Abstract
The coordinated regulation between cellular glucose uptake and endogenous glucose production is indispensable for the maintenance of constant blood glucose concentrations. The liver contributes significantly to this process by altering the levels of hepatic glucose release, through controlling the processes of de novo glucose production (gluconeogenesis) and glycogen breakdown (glycogenolysis). Various nutritional and hormonal stimuli signal to alter hepatic gluconeogenic flux, and suppression of this metabolic pathway during the postprandial state can, to a significant extent, be attributed to insulin. Here, we review some of the molecular mechanisms through which insulin modulates hepatic gluconeogenesis, thus controlling glucose production by the liver to ultimately maintain normoglycemia. Various signaling pathways governed by insulin converge at the level of transcriptional regulation of the key hepatic gluconeogenic genes PCK1 and G6PC, highlighting this as one of the focal mechanisms through which gluconeogenesis is modulated. In individuals with compromised insulin signaling, such as insulin resistance in type 2 diabetes, insulin fails to suppress hepatic gluconeogenesis, even in the fed state; hence, an insight into these insulin-moderated pathways is critical for therapeutic purposes.
Keywords: glycogenolysis, glucose, regulation, gluconeogenesis, insulin
Introduction
Glucose is a major metabolic fuel that serves the energetic demands of mammalian tissues. During periods of starvation, glucose can be generated through the gluconeogenesis pathway, which is highly evolutionarily conserved from microorganisms to vertebrates. In the human body, the liver is the main site of gluconeogenesis. Increased gluconeogenesis in the liver of patients with type 2 diabetes is considered a major contributor to hyperglycemia and subsequent diabetic organ damage. Insulin is a key hormone that inhibits gluconeogenesis, and insulin resistance is a hallmark of type 2 diabetes. Understanding the regulation of gluconeogenesis and the role of insulin signaling in this pathway is important to developing new therapies for type 2 diabetes. Here, we aim to depict the role of insulin in the framework of hepatic gluconeogenesis.
Gluconeogenesis contributes to hepatic glucose production
Hepatic glucose production is a sum of gluconeogenesis, which is the formation of glucose from pyruvate or other 3- or 4-carbon compounds, and glycogenolysis, which is the breakdown of glycogen to glucose. The main substrates of gluconeogenesis in humans are lactate, glycerol, alanine, and glutamine. Together, these account for 90% of gluconeogenic substrates; however, other amino acids and citric cycle intermediates can also serve as substrates for gluconeogenesis.1,2 Starting from lactate or an α-keto acid derived from amino acid breakdown, pyruvate can be generated for gluconeogenesis. Pyruvate is converted via carboxylation to oxaloacetate in the mitochondria. This reaction is stimulated by high levels of acetyl-CoA, which is produced via β-oxidation of fatty acids in the liver, and inhibited by high levels of ADP and glucose. After an intermediate step that allows oxaloacetate to leave the mitochondria via malate, oxaloacetate is decarboxylated and then phosphorylated to form phosphoenolpyruvate by phosphoenolpyruvate carboxykinase (PEPCK). This enzyme is a regulator of the rate of gluconeogenesis, and its transcription is targeted by multiple factors, including glucagon and insulin.3 After several steps of reverse glycolysis, fructose 1,6-bisphosphatase converts fructose 1,6-bisphosphate to fructose 6-phosphate. Fructose 2,6-bisphosphate and AMP inhibit this reaction, while citrate activates the fructose 1,6-bisphosphatase enzyme. Glucose-6-phosphate (G-6-P) is formed from fructose 6-phosphate by phosphoglucoisomerase. G-6-P can be used in other metabolic pathways or dephosphorylated by glucose-6-phosphatase (G-6-Pase) to form free glucose. Whereas free glucose can easily diffuse into and out of the cell, the phosphorylated form (G-6-P) cannot, providing a mechanism by which intracellular glucose levels are controlled. The final reaction of gluconeogenesis––the formation of glucose––occurs in the lumen of the endoplasmic reticulum, where G-6-P is hydrolyzed by G-6-Pase to produce glucose and release an inorganic phosphate. Like the two previous step, this step is a reversal of glycolysis, in which hexokinase catalyzes the conversion of glucose and ATP into G-6-P and ADP. Glucose is then shuttled into the cytoplasm by glucose transporters located in the endoplasmic reticulum membrane.
Enhanced hepatic glucose production leads to increased glucose release to the blood, which can cause hyperglycemia. Type 2 diabetes is characterized by persistent hyperglycemia. In patients with type 2 diabetes, gluconeogenesis has been identified as the primary source of glucose production, while glycogenolysis was found not to contribute.4 These findings underscore the importance of maintaining normal gluconeogenic rates to avoid disease pathophysiology. Insulin is a major hormone regulator of gluconeogenesis, so understanding its role in determining gluconeogenesis rates is essential to understanding the cause of and potential treatments for type 2 diabetes.
Insulin action on gluconeogenesis is both direct and indirect
A role for insulin in the regulation of hepatic glucose output is widely accepted. In healthy individuals, physiological hyperinsulinemia suppresses gluconeogenesis by 20%, while glycogenolysis is completely suppressed.5 Hyperglycemia alone suppresses hepatic glycogenolysis with only minimal effects on glycogen storage. Only the combination of hyperglycemia and hyperinsulinemia has a significant effect on hepatic glycogen synthesis.6 Thus, insulin plays a crucial role in hepatic glucose metabolism.
The dominant mechanism of insulin-mediated regulation of hepatic gluconeogenesis is not clear. Insulin exerts direct control of gluconeogenesis by acting on the liver, but also indirectly affects gluconeogenesis by acting on other tissues. The direct effect of insulin was demonstrated in fasted dogs, where portal plasma insulin suppressed hepatic glucose production. even without changes in glucagon or gluconeogenic precursors.7 However, in mouse models, insulin was found to have more potent effects on hepatic glucose production in vivo rather than in vitro.8–10 Moreover, indirect effects of insulin on extrahepatic tissues have been shown to be sufficient to maintain normal glucose metabolism, suggesting an important role for indirect insulin regulation of gluconeogenesis.11
Suppression of gluconeogenesis through indirect effects of insulin is known to involve multiple tissues and cell types, with pancreatic α cells, adipose tissue, skeletal muscle, and the brain exerting known effects on hepatic gluconeogenesis. In pancreatic α cells, insulin inhibits the secretion of glucagon, which can indirectly lead to suppression of hepatic glucose production by reducing hepatic glucagon signaling.12 Glucagon has effects on the transcriptional regulation of gluconeogenesis, primarily through the transcription factor CREB, but also through metabolite flux by affecting the activity of phosphofructokinase 1 (PFK1) in a PKA-dependent manner.13,14 Insulin decreased plasma glucagon levels in vivo and inhibited glucagon secretion from pancreatic αcells in vitro.15 However, in mice lacking the insulin receptor in the liver, insulin did not suppress glucagon secretion or hepatic glucose production, highlighting the importance of intrahepatic insulin signaling.16
Other indirect mechanisms by which insulin suppresses hepatic gluconeogenesis are through reducing gluconeogenic substrate release from adipose tissue and skeletal muscle or by acting on the brain. Insulin has inhibitory effects on lipolysis and proteolysis and thus decreases plasma levels of non-esterified fatty acids (NEFAs) and glycerol derived from adipose tissue, as well as amino acids from skeletal muscle.10,14 A reduction of free fatty acid delivery to the liver has been shown to decrease hepatic glucose output.17 However, NEFA failed to reduce hepatic glucose production in liver-specific insulin receptor gene knockout mice, suggesting that NEFAs are substrates that depend on intrahepatic insulin signaling to regulate hepatic gluconeogenesis.16 The role of the central nervous system in gluconeogenesis is complex and has been recently reviewed,18 but insulin has been found to inhibit gluconeogenesis by acting on the brain in an insulin receptor–dependent manner.19
The idea that extrahepatic insulin signaling can control hepatic glucose production (HGP) is supported by the fact that insulin can suppress HGP in mice where the canonical hepatic insulin signaling components Akt and FOXO1 are depleted.20 Moreover, acute depletion of the insulin receptor and FOXO1 in liver does not prevent insulin from suppressing HGP.21 It is important to mention, however, that in these experiments insulin can still signal through the IGF receptor, which might be sufficient to suppress HGP. A recent study further supports the idea that insulin’s main effect on HGP is through suppression of lipolysis in white adipose tissue.22 Here, intrahepatic acetyl-CoA levels were shown to be elevated in HFD-fed rodents, resulting in an increase in pyruvate carboxylase activity and gluconeogenesis. The increase in hepatic acetyl-CoA is a result of increased lipolysis due to insulin resistance in fat. In support of the importance of the fat lipolysis–hepatic acetyl-CoA axis in controlling HGP, reducing lipolysis by inhibition of adipose triglyceride lipase or by neutralizing interleukin (IL)-6, a cytokine known to promote lipolysis in fat, normalizes hepatic acetyl-CoA levels and pyruvate carboxylase activity as well as HGP.22
Insulin regulation of hepatic gluconeogenesis through transcriptional modulation
Insulin can regulate hepatic gluconeogenesis via transcription of genes involved in gluconeogenic control, including PCK1 and G6PC.23 Changes in transcription may not contribute to acute regulation of hepatic glucose output, but they determine the gluconeogenic capacity of the liver and may have long-term effects, especially in pathological states.
Whether transcriptional regulation of gluconeogenic enzymes can be observed in human livers is controversial. A recent study showed no induction of PCK1 and G6PC in liver biopsies of patients with type 2 diabetes.24 However, another study in patients found a clear correlation between insulin resistance and PCK1, G6PC, and FOXO1 mRNA levels.25 An explanation for these findings may be that lesions in nonalcoholic steatohepatitis are unequally distributed over the liver, and significant sampling errors might occur.26 Also, the degree of liver damage and other parameters, such as total body weight and tissue cross talk, may affect whether changes in gene expression are observed. Nonetheless, since gluconeogenesis is a primary driver of hepatic glucose production in type 2 diabetic patients4,27 and insulin affects gluconeogenesis through transcription, determining the insulin signaling pathways that alter gluconeogenic gene transcription and ultimately gluconeogenesis can contribute to our understanding of type 2 diabetes pathophysiology.
