Abstract
In this paper we describe a novel, dominant pleiotropic tomato (Lycopersicon esculentum)-ripening mutation, Cnr (colorless nonripening). This mutant occurred spontaneously in a commercial population. Cnr has a phenotype that is quite distinct from that of the other pleiotropic tomato-ripening mutants and is characterized by fruit that show greatly reduced ethylene production, an inhibition of softening, a yellow skin, and a nonpigmented pericarp. The ripening-related biosynthesis of carotenoid pigments was abolished in the pericarp tissue. The pericarp also showed a significant reduction in cell-to-cell adhesion, with cell separation occurring when blocks of tissue were incubated in water alone. The mutant phenotype was not reversed by exposure to exogenous ethylene. Crosses with other mutant lines and the use of a restriction fragment length polymorphism marker demonstrated that Cnr was not allelic with the pleiotropic ripening mutants nor, alc, rin, Nr, Gr, and Nr-2. The gene has been mapped to the top of chromosome 2, also indicating that it is distinct from the other pleiotropic ripening mutants. We undertook the molecular characterization of Cnr by examining the expression of a panel of ripening-related genes in the presence and absence of exogenous ethylene. The pattern of gene expression in Cnr was related to, but differed from, that of several of the other well-characterized mutants. We discuss here the possible relationships among nor, Cnr, and rin in a putative ripening signal cascade.
The ripening of a fruit imparts a variety of agronomically important characteristics to an otherwise unpalatable product. These include conversion of starch to sugars and changes in color, flavor, and texture. Ripening is a tightly controlled and highly programmed developmental event. Identifying the components of this developmental switch is important not only for manipulating this key plant process but also for understanding the regulation of plant development.
The biochemical and molecular basis of ripening in both climacteric and nonclimacteric fruit has been intensively studied. Genes involved in cell wall degradation, color change, ethylene synthesis, and perception have been cloned, and antisense techniques have been developed to manipulate these aspects of ripening (Gray et al., 1994; Wilkinson et al., 1995, 1997). Very little is known, however, about the regulatory genes specifically associated with ripening. Several single-gene mutations resulting in the reduction or almost complete elimination of ripening are known in tomato (Lycopersicon esculentum) fruit (Grierson, 1986). These mutant loci include rin (ripening-inhibitor; Robinson and Tomes, 1968), nor (nonripening; Tigchelaar et al., 1973), Nr (Never-ripe; Rick, 1956), Gr (Green-ripe; Kerr, 1958), Nr-2 (Never-ripe 2; Kerr, 1982), and alc (alcobaca; Kopeliovitch et al., 1981). These pleiotropic mutations are extremely rare and are likely to encode important regulatory genes. The Nr gene, which has been cloned, encodes a protein with homology to the Arabidopsis ethylene receptor ETR1 (Wilkinson et al., 1995), and the normal alleles residing at rin and nor are the subject of a map-based cloning program in one of the authors' laboratories (J.J.G.). The aim is to eventually understand the structures of the genes identified by these mutations and place their encoded proteins into a framework that will describe the molecular regulation of fruit ripening. It is unlikely that mutants are available for all of the steps in such a regulatory pathway and new ripening mutants are especially valuable. In the current paper we describe the molecular and genetic characterization of a novel, dominant pleiotropic tomato-ripening mutant, Cnr (colorless, nonripening).
MATERIALS AND METHODS
Tomato (Lycopersicon esculentum Mill. cvs Ailsa Craig and Liberto) fruit and the mutant line Cnr derived from cv Liberto were grown in a heated greenhouse using standard cultural practices with regular additions of N,P,K fertilizer and supplementary lighting when required. Plants were grown to three trusses. Fruits were harvested at the following stages: mature green (35 DPA), breaker, breaker plus 3 d, and breaker plus 7 d. Fruits of the Cnr mutant were also harvested at 35 DPA and then at 49, 52, and 56 DPA, which are equivalent to the ripening stages for the normal fruit. Seeds of Lycopersicon cheesmanii (LA483), Gr (LA2453), and Nr-2 (LA2455) were obtained from the Tomato Genetic Resource Center (Davis, CA). The seeds for the rin, nor, Nr, and y mutants were obtained from the Glasshouse Crops Research Institute collection at Horticulture Research International (Wellesbourne, UK).
Ethylene Production and Carotenoid Analysis
For measurement of ethylene production at each stage of ripening, three fruit from each stage were placed in a gas-tight 1-L glass jar at 20°C for 1 h, after which time a 1-mL sample of headspace was analyzed by GC (Ward et al., 1978). Each injection was repeated three times. For analysis of total carotenoid levels, pericarp tissue was freeze-dried and extracted with chloroform. Carotenoid content was then determined by the method of Wellburn (1994). Carotenoids were extracted from the pericarp of three individual Cnr and cv Ailsa Craig fruit at each ripening stage.
