression on GC/pre-GC B cells in a NSCLC tumor and matched non-tumoral distant lung, with percentages of cells expressing LLT1 among GC/pre-GC B cells in each compartment, n = 10. (C) Representative dot plots of LLT1 expression on different cell subsets in a NSCLC tumor, with percentages of cells expressing LLT1 among each cell subset in NSCLC tumors, n = 10. Mean percentages are indicated by red horizontal lines. P values were calculated using (A, C), One-way Anova, Kruskal-Wallis test, Dunn's Multiple Comparison Test and (B), unpaired t test. *p<0.05, **p<0.005.
Lung tumors are infiltrated with CD161 expressing CD4+ and CD8+ T cells accumulating within tumor stroma at the vicinity of TLS
As for LLT1, we thoroughly characterized CD161 expression in the tumor microenvironment of NSCLC patients. IHC staining indicated that CD161 was expressed in the stroma and at the vicinity of lymphoid aggregates (Fig. 1F–1G). By IF on serial sections, we confirmed CD161 expression in the T cell zone of the TLS (Fig. 4A–4G, 4I–4J), at the periphery of LLT1-expressing GC-B cells (Fig. 4F–4G), and within tumor stroma (Fig. 4H). CD161 expression was detected mostly on CD3+ T cells (Fig. 4D–4E, 4H–4J), and not on CD20+ B cells (Fig. 4C, 4G, 4I–4J), consistently with stainings of LN sections (Sup Fig. 5A–5B). No expression of CD161 was observed on CD21+ FDCs within TLS (Sup Fig. 5C–5F) or tonsils (Sup Fig. 5G), whatever the mAb clone used.
Figure 4.
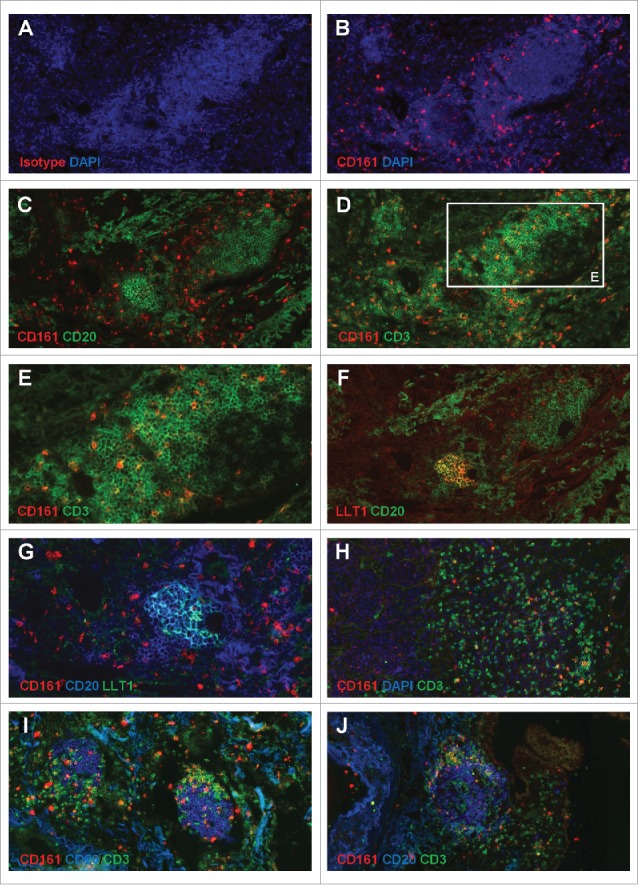
Accumulation of CD161-expressing CD3+ T cells within NSCLC tumor stroma and at the vicinity of TLS. (A-B) IF staining of a frozen section from NSCLC tumor using anti-human CD161 clone DX12 (B) or isotype control (A) associated with DAPI. (C-E, H) Double IF stainings of serial frozen sections from the same NSCLC tumor using anti-human CD161 clone DX12 associated with (C) anti-human CD20, (D-E, H) anti-human CD3, and (H) DAPI. (E) represents a higher magnification (x400) of area (white rectangle) in (D). (F) Double IF staining of a serial frozen section from the same NSCLC tumor using anti-human LLT1 clone 2F1 associated with anti-human CD20. (G) ImageJ-processed overlay of LLT1 IF staining from (F) and CD161/CD20 double IF staining from (C), at high magnification. (I-J) Triple IF stainings of frozen sections from two NSCLC tumors using anti-human CD161 clone DX12 associated with anti-human CD20 and anti-human CD3. (A-G, I-J) focus on TLS. (H) focus on tumor stroma and tumor nests. (A-D, F, H-J) magnification x200. (E, G) magnification x400.
Flow cytometry stainings showed that CD161 was expressed at the cell surface of a proportion of T cells and of the vast majority of NK cells in NSCLC patients (Sup Fig. 4 and Sup Fig. 6A). However, their distribution strongly varied with the anatomical site (Fig. 5A). In the blood of patients and healthy donors (HD), as well as in distant lung in NSCLC patients, NK cells encompassed about 50% of the CD161+ cells, followed by CD3+ CD4+ T cells (about 30%) and CD3+ CD8+ T cells (15–20%). In contrast, CD3+ CD4+ T cells were highly represented in NSCLC tumors and LN (48.7±3.1% and 79.5±7.9%, respectively), followed by CD3+CD8+ T cells (26.5±3.9% and 4±0.9%, respectively) and NK cells (18±2.6% and 11.3±6.4%, respectively). In each compartment, a small percentage of cells were CD3+ CD4− CD8−, accounting for TCR-γδ T cells (2.8±1.4% in tumor, n = 7, data not shown) and CD3− CD56− cells (3.3±0.5% in tumor), most likely being related to innate lymphoid cells (ILCs). Overall, these data identified a higher frequency of CD4+ and CD8+ T cells and a decreased frequency of NK cells among the CD161+ cell population in tumors compared to distant lung and blood.
Figure 5.
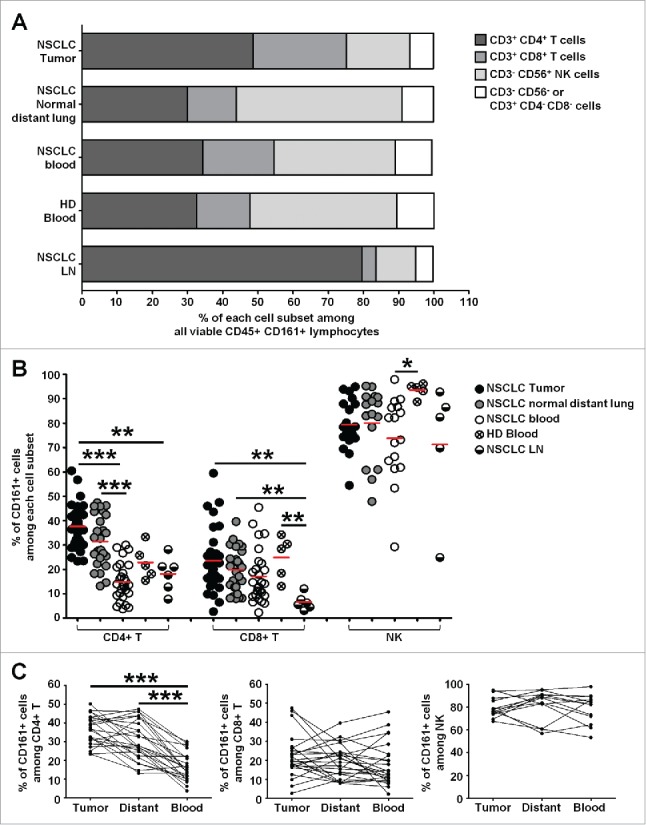
High frequencies of CD161-expressing CD4+ T cells in NSCLC tumors. (A) Histogram representing the mean percentages of each cell subset among viable CD45+ CD161+ lymphocytes, in each compartment. (B) Histogram representing the percentages of cells expressing CD161 among each cell subset, in each compartment, n = 33. Means are indicated by red horizontal lines. P values were calculated using One-way anova/Kruskal-Wallis/Dunn's test. (C) Graphs representing the percentages of cells expressing CD161 among each cell subset, in each compartment, in matched patients, n = 24. P values were calculated with One-way anova/Friedman/Dunn's test. *p<0.05, **p<0.005, ***p<0.001.
Interestingly, the frequency of CD161+ cells within CD4+ T cells was increased by two-fold in tumors compared to LN and blood from patients and HD. An increased frequency of CD161-expressing CD4+ T cells was also observed in tumor as compared to distant lung, although not significant (Fig. 5B, 5C). These TILs were mostly conventional T cells as we found a significantly lower frequency of FoxP3+ Tregs among CD161+ CD4+ TILs, together with lower frequencies of CD161+ cells among Tregs (Sup Fig. 7A, B). This is consistent with higher frequencies of CD127+ and lower frequencies of CD25+ cells among CD161+ CD4+ TILs (Sup Fig. 7C). PD-1bright CXCR5bright Tfh are CD4+ T cells located within GC, in close interaction with GC-B cells. In both NSCLC tumors and LN, we detected very low frequencies of CD161+ Tfh compared to non-Tfh (Sup Fig. 7F), in agreement with previous reports.15,22 This result correlated with IF staining showing no co-localization of PD1 expressed by Tfh and CD161 within TLS (Sup Fig. 7D–7E). These data suggest that the increased frequency of CD161+ TILs is more frequently associated with conventional CD4+ T cells.