Insulin activates several signaling pathways that regulate gluconeogenesis
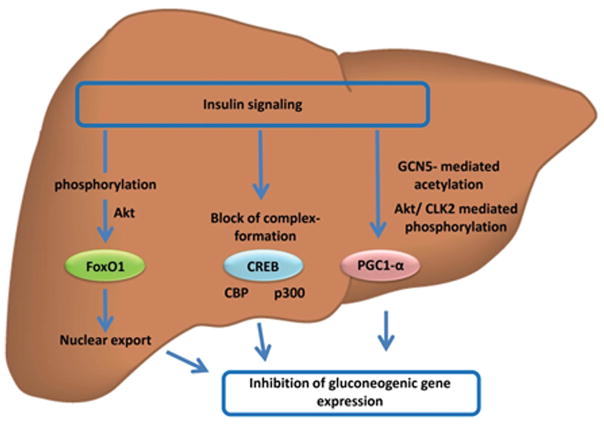
Insulin binds to and initiates signaling through the insulin receptor, which is a tyrosine kinase that is activated upon ligand binding. This activation leads to phosphorylation of a variety of intracellular substrates. Two major downstream pathways of insulin action are the PI3K and MAPK pathways.28 However, the activities of several other pathways are modulated by insulin action, with implications for transcriptional control of gluconeogenesis (Fig. 1).
Figure 1.
Regulation of gluconeogenic gene expression by hepatic insulin signaling. Insulin action regulates the activity of trancription factors controlling gluconeogenic gene expression. AKT-mediated phosphorylation leads to nuclear export of FOXO1. Inhibitory phosphorylation of CBP/p300 blocks trancription-complex formation of CREB. Modification of PGC-1α by GCN5-mediated acetylation or AKT/CLK2-mediated phosphorylation decreases PGC-1α transcriptional activity.
IRS1 and IRS2
Among the numerous downstream signaling components of the insulin receptor, the insulin receptor substrates (IRS) play a key role in the regulation of hepatic glucose production. Mice lacking the IRS2 protein exhibited diabetes-like symptoms and increased hepatic glucose production.29 Mice with knockout of Irs1 had decreased insulin sensitivity, but not diabetes per se.30 Double knockout of Irs1 and Irs2 in the liver leads to severe hyperglycemia, hyperinsulinemia, and induction of gluconeogenic genes, such as Pck1 and G6pc.31 However, the double knockout also had severe growth defects, so interpretation of data from this animal model must be cautious. Taken together, these data demonstrate that IRS proteins are important for insulin signal transduction, and that IRS1 and IRS2 can compensate for each other.
PI3K
The activation of IRS proteins results in the recruitment of the lipid kinase PI3K to the plasma membrane, where it phosphorylates PI-(4,5)-bisphosphate (PtdIns(4,5)P2/PIP2) to generate PtdIns(3,4,5)P3 (PIP3), an important second messenger of several growth factor receptors and mediators of PEPCK and G-6-Pase expression levels. Adenoviral overexpression of the dominant-negative mutant of the PI3K regulatory subunit p85α, which lacks the binding site for the PI3K catalytic subunit, increased Pck1 and G6pc gene expression, as well as hepatic glucose production in vivo.32 Pharmacological inhibition of PI3K also inhibited the insulin-mediated suppression of Pck1 and G6pc expression.33 Therefore, there is evidence that PI3K is a necessary downstream component of insulin-suppressed gluconeogenesis.
PDK1
Downstream of PI3K, the generation of PIP3 is known to increase the activity of 3-phosphatidylinositol-dependent kinase-1 (PDK1).28 Mice deficient for PDK1 in the liver have decreased glucose tolerance, decreased pyruvate tolerance, and fail to normalize blood glucose levels 2 h after insulin administration,34 probably because livers from these mice display a massive defect in glycogen storage. Furthermore, hepatic glucose output and gluconeogenic gene expression were not suppressed by feeding in these mice. Acute insulin-triggered Akt signaling was also impaired in mice lacking PDK1 in the liver. These findings suggest that PDK1 is important for insulin signaling but also plays a major role in glucose metabolism under physiological conditions.
Akt
PIP3 generated by PI3K allows Akt/PKB to bind and be phosphorylated by PDK1.35 Insulin-stimulated PI3K-mediated phosphorylation of Akt at Ser473 activates the kinase.36 Akt kinases control diverse functions, including cell growth, survival, proliferation, and metabolism. However, the mechanisms through which Akt activity is specified to particular cellular functions in response to extracellular stimuli are not fully understood. Studies of Akt isoform–specific gene knockout mice suggested that Akt signaling diversity might in part be due to the different functions of the three Akt family members AKT1, AKT2, and AKT3.37 Among the three, AKT2 appears to play the major role in glucose metabolism; however, redundancy in Akt functions also appear to occur. Akt2 knockout mice developed a type 2 diabetes–like phenotype, and cells derived from these mice had impaired glucose utilization, suggesting a central role for AKT2 in the maintenance of glucose homeostasis.38 Simultaneous deletion of Akt1 and Akt2 caused lethality shortly after birth,39 and Akt1 and Akt3 double-knockout mice were embryonic lethal.40 However, mice with knockout of Akt2 and Akt3 with a single functional allele of Akt1 (Akt1+/−Akt2−/−Akt3−/−) were viable despite reduced body weight and insulin and glucose intolerance.37 While knockout of Akt2 in mice caused insulin resistance and diabetes-like symptoms, including increased hepatic glucose production, AKT1 was dispensable for glucose homeostasis.38,39 Of note, a role for increased Pck1 and G6pc transcription was not demonstrated in these mouse models, suggesting regulation of acute glucose output in vivo rather than of chronic transcriptional changes. However, overexpression of Akt in hepatoma cells and primary hepatocyte cultures decreased Pck1 and G6pc gene transcription, demonstrating that Akt can regulate transcription of gluconeogenic enzymes.41
Several proteins have been identified to mediate suppression of Pck1 and G6pc by Akt, such as FOXO1, a master regulator of gluconeogenesis.42 Akt seems to be the key regulator of FOXO1 activity.43 Ablation of Akt1 and Akt2 in the livers of mice leads to insulin resistance and diabetes and increased expression of several FoOXO1 target genes.20 Pck1 and G6pc expression levels in the knockout livers were not different from that in the control livers during fasting, but the normal suppression of these genes after feeding was lost. Deletion of Foxo1 in these mice normalized the hyperglycemia, glucose intolerance, hyperinsulinemia, and the response to feeding despite defective insulin signaling.20 This study elegantly showcased the regulatory effect of the Akt–FOXO1 network on hepatic gluconeogenesis.
Activated Akt can phosphorylate and inhibit glycogen synthase kinase-3 (GSK-3), which inhibits glycogen synthase and therefore enhances the release of G-6-P from glycogen. However, whether this is a direct insulin-mediated effect remains unclear. Pharmacological inhibition of GSK-3 was shown to suppress Pck1 and G6pc transcription, but to a lesser extent than insulin.40 Furthermore, overexpression of GSK-3 had no effect on insulin-mediated suppression of Pck1 and G6pc transcription, suggesting different mechanisms of action for GSK-3 and insulin on Pck1 and G6pc expression.40
MAPK
Insulin is a potent simulator of the Raf/MEK/ERK1/2 pathway, and this pathway might be involved in the regulation of gluconeogenesis. Thus far, neither pharmacological inhibition nor overexpression of components of the Raf/MEK/ERK1/2 pathway have been shown to be efficient in suppressing Pck1 and G6pc transcription or hepatic glucose production.33,44,45 However, the downstream effector p38 has gained attention in the regulation of gluconeogenesis. Inhibition of p38 by siRNA or a pharmacological inhibitor reduced glycemia in mice and suppressed gluconeogenesis in liver, along with suppression of Pck1 and G6pc transcription.46 Activation of p38 by free fatty acids showed similar results on Pck1 and G6pc transcription.47,48 Mice lacking MAPK phosphatase 1 (MKP-1), a negative regulator of p38 and JNK activity, exhibited increased gluconeogenesis and hepatic insulin resistance.49 P38 inhibition also decreased transcription of peroxisome proliferator–activated receptor γ coactivator 1α (Ppargc1a) and Creb, suggesting transcriptional control of these major regulators of gluconeogenic gene expression.46 P38 can also activate PGC-1α by phosphorylation.50 Along with PGC-1α and CREB, p38 also phosphorylates and activates FOXO1, implicating a broader role for p38 in the transcriptional regulation of glucose homeostasis and gluconeogenesis.46,47,51 Additionally, insulin promotes glycogen synthesis through the activation of protein phosphatase-1.52 This is mediated through the Ras/MAPK pathway, and protein phosphatase-1 in turn activates glycogen synthase while simultaneously inactivating phosphorylase a and phosphorylase kinase to control glycogen metabolism.52
CDC-like kinase
CDC-like kinase 2 (CLK2) belongs to a large and highly conserved family of kinases. Among the CLKs, CLK1–4 play roles in mRNA splicing and nuclear recruitment of proteins.53 However, CLK2 is the only kinase known to be regulated by insulin and to have an effect on hepatic gluconeogenesis. CLK2 is directly phosphorylated and thereby stabilized via insulin-activated Akt.54 Overexpression of CLK2 decreased hepatic glucose output in mice and corrected hyperglycemia in db/db mice.55 On the contrary, in liver-specific Clk2 knockout mice, no diabetic phenotype was observed, suggesting a compensatory adaptive mechanism during chronic CLK2 deficiency.56 CLK2 potentially mediates its effects through modulation of the PP2A–phosphatase complex and phosphorylation of PGC-1α, leading to decreased transcriptional activity of PGC-1α.54,56
Cyclin-dependent kinases
Cyclin-dependent kinases (CDKs) are a family of protein kinases first characterized for their roles in the cell cycle.57 They are also involved in regulating transcription, mRNA processing, and differentiation.58 They are present in all known eukaryotes, and their regulatory function in the cell cycle has been evolutionarily conserved.59 The mechanism by which insulin affects the activity of CDKs is not completely understood. Reports suggested a role for insulin in the activity of CDK regulatory subunit 1 (CKS1), CDK4, and CDK5. CKS1 is upregulated by insulin via the insulin receptor to promote cell proliferation in insects, but no effect on gluconeogenesis was reported.60 Two independent reports recently showed the activation of CDK4 through refeeding and insulin in the liver. Both groups also observed profound effects on hepatic gluconeogenesis. This very likely occurs via the inhibition of PGC-1α transcriptional activity.61,62 The role of CDK5 in insulin signaling is unclear. CDK5 activity is increased by insulin, and this effect can be abolished by a PI3K inhibitor in primary adipocytes.63 In adipose tissue, activated CDK5 promotes adipogenesis through phosphorylation and activation of PPAR-γ.64 A role for CDK5 in regulating hepatic glucose production remains elusive.