Mechanical Tests for Fracture Energy and Analysis of Cell Separation
Measurements of fracture energy were made by the procedure described for potato tissue by Freeman et al. (1992). The force (F) required to propagate a preinitiated crack by driving a 0.28-mm stainless steel wire through 11-mm-diameter × 5-mm-thick discs of tissue was measured and converted to gross fracture energy (E) by the equation E = F/d, where d is the diameter of the disc.
For the cell-separation experiments, three individual Cnr, cv Ailsa Craig, or rin fruit were selected at each of the desired stages of development/ripeness, giving three replicates in each treatment. The fruits were peeled, and the pericarp was cut into 5-mm cubes. These cubes, with an approximate volume of 3 mL, were suspended in 9 mL of distilled water or 0.05 m CDTA, adjusted to pH 6.5, or in 20 units of pectinase enzyme from Aspergillus niger (Sigma) in 0.1 m sodium acetate buffer, pH 4.0, for 3 h at room temperature with gentle rocking. Cell separation was measured as the volume of separated cells after removal of the remaining tissue and settling for 2 h at 4°C. Cell separation in water and CDTA was calculated relative to 100% cell separation obtained after pectinase treatment.
RFLP and Linkage Analysis
Genomic DNA was purified according to the method of Fulton et al. (1995). Restriction digests, Southern blotting, and hybridization of the labeled probes to isolated plant genomic DNA were carried out using standard techniques (Sambrook et al., 1989). RFLP probes (provided by Dr. S. Tanksley, Cornell University, Ithaca, NY) were radiolabeled with 32P using the Bioline oligolabeling system (Bioline, London), according to the manufacturer's instructions. RFLP markers were mapped relative to the mutant locus as two-point data using the computer program MAPMAKER (Lander et al., 1987).
Radiolabeled Probes for RNA Analysis
Strand-specific, radiolabeled RNA probes for ACO1, PG, PSY1, and E8 were synthesized from linearized plasmid template DNA using either T3 or T7 RNA polymerase (Promega) according to the manufacturer's instructions. Radiolabeled ERT16 and 18S DNA probes were generated from a purified DNA insert with random primers according to the procedures described by Feinberg and Vogelstein (1983). A 189-bp fragment, corresponding to the 3′-untranslated region of the NR gene (Wilkinson et al., 1995), was amplified by PCR using the primers NR5 (5′-TAAATGACAAAAGGACAT-3′) and NR3 (5′-GTCAAAAGCTCGATGTAT-3′). The fragment was cloned using the TACloning kit (Invitrogen, NV Leek, The Netherlands), and the resulting plasmid clone generated a single-stranded, radiolabeled RNA probe, as described above. Details of the these cDNA clones were described by Gray et al. (1994) and Wilkinson et al. (1995).
RNA Gel-Blot Analysis and RNase Protection Assay
Total RNA was extracted from a pooled sample of pericarp from three individual mutant or normal fruit, as described by Hamilton et al. (1990). RNA gel-blot analysis was carried out using 10 μg of RNA according to the method of John et al. (1995). RNase protection assays were performed as described by Barry et al. (1996), with a few minor modifications. Fifty micrograms of total RNA was hybridized with radioactive probe in 50 μL of hybridization buffer. Digestion with 3 units of RNase ONE (Promega) for 3 h at 28°C removed the unhybridized RNA.
RESULTS
Isolation of the Mutant and Phenotype
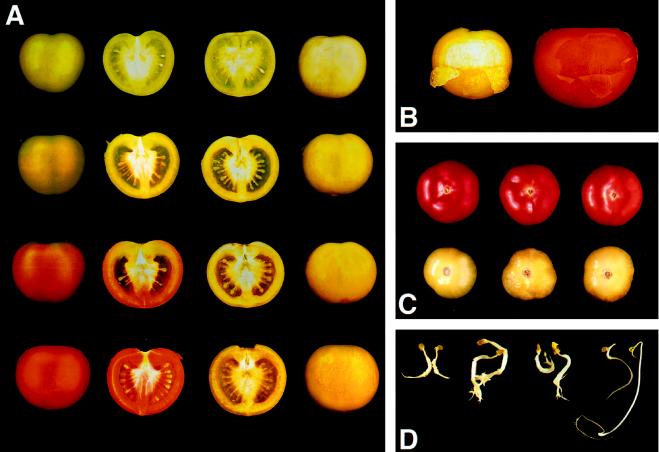
The mutant was isolated in 1993 as a single plant from a commercial planting of the F1 hybrid cv Liberto. It is characterized by fruit that fails to ripen, turns white when mature (40–50 DPA), remains very firm, and then develops a yellow skin. The underlying pericarp tissue remains completely nonpigmented/white (Fig. 1, A and B). Ripening was not restored by the application of exogenous ethylene at 100 μL L−1 to the mutant Cnr at 35 DPA (equivalent to mature-green fruit in cvs Ailsa Craig and Liberto) continuously for 4 d at 19°C (Fig. 1C). However, germinating seedlings showed the triple response in the presence of ethylene (Fig. 1D). To confirm that the yellow appearance of Cnr was due to pigmentation in the skin, a double mutant with a colorless epidermis (y) was generated. The fruit from this cross turned white/cream in color (data not shown).