The frequency of CD161+ cells within CD8+ T cells was also increased but moderately in tumors compared to distant lung and blood (Fig. 5B, 5C). The level of CD161 expression on CD8+ T cells is not homogenous and CD161bright cells have been described as Mucosal-Associated Invariant T (MAIT) cells.23 We therefore separately analyzed these CD8+ T cell populations and found a significant increase of the frequency of CD161+ cells between tumors and blood or LN when looking at CD161+/dim CD8+ T cells (Sup Fig. 6B–C). Besides, we found no influence of gender, histological subtype, chronic obstructive pulmonary disease (COPD) status, age and smoking exposure on the proportion of CD161+ cells within CD4+ T cells (Sup Fig. 8) or within CD8+ T cells (data not shown) in NSCLC tumors, distant lung and blood. A non-significant difference was however seen between late tumor stage III and earlier stages I and II. Moreover, a large majority of NK cells expressed CD161 in NSCLC patients (Fig. 5B, 5C), but the frequency of CD161+ cells within NK cells was significantly higher in blood of HD compared to patient's blood (Fig. 5B).
CD161 defines functional heterogeneity within T cells in the tumor microenvironment
To further characterize CD161-expressing T cells, we identified the genes most positively and negatively associated with KLRB1 encoding for CD161 within highly purified CD4+ and CD8+ TILs.
In CD4+ TILs (Fig. 6A), the genes most positively and significantly (R>0.6; FDR<0.05) correlated with KLRB1 were CCR2, RPS6, GZMA, CCL4 and CD96. Other genes such as GZMH or PPARG were also highly positively and significantly correlated with KLRB1 but they were expressed at lower levels within CD4+ TILs (Sup Fig. 9A). While CCR2 expression has been characterized has a marker of a stable population of EM CD4+ T cells,24 CCL4 production has been shown to be associated with a Th1 phenotype.25 RPS6 is part of activation pathways downstream TCR signaling,26 and CD96, also known as Tactile (T cell activation, increased late expression) plays a role in the adhesion of activated T and NK cells to their target cells and may also function in antigen presentation.27,28 KLRB1 gene was also positively and significantly (R>0.5; FDR<0.10) correlated with CTSH, ICAM3, CCND3, CYFIP2, AMICA1, DUSP6, CD81 and TNFSF8. ICAM3, AMICA1 (also known as JAML) and CYFIP2 are all three adhesion molecules playing a role in T cell-APC (antigen-presenting cells) interactions and/or leukocyte migration.29-31 CD81 on T cells associates with CD4 or CD8 and provides a co-stimulatory signal with CD3.32 TNFSF8, encodes for CD30L (CD153), which is expressed on activated T cells and acts as a co-stimulatory receptor.33 Among other genes positively correlated with KLRB1 (R>0.5; 0.10<FDR<0.15) we also found NUP107, ITGA6, SIGIRR, CD6, CD53 and CD40LG. Like CD96, CD6 is a cell adhesion molecule with co-stimulatory properties.34 SIGIRR (Single immunoglobulin and toll-interleukin 1 receptor (TIR) domain) is a negative regulator of IL-1 receptor, and was suggested to play a major role in suppression of Th17 differentiation.35,36 When considering all the genes positively correlated with KLRB1 (FDR<0.15), Cluego analysis37 identified several pathways associated with mononuclear cell migration or extravasation, and also with the immunological synapse (Fig. 6C). When genes negatively correlated with KLRB1 (FDR<0.15) were considered, Cluego analysis highlighted pathways linked to apoptosis (Fig. 6D).
Figure 6.

Heterogeneity of CD161-expressing CD4+ and CD8+ T cells within NSCLC tumors. (A-B) Correlation factors between KLRB1 gene expression and expression of genes from the “PanCancer Immune Profiling Panel” (NanoString Technologies) in highly purified intra-tumoral CD4+ T cells (A) or CD8+ T cells (B). Genes significantly correlated with KLRB1 (FDR < 0.05, 0.05 < FDR < 0.10, and up to 0.10 < FDR < 0.15) are shown. Vertical dashed lines represent limits for correlation factors <-0.5 and >+0.5. (C-D) Potential associations between genes positively (C) and negatively (D) correlated with KLRB1 in CD4+ T cells (FDR < 0.15) with corresponding pathways determined using ClueGo software.
Regarding CD8 TILs (Fig. 6B), only few genes were positively correlated with KLRB1 gene, including CCL20, NT5E, IL17A and NCR1. Positive correlation between KLRB1 gene and IL-17A gene but also CCL20 would suggest the presence within the tumor microenvironment of previously described CD161-expressing IL-17-producing CD8 T cells.38 However, the low number of genes positively or negatively correlated with KLRB1 in CD8 TILs did not allow ClueGo analysis to highlight any functional pathways.
Overall, these gene expression profiles revealed a link between CD161 and T-cell activation, co-stimulation and differentiation, and strongly suggest that CD161 may actively participate to tumor immunosurveillance.
Tumor infiltrating CD161+ T cells display an effector-memory phenotype and predominantly secrete IFN-γ
Using CD45RA and CCR7 cell surface markers (Sup Fig. 10A), we found that most CD161-expressing CD4+ and CD8+ T cells displayed an EM phenotype within tumor and non-tumoral distant lung, a feature shared with their CD161-negative counterparts (Fig. 7A). Besides, CD4+ T cells also displayed a central-memory (CM) phenotype in both tumor and distant lung, while their frequency was much higher in the blood of patients and HD. Regarding CD8+ CD161+ and – T cells, in addition to EM, a small proportion of cells displayed an effector phenotype (EMRA) in tumor, and this subset was even more represented in normal distant lung and in the blood of patients and HD. Of note, whatever the considered anatomical site, extremely low frequencies of naive cells could be observed among the CD161-expressing CD4+ and CD8+ T cells as compared to their CD161-negative counterparts. In agreement with this finding, we detected lower frequencies of CD161+ cells among CD62L-expressing CD4+ T cells, which include naive and CM cells, as compared to CD62L-negative CD4+ T cells, which include EM and effector cells (Sup Fig. 10B).
Figure 7.

Higher frequencies of CD161-expressing central-memory CD4+ T cells in NSCLC tumors as compared to distant lung and peripheral blood. (A) Rings illustrating the mean percentages of naive, effector memory (EM), central memory (CM) and effectors (EMRA) subsets within CD161− CD4+ or CD161+ CD4+ T cells (left panels), or within CD161− CD8+ or CD161+ CD8+ T cells (right panels), in each compartment, n = 12. (B) Histograms representing the percentages of CD161 expression within each CD4+ (left panel) and CD8+ (right panel) T cell subsets in each compartment, n = 12. Means are indicated by red horizontal lines. *p <0.05 (One-way anova/Kruskal-Wallis/Dunn's test). (C) Mean percentages of CD161 expression within each CD4+ (left panel) and CD8+ (right panel) T cell subsets in matched compartments, n = 11. *p<0.05, **p<0.005 (One-way anova/Friedman/Dunn's test).
Interestingly, we found a significant increase in the frequency of CD161-expressing CM CD4+ and CD8+ TILs compared to distant lung and blood (Fig. 7B, 7C), which suggests that they have been shaped by the tumor microenvironment.
To gain insights into the functions of CD161-expressing CD4+ and CD8+ T cells within NSCLC tumors, we assessed their cytotoxic potential and their cytokine secretion (Fig. 8). We detected lower frequencies of Granzyme B+ CD161+ CD4+ and CD8+ T cells, indicating decreased potential cytotoxicity. By contrast, the frequencies of IFN-γ-secreting cells and of TNF-α-secreting cells were significantly higher among CD161+ CD4+ TILs compared to their matched CD161− CD4+ TILs. No differences were observed for CD8+ T cells, and only very low frequencies of IL-17-producing CD4+ or CD8+ T cells were detected, with no significant differences between the CD161+ and the CD161− fractions (data not shown). These data support a more prominent Th1-oriented differentiation of the CD161-expressing CD4+ TILs.
Figure 8.

CD161-expressing CD4+ and CD8+ T cell phenotypes in NSCLC tumors. (A-B) Histograms represent the percentages of cells producing Granzyme B (GzB), IFN-γ or TFN-α among PMA/Ionomycin-stimulated CD161− or CD161+ CD4+ T cells (A), and CD161− or CD161+ CD8+ T cells (B) in NSCLC tumors, n = 18. (C-D) Upper heat maps illustrate the percentages of cells producing Granzyme B (GzB), IFN-γ or TFN-α among PMA/Ionomycin-stimulated CD4+ CD161− T cells and CD4+ CD161+ T cells (A) or CD8+ CD161− T cells and CD8+ CD161+ T cells (B) in NSCLC tumors, n = 18. Lower heat maps illustrate the percentages of cells expressing CD38 (n = 14), CD69 (n = 11), CD96 (n = 9), CD30L (n = 9), CD40L (n = 11), OX40 (n = 11), 4-1BB (n = 11), CD27 (n = 13), PD-1 (n = 15), Tim-3 (n = 13) or CD103 (n = 12) among CD4+ CD161− T cells and CD4+ CD161+ T cells (A), or CD8+ CD161− T cells and CD8+ CD161+ T cells (B) in NSCLC tumors. P values were calculated using Wilcoxon (paired, non-gaussian) test between CD161− and CD161+ cells. *p<0.05, **p<0.005, ***p<0.001.