Transcriptional regulation of gluconeogenesis by insulin
FOXO1
In mammals, the Forkhead protein family comprises four proteins, FOXO1, FOXO3 (FOXO3a), FOXO4, and FOXO6. Among these, FOXO6 shows a high specificity to neuronal localization, while the other three factors are widely distributed and are present in most tissues. There seems to be a considerable overlap in the transcriptional targets of the first three, and there is also evidence that each of these can compensate for loss of the others to some degree.65,66 While Foxo1 knockout is embryonically lethal owing a the failure of angiogenesis, Foxo3 knockout produces premature ovarian failure, and Foxo4 knockout has no obvious phenotype.28 Triple conditional knockouts resulted in lymphomas, hemangiomas, and angiosarcomas, which did not occur with double-knockout combinations.65 FOXO1 specifically plays a major role in the regulation of insulin sensitivity.66 FOXO1 transcriptional activity is regulated by a complex array of posttranslational modifications (PTMs). With regards to the gluconeogenic function of FOXO1, the primary regulatory event is Akt-mediated phosphorylation of three conserved residues, one threonine and two serines (Thr24, Ser253, and Ser316 in Mus musculus), that results in binding to 14-3-3 proteins and nuclear export of FOXO1.66
Mice with specific deletion of Foxo1 in the liver show reduced fasting glucose and gluconeogenic gene expression, and glucose clamp studies demonstrate that Foxo1 ablation impairs fasting- and cAMP-induced glycogenolysis and gluconeogenesis. Furthermore, Foxo1 deletion prevents neonatal diabetes and steatohepatitis in insulin receptor gene knockout mice.67 When FOXO1 is constitutively expressed in the murine liver, fasting blood glucose increases.68 There is strong evidence that FOXO1 directly regulates Pck1 and G6pc transcription. FOXO1 has been reported to bind to the insulin-responsive sequences of the Pck1 and G6pc promoters in vitro.69,70 The importance of Akt phosphorylation on FOXO1 regulation is striking. In the fed state, insulin signaling activates PI3K and subsequently Akt. Akt then phosphorylates FOXO1, leading to its inactivation though its export from the nucleus, with subsequent suppression of gluconeogenesis.20 Hyperglycemia following Akt gene deletion can be corrected by concomitant hepatic deletion of Foxo1.20,38 These data indicate that the regulation of hepatic glucose metabolism and the maintenance of glucose homeostasis are strongly mediated though the Akt–FOXO1 axis. In this context, the main function of insulin-mediated signaling is to counteract FOXO1 and thus reduce glucose production during the fed state. However, FOXO1 does not inhibit the insulin-mediated upregulation of anabolic processes, such as glycogen and lipid synthesis.20
As mentioned briefly earlier, the activity of FOXO1 is also modulated by processes other than Akt-mediated phosphorylation. Its acetylation and deacetylation provide a second tier of regulation, and deacetylation is generally considered to enhance FOXO1 activity to promote gluconeogenesis. FOXO1 can be deacetylated by SIRT1 under conditions of cellular stress, in the process overriding the nuclear exclusion effect of Akt and causing nuclear retention and expression of FOXO1 target genes, including Pck1 and G6pc.71 Class IIa HDACs can also contribute to deacetylation of FOXO1 and have been shown to be positive regulators of hepatic FOXO1 in response to glucagon signaling during fasting.72 Once these HDACs are activated through AMPK-dependent phosphorylation, they translocate to the nucleus, where they deacetylate and activate FOXO1, inducing transcription of gluconeogenic genes.66,71
Several other mechanisms of modulating FOXO1 activity, such as regulation of its glycosylation, ubiquitination, and proteosomal degradation, have recently been described. The transcription factor XBP-1, involved in the unfolded protein response, has been shown to increase insulin sensitivity. This activity is independent of its transcriptional effects, and may be mediated through its indirect interaction with FOXO1, acting as a chaperone to direct it toward proteosomal degradation.73 The herpesvirus-associated ubiquitin-specific protease 7 (USP7; also known as herpesvirus-associated ubiquitin-specific protease (HAUSP)) is capable of mono-deubiquinating FOXO1, resulting in suppression of FOXO1 transcriptional activity through decreased FOXO1 occupancy on the promoters of gluconeogenic genes.74 Another mechanism that can be utilized to modulate FOXO1 activity is through the glucose-derived O-linked β-N-acetylglucosamine (O-GlcNAc) modification. In diabetes, this specific modification is increased on hepatic FOXO1. It regulates FOXO1 activation in response to glucose, resulting in increased expression of gluconeogenic genes, while concomitantly inducing expression of genes involved in the ROS detoxification pathways.75,76 Paradoxically, it is induced by hyperglycemia and appears to result from PGC-1α binding to O-GlcNAc transferase and targeting it to nuclear FOXO1.77 Other signaling pathways, such as thyroid hormone signaling, retinoid signaling, and p53-regulated pathways, have been linked to modulation of the FOXO1 transcriptional activity on gluconeogenic genes.55,78,79 The physiological relevance of these findings has yet to be determined.
CREB
CREB was first identified as a transcription factor that binds to cyclic AMP–binding element (CRE) on the promoter of the somatostatin gene.80 Mice that express a dominant-negative inhibitor of CREB in the liver (alb-ACREB transgenic mice) showed decreased blood glucose levels and decreased expression of gluconeogenic genes, emphasizing the role of CREB in hepatic gluconeogenesis as a progluconeogenic factor.81 Since then, the role of CREB as a crucial transcription factor in liver metabolism has been studied extensively. The key event in CREB activation is its phosphorylation at Ser133 following an increase in intracellular cAMP levels and subsequent activation of PKA. Activated PKA translocates to the nucleus and phosphorylates CREB, which is critical for the interaction of CREB with CBP/p300 to promote its transcriptional activity.13 Apart from PKA, other kinases have been reported to phosphorylate CREB, including calmodulin-dependent kinases and MAPK kinases, such as p38.13 Reversal of CREB phosphorylation can be mediated through PP1 and PP2 phosphatases, with potential tissue- and cell-specific regulatory patterns.13 However, the interaction of CREB with its coactivators, rather than the phosphorylation of Ser133, appears to be the critical event that promotes its transcriptional activity.13 CBP and p300 are closely related histone acetyl transferase orthologs that catalyze the transfer of the acetyl group from acetyl CoA to lysine residues of histone or non-histone proteins. The formation of a transcriptional complex of CREB and CBP/p300 seems to be critical for the selective transcriptional induction of cAMP-responsive genes.13,82 Insulin has also been found to inhibit gluconeogenesis by selectively disrupting the CREB–CBP interaction. Refeeding triggers the phosphorylation of CBP at Ser436 by the atypical PKC-ι/γ (aPKCι/γ). This modification on CBP appears to block binding of CREB that is phosphorylated at Ser133. However, the regulatory Ser436 site in CBP is not conserved in p300, suggesting that CBP and p300 perform distinct roles in the liver.13,83 CBP/p300 can additionally be phosphorylated and thereby inactivated by salt-inducible kinase (SIK).84 It has also been reported that active CBP/p300 can stabilize CRTC2 through acetylation of a lysine residue, thereby increasing its activity.85 The CRTC family consists of three members (CRTC1, CRTC2, and CRTC3) that have similar modular structures: they all contain an N-terminal CREB-binding domain (CBD), a central regulatory domain (REG), a splicing domain (SD), and a C-terminal transactivation domain (TAD).13 In the basal state, CRTCs are sequestered in the cytoplasm through phosphorylation-dependent interactions with 14-3-3 proteins, such as SIK.13 Exposure to cAMP and calcium, but not other signals, triggers the calcineurin-mediated dephosphorylation and nuclear translocation of CRTCs, which then bind to CREB over relevant gene promoters. Among the different family members, CRTC2 is highly expressed in the liver. In fasting conditions, PKA-dependent inactivation of SIK leads to dephosphorylation of CRTC2 and its nuclear translocation.86 Mice with a Crtc2 knockout have decreased circulating glucose concentrations during fasting, lowered circulating blood glucose concentrations, and improved insulin sensitivity in the context of diet-induced obesity.87 Hyperglycemia affects the activity of CRTC2 by enhancing O-glycosylation, resulting in reduced phosphorylation at Ser171, nuclear retention, and subsequent increased activity.13,88
CREBH