Figure 1.
A, Ripening of cv Ailsa Craig (left) and homozygous Cnr fruit (right) in a cv Liberto background. Fruit are shown at various DPA corresponding in the wild-type fruit to mature-green, breaker, breaker-plus-3-d, and breaker-plus-7-d stages. B, Skin peeled back on Cnr fruit 56 DPA (left) and red-ripe cv Ailsa Craig (right) to reveal colorless flesh in the mutant. C, Effect of exposure to 100 μL L−1 ethylene for 4 d and then air alone for an additional 5 d at 19°C on the appearance of cv Ailsa Craig (top) and Cnr (bottom) fruit. D, Seedlings from (left to right) Cnr, cv Ailsa Craig, and cv Liberto displaying sensitivity to ethylene, in contrast to those of the Nr mutant.
Allelism Tests with Other Pleiotropic Tomato-Ripening Mutants and Genetic Mapping of Cnr
The original mutant was selfed to produce a homozygous line in the F3 generation, which was crossed to L. esculentum cv Ailsa Craig. We scored 99 of the F2 plants for their ripening phenotype (Table I). Mutant plants could not be separated into heterozygous and homozygous classes based on the severity of the phenotype. A χ2 test for goodness-of-fit to a ratio of 3:1 (mutant:wild type) gave a test statistic of 1.22, which is not significant at the 5% level. Thus, we concluded that Cnr is a dominant, nuclear-encoded mutation.
Table I.
Test for dominance and allelism
| Test and Cross | Phenotypic Class | Expected Ratio | |||
|---|---|---|---|---|---|
| Dominance | Cnr | +a | |||
| Cnr/Cnr × +/+ F2 self | 79 | 20 | 3:1 | ||
| Allelism | Cnr, + | +, + | Cnr, mutb | +, mut | |
| Cnr/Cnr × rin/rin F2 self | 14 | 2 | 1 | 3 | 9:3:3:1 |
| rin/rin × Cnr/Cnr F2 self | 4 | 8 | 6 | 2 | 9:3:3:1 |
| Nr/Nr × Cnr/Cnr BCc(male) | 8*d | 7 | 5 | 2:1:1 | |
| Nr/Nr × Cnr/Cnr BC (female) | 12* | 5 | 3 | 2:1:1 | |
| Nr-2/Nr-2 × Cnr/Cnr BC (male) | |||||
| Fruit Ae | 22* | 6 | 6 | 2:1:1 | |
| Fruit B | 9* | 1 | 3 | 2:1:1 | |
| Gr/Gr × Cnr/Cnr BC (male) | |||||
| Fruit A | 12* | 5 | 7 | 2:1:1 | |
| Fruit B | 12* | 7 | 6 | 2:1:1 | |
+, Wild-type phenotype.
mut, The phenotype of the ripening mutant tested in the cross.
BC indicates a back-cross of the F1 parent to either male or female wild type. In allelism tests, expected ratios assume independent segregation.
Asterisk indicates that the Cnr, + phenotype could not be distinguished from the Cnr, mut double-mutant phenotype, and so the two classes were pooled. These two classes could be distinguished only in the case of rin because of the macrocalyx phenotype tightly linked to the rin mutation.
Fruit A or B indicates progeny from two single fruit.