CD4+ TILs expressing CD161 are more activated and not terminally exhausted
Next, we investigated whether activation markers, co-stimulatory receptors and ICP molecules could be differentially associated with CD161 expression (Fig. 8C–8D, Sup Fig. 11 and Sup Fig. 12). When evaluating activation markers, we did not observe significant differences regarding CD38 expression but found that both CD161+ CD4+ and CD161+ CD8+ T cells expressed more frequently CD69 as compared to their CD161− counterparts.
Gene expression analysis has revealed a positive association between KLRB1 gene and genes coding for CD96, CD30L and CD40L among CD4+ TILs (Fig. 6A). By flow cytometry (Fig. 8C–8D, Sup Fig. 11 and Sup Fig. 12), we also detected significantly higher frequencies of CD96 and CD30L-expressing cells in CD161+ CD4+ T cells compared to their CD161− counterparts, while no differences were found for CD40L. Of note, very low percentages of cells expressed CD30L and CD40L. CD30L and its receptor CD30 belong to the TNFR/TNF-Ligand family of TCR co-stimulating receptors. This family also includes OX40/OX40L, GITR/GITRL, 4-1BB/4-1BBL and CD27/CD70 receptor/ligand pairs, found on activated T cells and APCs.39 We detected significantly higher frequencies of OX40+ CD4+ T cells and significantly lower frequencies of CD4+ T cells expressing 4-1BB and CD27 in the CD161+ versus the CD161− fraction. These co-stimulatory receptors were not differentially expressed on CD8+ T cells. Very interestingly, significantly higher frequencies of both CD161+ CD4+ T cells and CD161+ CD8+ T cells expressed PD-1 as compared to their CD161− counterparts (Fig. 8C–8D, Sup Fig. 11 and Sup Fig. 12). In agreement with Sup Fig. 7F, PD-1-expressing CD161+ CD4+ T cells showed lower frequencies of PD-1high CXCR5high Tfh cells (data not shown), highlighting that the increased frequencies of PD-1-expressing cells is mostly observed among non-Tfh CD161+ CD4+ T cells. When analy-zing Tim-3+ cells, we found a lower frequency among CD161+ CD4+ T cells together with a higher frequency among CD161+ CD8+ T cells, compared to their respective CD161− counterparts. Finally, when looking at the residency marker CD103, significantly higher frequencies of cells were observed in the CD161+ CD8+ T cell fraction, but no differences could be observed among CD4+ T cells. Thus, while CD161+ CD8+ T cells express markers associated with T-cell exhaustion6 similarly to their CD161− counterparts, CD161+ CD4+ T cells display a phenotype which is more activated and less dysfunctional than their CD161− counterparts, suggesting a functional diversity within CD4+ TILs.
CLEC2D and KLRB1 are associated with favorable clinical outcome in NSCLC
Lastly, we assessed the clinical impact of KLRB1 and CLEC2D, respectively coding for CD161 receptor and its ligand LLT1. We processed 730 publically-available transcriptomic profiles of NSCLC tumors, originating from 8 unique studies and annotated for overall survival. By performing a meta-analysis over these individual series, we found a positive association between CLEC2D expression and favorable clinical outcome in most of the NSCLC cohorts we analyzed (p = 0.044, HR = 0.86) (Fig. 9A). In agreement with Gentles et al.,20 we also observed an association between KLRB1 expression and favorable clinical outcome in the majority of these cohorts (p = 0.016, HR = 0.88) (Fig. 9B).
Figure 9.

Correlation of CLEC2D and KLRB1 with clinical outcome. (A-B) Forest plot depicting the meta-analysis of (A) CLEC2D and (B) KLRB1 gene expression associated with better clinical outcome in different cohorts of NSCLC patients as indicated. Global p values are indicated. The width of the diamond shows the confidence intervals for each Hazard ratio. HR, Hazard Ratio. (C-D) Kaplan-Meier curves of overall survival among the GSE50081 cohort, according to CLEC2D gene expression combined with estimated B cell density (C), or KLRB1 gene expression combined with estimated cytotoxic cell density (D), after transformation of each variable into binary (high or low) variables (median cut-off value).
To exclude that the favorable prognostic value of KLRB1 could only reflect tumor infiltration by immune cell populations classically associated with anti-tumor response and favorable clinical outcome, namely CD8+ T cells and NK cells, we used the “Microenvironment Cell Populations-counter” (MCP-counter) method developed in our group,40 to determine the level of tumor infiltration by cytotoxic cells (CD8+ T cells and NK cells) and B cells, from GSE50081 transcriptomic dataset. After transformation of each variable into binary variables (high or low) according to a median cut-off value, we performed Kaplan-Meier analyses combining CLEC2D and estimated B cell density (Fig. 9C), and KLRB1 and estimated cytotoxic cell density (Fig. 9D). Results indicate that high expression of both KLRB1 and CLEC2D are significantly associated with better clinical outcome, even after accounting for tumor infiltration in cytotoxic cells or B cells (Fig. 9C, 9D and Sup Table 2), and that the prognostic value of KLRB1 and CLEC2D is thus not solely due to tumor infiltration by these immune cells.
These findings are consistent with the observed association between CD161 expression and IFN-γ production by T cells and suggest a role for LLT1/CD161 in NSCLC tumor immunosurveillance.
Discussion
By performing an exhaustive analysis of retrospective and prospective cohorts of NSCLC patients, we show here that CD161 and its ligand LLT1 are expressed in the lung tumor microenvironment. Importantly, both CLEC2D (coding for LLT1) and KLRB1 (coding for CD161) gene expressions are associated with a favorable clinical outcome in NSCLC, independently of the level of infiltration by B cells or cytotoxic cells.
We observed that LLT1 expression is restricted to immune cells within NSCLC tumors and is absent from non-tumoral distant lung and peripheral blood. LLT1 is also not expressed in HPAP samples, ruling out a sole link with inflammation of the lung. Importantly, LLT1 is expressed at the cell surface of GC-B cells within TLS. This is consistent with our previous work identifying LLT1 as a marker of GC-B cells within SLOs and of GC-derived B cell lymphomas.15 The presence of TLS in tumors has been associated with better clinical outcome and anti-tumor immune responses in a large variety of cancers.5 Our observation makes LLT1 a new hallmark of active TLS in the tumor microenvironment.
Expression of its receptor CD161 can similarly be observed in the tumor microenvironment, and also in normal lung and blood. In tumor, CD161 is found within tumor stroma and tumor nests, as well as in the T-cell area of TLS. It is not found in the GC area of TLS and is not expressed on CD21+ FDCs, as opposed to what was previously reported.22 The lack of clear co-localization in established tumors may relate to dynamic processes in which LLT1/CD161 could play a role, such as the initial cross-talk between T cells and activated B cells and/or neogenesis of TLS. Interestingly, the distribution among CD161-expressing cells differs in tumors compared to distant non-tumoral lung and peripheral blood. While NK cells are the predominant CD161+ population in distant lung and blood, CD4+ and CD8+ T cells become the most abundant CD161+ cells in tumors. In agreement with Platonova et al.,41 we report that most NK cells express CD161 and with similar frequencies in tumors, distant non-tumoral lung and peripheral blood. No clear colocalization of NK cells with LLT1-expressing GC-B cells is seen, suggesting that LLT1/CD161 interaction may not occur at this stage of the disease. This would also be consistent with Platonova et al.41 who found NK cells within NSCLC tumors, but mainly concentrated at the invasive margin of the tumor, rarely in contact with tumor cells, and never in TLS.
By contrast to NK cells expressing CD161 at all anatomical sites, we report higher frequencies of CD161-expressing CD4+ and to a lesser extent of CD161+ CD8+ T cells within NSCLC tumors, as compared to matched peripheral blood and distant non-tumoral lung. Statistical significance was reached with blood but not with distant lung which suggests that this tissue is strongly imprinted by the ongoing immune response within the tumor, as also observed in the context of COPD.42 Iliopoulou et al. similarly observed increased frequencies of CD161+ CD4+ CD56− T cells in blood and in malignant effusions of patients with breast, ovarian, lung, colon, pancreas and stomach cancers, compared to blood from HD, as well as in few breast tumors where the frequency was even higher.43 They notably found a positive correlation between the stage of the disease and the frequencies of CD161+ CD4+ CD56− T cells in the blood of cancer patients. Likewise, our study revealed a positive relationship between CD161+ CD4+ T cell frequencies in tumor, distant lung and blood of NSCLC patients and cancer progression (stages I-II versus III), even if statistical significance was not reached. This is in favor of a direct link between the frequencies of circulating/intratumoral CD161+ CD4+ T cells and disease progression.