CREBH is an endoplasmic reticulum (ER)-bound transcription factor that is controlled by regulated intramembrane proteolysis (RIP) of the ER, which is known to maintain sterol homeostasis and to mediate the unfolded protein response (UPR). It is mainly expressed in the liver and is activated upon acute ER stress, when it promotes the transcription of acute phase response genes, such as Crp (C-reactive protein) or Sap (serum amyloid P-component). CREBH has been reported to induce the transcription of gluconeogenic genes, and acute knockdown of CREBH reduces fasting blood glucose levels in mice.89 In mice with hepatic knockout of CREBH, a decrease in hepatic lipid content was observed in those on normal chow diet, while those on a high-fat diet displayed increased steatosis, suggesting that CREBH is involved in lipogenesis and lipolysis; however, no abnormalities in blood glucose were observed.90 The same study reported that insulin treatment promotes cleavage and thereby activation of CREBH.90
PGC-1α
PGC-1α was initially identified in brown adipocytes as a coactivator of the transcription factor PPAR-γ, where it is critical for the control of thermogenesis and mitochondrial biogenesis.91,92 The family of PGC-1 transcriptional coactivators includes various isoforms of PGC-1α as well as PGC-1β and PGC-1–related co-activator (PRC). They share similar structural features, comprising an N-terminal activation domain, a central regulatory domain, and a C-terminal RNA-binding domain.93 Following PPAR-γ, PGC-1α has been shown to interact with various transcription factors, such as PPAR-α, PPAR-δ, FOXO1, and SREBP, and nuclear receptors, such as estrogen related receptors (ERRs), hepatocyte nuclear factor 4α (HNF4α), and glucocorticoid receptor (GR).93 In addition, PGC-1α interacts with several other proteins, including HAT domain–containing proteins, such as CBP and p300, via the N-terminal domain, and the mediator complex via the C-terminal region.94,95 Knockdown or knockout of Ppargc1a in primary hepatocytes results in decreased glucose production and reduced expression of gluconeogenic genes, while Ppargc1a null mice show increased gluconeogenic gene expression insensitive to normal feeding controls.96 Overexpression of PGC-1α in the liver of mice promotes the expression of gluconeogenic genes by enhancing the activity of various transcription factors, such as CREB, HNF4α, and FOXO1.81,97,98 Insulin-stimulated Akt-mediated phosphorylation of FOXO1 obstructs its interaction with PGC-1α and interferes with the activation of hepatic gluconeogenic gene expression in mice.97 Insulin can also regulate PGC-1α activity via the alteration of its acetylation and phosphorylation status. Acetylation of hepatic PGC-1α is controlled by counteracting effects of the GCN5 acetyltransferase and the SIRT1 deacetylase, and this PTM impairs the ability of PGC-1α to promote gluconeogenic gene expression.99,100 Insulin activates the cyclin D1/cyclin-dependent kinase 4 complex, which increases GCN5 acetyltransferase activity through phosphorylation, to promote the acetylation of PGC-1α that results in the suppression of hepatic glucose production.62 The transcriptional co-regulator CITED2 (CBP- and p300-interacting transactivator with glutamic acid– and aspartic acid–rich COOH-terminal domain 2) activates PGC-1α by decreasing its acetylation through blocking its interaction with GCN5, resulting in increased gluconeogenic gene expression; however, this event is negatively regulated by insulin in an Akt-dependent manner.101 Additionally, activation of Akt through insulin signaling enables it to phosphorylate PGC-1α directly at Ser570, thereby inhibiting its ability to be recruited to gluconeogenic gene promoters.102 The insulin-regulated CLK2 phosphorylates PGC-1α in its serine- and arginine-rich (SR) domain, impairing its ability to function as a transcriptional coactivator, specifically toward FOXO1.55 The insulin-inducible corepressor small heterodimer partner–interacting leucine zipper protein (SMILE) directly competes with PGC-1α for HNF4α to suppress its hepatic transcriptional activity and gluconeogenesis.103
Glucocorticoid receptor
GR has been linked to hepatic gluconeogenesis for a long time. It is activated by endogenous cortisol as well as exogenous corticosteroids. Upon fasting, increased secretion of glucocorticoids activates GR in the liver, which promotes transcription of gluconeogenic genes.104 Even though insulin does not directly interact with GR, the transcriptional activity of GR is important for modulating insulin-mediated signaling. GR increases the transcription of Mapk14 (p38 MAPK) and thereby counteracts the insulin-mediated effects on gluconeogenesis. Glucocorticoids also diminish IR and IRS1 phosphorylation in response to insulin in liver.105
Sterol response element–binding protein
Members of the sterol response element–binding protein (SREBP) family were initially characterized as transcription factors that are activated by low cholesterol levels in the cell.106 The SREBP-1c isoform in the liver has been demonstrated to be regulated by nutritional stimuli in vivo, such as a carbohydrate-rich diet, and insulin has been found to rapidly stimulate the transcription of Srebp1 in hepatocytes and adipose and muscle tissue. The effect of insulin on the expression level of SREBP-1c protein in hepatocytes appears to be mediated through activation of PI3K. In addition, by employing a version of Akt that can be conditionally regulated, studies in hepatoma cells provide evidence that activation of Akt is sufficient to mimic this biological response to insulin.107 While mice with siRNA-mediated knockdown of Srebp1 show decreased gluconeogenic gene expression, no effect was observed on fasting glucose levels.108 A suggested mechanism of SREBP-1 action on gluconeogenesis is its interference with the HNF4α-mediated recruitment of PGC-1α to gluconeogenic gene promoters.109 Coimmunoprecipitation experiments demonstrate that these two transcription factors (SREBP-1 and HNF4α) directly interact through the transactivation domain of SREBP and the ligand binding/AF2 domains of HNF4α.109 In the same study, SREBP-1 does not bind the Pck1 promoter, supporting an indirect effect of SREBP-1 on gluconeogenic gene expression, possibly through altering the localization of HNF4α and PGC-1α.109 Overall, the role of SREBP-1 in gluconeogenesis appears to be moderate, and the main functional role lies in the regulation of lipid metabolism.
STAT3
STAT3 is a member of the signal transducer and activator of transcription (STAT) protein family. STAT3 is phosphorylated by receptor-associated Janus kinases (JAKs) in response to cytokines and growth factors. Activated STAT3 proteins form homo- or heterodimers, and translocate to the cell nucleus, where they act as transcription activators.110 In the liver, STAT activation is mainly linked to inflammation and cancer. Interestingly, STAT3 has the ability to directly bind to the promoters of Pck1 and G6pc to inhibit the promoter activity of these gluconeogenic genes.111 In mice, liver-specific deletion of Stat3 increases the expression of Pck1, G6pc, and Ppargc1a. Conversely, liver-specific overexpression of a constitutively active form of STAT3 decreases hepatic glucose production and blood glucose levels in diabetic mice.112 A significant mechanism through which insulin is able to modulate STAT3 activity is its hypothalamic action. Insulin action in the hypothalamus stimulates IL-6 production in the liver, and IL-6 in turn suppresses gluconeogenesis by activating STAT3.113 Another point of cross talk between insulin and STAT3 is through GSK-3β. STAT3 suppresses the expression of GSK-3β, a negative regulator of the insulin signaling pathway. Mice lacking STAT3 in the liver do not exhibit a physiological decrease of Gsk3b mRNA or GSK-3β protein levels in response to feeding, which suggests that these mice display an impaired response to insulin.114
DAX1
Dosage-sensitive sex reversal, adrenal hypoplasia congenita critical region on the X-chromosome, gene 1 (DAX1) is an atypical nuclear receptor that lacks a classical DNA-binding domain. It was initially described in hypogonadotropic hypogonadism and is mainly expressed in adrenal glands, the pituitary gland, and the testes.115 DAX1 has been shown to be expressed in the liver, and its expression is increased by insulin and SIK1, whereas it is decreased in diabetic and high-fat diet–fed mice.116 It has been shown that DAX1 blocks the association of HNF4α and PGC-1α with the gluconeogenic gene promoters, suggesting a novel mechanism for feeding-induced repression of gluconeogenesis.116 However, the mechanism through which insulin or insulin-related signaling increases DAX1 is not clear. Current data suggest an increase in Dax1 transcription. In testicular Leydig cells, Dax1 transcription is increased by insulin in an Akt-dependent manner. However, high-fat diet suppresses hepatic DAX1 levels in mice, suggesting that DAX1 regulation is distinct in different tissues.116,117
Insulin signaling and its impact on hepatic gluconeogenesis in diabetes and obesity
In obesity and diabetes, tissue homeostasis is disturbed in many compartments, such as skeletal muscle, adipose tissue, liver, kidney, and connective tissues. Eventually, these changes lead to tissue damage and remodeling, as seen in liver fibrosis, kidney fibrosis, or atherosclerosis. Additionally, these changes are major drivers of insulin resistance and thereby fuel a vicious cycle leading to organ damage, morbidity, and mortality.