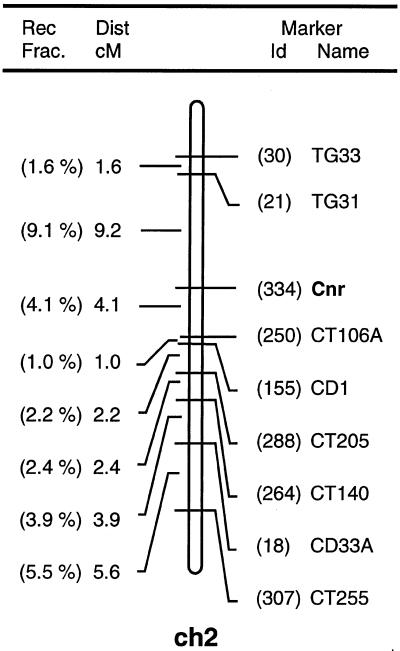
Additional crosses, including reciprocal crosses, with the pleiotropic ripening mutants Nr, rin, Nr-2, and Gr, all yielded wild-type plants in the F2 self- or back-cross progeny (Table I), demonstrating that Cnr is nonallelic to these loci and further supporting the dominant nuclear nature of the mutation. In all cases, except for Cnr × rin, the ripening characteristics of the double mutants were not visually distinct from Cnr alone. The phenotype of Cnr was easily distinguished from the other mutants by the lack of pigment in the pericarp. To test whether Cnr was allelic with the two additional pleiotropic ripening mutants nor and alc, an RFLP analysis was performed. Tigchelaar and Barman (1985) concluded that nor and alc are allelic, although two tightly linked loci cannot be excluded. The RFLP marker CT16, which maps 0.9 centimorgan from nor (Giovannoni et al., 1995) was used to follow the segregation pattern from a cross between Cnr (in L. esculentum background) and L. cheesmanii in an F2 population. Of the 21 F2 plants analyzed, 13 showed recombination between Cnr and CT16, indicating that these two loci segregate independently (Table II). It follows that Cnr cannot be allelic with nor or alc. Using the F2 mapping population, we carried out additional experiments to map the Cnr gene. Initially, the least ambiguous class of homozygous, wild-type F2 families was used to map the position of the gene. Subsequently, as linked markers were identified, confirmation of the map position was sought using heterozygous and homozygous mutant families, and linkage was observed with molecular markers at the top of chromosome 2 (Fig. 2). Thus, Cnr was located in an interval between the RFLP markers TG31 and CT106A. Nr, rin, nor, alc, and Nr-2 all map to chromosomes other than 2 (Gray et al., 1994). No map location could be found in the literature for Gr.
Table II.
Test for independent segregation of Cnr and CT16 in the cross L. esculentum, (Cnr/Cnr) × L. cheesmanii, (+/+) F2 self
c and e are the CT16 alleles from L. cheesemaii and L. esculentum, respectively.
Classes that show recombination between Cnr and CT16.
Figure 2.
Linkage map of tomato chromosome 2 from the analysis of F2 progeny from the cross L. esculentum (Cnr/Cnr) × L. cheesmannii (+/+). Rec Frac., Recombination fraction; Dist, distance; cM, centimorgan.
Biochemical and Molecular Characterization of Ripening Fruit
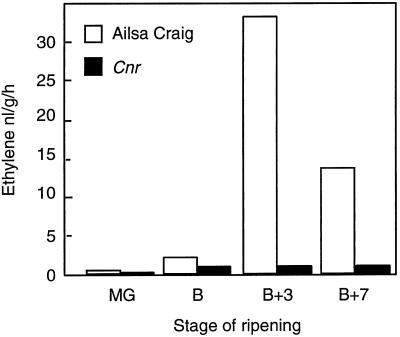
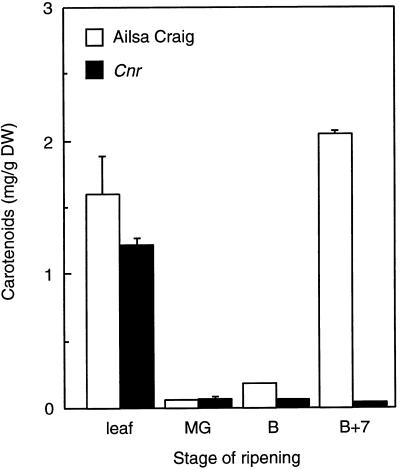
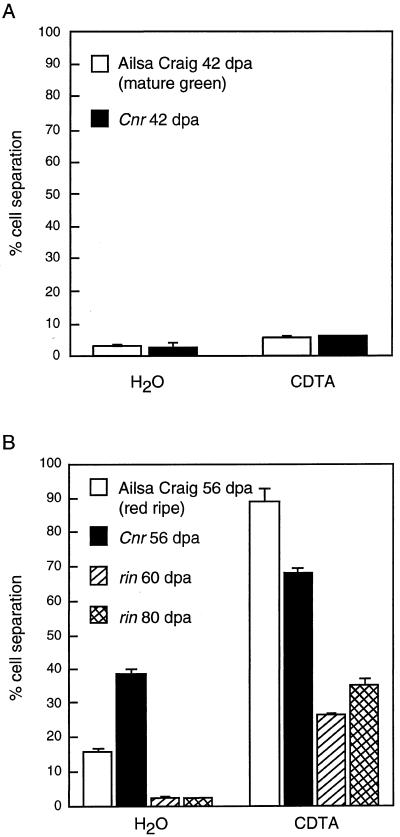
Ethylene production in the fruit of both cvs Liberto and Ailsa Craig showed a climacteric rise as the fruit ripened. A much reduced level of ethylene production was apparent in Cnr, as illustrated in Figure 3. The ethylene measurements were repeated on two separate occasions with different batches of fruit, and similar results were obtained. A distinctive feature of Cnr was the lack of pigmentation in the pericarp tissue. Analysis of the total carotenoid content revealed that ripening-related production of these compounds was absent in the Cnr fruit. However, similar levels of total colored carotenoids were present in the mature, unripe fruit and leaves of Cnr, in comparison with wild-type plants (Fig. 4). On handling the Cnr fruit, we found that they were very firm in comparison with the wild type, and mechanical tests demonstrated that they exhibited a higher fracture energy (Table III), although the pericarp tissues often had a mealy appearance when cut. This apparent