These results lead us to speculate that the presence of TLS and the enrichment of the tumor microenvironment in CD161-expressing CD4+ and CD8+ T cells reflect an ongoing adaptive immune response. We previously proposed that engagement of CD161 on NK cells and T cells by its ligand LLT1 on APCs may favor the switch from innate to adaptive immunity, based on their capacity to inhibit NK cell cytotoxicity and at the same time to co-stimulate proliferation and IFN-γ/IL-17 production by T cells.10,13 This dual role on NK cells and T cells may be linked to the involvement of acid sphingomyelinase (aSMase), a membrane lipid hydrolase that generates ceramide by hydrolyzing sphingomyelin, in the signaling pathway downstream CD161.44,45
Because of their enrichment in tumors, the phenotype of NSCLC-infiltrating CD161+ CD4+ and CD161+ CD8+ T cells was further characterized. We observed that within NSCLC tumors, CD161-expressing T cells are predominantly EM and some are CM cells. This observation is consistent with previously published studies.43,46,47 This is also in agreement with the positive correlation between CCR2 and KLRB1 gene expression among CD4+ TILs. Indeed, CCR2 identifies a stable population of EM T cells equipped for rapid recall response, with increased memory character, more prone to proliferate, to produce cytokines such as IFN-γ and TNF-α, and expressing less CD25 and FoxP3 than CCR2− EM cells.24 CM cells and EM cells differ according to their trafficking, proliferative capacity, and cytokine profile. While EM cells express adhesion molecules facilitating their entry into non-lymphoid tissues, display lineage-specific transcriptional programs and produce appropriate cytokines upon rechallenge, CM cells traffic between the SLOs via the blood, have a great proliferative potential and produce IL-2 rather than lineage-specific cytokines upon re-challenge.48 The finding that higher frequencies of CM cells express CD161 in the tumor as compared to the distant non-tumoral lung and the peripheral blood would suggest that CD161 may play a role in the homing of CM T cells to TLS, where it would be able to encounter its ligand LLT1 on B cells. Interestingly, gene expression analysis also revealed a positive association between KLRB1 gene and several genes involved in cell adhesion and/or leukocyte migration, consistent with a previous study proposing the involvement of CD161 in the process of trans-endothelial migration.49
On the functional level, we found significantly higher frequencies of IFN-γ- and TNF-α-producing cells among the CD161+ CD4+ TILs as compared to their matched CD161− counterparts. This reflects an IL-12/Th1 oriented phenotype,19 and is consistent with the association of CLEC2D and KLRB1 with a favorable clinical outcome. Interestingly, our findings are also in agreement with the recent report by Welters et al,47 linking a better control of oropharyngeal cancer with a dense infiltration of tumors with IFN-γ/IL-17 oriented CD4+ and CD8+ CD161+ T cells. In our study, we detected very low frequencies of IL-17-producing CD4+ T cells, with no differences between the CD161+ and the CD161− fraction. The predominance of a Th1-oriented response is in agreement with the finding that among CD4+ TILs, KLRB1 gene is positively correlated with SIGIRR. SIGIRR is a negative regulator of IL-1 receptor. IL-1R was shown to be expressed by a subset of “non-classical” Th1 clones derived from Th17 cells and characterized by the expression of CD161.50,51 IL-1-mediated signaling in T cells is essential for Th17 cell differentiation. Yet, Gulen et al. showed that SIGIRR suppresses Th17 cell proliferation via inhibition of the interleukin-1 receptor pathway.35 Within CD8+ T cells, KLRB1 gene was associated with IL-17A, thus linking Tc17 with CD161, as previously described.12,52 Besides conventional CD4+ T cells, we found a significantly lower frequency of CD161+ Tregs in NSCLC tumors as compared to CD161− Tregs. CD161+ Tregs have been shown to display immune-regulatory functions, similarly to their CD161− Treg counterparts.43,53,54 However, these CD161+ Tregs were also shown to display a high pro-inflammatory potential, as they produced higher levels of IL-17, IFN-γ and IL-2 after PMA/Ionomycin stimulation and expressed higher percentages of CCR6, IL-23R, CD45RO, RORC and T-bet than CD161− Tregs. It remains to define the exact role of these CD161+ Tregs in the context of NSCLC tumors.
Because T cells in the tumor microenvironment have been shown to display either anti-tumoral functions or dysfunctional/exhausted phenotypes, many studies have attempted to delineate their phenotype. Among co-stimulatory receptors and ICP molecules upregulated on both CD161+ CD4+ and CD161+ CD8+ T cells, we found the activation marker CD69 and the well-known inhibitory co-receptor PD-1. Expression of PD-1 has been associated with Tim-3 on exhausted CD8+ T cells.6,55 We found a significantly higher frequency of Tim-3+ cells among CD161+ CD8+ T cells, suggesting that some CD161-expressing CD8+ T cells could be dysfunctional. By contrast, CD161+ CD4+ T cells expressed significantly lower frequencies of Tim-3 as compared to their CD161− counterparts, suggesting that they rather retain high functional capacities than being terminally exhausted. Consistently with gene expression analysis, a higher frequency of CD96, CD30L, and OX40-expressing cells and a lower frequency of 4-1BB+ cells were detected among CD161+ CD4+ T cells compared to CD161− CD4+ T cells. OX40, CD30 and 4-1BB are known to synergize with TCR-CD3 to promote cell cycle progression, cytokine production and T cell survival.39 Their expression is induced on T cells following activation, is predominant on effector T cells, and is maintained on memory T cells. In pre-clinical studies, the use of agonistic antibodies of these co-receptors resulted in a marked increase of antigen-specific CTL and CD4+ T cell responses, and the generation of memory T cells.39,56 Similarly, expression of these co-receptors on immune cells in the tumor microenvironment is usually associated with good prognosis.57 Their ligands, OX40L, CD30L and 4-1BBL, are mainly expressed by activated/mature APCs but can also be expressed by activated T cells.6,39 The finding that within CD4+ TILs, CD161 is more frequently co-expressed with OX40 but not with 4-1BB may be explained by the fact that 4-1BB and OX40 are expressed sequentially on CD4+ T cells. Dawicki et al. showed that while 4-1BB is detected on CD4+ T cells early after immunization, OX40 is not highly expressed until after 4-1BB levels have dropped.58 They showed that OX40/OX40L interactions are critical for optimal CD4+ T cell responses, maintenance of CD4+ T cells late in the primary response and for triggering CD4+ T cell expansion upon rechallenge, while 4-1BB/4-1BBL interactions have a more moderated impact. Co-expression of CD161 with OX40, and restriction of CD161 to EM and CM T cells, lead us to hypothesize that CD161 may have a similar function as OX40 in the establishment of a robust CD4 memory pool that can vigorously respond upon rechallenge.
Similarly to CD161, CD30 expression has been mainly reported on CD45RO+ memory T cells, while like LLT1, CD30L is expressed on activated B cells. CD30L gene expression level is higher on GC-B cells than memory B cells, and CD30L has been reported as constitutively expressed on GC-derived cell lines such as Burkitt's lymphomas.59 Similarly to LLT1, CD30L can also be expressed on activated T cells. However, despite the detection by IHC of LLT1 expression by CD3+ T cells in the tumor stroma, we could not observe any LLT1 expression on the cell surface of CD3+ T cells by flow cytometry, and we similarly observed very low frequencies of TILs expressing CD30L on the cell surface. This suggests that LLT1/CD161 and CD30L/CD30 pairs may have a similar regulation of their expression. Bi-directional co-signaling has been described for many TNFR/TNF-Ligand, such as OX40L that would enhance antibody production by plasma cells,60 or CD30L engagement on B cells that would inhibit class-switch antibody to modulate the magnitude of IgG, IgA or IgE production.61 Whether LLT1 signals within B cells but also within T cells, after potential trans or cis interactions with CD161, and the consequences of this signaling, remain to be elucidated.
In agreement with our previous report in tonsils and NHLs,15 we observed a lower expression of CD161 on the cell surface of CXCR5high PD-1high Tfh cells, compared to CXCR5neg T cells. KLRB1 seems to be transcribed as it was detected by RNAseq on Tfh purified from tonsils but the level of transcription was significantly lower than effector T cells.62 Similarly, a recent study22 confirmed lower KLRB1 transcripts and surface expression of CD161 on Tfh cells. Because CD161 is known to be downregulated after engagement with LLT1,10 a lower expression of CD161 on Tfh may suggest that this ligand/receptor pair has been involved in B/Tfh interactions within GCs and may contribute to GC development and maintenance. As CD30L and OX40 were both shown to play a major role in Tfh survival and GC development through the maintenance of CD4 T cell memory,63 one could speculate that LLT1/CD161 are also active players in the control of GC development. Also, several mechanisms of post-transcriptional and/or epigenetic regulation of gene expression have been shown to be particularly important for limiting the numbers of Tfh cells and preventing autoimmunity. Among them, the RNA-binding proteins ROQUIN-1 and ROQUIN-2 are able to bind to ICOS, OX40, IL-6 and TNF mRNAs and to induce their degradation.64 KLRB1 mRNA within Tfh could be submitted to the same type of regulation, explaining its lower expression on Tfh cells. An interesting model has been proposed, that under limiting IL-2 conditions, exposure to IL-21 and interaction with B cells, CD4+ T cells differentiate into a plastic population of Tfh-like cells that give rise to Tfh memory or CM T cells. Upon subsequent exposure to antigens, Tfh, CM and EM T cell pools may then differentiate into Th1 effectors and Tfh effectors.48 Thus, we suggest that LLT1 expression on B cells and CD161 expression on CM CD4+ T cells may participate in re-constituting the pool of Tfh memory and CM T cells for subsequent re-challenge. Our observation that CD62L+ CD4+ T cells express lower frequencies of CD161, together with our previous observation that CD62L+ T cells within NSCLC tumors selectively localize inside TLS,19 are also strongly in favor of potential interactions between CD161-expressing CM CD4+ T cells and LLT1-expressing B cells within TLS, accompanied by CD161 downregulation.
In conclusion, LLT1 and CD161 appear as novel promising co-receptors that are worth exploring further in the cancer context. Higher frequencies of IFN-γ-producing T cells and lower frequencies of Tregs among the CD161+ CD4+ T cell fraction is consistent with the positive impact of high KLRB1 and CLEC2D gene expression on NSCLC patient survival. Restricted expression of CD161 on CD4+ and CD8+ EM T cells is in favor of its implication in rapid recall responses, similarly to OX40. The fact that CD161 expression on CD4+ TILs is associated with an activated but not terminally exhausted phenotype is also predictive of their functional relevance. The putative role of CD161/LLT1 axis in Tfh biology and in GC development within TLS further emphasizes the importance of TLS and their positive impact on NSCLC patient survival. Thus, LLT1 and CD161 represent promising new targets for immunotherapy strategies, in order to boost ongoing immune responses that are taking place in cancer-associated TLS.