Inflammation is a widely recognized occurrence in obesity and diabetes and has been linked to the pathogenesis of diabetes, mainly through impaired insulin signaling.118 Chronic as well as acute inflammation have been clearly linked to insulin resistance and elevated levels of cytokines, such as TNF-α, and have been described and extensively studied in this context.119,120 TNF-α signaling can activate various intracellular pathways, such as those involving JNK and IKK, and this inhibits insulin receptor signaling through serine phosphorylation of IRS1.121 In contrast to the activating tyrosine phosphorylation, this modification leads to inactivity of IRS1 and eventual degradation of the protein.122 Serine phosphorylation of IRS1 can come about as a result of several events, including through mTOR, S6 kinase, PKC-θ, and JNK signaling.123
Disturbed hepatic lipid metabolism is also known to affect insulin signaling and hepatic glucose production. Several studies support a role for hepatic diacylglycerol (DAG) accumulation and PKC-ε activation as factors for impaired hepatic insulin action. DAG has been shown to promote PKC activation with a subsequent decrease in insulin receptor tyrosine kinase activity. Mice lacking PKC are protected from diet-induced insulin resistance, even though hepatic lipid content is increased.124 In humans, FOXO1 expression and transcriptional activity are increased in nonalcoholic steatohepatitis (NASH) liver, emphasizing a role for inflammation and disturbed lipid metabolism in the dysregulation of hepatic gluconeogenesis.25
Non-alcoholic fatty liver disease (NAFLD) is a growing subclass of liver disease, as it is closely related to obesity, metabolic syndrome, and diabetes. Patients diagnosed with hepatic steatosis have an increased risk for cardiovascular disease, as well as for diabetes and related conditions.125 An important trigger may be hepatic insulin resistance, as it may be sufficient to promote dyslipidemia and atherosclerosis.126 In addition, excess lipid accumulation in the liver has negative effects on the progression of liver disease. Interestingly, hepatic insulin signaling is impaired, leading to not only an increase in HGP but also an ongoing hepatic lipid production that results in hepatic steatosis, steatohepatitis, and cirrhosis. The phenomenon where impaired insulin signaling blocks one of its actions (decrease of HGP) while promoting another action (lipogenesis) is termed selective insulin resistance. A potential mechanism leading to this paradox in insulin action in the brain and the adipose tissue, which results in increased lipolysis and excess triglyceride synthesis in the liver, has been recently reviewed elsewhere.127
Additionally, insulin action in the central nervous system can indirectly regulate metabolism in the liver. Insulin signaling in the brain through hypothalamic PI3K and ATP-sensitive potassium channels has been suggested to suppress Pck1 and G6pc expression in the liver, which results in the blunting of gluconeogenesis; activation of the hepatic IL-6/STAT3 pathway has been demonstrated to be mechanistically involved in this process.112,128–130 Insulin-mediated FOXO1 nuclear exclusion through the PI3K–Akt axis in pro-opiomelanocortin (POMC) neurons has also been suggested to suppress basal HGP.131 However, a study by Ramnanan et al. indicated that, in dogs, regulation of hepatic gluconeogenesis mediated by the action of insulin on the brain is debatable.132 Scherer et al. additionally showed that insulin signaling in the rat brain is able to modulate triglyceride secretion and lipid content in the liver.133 Thus, we highlighted the promise of potentially targeting insulin resistance in the central nervous system for the treatment of type 2 diabetes.134
Concluding remarks
An intact insulin signaling system is paramount to maintaining blood glucose levels within a narrow normal glycemic range during periods of fasting or excess nutrient availability, and this is achieved in particular via regulation of metabolic flux through the hepatic gluconeogenic pathway. This review summarizes some of the mechanisms through which insulin can modulate hepatic gluconeogenesis. A significant node of control involves the transcriptional regulation of expression of the key hepatic gluconeogenic genes Pck1 and G6pc, which occurs primarily through the transcription factor FOXO1 and nuclear receptor HNF4α and their transcriptional coactivator PGC-1α. Understanding these regulatory pathways is of extreme importance in terms of identifying potential targets and devising treatments for disorders involving insulin resistance and dysregulated hepatic gluconeogenesis, such as in type 2 diabetes.
Table 1.
Extrahepatic effects of insulin that regulate hepatic gluconeogenesis.
| Organ/tissue | Action | Effect on liver | Reference |
|---|---|---|---|
|
| |||
| Pancreatic alpha cells | Secretion of glucagon ↓ | Transcriptional regulation of gluconeogenic genes | 12–14, 16 |
| White adipose tissue | Lipolysis ↓ | Reduction of free fatty acid delivery to the liver | 10,14,17 |
| Skeletal muscle | Proteolysis ↓ | Reduction of amino acid flux to the liver | 10,14 |
| Central nervous system | Pleiotropic manner | Multiple effects | 18 |
Table 2.
Animal models of targets of insulin signaling
| Insulin target | Phenotype of (liver-specific) KO mice | Potential mechanism of action | Reference |
|---|---|---|---|
|
| |||
| IRS1, IRS2 | Hyperglycemia, hyperinsulinemia | Induction of gluconeogenic genes | 29–31 |
| PI3K (p110-α and p110-β subunits) | Diet-induced liver steatosis ↓ Glucose intolerance (p110-α) ↑ |
Insulin-induced Akt phosphorylation at Ser473 ↓ | 135 |
| PDK1 | Glucose tolerance ↓ Pyruvate tolerance ↓ Insulin sensitivity ↑ |
Disruption of acute AKT-dependent insulin signaling | 34 |
| Akt2 | Insulin resistance ↑ Hepatic glucose production ↑ |
Increased expression of FOXO1 target genes | 20, 38, 39 |
| p38 MAPK | Glycemia ↓ Gluconeogenesis ↓ |
Decreased transcription of Pparg, Ppargc1a | 46 |
| FoxO1 | Fasting glucose ↓ Gluconeogenic gene expression ↓ |
Direct regulation of G6pc and Pck1 transcription | 67–70 |
| PGC-1α | Gluconeogenic gene expression ↑ Loss of feeding-induced regulation |
Regulation of G6pc and Pck1 transcription | 96 |
Footnotes
Competing Interests
The author declares no competing interests
References
- 1.Gerich JE, Meyer C, Woerle HJ, Stumvoll M. Renal gluconeogenesis: its importance in human glucose homeostasis. Diabetes care. 2001;24:382–391. doi: 10.2337/diacare.24.2.382. [DOI] [PubMed] [Google Scholar]
- 2.Garrett Reginald H, CMG . Principles of Biochemistry with a Human Focus. USA: Brooks/Cole, Thomson Learning; 2002. [Google Scholar]
- 3.O’Brien RM, Lucas PC, Forest CD, Magnuson MA, Granner DK. Identification of a sequence in the PEPCK gene that mediates a negative effect of insulin on transcription. Science. 1990;249:533–537. doi: 10.1126/science.2166335. [DOI] [PubMed] [Google Scholar]
- 4.Magnusson I, Rothman DL, Katz LD, Shulman RG, Shulman GI. Increased rate of gluconeogenesis in type II diabetes mellitus. A 13C nuclear magnetic resonance study. The Journal of clinical investigation. 1992;90:1323–1327. doi: 10.1172/JCI115997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gastaldelli A, Toschi E, Pettiti M, Frascerra S, Quinones-Galvan A, Sironi AM, Natali A, Ferrannini E. Effect of physiological hyperinsulinemia on gluconeogenesis in nondiabetic subjects and in type 2 diabetic patients. Diabetes. 2001;50:1807–1812. doi: 10.2337/diabetes.50.8.1807. [DOI] [PubMed] [Google Scholar]
- 6.Petersen KF, Laurent D, Rothman DL, Cline GW, Shulman GI. Mechanism by which glucose and insulin inhibit net hepatic glycogenolysis in humans. The Journal of clinical investigation. 1998;101:1203–1209. doi: 10.1172/JCI579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edgerton DS, Lautz M, Scott M, Everett CA, Stettler KM, Neal DW, Chu CA, Cherrington AD. Insulin’s direct effects on the liver dominate the control of hepatic glucose production. The Journal of clinical investigation. 2006;116:521–527. doi: 10.1172/JCI27073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williamson JR, Wright PH, Malaisse WJ, Ashmore J. Control of gluconeogenesis by acetyl CoA in rats treated with glucagon and anti-insulin serum. Biochemical and biophysical research communications. 1966;24:765–770. doi: 10.1016/0006-291x(66)90391-3. [DOI] [PubMed] [Google Scholar]
- 9.Mondon CE, Mortimore GE. Effects of insulin on amino acid release and urea formation in perfused rat liver. The American journal of physiology. 1967;212:173–178. doi: 10.1152/ajplegacy.1967.212.1.173. [DOI] [PubMed] [Google Scholar]
- 10.Girard J. Insulin’s effect on the liver: “direct or indirect?” continues to be the question. The Journal of clinical investigation. 2006;116:302–304. doi: 10.1172/JCI27743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.IOS, Zhang W, Wasserman DH, Liew CW, Liu J, Paik J, DePinho RA, Stolz DB, Kahn CR, Schwartz MW, Unterman TG. FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nature communications. 2015;6:7079. doi: 10.1038/ncomms8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravier MA, Rutter GA. Glucose or insulin, but not zinc ions, inhibit glucagon secretion from mouse pancreatic alpha-cells. Diabetes. 2005;54:1789–1797. doi: 10.2337/diabetes.54.6.1789. [DOI] [PubMed] [Google Scholar]