mealiness was reflected in altered cell-to-cell adhesion properties in Cnr. Preliminary experiments showed that, unlike wild-type fruit, significant cell separation occurred in the Cnr pericarp tissue when it was left in water for a few hours. To obtain a more accurate comparison of cell-to-cell adhesion in Cnr and wild-type fruit, blocks of pericarp were floated in water alone or in a solution containing the Ca chelator CDTA. The degree of cell separation was then compared with the 100% cell separation obtained after pectinase treatment. In unripe fruit there was limited cell separation apparent in the Cnr and the wild-type tissue either in water or in CDTA. However, blocks of Cnr pericarp from 56-d-old fruit (equivalent to red-ripe, wild-type fruit) incubated in water alone showed extensive cell separation (Fig. 5). In contrast, although the Ca chelator CDTA caused more extensive cell separation than water in all of the fruit, the effects on Cnr were less pronounced than on the wild-type material (Fig. 5B). In rin fruit, even at 80 DPA, minimal cell separation occurred in water, although loss of cell adhesion was enhanced by CDTA (Fig. 5B).
Figure 3.
Ethylene production by cv Ailsa Craig fruit at mature-green (MG), breaker (B), breaker-plus-3-d (B+3), and breaker-plus-7-d (B+7) stages and Cnr fruit at equivalent ages postanthesis. Three fruit were placed in a gas-tight jar, and ethylene was sampled in the headspace after 1 h. Values shown are the means of three injections on the GC.
Figure 4.
Total carotenoid content of cv Ailsa Craig and Cnr leaf, mature-green- (MG), breaker- (B), and breaker-plus-7-d (B+7)-stage pericarp tissue. Vertical bars = se; n = 3. DW, Dry weight.
Table III.
Tissue strength in red-ripe fruit of cvs Ailsa Craig and Liberto and Cnr fruit at an equivalent time postanthesis (65 DPA)
| Fruit Type | Pericarp | Locule |
|---|---|---|
| gross fracture energy, Jm2 | ||
| cv Ailsa Craig | 66a | 0.6a |
| cv Liberto | 90a | 0.8a |
| Cnr | 335b | 13.0b |
Means are six individual fruit (pericarp) or four individual fruit (locule), with two replicates per fruit. Data analysis was by analysis of variance after logarithmic transformation. For each column the same letter denotes no significant difference at P < 0.01.
Figure 5.
Cell separation in pericarp tissue in unripe (A) and ripe (B) cv Ailsa Craig and Cnr (from fruit of an equivalent age) and from 60- and 80-DPA (dpa) rin fruits incubated in water or CDTA for 3 h at room temperature. Values were calculated relative to 100% cell separation obtained after pectinase treatment. Vertical bars = se; n = 3.
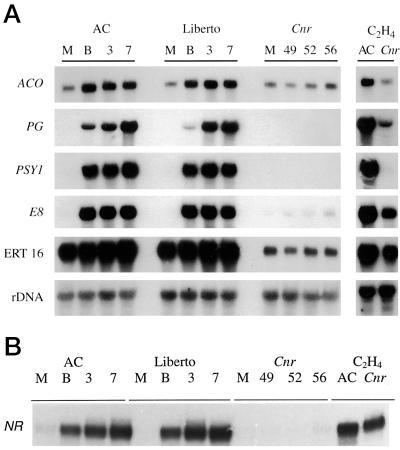
We followed changes in the expression of a panel of ripening-related genes in cvs Ailsa Craig and Liberto and Cnr in the presence and absence of exogenous ethylene (Fig. 6). ACO (ACC oxidase), PG (polygalacturonase), PSY1 (phytoene synthase), E8, ERT16, a clone encoding an ABA stress-related protein, and NR (an ethylene receptor) were chosen, because these messages are all up-regulated or present during normal ripening. The pattern of expression of these genes in Cnr could be grouped into four broad categories: (a) no detectable message and expression not restored by ethylene, e.g. PSY1, (b) transcripts detectable only after exposure of the fruit to ethylene, e.g. PG, (c) low levels of message in untreated fruit and enhanced expression in the presence of ethylene, e.g. E8, NR, and ERT16; and (d) low levels of message in untreated fruit that is not enhanced by ethylene treatment, e.g. ACO.