Materials and methods
NSCLC patients
Fresh, Formalin-Fixed Paraffin-Embedded (FFPE) and frozen samples were obtained from NSCLC patients at the Institut Mutualiste Montsouris (IMM) or Hôpitaux Universitaires Paris Centre (HUPC) (Paris, France). Pre-operative evaluation of patients included lung, brain and adrenal computed tomography scanning and liver ultrasound echography. They all underwent complete surgical resection of their tumors, including multilevel lymph node sampling or lymphadenectomy. Patients with an Eastern Cooperative Oncology Group performance status ≤ 1 were eligible.
A first retrospective cohort of 32 untreated NSCLC patients (HUPC) and a second retrospective cohort of 20 NSCLC patients treated with neo-adjuvant chemotherapy (HUPC) were selected for comparison and analyzed by IHC for LLT1 and CD21 expression within B-cell follicles.
A first prospective flow cytometry study was performed on 33 untreated patients with NSCLC (Table 1) (IMM or HUPC). Fresh tumor samples were retrieved from NSCLC patients, associated when possible with distant lung samples, draining LN and/or peripheral blood. A second prospective study was performed for Nanostring analysis, on 31 untreated patients with NSCLC (Table 2) (IMM or HUPC). Only fresh tumor samples were obtained from these NSCLC patients. Patients who received neoadjuvant chemotherapy or radiotherapy were ineligible. The protocol was approved by local ethic and human investigation committee (n° 2008–133 and 2012–0612) and by the Assistance Publique-Hôpitaux de Paris (AP-HP), in application with the article L.1121-1 of French law. A written informed consent was obtained from patients prior to inclusion in the studies. When possible, serial sections of the corresponding FFPE NSCLC tumors were obtained for patients from these two prospective cohorts, and analyzed by IHC for LLT1 and/or CD20/CD21 expression.
Table 1.
Clinical and pathological features of the 33 untreated patients with NSCLC used in flow cytometry analysis.
| Characteristics | Number (%) | |
|---|---|---|
| Gender | ||
| Male | 20 (61%) | |
| Female | 13 (39%) | |
| Age | ||
| Mean (years) +/- SEM | 70 +/- 9 | |
| Range | 52–90 | |
| Smoking History | ||
| Past/current | 26 (79%) | |
| Never smokers | 3 (9%) | |
| ND | 4 (12%) | |
| Pack-years (years) +/- SEM | 35 +/- 17 | |
| Range | 0–60 | |
| Histological subtype | ||
| Adenocarcinoma | 18 (55%) | |
| Squamous cell carcinoma | 14 (42%) | |
| Sarcomatoid carcinoma | 1 (3%) | |
| PTNM stage | ||
| I | 13 (39%) | |
| II | 13 (39%) | |
| III | 7 (22%) |
Pathologic staging of lung cancer was determined according to the new TNM staging classification.66 Histological subtypes were determined according to the classification of the WHO.67 Patients did not receive any pre-operative chemotherapy nor radiotherapy.
Table 2.
Clinical and pathological features of the 31 untreated patients with NSCLC used in gene expression analysis.
| Characteristics | Number (%) |
|---|---|
| Gender | |
| Male | 13 (42%) |
| Female | 18 (58%) |
| Age | |
| Mean (years) +/- SEM | 68 +/- 9 |
| Range | 51–85 |
| Smoking History | |
| Past/current | 23 (74%) |
| Never smokers | 4 (13%) |
| ND | 4 (13%) |
| Pack-years (years) +/- SEM | 31 +/- 23 |
| Range | 0–80 |
| Histological subtype | |
| Adenocarcinoma | 16 (52%) |
| Squamous cell carcinoma | 14 (45%) |
| Large-cell Neuroendocrine carcinoma | 1 (3%) |
| PTNM stage | |
| I | 14 (45%) |
| II | 11 (36%) |
| III | 6 (19%) |
Pathologic staging of lung cancer was determined according to the new TNM staging classification.66 Histological subtypes were determined according to the classification of the WHO.67 Patients did not receive any pre-operative chemotherapy nor radiotherapy.
Control tissues and primary cells
Frozen sections of NSCLC LN, obtained from IMM hospital, and FFPE sections of hyper pulmonary arterial pressure (HPAP) lung tissue or tonsil, obtained from HUPC and Geneticist Inc (CA, USA), respectively, were used as control for IHC and IF stainings.
Tonsil cells were obtained from discarded benign tonsils after informed consent from patients undergoing routine tonsillectomies at the Lenval Hospital, Nice.
Peripheral blood was obtained from healthy volunteers at the “Etablissement Français du Sang” (EFS, Paris, France, n 12/EFS/079-E2075).
Immunohistochemistry
Sections of FFPE NSCLC tumors were stained as previously described17 using mouse (Mo) anti-human CD20 and Mo anti-human CD21 (Sup Table 1).
For determining the number of LLT1-positive and CD21-positive B-cell follicles, two independent IHC stainings (anti-human LLT1 and anti-human CD20/antihuman CD21) were performed on serial and non-serial FFPE NSCLC tumor sections for each patient.
IHC staining for LLT1 was performed as previously described15 with Mo anti-LLT1 clone 2F1 mAb. Briefly, FFPE tissue sections were deparaffinized and rehydrated in successive baths of xylene, ethanol and water followed by heat-induced epitope retrieval in Tris/EGTA buffer pH 9. Endogenous peroxidase activity and biotin were blocked by H2O2 (0.5% v/v) and avidin–biotin blocking kit (Vector), respectively. Unspecific protein-binding was blocked with 10% donkey serum (Do), 3% human serum, 3% bovine serum albumin, 3% skim milk in Tris Buffer Saline (TBS). Sections were washed in TBS with 0.05% Tween20 between steps. FFPE sections were incubated with Mo anti-LLT1 mAb (2F1) or Mo IgG1 isotype control (BD Biosciences) at 0.1 µg/ml. Antibody binding was detected with biotinylated Do anti-Mo IgG1 secondary antibody (Jackson Immunoresearch) at 0.3 µg/ml and visualized using avidin-biotin-HRP complexes (PK-6100, Vector) in combination with the highly sensitive detection method Tyramid Signal Amplification (TSA), according to manufacturer's instructions (NEL700A, Perkin Elmer) (Sup Table 1). Nuclei were counterstained with hematoxylin.
IHC staining for CD161 was performed with Mo anti-CD161 clone DX12 (BD Biosciences) on frozen tissue sections fixed in acetone. Endogenous peroxidase activity and unspecific protein-binding were blocked as described above. Mo anti-CD161 or Mo IgG1 isotype control (BD Biosciences) were used at 1 µg/ml. Antibody binding was detected with biotinylated Do anti-Mo IgG1 secondary antibody (Jackson Immunoresearch) at 0.3 µg/ml and visualized as described above (Sup Table 1).
Immunostained slides were scanned using NanoZoomer 2.0HT slide scanner (Hamamatsu). Pictures were captured with NDP.View2 software.
Immunofluorescence
FFPE tissue sections were deparaffinized, pre-treated as described above, and incubated with Mo anti-LLT1 mAb (2F1) or Mo IgG1 isotype control (BD Biosciences) at 0.1 µg/ml followed by secondary biotinylated Do anti-Mo IgG1 (Jackson Immunoresearch) at 0.3 µg/ml. Antibody binding was fluorescently stained using avidin-biotin-HRP complexes (PK-6100, Vector) in combination with the highly sensitive detection method Tyramid Signal Amplification (TSA), as described above, and with AF647-conjugated streptavidin (Jackson Immunoresearch) (Sup Table 1). Nuclei were visualized using DAPI.
IF staining for CD161 was performed as described above, on frozen tissue sections fixed in acetone, using Mo anti-CD161 clone DX12 (BD Biosciences) at 1 µg/ml, Mo anti-CD161 clone B199.2 (LSBio) at 2 µg/ml or Mo IgG1 isotype control (BD Biosciences), followed by secondary biotinylated Do anti-Mo IgG1 (Jackson Immunoresearch) at 0.3 µg/ml. Antibody binding was fluorescently stained using AF647-conjugated streptavidin (Jackson Immunoresearch), as described above (Sup Table 1). Nuclei were visualized using DAPI.
For double and triple-IF stainings, LLT1 or CD161 staining was repeated as described above, followed by application of primary antibodies and appropriate fluorochrome-conjugated secondary antibodies for the following immune cell subsets: B cells (anti-human CD20 or anti-human CD19), T cells (anti-human CD3), NK cells (anti-human NKp46), mature DCs (anti-human DC-LAMP), macrophages (anti-human CD68), FDCs (anti-human CD21), and Tfh cells (anti-human PD-1) (Sup Table 1). Nuclei were visualized using DAPI. Immunostained slides were scanned using Axio Scan.Z1 and pictures were captured with Zen 2012 (blue edition) (Zeiss).
A virtual multiplexing method based on image registration using ImageJ software was used in Fig. 4G, for visualizing the localization of LLT1-expressing cells and CD161-expressing cells together with CD20-expressing cells in the same virtual area.
Methods for cell quantification
The number of LLT1-positive or CD21-positive B-cell follicles was determined manually, across the whole tumor section, on NDP.View2 software.