- 13.Altarejos JY, Montminy M. CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nature reviews. Molecular cell biology. 2011;12:141–151. doi: 10.1038/nrm3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharabi K, Tavares CD, Rines AK, Puigserver P. Molecular pathophysiology of hepatic glucose production. Molecular aspects of medicine. 2015;46:21–33. doi: 10.1016/j.mam.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawamori D, Kurpad AJ, Hu J, Liew CW, Shih JL, Ford EL, Herrera PL, Polonsky KS, McGuinness OP, Kulkarni RN. Insulin signaling in alpha cells modulates glucagon secretion in vivo. Cell metabolism. 2009;9:350–361. doi: 10.1016/j.cmet.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher SJ, Kahn CR. Insulin signaling is required for insulin’s direct and indirect action on hepatic glucose production. The Journal of clinical investigation. 2003;111:463–468. doi: 10.1172/JCI16426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis GF, Vranic M, Harley P, Giacca A. Fatty acids mediate the acute extrahepatic effects of insulin on hepatic glucose production in humans. Diabetes. 1997;46:1111–1119. doi: 10.2337/diab.46.7.1111. [DOI] [PubMed] [Google Scholar]
- 18.Morton GJ, Schwartz MW. Leptin and the central nervous system control of glucose metabolism. Physiological reviews. 2011;91:389–411. doi: 10.1152/physrev.00007.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buettner C, Patel R, Muse ED, Bhanot S, Monia BP, McKay R, Obici S, Rossetti L. Severe impairment in liver insulin signaling fails to alter hepatic insulin action in conscious mice. The Journal of clinical investigation. 2005;115:1306–1313. doi: 10.1172/JCI23109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu M, Wan M, Leavens KF, Chu Q, Monks BR, Fernandez S, Ahima RS, Ueki K, Kahn CR, Birnbaum MJ. Insulin regulates liver metabolism in vivo in the absence of hepatic Akt and Foxo1. Nature medicine. 2012;18:388–395. doi: 10.1038/nm.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Titchenell PM, Chu Q, Monks BR, Birnbaum MJ. Hepatic insulin signalling is dispensable for suppression of glucose output by insulin in vivo. Nature communications. 2015;6:7078. doi: 10.1038/ncomms8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perry RJ, Camporez JP, Kursawe R, Titchenell PM, Zhang D, Perry CJ, Jurczak MJ, Abudukadier A, Han MS, Zhang XM, Ruan HB, Yang X, Caprio S, Kaech SM, Sul HS, Birnbaum MJ, Davis RJ, Cline GW, Petersen KF, Shulman GI. Hepatic acetyl CoA links adipose tissue inflammation to hepatic insulin resistance and type 2 diabetes. Cell. 2015;160:745–758. doi: 10.1016/j.cell.2015.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barthel A, Schmoll D. Novel concepts in insulin regulation of hepatic gluconeogenesis. American journal of physiology. Endocrinology and metabolism. 2003;285:E685–692. doi: 10.1152/ajpendo.00253.2003. [DOI] [PubMed] [Google Scholar]
- 24.Samuel VT, Beddow SA, Iwasaki T, Zhang XM, Chu X, Still CD, Gerhard GS, Shulman GI. Fasting hyperglycemia is not associated with increased expression of PEPCK or G6Pc in patients with Type 2 Diabetes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12121–12126. doi: 10.1073/pnas.0812547106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valenti L, Rametta R, Dongiovanni P, Maggioni M, Fracanzani AL, Zappa M, Lattuada E, Roviaro G, Fargion S. Increased expression and activity of the transcription factor FOXO1 in nonalcoholic steatohepatitis. Diabetes. 2008;57:1355–1362. doi: 10.2337/db07-0714. [DOI] [PubMed] [Google Scholar]
- 26.Ratziu V, Charlotte F, Heurtier A, Gombert S, Giral P, Bruckert E, Grimaldi A, Capron F, Poynard T, Group LS. Sampling variability of liver biopsy in nonalcoholic fatty liver disease. Gastroenterology. 2005;128:1898–1906. doi: 10.1053/j.gastro.2005.03.084. [DOI] [PubMed] [Google Scholar]
- 27.Rizza RA. Pathogenesis of fasting and postprandial hyperglycemia in type 2 diabetes: implications for therapy. Diabetes. 2010;59:2697–2707. doi: 10.2337/db10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lizcano JM, Alessi DR. The insulin signalling pathway. Current biology : CB. 2002;12:R236–238. doi: 10.1016/s0960-9822(02)00777-7. [DOI] [PubMed] [Google Scholar]
- 29.Withers DJ, Gutierrez JS, Towery H, Burks DJ, Ren JM, Previs S, Zhang Y, Bernal D, Pons S, Shulman GI, Bonner-Weir S, White MF. Disruption of IRS-2 causes type 2 diabetes in mice. Nature. 1998;391:900–904. doi: 10.1038/36116. [DOI] [PubMed] [Google Scholar]
- 30.Tamemoto H, Kadowaki T, Tobe K, Yagi T, Sakura H, Hayakawa T, Terauchi Y, Ueki K, Kaburagi Y, Satoh S, et al. Insulin resistance and growth retardation in mice lacking insulin receptor substrate-1. Nature. 1994;372:182–186. doi: 10.1038/372182a0. [DOI] [PubMed] [Google Scholar]
- 31.Dong X, Park S, Lin X, Copps K, Yi X, White MF. Irs1 and Irs2 signaling is essential for hepatic glucose homeostasis and systemic growth. The Journal of clinical investigation. 2006;116:101–114. doi: 10.1172/JCI25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyake K, Ogawa W, Matsumoto M, Nakamura T, Sakaue H, Kasuga M. Hyperinsulinemia, glucose intolerance, and dyslipidemia induced by acute inhibition of phosphoinositide 3-kinase signaling in the liver. The Journal of clinical investigation. 2002;110:1483–1491. doi: 10.1172/JCI15880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agati JM, Yeagley D, Quinn PG. Assessment of the roles of mitogen-activated protein kinase, phosphatidylinositol 3-kinase, protein kinase B, and protein kinase C in insulin inhibition of cAMP-induced phosphoenolpyruvate carboxykinase gene transcription. The Journal of biological chemistry. 1998;273:18751–18759. doi: 10.1074/jbc.273.30.18751. [DOI] [PubMed] [Google Scholar]
- 34.Mora A, Lipina C, Tronche F, Sutherland C, Alessi DR. Deficiency of PDK1 in liver results in glucose intolerance, impairment of insulin-regulated gene expression and liver failure. The Biochemical journal. 2005;385:639–648. doi: 10.1042/BJ20041782. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Hemmings BA, Restuccia DF. The PI3K-PKB/Akt pathway. Cold Spring Harbor perspectives in biology. 2015;7:261–269. doi: 10.1101/cshperspect.a026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brozinick JT, Jr, Birnbaum MJ. Insulin, but not contraction, activates Akt/PKB in isolated rat skeletal muscle. The Journal of biological chemistry. 1998;273:14679–14682. doi: 10.1074/jbc.273.24.14679. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez E, McGraw TE. The Akt kinases: isoform specificity in metabolism and cancer. Cell cycle. 2009;8:2502–2508. doi: 10.4161/cc.8.16.9335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB, 3rd, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 39.Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. The Journal of biological chemistry. 2001;276:38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 40.Lochhead PA, Coghlan M, Rice SQ, Sutherland C. Inhibition of GSK-3 selectively reduces glucose-6-phosphatase and phosphatase and phosphoenolypyruvate carboxykinase gene expression. Diabetes. 2001;50:937–946. doi: 10.2337/diabetes.50.5.937. [DOI] [PubMed] [Google Scholar]
- 41.Liao J, Barthel A, Nakatani K, Roth RA. Activation of protein kinase B/Akt is sufficient to repress the glucocorticoid and cAMP induction of phosphoenolpyruvate carboxykinase gene. The Journal of biological chemistry. 1998;273:27320–27324. doi: 10.1074/jbc.273.42.27320. [DOI] [PubMed] [Google Scholar]
- 42.Schmoll D, Walker KS, Alessi DR, Grempler R, Burchell A, Guo S, Walther R, Unterman TG. Regulation of glucose-6-phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. The Journal of biological chemistry. 2000;275:36324–36333. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- 43.Tzivion G, Dobson M, Ramakrishnan G. FoxO transcription factors; Regulation by AKT and 14–3-3 proteins. Biochimica et biophysica acta. 2011;1813:1938–1945. doi: 10.1016/j.bbamcr.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 44.Gabbay RA, Sutherland C, Gnudi L, Kahn BB, O’Brien RM, Granner DK, Flier JS. Insulin regulation of phosphoenolpyruvate carboxykinase gene expression does not require activation of the Ras/mitogen-activated protein kinase signaling pathway. The Journal of biological chemistry. 1996;271:1890–1897. doi: 10.1074/jbc.271.4.1890. [DOI] [PubMed] [Google Scholar]
- 45.Sutherland C, Waltner-Law M, Gnudi L, Kahn BB, Granner DK. Activation of the ras mitogen-activated protein kinase-ribosomal protein kinase pathway is not required for the repression of phosphoenolpyruvate carboxykinase gene transcription by insulin. The Journal of biological chemistry. 1998;273:3198–3204. doi: 10.1074/jbc.273.6.3198. [DOI] [PubMed] [Google Scholar]
- 46.Cao W, Collins QF, Becker TC, Robidoux J, Lupo EG, Jr, Xiong Y, Daniel KW, Floering L, Collins S. p38 Mitogen-activated protein kinase plays a stimulatory role in hepatic gluconeogenesis. The Journal of biological chemistry. 2005;280:42731–42737. doi: 10.1074/jbc.M506223200. [DOI] [PubMed] [Google Scholar]
- 47.Collins QF, Xiong Y, Lupo EG, Jr, Liu HY, Cao W. p38 Mitogen-activated protein kinase mediates free fatty acid-induced gluconeogenesis in hepatocytes. The Journal of biological chemistry. 2006;281:24336–24344. doi: 10.1074/jbc.M602177200. [DOI] [PubMed] [Google Scholar]
- 48.Liu HY, Collins QF, Xiong Y, Moukdar F, Lupo EG, Jr, Liu Z, Cao W. Prolonged treatment of primary hepatocytes with oleate induces insulin resistance through p38 mitogen-activated protein kinase. The Journal of biological chemistry. 2007;282:14205–14212. doi: 10.1074/jbc.M609701200. [DOI] [PubMed] [Google Scholar]
- 49.Lawan A, Zhang L, Gatzke F, Min K, Jurczak MJ, Al-Mutairi M, Richter P, Camporez JP, Couvillon A, Pesta D, Roth Flach RJ, Shulman GI, Bennett AM. Hepatic mitogen-activated protein kinase phosphatase 1 selectively regulates glucose metabolism and energy homeostasis. Molecular and cellular biology. 2015;35:26–40. doi: 10.1128/MCB.00503-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Puigserver P, Rhee J, Lin J, Wu Z, Yoon JC, Zhang CY, Krauss S, Mootha VK, Lowell BB, Spiegelman BM. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Molecular cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- 51.Asada S, Daitoku H, Matsuzaki H, Saito T, Sudo T, Mukai H, Iwashita S, Kako K, Kishi T, Kasuya Y, Fukamizu A. Mitogen-activated protein kinases, Erk and p38, phosphorylate and regulate Foxo1. Cellular signalling. 2007;19:519–527. doi: 10.1016/j.cellsig.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 52.Ragolia L, Begum N. Protein phosphatase-1 and insulin action. Molecular and cellular biochemistry. 1998;182:49–58. [PubMed] [Google Scholar]