Figure 6.
A, Ripening-related gene expression by RNA gel-blot analysis. Total RNA was isolated from the fruit of cv Ailsa Craig (AC), cv Liberto, and the mutant Cnr, and both Cnr and cv Ailsa Craig after treatment with 100 μL/L exogenous ethylene at mature-green (M), breaker (B), breaker-plus-3-d (3), and breaker-plus-7-d (7) stages. Cnr fruit were picked at equivalent DPA, as described in Methods. Ten micrograms of RNA was loaded per lane and hybridized with previously characterized ripening-related genes (Gray et al., 1992, and refs. therein; Picton et al., 1993). B, Analysis of NR gene expression in Cnr by the RNase protection assay. Fifty micrograms of total RNA from cvs Ailsa Craig, Liberto, and Cnr from stages described in A was hybridized with a radiolabeled NR gene fragment, and RNase protection assay analysis was performed as described in Methods.
DISCUSSION
Cnr is a spontaneous mutation isolated from a commercial population of plants. The mutant has a phenotype that is quite distinct from the other known pleiotropic mutants at both the physiological and molecular levels. Our allelism tests and genetic mapping experiments indicated that Cnr is a novel, dominant pleiotropic ripening mutant.
The most readily apparent feature that distinguishes Cnr from the other pleiotropic ripening mutants is its lack of pigment in the pericarp tissue. The yellow color of the intact fruit comes from pigmentation in the skin, as demonstrated by the cross between Cnr and the colorless fruit epidermis mutant y. Analysis of the carotenoid composition of the Cnr fruit pericarp indicated that the biosynthesis of these pigments is completely inhibited in the pericarp of the mutant during ripening, although unripe fruit and nonfruit tissues appeared to be unaffected by the mutation. Other pleiotropic tomato-ripening mutants, e.g. Nr, nor, and rin, all show some carotenoid production in the pericarp, although the synthesis of lycopene is reduced, delayed, or absent, depending on the mutation (Tigchelaar et al., 1978). In Cnr, PSY1 gene expression is absent, even in the presence of ethylene (Fig. 6A), whereas in normal tomatoes it is up-regulated during ripening (see Fray and Grierson, 1993, and refs. therein). The protein product of PSY1 is phytoene synthase, which catalyzes the formation of phytoene, the first C40 carotene intermediate in carotenoid biosynthesis. This is an essential step in the production of the carotenoids, which give the fruit its red color. The product of PSY1 expression is responsible for the formation of carotenoids in the fruit, whereas a functionally distinct phytoene synthase is active in green tissues (Bramley et al., 1992; Fray and Grierson, 1993; Fraser et al., 1994). The absence of PSY1 expression in Cnr can probably explain the lack of carotenoids in the pericarp of the fruit.
Another striking feature of Cnr fruit is their altered cell adhesion, which is apparent when the fruit have reached an age equivalent to that of the red-ripe cvs Ailsa Craig and Liberto. The gross energy needed for mechanical fracture of Cnr tissue was greater than that needed for the red-ripe wild type, but Cnr pericarp cells appeared to separate more readily in water, where turgor may provide the cell-separation forces (Jarvis, 1998). Experiments to compare the cell adhesion in Cnr with wild-type pericarp tissue by incubation in water or the Ca chelator CDTA (Fig. 5) showed that in unripe fruit these solutions had very limited ability to induce cell separation and therefore to solubilize the cell wall components responsible for cell adhesion. However, CDTA was effective at inducing substantial cell separation in ripe wild-type fruit, which indicates that Ca probably plays an important role in cell adhesion in these fruit, most likely through its association with pectic polysaccharides (Carpita and Gibeaut, 1993). In contrast, water alone induced marked cell separation in Cnr fruit of an equivalent age, indicating that a proportion of the wall components involved in cell adhesion in the mutant are readily soluble. These unusual physical properties of Cnr deserve further study, and investigations are now underway on the type and nature of the pectic components in the cell walls of this mutant (C. Orfila, G.B. Seymour, A.J. Thompson, and J.P. Knox, unpublished data). That wall hydrolase activity is altered in Cnr is indicated from measurements of PG mRNA, which was detected only in the fruit that was exposed to exogenous ethylene. Reduced levels of wall hydrolases and modified tissue composition are likely to account for the altered textural properties of the fruit.