The density of TLS-B cells was determined automatically, across the whole tumor section, using Calopix software (Tribvn), and expressed as a percentage:
[Total surface of CD20+ B-cell follicles (µm2) / Total surface of tumor area (µm2)]* 100.
Flow cytometry
Fresh NSCLC-derived tissues were mechanically dissociated and digested in the presence of non-enzymatic Cell Recovery Solution (Corning) for 1 h at 4°C. The cell suspension was then filtered through a 70 µm filter and mononuclear cells were isolated by centrifugation over a Ficoll Hypaque density gradient. Multiple stainings on isolated mononuclear cells were performed as previously described.15 Briefly, cells were incubated in PBS 0.5 mM EDTA, 10% normal mouse serum and 100 µg/ml human Ig for 30 min on ice prior to incubation with biotinylated anti-LLT1 (4F68) mAb or biotinylated Mo IgG1 isotype control (BD Biosciences), followed by PE-CF594-conjugated streptavidin (BD Biosciences) and additional fluorochrome-labeled antibodies to CD45, CD19, CD38, CD3, CD4, CD8, CD56, CD11c, CD14, CD123, CD10, CD35, CD21, TCR-γδ, CD161 (or isotype control), CD45RA, CCR7, CD25, CD127, CD62L, CXCR5, PD-1, CD69, OX40, 4-1BB, Tim-3, CD96, CD30L, CD40L, CD27, CD28, GITR, ICOS and CD103 (Sup Table 1).
For intranuclear FoxP3 staining, the anti-FoxP3 staining set (eBioscience) was used according to the manufacturer's instructions.
For intracellular cytokine staining, cells were stimulated for 4 h with phorbol 12-myristate 13-acetate (PMA) and ionomycin (Sigma-Aldrich) in the presence of brefeldin A and monensin (BD Pharmingen). After cell surface staining with appropriate lineage markers, including anti-CD161 or isotype control, cells were permeabilized using the Fixation/Permeabilization Solution (BD Biosciences) and stained with fluorochrome-labeled antibodies to IFN-γ, TNF-α, IL-17 and Granzyme B (Sup Table 1).
Cells were fixed in PBS 0.5% formaldehyde before analysis on a LSR Fortessa cell analyzer (BD Biosciences). Dead cells were excluded based on a viability marker (Sup Table 1). Results were subsequently analyzed using DIVA (BD Biosciences) and/or FlowJo software (TreeStar, Inc).
Cell sorting and NanoString-based gene expression profiling
Fresh NSCLC tumors were mechanically dissociated and digested in the presence of DNAse I (Sigma Aldrich) and collagenase A (Roche) for 1 h at 37°C. The cell suspension was then filtered through a 70 µm filter, and mononuclear cells were isolated by centrifugation over a Ficoll Hypaque density gradient. CD3+ CD4+ and CD3+ CD8+ T cells were sorted using a FACS Aria III cell sorter (BD Biosciences) among alive cells (gated DAPI-), non-epithelial cells (gated CD227-, epithelial antigen-, pan-cytokeratins-), and hematopoietic cells (CD45+) (Sup Table 1). Purity was >98%.
Total RNA was extracted using RNeasy Mini Kit (Qiagen SAS, Courtaboeuf, France), including DNAse treatment, according to the manufacturer's instructions. RNA quantity and quality were determined with RNA 6000 Nano Kit on a 2100 Bioanalyzer (Agilent Technologies SAS, Les Ulis, France). Digital multiplexed gene expression analysis using the NanoString nCounter system (PanCancer Immune Profiling Panel, NanoString Technologies) was performed on 4 ng of total RNA from each sample according to the manufacturer's instructions. Pre-amplification was performed using Taqman PreAmp 2x and MTE primers (NanoString Technologies) according to the manufacturer's instructions. Genes with geomean counts before normalization below a threshold determined on background, i.e. inferior to 20 geomean counts, were excluded from subsequent analysis. Normalization of raw data was performed using the nSolver software (NanoString Technologies) based on the 10 most relevant housekeeping genes. To investigate the biological role of the genes significantly correlated with KLRB1 gene, we used ClueGO.37 ClueGO integrates Gene Ontology (GO) terms as well as KEGG/BioCarta pathways and creates a functionally organized GO/pathway term network. Terms found in the GO interval of 3 to 8, with at least three genes from the initial list, were selected.
Survival analysis of the expression of KLRB1 and CLEC2D genes
We curated publically-available transcriptomic data from cohorts of NSCLC patients with annotation for overall survival, from three gene expression platforms (Affymetrix 133A, Plus 2.0, and Illumina HiSeq). Samples analyzed (n = 1760) are listed in additional online data supplement. For Affymetrix platforms, we used the fRMA-normalized expression of the probeset 214470_at to represent KLRB1 and 220132_s_at for CLEC2D.65 For the RNAseq data, we used FPKM values corresponding to each gene. For each gene we then fitted Cox proportional-hazards models for each individual series using the R package survival (version 2.41.3). We then performed a random-effects meta-analysis using the R package meta (version 4.8-2).
For Kaplan-Meier analyses, we used the “Microenvironment Cell Populations-counter” (MCP-counter) method,40 to determine the level of tumor infiltration by cytotoxic cells (CD8+ T cells and NK cells) and B cells, from GSE50081 transcriptomic data. We transformed each variable into binary variables (high or low) according to a median cut-off value and performed Kaplan Meier analyses combining KLRB1 and estimated cytotoxic cell density, or CLEC2D and estimated B cell density. Hazard Ratios and associated p values were determined using the log-rank test.
Statistical analysis
In IHC experiment, a spearman test was applied to determine the correlation factor between the number of LLT1-positive and the number of CD21-positive B-cell follicles (GraphPad Prism 5 software).
In flow cytometry experiments, depending on data distribution (Shapiro normality test) and matched or not matched observations, a Kruskal-Wallis test, an unpaired t test, a Friedman test, a Mann Whitney test, or a Wilcoxon (paired, non-gaussian) test, with appropriate post-hoc comparison, was used to compare quantitative variables across the different groups (GraphPad Prism 5 software). A p value < 0.05 was considered statistically significant.
In Nanostring analysis, a spearman test was applied to determine the correlation factors between KLRB1 gene and all the genes from the PanCancer Immune Profiling Panel. Benjamini–Hochberg correction was applied to all the associated p values to determine the false discovery rates (FDRs) (R software). Only FDRs < 0.10 were considered statistically significant, but FDRs < 0.15 were still considered for subsequent ClueGO analyses.
Supplementary Material
Funding Statement
This work was supported by the “Institut National de la Santé et de la Recherche Médicale (INSERM), Centre National de la Recherche Scientifique (CNRS), Pierre and Marie Curie University, Paris-Descartes University, the Labex Immuno-Oncology (LAXE62_9UMRS872 Fridman), Fondation ARC, Cancéropole PACA, Agence Nationale de la Recherche (ANR) through the “Investments for the Future” LABEX SIGNALIFE (ANR-11-LABX-0028-01).
Abbreviations
- ADC
Adenocarcinoma
- APC
Antigen-Presenting Cell
- CM
Central-Memory
- COPD
Chronic Obstructive Pulmonary Disease
- DC
Dendritic Cell
- EM
Effector-Memory
- FDC
Follicular Dendritic Cell
- FDR
False Discovery Rate
- FFPE
Formalin-Fixed Paraffin-Embedded
- GC
Germinal Center
- HD
Healthy Donor
- HR
Hazard Ratio
- ICP
Immune Check Point
- IF
Immunofluorescence
- IHC
Immunohistochemistry
- ILC
Innate Lymphoid Cell
- LN
Lymph Node
- mAb
monoclonal Antibody
- MAIT
Mucosal-Associated Invariant T cell
- NSCLC
Non-Small Cell Lung Cancer
- NHL
Non-Hodgkin Lymphoma
- NK
Natural Killer
- PBMC
Peripheral Blood Mononuclear Cell
- PC
Plasma Cell
- pDC
plasmacytoid Dendritic Cell
- PMA
phorbol 12-myristate 13-acetate
- SCC
Squamous Cell Carcinoma
- SLO
Secondary Lymphoid Organ
- Tfh
T follicular helper cell
- Th
T helper cell
- TIL
Tumor-Infiltrating Lymphocyte
- TSA
Tyramid Signal Amplification
- TLS
Tertiary Lymphoid Structure
- Treg
Regulatory T cell.
Disclosure of potential conflicts of interest
The authors disclose no potential conflicts of interest.
Acknowledgments
We thank Dr Myriam Lawand and Dr Frédéric Brau for advices as well as Dr Estelle Pécheux-Devêvre, Dr Hélène Fohrer-Ting and Mr. Christophe Klein from the platform “Centre d'Imagerie Cellulaire et de Cytométrie” (Cordeliers Research Center, Paris, France) for technical support.