- 53.Araki S, Dairiki R, Nakayama Y, Murai A, Miyashita R, Iwatani M, Nomura T, Nakanishi O. Inhibitors of CLK protein kinases suppress cell growth and induce apoptosis by modulating pre-mRNA splicing. PloS one. 2015;10:e0116929. doi: 10.1371/journal.pone.0116929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodgers JT, Vogel RO, Puigserver P. Clk2 and B56beta mediate insulin-regulated assembly of the PP2A phosphatase holoenzyme complex on Akt. Molecular cell. 2011;41:471–479. doi: 10.1016/j.molcel.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodgers JT, Haas W, Gygi SP, Puigserver P. Cdc2-like kinase 2 is an insulin-regulated suppressor of hepatic gluconeogenesis. Cell metabolism. 2010;11:23–34. doi: 10.1016/j.cmet.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tabata M, Rodgers JT, Hall JA, Lee Y, Jedrychowski MP, Gygi SP, Puigserver P. Cdc2-like kinase 2 suppresses hepatic fatty acid oxidation and ketogenesis through disruption of the PGC-1alpha and MED1 complex. Diabetes. 2014;63:1519–1532. doi: 10.2337/db13-1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suryadinata R, Sadowski M, Sarcevic B. Control of cell cycle progression by phosphorylation of cyclin-dependent kinase (CDK) substrates. Bioscience reports. 2010;30:243–255. doi: 10.1042/BSR20090171. [DOI] [PubMed] [Google Scholar]
- 58.Hydbring P, Malumbres M, Sicinski P. Non-canonical functions of cell cycle cyclins and cyclin-dependent kinases. Nature reviews. Molecular cell biology. 2016;17:280–292. doi: 10.1038/nrm.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morgan DO. The cell cycle : principles of control. New Science Press ; Sinauer Associates; London Sunderland, MA: 2007. [Google Scholar]
- 60.Liu CY, Zhao WL, Wang JX, Zhao XF. Cyclin-dependent kinase regulatory subunit 1 promotes cell proliferation by insulin regulation. Cell cycle. 2015;14:3045–3057. doi: 10.1080/15384101.2015.1053664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhalla K, Liu WJ, Thompson K, Anders L, Devarakonda S, Dewi R, Buckley S, Hwang BJ, Polster B, Dorsey SG, Sun Y, Sicinski P, Girnun GD. Cyclin D1 represses gluconeogenesis via inhibition of the transcriptional coactivator PGC1alpha. Diabetes. 2014;63:3266–3278. doi: 10.2337/db13-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee Y, Dominy JE, Choi YJ, Jurczak M, Tolliday N, Camporez JP, Chim H, Lim JH, Ruan HB, Yang X, Vazquez F, Sicinski P, Shulman GI, Puigserver P. Cyclin D1-Cdk4 controls glucose metabolism independently of cell cycle progression. Nature. 2014;510:547–551. doi: 10.1038/nature13267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lalioti V, Muruais G, Dinarina A, van Damme J, Vandekerckhove J, Sandoval IV. The atypical kinase Cdk5 is activated by insulin, regulates the association between GLUT4 and E-Syt1, and modulates glucose transport in 3T3-L1 adipocytes. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4249–4253. doi: 10.1073/pnas.0900218106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choi JH, Banks AS, Estall JL, Kajimura S, Bostrom P, Laznik D, Ruas JL, Chalmers MJ, Kamenecka TM, Bluher M, Griffin PR, Spiegelman BM. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARgamma by Cdk5. Nature. 2010;466:451–456. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tikhanovich I, Cox J, Weinman SA. Forkhead box class O transcription factors in liver function and disease. Journal of gastroenterology and hepatology. 2013;28(Suppl 1):125–131. doi: 10.1111/jgh.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell metabolism. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 68.Matsumoto M, Han S, Kitamura T, Accili D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. The Journal of clinical investigation. 2006;116:2464–2472. doi: 10.1172/JCI27047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ayala JE, Streeper RS, Desgrosellier JS, Durham SK, Suwanichkul A, Svitek CA, Goldman JK, Barr FG, Powell DR, O’Brien RM. Conservation of an insulin response unit between mouse and human glucose-6-phosphatase catalytic subunit gene promoters: transcription factor FKHR binds the insulin response sequence. Diabetes. 1999;48:1885–1889. doi: 10.2337/diabetes.48.9.1885. [DOI] [PubMed] [Google Scholar]
- 70.Hall RK, Yamasaki T, Kucera T, Waltner-Law M, O’Brien R, Granner DK. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. The Journal of biological chemistry. 2000;275:30169–30175. doi: 10.1074/jbc.M004898200. [DOI] [PubMed] [Google Scholar]
- 71.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. The Journal of biological chemistry. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 72.Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhou Y, Lee J, Reno CM, Sun C, Park SW, Chung J, Lee J, Fisher SJ, White MF, Biddinger SB, Ozcan U. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nature medicine. 2011;17:356–365. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall JA, Tabata M, Rodgers JT, Puigserver P. USP7 attenuates hepatic gluconeogenesis through modulation of FoxO1 gene promoter occupancy. Molecular endocrinology (Baltimore, Md ) 2014;28:912–924. doi: 10.1210/me.2013-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Housley MP, Rodgers JT, Udeshi ND, Kelly TJ, Shabanowitz J, Hunt DF, Puigserver P, Hart GW. O-GlcNAc regulates FoxO activation in response to glucose. The Journal of biological chemistry. 2008;283:16283–16292. doi: 10.1074/jbc.M802240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuo M, Zilberfarb V, Gangneux N, Christeff N, Issad T. O-glycosylation of FoxO1 increases its transcriptional activity towards the glucose 6-phosphatase gene. FEBS letters. 2008;582:829–834. doi: 10.1016/j.febslet.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 77.Housley MP, Udeshi ND, Rodgers JT, Shabanowitz J, Puigserver P, Hunt DF, Hart GW. A PGC-1alpha-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. The Journal of biological chemistry. 2009;284:5148–5157. doi: 10.1074/jbc.M808890200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shin DJ, Joshi P, Hong SH, Mosure K, Shin DG, Osborne TF. Genome-wide analysis of FoxO1 binding in hepatic chromatin: potential involvement of FoxO1 in linking retinoid signaling to hepatic gluconeogenesis. Nucleic acids research. 2012;40:11499–11509. doi: 10.1093/nar/gks932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang P, Tu B, Wang H, Cao Z, Tang M, Zhang C, Gu B, Li Z, Wang L, Yang Y, Zhao Y, Wang H, Luo J, Deng CX, Gao B, Roeder RG, Zhu WG. Tumor suppressor p53 cooperates with SIRT6 to regulate gluconeogenesis by promoting FoxO1 nuclear exclusion. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:10684–10689. doi: 10.1073/pnas.1411026111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 81.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 82.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhou XY, Shibusawa N, Naik K, Porras D, Temple K, Ou H, Kaihara K, Roe MW, Brady MJ, Wondisford FE. Insulin regulation of hepatic gluconeogenesis through phosphorylation of CREB-binding protein. Nature medicine. 2004;10:633–637. doi: 10.1038/nm1050. [DOI] [PubMed] [Google Scholar]
- 84.Dentin R, Liu Y, Koo SH, Hedrick S, Vargas T, Heredia J, Yates J, 3rd, Montminy M. Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature. 2007;449:366–369. doi: 10.1038/nature06128. [DOI] [PubMed] [Google Scholar]
- 85.Liu Y, Dentin R, Chen D, Hedrick S, Ravnskjaer K, Schenk S, Milne J, Meyers DJ, Cole P, Yates J, 3rd, Olefsky J, Guarente L, Montminy M. A fasting inducible switch modulates gluconeogenesis via activator/coactivator exchange. Nature. 2008;456:269–273. doi: 10.1038/nature07349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Koo SH, Flechner L, Qi L, Zhang X, Screaton RA, Jeffries S, Hedrick S, Xu W, Boussouar F, Brindle P, Takemori H, Montminy M. The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature. 2005;437:1109–1111. doi: 10.1038/nature03967. [DOI] [PubMed] [Google Scholar]
- 87.Wang Y, Inoue H, Ravnskjaer K, Viste K, Miller N, Liu Y, Hedrick S, Vera L, Montminy M. Targeted disruption of the CREB coactivator Crtc2 increases insulin sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:3087–3092. doi: 10.1073/pnas.0914897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dentin R, Hedrick S, Xie J, Yates J, 3rd, Montminy M. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 2008;319:1402–1405. doi: 10.1126/science.1151363. [DOI] [PubMed] [Google Scholar]
- 89.Lee MW, Chanda D, Yang J, Oh H, Kim SS, Yoon YS, Hong S, Park KG, Lee IK, Choi CS, Hanson RW, Choi HS, Koo SH. Regulation of hepatic gluconeogenesis by an ER-bound transcription factor, CREBH. Cell metabolism. 2010;11:331–339. doi: 10.1016/j.cmet.2010.02.016. [DOI] [PubMed] [Google Scholar]
- 90.Zhang C, Wang G, Zheng Z, Maddipati KR, Zhang X, Dyson G, Williams P, Duncan SA, Kaufman RJ, Zhang K. Endoplasmic reticulum-tethered transcription factor cAMP responsive element-binding protein, hepatocyte specific, regulates hepatic lipogenesis, fatty acid oxidation, and lipolysis upon metabolic stress in mice. Hepatology (Baltimore, Md ) 2012;55:1070–1082. doi: 10.1002/hep.24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 92.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 93.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell metabolism. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 94.Puigserver P, Adelmant G, Wu Z, Fan M, Xu J, O’Malley B, Spiegelman BM. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 95.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Molecular cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 96.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 97.Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 98.Rhee J, Inoue Y, Yoon JC, Puigserver P, Fan M, Gonzalez FJ, Spiegelman BM. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell metabolism. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 100.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 101.Sakai M, Matsumoto M, Tujimura T, Yongheng C, Noguchi T, Inagaki K, Inoue H, Hosooka T, Takazawa K, Kido Y, Yasuda K, Hiramatsu R, Matsuki Y, Kasuga M. CITED2 links hormonal signaling to PGC-1[alpha] acetylation in the regulation of gluconeogenesis. Nature medicine. 2012;18:612–617. doi: 10.1038/nm.2691. [DOI] [PubMed] [Google Scholar]