Cnr fruit did not exhibit the characteristic climacteric rise in ethylene production at the onset of ripening; these data are consistent with reduced levels of ACO gene expression in Cnr. Although ripening in Cnr is not restored by the addition of exogenous ethylene, the mutant is not completely insensitive to ethylene, as indicated by the characteristic triple response in dark-grown seedlings and the accumulation of some ethylene- and ripening-related mRNAs in the fruit after ethylene treatment (Figs. 1 and 6). In these respects, Cnr very closely resembles rin and nor (Knapp et al., 1989; Lanahan et al., 1994; Yen et al., 1995) and does not possess the ethylene-insensitive phenotype characteristic of Nr (Lanahan et al., 1994; Yen et al., 1995). Therefore, based on these criteria, Cnr, like rin and nor (Yen et al., 1995), can be presumed to act upstream of ethylene biosynthesis during the regulation of ripening.
Cnr, nor, and rin may act together in a cascade or independently in a multibranched regulatory network to control ripening, or they may represent developmental components leading to fruit tissue correctly primed for ripening. To begin to understand the relationship of Cnr to the other mutants, we have examined the expression of a number of ripening-related genes in the presence and absence of exogenous ethylene. A number of differences in the molecular fingerprints of the mutants were observed. For example, in mature nor fruit, PSY1 and E8 mRNA are lacking and expression of these genes is not restored by exposing the fruit to ethylene (Yen et al., 1995; J.J. Giovannoni, unpublished data). In mature rin, PSY1 and E8 are expressed at low levels, and the transcripts are up-regulated by ethylene (Knapp et al., 1989). In Cnr there is no evidence for PSY1 expression in either the presence or absence of exogenous ethylene, but the E8 message is partially restored by ethylene treatment. Similarly, PG transcripts are absent in nor and rin in the presence and absence of ethylene (Knapp et al., 1989; Yen et al., 1995) but are partially restored by ethylene in Cnr. ACO transcripts are enhanced by ethylene treatment in rin fruit (Knapp et al., 1989) but are unaffected by ethylene in Cnr. These observations suggest that nor may have a more global effect on ethylene/ripening-related gene expression than Cnr and rin, whereas Cnr and rin may differentially regulate different subsets of ripening genes in response to ethylene. In this model nor is upstream of rin and Cnr, because ethylene/ripening-related gene expression appears to be more extensively inhibited in this mutant. However, further work will almost certainly show that these genes act as part of a complex, multibranched regulatory network. The real test of this and other models awaits the cloning of the normal alleles for nor, rin, and Cnr. The pleiotropic effects of the Cnr mutation indicate that the gene has a regulatory function, but its identity may be difficult to establish using only biochemical and molecular assays. Current efforts are focused on isolation of the Cnr gene by more precisely refining the map position of this locus and by identifying tomato bacteria artificial chromosome clones that hybridize to RFLP- or amplified restriction fragment polymorphism-based flanking markers.
ACKNOWLEDGMENTS
We thank Rachel Edwards for expert technical assistance, John Maxam-Smith (Practical Plant Genetics, Chichester, UK) for help with performing the crosses, and Steve Tanksley (Cornell University, Ithaca, NY) for the use of the RFLP markers. We are grateful to Peter Bramley and Paul Fraser (University of London) and to Emma Schofield (Zeneca Plant Science, Bracknell, Berkshire, UK) for help with the carotenoid analysis and for useful discussions.
Abbreviations:
- CDTA
cyclohexane diamine tetraacetic acid
- DPA
days postanthesis
- RFLP
restriction fragment-length polymorphism
Footnotes
This work was supported by the United Kingdom Biotechnology and Biological Sciences Research Council.
LITERATURE CITED
- Barry CS, Blume B, Bouzayen M, Cooper W, Hamilton AJ, Grierson D. Differential expression of the 1-aminocyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 1996;9:525–535. doi: 10.1046/j.1365-313x.1996.09040525.x. [DOI] [PubMed] [Google Scholar]
- Bramley PM, Teulieres C, Blain I, Bird CR, Schuch W. Biochemical characterization of transgenic tomato plants in which carotenoid synthesis has been inhibited through expression of antisense RNA to pTom5. Plant J. 1992;2:343–349. [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of the primary walls in flowering plants: consistency of molecular structure with physical properties of walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific radioactivity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fraser PD, Truesdale M, Bird CR, Schuch W, Bramley PM. Carotenoid biosynthesis during tomato fruit development. Plant Physiol. 1994;105:405–413. doi: 10.1104/pp.105.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fray R, Grierson D. Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol Biol. 1993;22:589–602. doi: 10.1007/BF00047400. [DOI] [PubMed] [Google Scholar]