References
- 1.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: From immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–8. doi: 10.1038/ni1102-991. PMID:12407406. [DOI] [PubMed] [Google Scholar]
- 2.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. PMID:22419253. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. PMID:21376230. [DOI] [PubMed] [Google Scholar]
- 4.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, et al.. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–95. doi: 10.1016/j.immuni.2013.10.003. PMID:24138885. [DOI] [PubMed] [Google Scholar]
- 5.Dieu-Nosjean M-C, Giraldo NA, Kaplon H, Germain C, Fridman WH, Sautès-Fridman C. Tertiary lymphoid structures, drivers of the anti-tumor responses in human cancers. Immunol Rev. 2016;271:260–75. doi: 10.1111/imr.12405. PMID:27088920. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. 2013;13:227–42. doi: 10.1038/nri3405. PMID:23470321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: Toward combination strategies with curative potential. Cell. 2015;161:205–14. doi: 10.1016/j.cell.2015.03.030. PMID:25860605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dempke WCM, Fenchel K, Uciechowski P, Dale SP. Second- and third-generation drugs for immuno-oncology treatment-The more the better? Eur J Cancer Oxf Engl 1990. 2017;74:55–72. [DOI] [PubMed] [Google Scholar]
- 9.Fridman WH, Zitvogel L, Sautès-Fridman C, Kroemer G. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol. 2017;14(12):717–734. doi: 10.1038/nrclinonc.2017.101. PMID:28741618. [DOI] [PubMed] [Google Scholar]
- 10.Aldemir H, Prod'homme V, Dumaurier M-J, Retiere C, Poupon G, Cazareth J, Bihl F, Braud VM. Cutting edge: Lectin-like transcript 1 is a ligand for the CD161 receptor. J Immunol Baltim Md 1950. 2005;175:7791–5. [DOI] [PubMed] [Google Scholar]
- 11.Rosen DB, Bettadapura J, Alsharifi M, Mathew PA, Warren HS, Lanier LL. Cutting edge: Lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J Immunol Baltim Md 1950. 2005;175:7796–9. [DOI] [PubMed] [Google Scholar]
- 12.Fergusson JR, Fleming VM, Klenerman P. CD161-expressing human T cells. Front Immunol. 2011;2:36. doi: 10.3389/fimmu.2011.00036. PMID:22566826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Germain C, Meier A, Jensen T, Knapnougel P, Poupon G, Lazzari A, Neisig A, Håkansson K, Dong T, Wagtmann N, et al.. Induction of lectin-like transcript 1 (LLT1) protein cell surface expression by pathogens and interferon-γ contributes to modulate immune responses. J Biol Chem. 2011;286:37964–75. doi: 10.1074/jbc.M111.285312. PMID:21930700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germain C, Bihl F, Zahn S, Poupon G, Dumaurier M-J, Rampanarivo HH, Padkjær SB, Spee P, Braud VM. Characterization of alternatively spliced transcript variants of CLEC2D gene. J Biol Chem. 2010;285:36207–15. doi: 10.1074/jbc.M110.179622. PMID:20843815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Germain C, Guillaudeux T, Galsgaard ED, Hervouet C, Tekaya N, Gallouet A-S, Fassy J, Bihl F, Poupon G, Lazzari A, et al.. Lectin-like transcript 1 is a marker of germinal center-derived B-cell non-Hodgkin's lymphomas dampening natural killer cell functions. Oncoimmunology. 2015;4:e1026503. doi: 10.1080/2162402X.2015.1026503. PMID:26405582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dieu-Nosjean M-C, Antoine M, Danel C, Heudes D, Wislez M, Poulot V, Rabbe N, Laurans L, Tartour E, de Chaisemartin L, et al.. Long-term survival for patients with non–small-cell lung cancer with intratumoral lymphoid structures. J Clin Oncol. 2008;26:4410–7. doi: 10.1200/JCO.2007.15.0284. PMID:18802153. [DOI] [PubMed] [Google Scholar]
- 17.Germain C, Gnjatic S, Tamzalit F, Knockaert S, Remark R, Goc J, Lepelley A, Becht E, Katsahian S, Bizouard G, et al.. Presence of B cells in tertiary lymphoid structures is associated with a protective immunity in patients with lung cancer. Am J Respir Crit Care Med. 2014;189:832–44. doi: 10.1164/rccm.201309-1611OC. PMID:24484236. [DOI] [PubMed] [Google Scholar]
- 18.Germain C, Gnjatic S, Dieu-Nosjean M-C. Tertiary lymphoid structure-associated B cells are key players in anti-tumor immunity. Front Immunol. 2015;6:67. doi: 10.3389/fimmu.2015.00067. PMID:25755654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goc J, Germain C, Vo-Bourgais TKD, Lupo A, Klein C, Knockaert S, de Chaisemartin L, Ouakrim H, Becht E, Alifano M, et al.. Dendritic cells in tumor-associated tertiary lymphoid structures signal a Th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ T cells. Cancer Res. 2014;74:705–15. doi: 10.1158/0008-5472.CAN-13-1342. PMID:24366885. [DOI] [PubMed] [Google Scholar]
- 20.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al.. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–45. doi: 10.1038/nm.3909. PMID:26193342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.JØ Bohnhorst, MB Bjørgan, Thoen JE, Natvig JB, Thompson KM. Bm1-Bm5 classification of peripheral blood B cells reveals circulating germinal center founder cells in healthy individuals and disturbance in the B cell subpopulations in patients with primary Sjögren's syndrome. J Immunol Baltim Md 1950. 2001;167:3610–8. [DOI] [PubMed] [Google Scholar]
- 22.Llibre A, López-Macías C, Marafioti T, Mehta H, Partridge A, Kanzig C, Rivellese F, Galson JD, Walker LJ, Milne P, et al.. LLT1 and CD161 Expression in Human Germinal Centers Promotes B Cell Activation and CXCR4 Downregulation. J Immunol Baltim Md 1950. 2016;196:2085–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martin E, Treiner E, Duban L, Guerri L, Laude H, Toly C, Premel V, Devys A, Moura IC, Tilloy F, et al.. Stepwise development of MAIT cells in mouse and human. PLoS Biol. 2009;7:e54. doi: 10.1371/journal.pbio.1000054. PMID:19278296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang HH, Song K, Rabin RL, Hill BJ, Perfetto SP, Roederer M, Douek DC, Siegel RM, Farber JM. CCR2 identifies a stable population of human effector memory CD4+ T cells equipped for rapid recall response. J Immunol Baltim Md 1950. 2010;185:6646–63. [DOI] [PubMed] [Google Scholar]
- 25.Schrum S, Probst P, Fleischer B, Zipfel PF. Synthesis of the CC-chemokines MIP-1alpha, MIP-1beta, and RANTES is associated with a type 1 immune response. J Immunol Baltim Md 1950. 1996;157:3598–604. [PubMed] [Google Scholar]
- 26.Salmond RJ, Emery J, Okkenhaug K, Zamoyska R. MAPK, phosphatidylinositol 3-kinase, and mammalian target of rapamycin pathways converge at the level of ribosomal protein S6 phosphorylation to control metabolic signaling in CD8T cells. J Immunol Baltim Md 1950. 2009;183:7388–97. [DOI] [PubMed] [Google Scholar]
- 27.Wang PL, O'Farrell S, Clayberger C, Krensky AM. Identification and molecular cloning of tactile. A novel human T cell activation antigen that is a member of the Ig gene superfamily. J Immunol Baltim Md 1950. 1992;148:2600–8. [PubMed] [Google Scholar]
- 28.Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155). J Immunol Baltim Md 1950. 2004;172:3994–8. [DOI] [PubMed] [Google Scholar]
- 29.Springer TA. Adhesion receptors of the immune system. Nature. 1990;346:425–34. doi: 10.1038/346425a0. PMID:1974032. [DOI] [PubMed] [Google Scholar]
- 30.Hogg N, Landis RC. Adhesion molecules in cell interactions. Curr Opin Immunol. 1993;5:383–90. doi: 10.1016/0952-7915(93)90057-Y. PMID:7688516. [DOI] [PubMed] [Google Scholar]
- 31.Mayne M, Moffatt T, Kong H, McLaren PJ, Fowke KR, Becker KG, Namaka M, Schenck A, Bardoni B, Bernstein CN, et al.. CYFIP2 is highly abundant in CD4+ cells from multiple sclerosis patients and is involved in T cell adhesion. Eur J Immunol. 2004;34:1217–27. doi: 10.1002/eji.200324726. PMID:15048733. [DOI] [PubMed] [Google Scholar]
- 32.Witherden DA, Boismenu R, Havran WL. CD81 and CD28 costimulate T cells through distinct pathways. J Immunol Baltim Md 1950. 2000;165:1902–9. [DOI] [PubMed] [Google Scholar]
- 33.Wiley SR, Goodwin RG, Smith CA. Reverse signaling via CD30 ligand. J Immunol Baltim Md 1950. 1996;157:3635–9. [PubMed] [Google Scholar]