- 102.Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 103.Lee JM, Seo WY, Han HS, Oh KJ, Lee YS, Kim DK, Choi S, Choi BH, Harris RA, Lee CH, Koo SH, Choi HS. Insulin-Inducible SMILE Inhibits Hepatic Gluconeogenesis. Diabetes. 2016;65:62–73. doi: 10.2337/db15-0249. [DOI] [PubMed] [Google Scholar]
- 104.Zinker B, Mika A, Nguyen P, Wilcox D, Ohman L, von Geldern TW, Opgenorth T, Jacobson P. Liver-selective glucocorticoid receptor antagonism decreases glucose production and increases glucose disposal, ameliorating insulin resistance. Metabolism: clinical and experimental. 2007;56:380–387. doi: 10.1016/j.metabol.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 105.Saad MJ, Folli F, Kahn JA, Kahn CR. Modulation of insulin receptor, insulin receptor substrate-1, and phosphatidylinositol 3-kinase in liver and muscle of dexamethasone-treated rats. The Journal of clinical investigation. 1993;92:2065–2072. doi: 10.1172/JCI116803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang X, Sato R, Brown MS, Hua X, Goldstein JL. SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell. 1994;77:53–62. doi: 10.1016/0092-8674(94)90234-8. [DOI] [PubMed] [Google Scholar]
- 107.Foufelle F, Ferre P. New perspectives in the regulation of hepatic glycolytic and lipogenic genes by insulin and glucose: a role for the transcription factor sterol regulatory element binding protein-1c. The Biochemical journal. 2002;366:377–391. doi: 10.1042/BJ20020430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ruiz R, Jideonwo V, Ahn M, Surendran S, Tagliabracci VS, Hou Y, Gamble A, Kerner J, Irimia-Dominguez JM, Puchowicz MA, DePaoli-Roach A, Hoppel C, Roach P, Morral N. Sterol regulatory element-binding protein-1 (SREBP-1) is required to regulate glycogen synthesis and gluconeogenic gene expression in mouse liver. The Journal of biological chemistry. 2014;289:5510–5517. doi: 10.1074/jbc.M113.541110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yamamoto T, Shimano H, Nakagawa Y, Ide T, Yahagi N, Matsuzaka T, Nakakuki M, Takahashi A, Suzuki H, Sone H, Toyoshima H, Sato R, Yamada N. SREBP-1 interacts with hepatocyte nuclear factor-4 alpha and interferes with PGC-1 recruitment to suppress hepatic gluconeogenic genes. The Journal of biological chemistry. 2004;279:12027–12035. doi: 10.1074/jbc.M310333200. [DOI] [PubMed] [Google Scholar]
- 110.Lim CP, Cao X. Structure, function, and regulation of STAT proteins. Molecular bioSystems. 2006;2:536–550. doi: 10.1039/b606246f. [DOI] [PubMed] [Google Scholar]
- 111.Ramadoss P, Unger-Smith NE, Lam FS, Hollenberg AN. STAT3 targets the regulatory regions of gluconeogenic genes in vivo. Molecular endocrinology (Baltimore, Md ) 2009;23:827–837. doi: 10.1210/me.2008-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Inoue H, Ogawa W, Ozaki M, Haga S, Matsumoto M, Furukawa K, Hashimoto N, Kido Y, Mori T, Sakaue H, Teshigawara K, Jin S, Iguchi H, Hiramatsu R, LeRoith D, Takeda K, Akira S, Kasuga M. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nature medicine. 2004;10:168–174. doi: 10.1038/nm980. [DOI] [PubMed] [Google Scholar]
- 113.Inoue H, Ogawa W, Asakawa A, Okamoto Y, Nishizawa A, Matsumoto M, Teshigawara K, Matsuki Y, Watanabe E, Hiramatsu R, Notohara K, Katayose K, Okamura H, Kahn CR, Noda T, Takeda K, Akira S, Inui A, Kasuga M. Role of hepatic STAT3 in brain-insulin action on hepatic glucose production. Cell metabolism. 2006;3:267–275. doi: 10.1016/j.cmet.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 114.Moh A, Zhang W, Yu S, Wang J, Xu X, Li J, Fu XY. STAT3 sensitizes insulin signaling by negatively regulating glycogen synthase kinase-3 beta. Diabetes. 2008;57:1227–1235. doi: 10.2337/db06-1582. [DOI] [PubMed] [Google Scholar]
- 115.Iyer AK, McCabe ER. Molecular mechanisms of DAX1 action. Molecular genetics and metabolism. 2004;83:60–73. doi: 10.1016/j.ymgme.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 116.Nedumaran B, Hong S, Xie YB, Kim YH, Seo WY, Lee MW, Lee CH, Koo SH, Choi HS. DAX-1 acts as a novel corepressor of orphan nuclear receptor HNF4alpha and negatively regulates gluconeogenic enzyme gene expression. The Journal of biological chemistry. 2009;284:27511–27523. doi: 10.1074/jbc.M109.034660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ahn SW, Gang GT, Kim YD, Ahn RS, Harris RA, Lee CH, Choi HS. Insulin directly regulates steroidogenesis via induction of the orphan nuclear receptor DAX-1 in testicular Leydig cells. The Journal of biological chemistry. 2013;288:15937–15946. doi: 10.1074/jbc.M113.451773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Johnson AM, Olefsky JM. The origins and drivers of insulin resistance. Cell. 2013;152:673–684. doi: 10.1016/j.cell.2013.01.041. [DOI] [PubMed] [Google Scholar]
- 119.Gregor MF, Hotamisligil GS. Inflammatory mechanisms in obesity. Annual review of immunology. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 120.Shangraw RE, Jahoor F, Miyoshi H, Neff WA, Stuart CA, Herndon DN, Wolfe RR. Differentiation between septic and postburn insulin resistance. Metabolism: clinical and experimental. 1989;38:983–989. doi: 10.1016/0026-0495(89)90010-3. [DOI] [PubMed] [Google Scholar]
- 121.Qiao LY, Zhande R, Jetton TL, Zhou G, Sun XJ. In vivo phosphorylation of insulin receptor substrate 1 at serine 789 by a novel serine kinase in insulin-resistant rodents. The Journal of biological chemistry. 2002;277:26530–26539. doi: 10.1074/jbc.M201494200. [DOI] [PubMed] [Google Scholar]
- 122.Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Current biology : CB. 2004;14:1650–1656. doi: 10.1016/j.cub.2004.08.026. [DOI] [PubMed] [Google Scholar]
- 123.Draznin B. Molecular mechanisms of insulin resistance: serine phosphorylation of insulin receptor substrate-1 and increased expression of p85alpha: the two sides of a coin. Diabetes. 2006;55:2392–2397. doi: 10.2337/db06-0391. [DOI] [PubMed] [Google Scholar]
- 124.Samuel VT, Shulman GI. The pathogenesis of insulin resistance: integrating signaling pathways and substrate flux. The Journal of clinical investigation. 2016;126:12–22. doi: 10.1172/JCI77812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Expert Panel on Detection E and Treatment of High Blood Cholesterol in Adults. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III) Jama. 2001;285:2486–2497. doi: 10.1001/jama.285.19.2486. [DOI] [PubMed] [Google Scholar]
- 126.Biddinger SB, Hernandez-Ono A, Rask-Madsen C, Haas JT, Aleman JO, Suzuki R, Scapa EF, Agarwal C, Carey MC, Stephanopoulos G, Cohen DE, King GL, Ginsberg HN, Kahn CR. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell metabolism. 2008;7:125–134. doi: 10.1016/j.cmet.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ferris HA, Kahn CR. Unraveling the Paradox of Selective Insulin Resistance in the Liver: the Brain-Liver Connection. Diabetes. 2016;65:1481–1483. doi: 10.2337/dbi16-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gelling RW, Morton GJ, Morrison CD, Niswender KD, Myers MG, Jr, Rhodes CJ, Schwartz MW. Insulin action in the brain contributes to glucose lowering during insulin treatment of diabetes. Cell metabolism. 2006;3:67–73. doi: 10.1016/j.cmet.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 129.Myers MG., Jr Role reversal: brain insulin and liver STAT3. Cell metabolism. 2006;3:231–232. doi: 10.1016/j.cmet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 130.Obici S, Zhang BB, Karkanias G, Rossetti L. Hypothalamic insulin signaling is required for inhibition of glucose production. Nature medicine. 2002;8:1376–1382. doi: 10.1038/nm1202-798. [DOI] [PubMed] [Google Scholar]
- 131.Shi X, Zhou F, Li X, Chang B, Li D, Wang Y, Tong Q, Xu Y, Fukuda M, Zhao JJ, Li D, Burrin DG, Chan L, Guan X. Central GLP-2 enhances hepatic insulin sensitivity via activating PI3K signaling in POMC neurons. Cell metabolism. 2013;18:86–98. doi: 10.1016/j.cmet.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ramnanan CJ, Saraswathi V, Smith MS, Donahue EP, Farmer B, Farmer TD, Neal D, Williams PE, Lautz M, Mari A, Cherrington AD, Edgerton DS. Brain insulin action augments hepatic glycogen synthesis without suppressing glucose production or gluconeogenesis in dogs. The Journal of clinical investigation. 2011;121:3713–3723. doi: 10.1172/JCI45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Scherer T, Lindtner C, O’Hare J, Hackl M, Zielinski E, Freudenthaler A, Baumgartner-Parzer S, Todter K, Heeren J, Krssak M, Scheja L, Furnsinn C, Buettner C. Insulin Regulates Hepatic Triglyceride Secretion and Lipid Content via Signaling in the Brain. Diabetes. 2016;65:1511–1520. doi: 10.2337/db15-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hallschmid M, Schultes B. Central nervous insulin resistance: a promising target in the treatment of metabolic and cognitive disorders? Diabetologia. 2009;52:2264–2269. doi: 10.1007/s00125-009-1501-x. [DOI] [PubMed] [Google Scholar]
- 135.Chattopadhyay M, Selinger ES, Ballou LM, Lin RZ. Ablation of PI3K p110-alpha prevents high-fat diet-induced liver steatosis. Diabetes. 2011;60:1483–1492. doi: 10.2337/db10-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]