- Freeman M, Jarvis MC, Duncan HJ. The textural analysis of cooked potato: three simple methods for determining texture. Potato Res. 1992;35:103–109. [Google Scholar]
- Fulton T, Julapark C, Tanksley S. Miniprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep. 1995;13:207–209. [Google Scholar]
- Giovannoni JJ, Noensie EN, Ruezinsky DM, Lu X, Tracy SL, Ganal MW, Martin GB, Pillen K, Alpert K, Tanksley SD. Molecular genetic analysis of the ripening inhibitor and non-ripening loci of tomato: a first step in the genetic map-based cloning of fruit ripening genes. Mol Gen Genet. 1995;248:195–206. doi: 10.1007/BF02190801. [DOI] [PubMed] [Google Scholar]
- Gray JE, Picton S, Giovannoni JJ, Grierson D. The use of transgenic and naturally occurring mutants to understand and manipulate tomato fruit ripening. Plant Cell Environ. 1994;17:557–571. [Google Scholar]
- Grierson D. Molecular biology of fruit ripening. Oxf Surv Plant Mol Cell Biol. 1986;3:363–383. [Google Scholar]
- Hamilton AJ, Lycett GW, Grierson D. Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature. 1990;346:284–287. [Google Scholar]
- Jarvis MC. Intercellular separation forces generated by intracellular pressure. Plant Cell Environ. 1998;21:1307–1310. [Google Scholar]
- John I, Drake R, Farrell A, Cooper W, Lee P, Horton P, Grierson D. Delayed leaf senescence in ethylene-deficient ACC-oxidase antisense tomato plants: molecular and physiological analysis. Plant J. 1995;7:483–490. [Google Scholar]
- Kerr EA. Mutations of chlorophyll retention in ripe fruit. Rep Tomato Genet Coop. 1958;8:22. [Google Scholar]
- Kerr EA. Never ripe-2 (Nr-2) a slow ripening mutant resembling Nr and Gr. Rep Tomato Genet Coop. 1982;32:33. [Google Scholar]
- Knapp J, Moureau P, Schuch W, Grierson D. Organisation and expression of polygalacturonase and other ripening genes in Ailsa Craig ‘Neverripe ’ and ‘ripening inhibitor’ tomato mutants. Plant Mol Biol. 1989;12:105–116. doi: 10.1007/BF00017453. [DOI] [PubMed] [Google Scholar]
- Kopeliovitch E, Rabinowitch HD, Mizrahi Y, Kedar N. Mode of inheritance of alcobaca, a tomato ripening mutant. Euphytica. 1981;30:223–225. [Google Scholar]
- Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ. The Never ripe mutation blocks ethylene perception in tomato. Plant Cell. 1994;6:521–530. doi: 10.1105/tpc.6.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ER, Green P, Abrahamson J, Barlow A, Daly M, Lincoln SE, Newburg L. MAPMAKER: an interactive package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- Rick CM. New mutants. Rep Tomato Genet Coop. 1956;6:22–23. [Google Scholar]
- Robinson R, Tomes M. Ripening inhibitor: a gene with multiple effects on ripening. Rep Tomato Genet Coop. 1968;18:36–37. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Tigchelaar EC, Barman RJ. Allelism of the alcobaca ripening mutant and nor. Rep Tomato Genet Coop. 1985;35:20. [Google Scholar]
- Tigchelaar EC, McGlasson WB, Buescher RW. Genetic regulation of tomato fruit ripening. HortScience. 1978;13:508–513. [Google Scholar]
- Tigchelaar EC, Tomes M, Kerr E, Barman R. A new fruit ripening mutant, non-ripening (nor) Rep Tomato Genet Coop. 1973;23:33. [Google Scholar]
- Ward TM, Wright M, Roberts JA, Self R, Osborne DJ. Analytical procedures for the assay and identification of ethylene. In: Hillman JR, editor. Isolation of Plant Growth Substances, Vol 4. Cambridge, UK: Cambridge University Press; 1978. pp. 135–151. [Google Scholar]
- Wellburn AR. The spectral determination of chlorophyll-A and chlorophyll-B, as well as total carotenoids using various solvents with spectrophotometers of different resolution. J Plant Physiol. 1994;144:301–313. [Google Scholar]
- Wilkinson JQ, Lanahan MB, Clark DG, Bleeker AB, Chang C, Meyerowitz EM, Klee HJ. A dominant mutant receptor from Arabidopsis confers ethylene insensitivity in heterologous plants. Nat Biotechnol. 1997;15:444–447. doi: 10.1038/nbt0597-444. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Yen H-C, Giovannoni JJ, Klee HJ. An ethylene-inducible component of signal transduction encoded by Never-ripe. Science. 1995;270:1807–1809. doi: 10.1126/science.270.5243.1807. [DOI] [PubMed] [Google Scholar]
- Yen H-C, Lee S, Tanksley SD, Lanahan MB, Klee HJ, Giovannoni JJ. The tomato Never-ripe locus regulates ethylene-inducible gene expression and is linked to a homolog of the Arabidopsis ETR1 gene. Plant Physiol. 1995;107:1343–1353. doi: 10.1104/pp.107.4.1343. [DOI] [PMC free article] [PubMed] [Google Scholar]