- 34.Alonso-Ramirez R, Loisel S, Buors C, Pers J-O, Montero E, Youinou P, Renaudineau Y. Rationale for targeting CD6 as a treatment for autoimmune diseases. Arthritis. 2010;2010:130646. doi: 10.1155/2010/130646. PMID:22076177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gulen MF, Kang Z, Bulek K, Youzhong W, Kim TW, Chen Y, Altuntas CZ, Bak-Jenson K, McGeachy MJ, Do JS, et al.. The receptor SIGIRR suppresses Th17 cell proliferation via inhibition of the interleukin-1 receptor pathway and mTOR kinase activation. Immunity 2010;32:54–66. doi: 10.1016/j.immuni.2009.12.003. PMID:20060329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garlanda C, Dinarello CA, Mantovani A. The interleukin-1 family: Back to the future. Immunity. 2013;39:1003–18. doi: 10.1016/j.immuni.2013.11.010. PMID:24332029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman W-H, Pagès F, Trajanoski Z, Galon J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinforma Oxf Engl. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srenathan U, Steel K, Taams LS. IL-17+ CD8+ T cells: Differentiation, phenotype and role in inflammatory disease. Immunol Lett. 2016;178:20–6. doi: 10.1016/j.imlet.2016.05.001. PMID:27173097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Croft M. Co-stimulatory members of the TNFR family: Keys to effective T-cell immunity? Nat Rev Immunol. 2003;3:609–20. doi: 10.1038/nri1148. PMID:12974476. [DOI] [PubMed] [Google Scholar]
- 40.Becht E, Giraldo NA, Lacroix L, Buttard B, Elarouci N, Petitprez F, Selves J, Laurent-Puig P, Sautès-Fridman C, Fridman WH, et al.. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. PMID:27765066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Platonova S, Cherfils-Vicini J, Damotte D, Crozet L, Vieillard V, Validire P, André P, Dieu-Nosjean M-C, Alifano M, Régnard J-F, et al.. Profound coordinated alterations of intratumoral NK cell phenotype and function in lung carcinoma. Cancer Res. 2011;71:5412–22. doi: 10.1158/0008-5472.CAN-10-4179. PMID:21708957. [DOI] [PubMed] [Google Scholar]
- 42.Mark NM, Kargl J, Busch SE, Yang GHY, Metz HE, Zhang H, Hubbard JJ, Pipavath SNJ, Madtes DK, Houghton AM. COPD alters immune cell composition and immune checkpoint inhibitor efficacy in NSCLC. Am J Respir Crit Care Med. 2017. doi: 10.1164/rccm.201704-0795OC. PMID:28934595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iliopoulou EG, Karamouzis MV, Missitzis I, Ardavanis A, Sotiriadou NN, Baxevanis CN, Rigatos G, Papamichail M, Perez SA. Increased frequency of CD4+ cells expressing CD161 in cancer patients. Clin Cancer Res Off J Am Assoc Cancer Res. 2006;12:6901–9. doi: 10.1158/1078-0432.CCR-06-0977. [DOI] [PubMed] [Google Scholar]
- 44.Pozo D, Valés-Gómez M, Mavaddat N, Williamson SC, Chisholm SE, Reyburn H. CD161 (human NKR-P1A) signaling in NK cells involves the activation of acid sphingomyelinase. J Immunol Baltim Md 1950. 2006;176:2397–406. [DOI] [PubMed] [Google Scholar]
- 45.Bai A, Robson S. Beyond ecto-nucleotidase: CD39 defines human Th17 cells with CD161. Purinergic Signal. 2015;11:317–9. doi: 10.1007/s11302-015-9457-4. PMID:26059452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takahashi T, Dejbakhsh-Jones S, Strober S. Expression of CD161 (NKR-P1A) defines subsets of human CD4 and CD8T cells with different functional activities. J Immunol Baltim Md 1950. 2006;176:211–6. [DOI] [PubMed] [Google Scholar]
- 47.Welters MJP, Ma W, Santegoets SJ, Goedemans R, Ehsan I, Jordanova KS, van Ham VJ, van Unen V, Koning F, van Egmond SI, et al.. Intratumoral HPV16-specific T-cells constitute a type 1 oriented tumor microenvironment to improve survival in HPV16-driven oropharyngeal cancer. Clin Cancer Res. 2017. doi: 10.1158/1078-0432.CCR-17-2140. PMID:29018052. [DOI] [PubMed] [Google Scholar]
- 48.Gasper DJ, Tejera MM, Suresh M. CD4T-cell memory generation and maintenance. Crit Rev Immunol. 2014;34:121–46. doi: 10.1615/CritRevImmunol.2014010373. PMID:24940912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poggi A, Costa P, Zocchi MR, Moretta L. Phenotypic and functional analysis of CD4+ NKRP1A+ human T lymphocytes. Direct evidence that the NKRP1A molecule is involved in transendothelial migration. Eur J Immunol. 1997;27:2345–50. doi: 10.1002/eji.1830270932. PMID:9341779. [DOI] [PubMed] [Google Scholar]
- 50.Maggi L, Santarlasci V, Capone M, Rossi MC, Querci V, Mazzoni A, Cimaz R, De Palma R Liotta F, Maggi E, et al.. Distinctive features of classic and nonclassic (Th17 derived) human Th1 cells. Eur J Immunol. 2012;42:3180–8. doi: 10.1002/eji.201242648. PMID:22965818. [DOI] [PubMed] [Google Scholar]
- 51.Cosmi L, De Palma R Santarlasci V, Maggi L, Capone M, Frosali F, Rodolico G, Querci V, Abbate G, Angeli R, et al.. Human interleukin 17-producing cells originate from a CD161+CD4+ T cell precursor. J Exp Med. 2008;205:1903–16. doi: 10.1084/jem.20080397. PMID:18663128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fergusson JR, Smith KE, Fleming VM, Rajoriya N, Newell EW, Simmons R, Marchi E, Björkander S, Kang Y-H, Swadling L, et al.. CD161 defines a transcriptional and functional phenotype across distinct human T cell lineages. Cell Rep. 2014;9:1075–88. doi: 10.1016/j.celrep.2014.09.045. PMID:25437561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pesenacker AM, Bending D, Ursu S, Wu Q, Nistala K, Wedderburn LR. CD161 defines the subset of FoxP3+ T cells capable of producing proinflammatory cytokines. Blood. 2013;121:2647–58. doi: 10.1182/blood-2012-08-443473. PMID:23355538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Afzali B, Mitchell PJ, Edozie FC, Povoleri GAM, Dowson SE, Demandt L, Walter G, Canavan JB, Scotta C, Menon B, et al.. CD161 expression characterizes a subpopulation of human regulatory T cells that produces IL-17 in a STAT3-dependent manner. Eur J Immunol. 2013;43:2043–54. doi: 10.1002/eji.201243296. PMID:23677517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207:2187–94. doi: 10.1084/jem.20100643. PMID:20819927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aspeslagh S, Postel-Vinay S, Rusakiewicz S, Soria J-C, Zitvogel L, Marabelle A. Rationale for anti-OX40 cancer immunotherapy. Eur J Cancer Oxf Engl 1990. 2016;52:50–66. [DOI] [PubMed] [Google Scholar]
- 57.Weixler B, Cremonesi E, Sorge R, Muraro MG, Delko T, Nebiker CA, Däster S, Governa V, Amicarella F, Soysal SD, et al.. OX40 expression enhances the prognostic significance of CD8 positive lymphocyte infiltration in colorectal cancer. Oncotarget. 2015;6:37588–99. doi: 10.18632/oncotarget.5940. PMID:26439988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dawicki W, Bertram EM, Sharpe AH, Watts TH. 4-1BB and OX40 act independently to facilitate robust CD8 and CD4 recall responses. J Immunol Baltim Md 1950. 2004;173:5944–51. [DOI] [PubMed] [Google Scholar]
- 59.Kennedy MK, Willis CR, Armitage RJ. Deciphering CD30 ligand biology and its role in humoral immunity. Immunology. 2006;118:143–52. doi: 10.1111/j.1365-2567.2006.02354.x. PMID:16771849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morimoto S, Kanno Y, Tanaka Y, Tokano Y, Hashimoto H, Jacquot S, Morimoto C, Schlossman SF, Yagita H, Okumura K, et al.. CD134L engagement enhances human B cell Ig production: CD154/CD40, CD70/CD27, and CD134/CD134L interactions coordinately regulate T cell-dependent B cell responses. J Immunol Baltim Md 1950. 2000;164:4097–104. [DOI] [PubMed] [Google Scholar]
- 61.Cerutti A, Schaffer A, Goodwin RG, Shah S, Zan H, Ely S, Casali P. Engagement of CD153 (CD30 ligand) by CD30+ T cells inhibits class switch DNA recombination and antibody production in human IgD+ IgM+ B cells. J Immunol Baltim Md 1950. 2000;165:786–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weinstein JS, Lezon-Geyda K, Maksimova Y, Craft S, Zhang Y, Su M, Schulz VP, Craft J, Gallagher PG. Global transcriptome analysis and enhancer landscape of human primary T follicular helper and T effector lymphocytes. Blood. 2014;124:3719–29. doi: 10.1182/blood-2014-06-582700. PMID:25331115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim M-Y, Gaspal FMC, Wiggett HE, McConnell FM, Gulbranson-Judge A, Raykundalia C, Walker LSK, Goodall MD, Lane PJL. CD4(+)CD3(-) accessory cells costimulate primed CD4T cells through OX40 and CD30 at sites where T cells collaborate with B cells. Immunity 2003;18:643–54. doi: 10.1016/S1074-7613(03)00110-9. PMID:12753741. [DOI] [PubMed] [Google Scholar]
- 64.Jiang SH, Shen N, Vinuesa CG. Posttranscriptional T cell gene regulation to limit Tfh cells and autoimmunity. Curr Opin Immunol. 2015;37:21–7. doi: 10.1016/j.coi.2015.09.003. PMID:26432764. [DOI] [PubMed] [Google Scholar]
- 65.McCall MN, Bolstad BM, Irizarry RA. Frozen robust multiarray analysis (fRMA). Biostat Oxf Engl. 2010;11:242–53. doi: 10.1093/biostatistics/kxp059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Detterbeck FC, Boffa DJ, Tanoue LT. THe new lung cancer staging system. CHEST J. 2009;136:260–71. doi: 10.1378/chest.08-0978. [DOI] [PubMed] [Google Scholar]
- 67.Brambilla E, Travis WD, Colby TV, Corrin B, Shimosato Y. The new World Health Organization classification of lung tumours. Eur Respir J. 2001;18:1059–68. doi: 10.1183/09031936.01.00275301. PMID:11829087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


