ABSTRACT
LncRNA in non-homologous end joining (NHEJ) pathway 1 (LINP1) is an lncRNA which promotes therapeutic resistance in triple-negative breast cancer (TNBC). However, the expression and function of LINP1 in cervical cancer is not yet well-understood. In this study, we evaluated the expression levels of LINP1 in tumor tissues and cell lines of cervical cancer. We found that LINP1 associates with NHEJ proteins (Ku80 and DNA-PKcs). LINP1 translocates from cytosol to nucleus in response to irradiation. In addition, LINP1 knockdown significantly increases the levels of cleaved caspase3 and PARP, leading to enhanced cell apoptosis after ionizing radiation (IR). LINP1-knockdown cells showed delayed repairs of DNA double-strand breaks (DSBs) after IR. Finally, LINP1 knockdown increases radiosensitivity of Hela S3 cells. These results suggest that LINP1 facilitates DSBs repair through NHEJ pathway and may thus serve as a prognostic marker and a potential target for the therapy of cervical cancer.
KEYWORDS: LINP1, long non-coding RNA, cervical cancer, radiotherapy, radiation resistance
Introduction
Although human papillomavirus (HPV) vaccines are available and effective screening methods have been established to reduce the incidence and mortality, cervical cancer (CC) remains the second most common cancer and third leading cause of death from cancer among females in developing countries [1]. Treatment of cervical cancer may vary within some combination of surgery, chemotherapy, radiotherapy and targeted therapy, depending mainly on the individual stages defined by Féderation Internationale de Gynécologie et d'Obstétrique (FIGO). According to National Comprehensive Cancer Network (NCCN) 2016, radiotherapy can be performed in all stages of patients including: as a treatment for early stage patients when surgery is out of option; combined with chemotherapy or as an adjuvant therapy after hysterectomy in larger early-stage and advanced-stage tumors; as palliative therapy to relieve symptoms caused by the cancer and improve quality of life in late-stage patients. However, some patients respond less well to radiation therapy and have higher incidences of relapse and treatment failure [2-4]. Therefore, questions remain to be answered about which factors are accounted for the radioresistance and poor outcomes of CC.
Long non-coding RNAs (lncRNAs) are defined as transcripts that are longer than 200 nucleotides with extremely limited protein coding capability [5,6]. Recent studies demonstrated that lncRNAs regulate gene expression and protein functions by recruiting epigenetic modification complexes to a chromosome or a particular gene region [7,8], acting as scaffolds to facilitate specific protein components into its position to form unique functional complexes [9,10]. Several studies have demonstrated the aberrant expression of lncRNAs in human cancer [11,12]. For example, ROR serves as a decoy that blocks the recruitment of G9A methyltransferase, abolishes histone H3K9 modification of the TESC promoter and induces tumorigenesis and metastasis [13]. LncRNA Malat1 (metastasis-associated lung adenocarcinoma transcript 1) up-regulation has been identified in a broad spectrum of tumor types [14-16], and its level may be even higher in the metastatic lesions compared with the primary tumor [17]. In a mouse model of breast cancer, Malat1 loss using antisense Oligonucleotides (ASOs) induced a dramatic differentiation of the primary tumor and a significant reduction in lung metastasis [17]. Notably, Zhang et al. recently discovered an lncRNA termed lncRNA in non-homologous end joining (NHEJ) pathway 1 (LINP1) in TNBC [18,19]. LINP1 functions as a scaffold interacting with Ku80 and DNA-PKcs, key proteins in NHEJ pathway [20], thereby promoting NHEJ-mediated DNA repair activity [18,21]. Considering the importance of LINP1 in DNA damage repair progression and the possibility it may help to classify radioresistant tumors and serve as a treatment target, we investigated its expression and functions in cervical cancers.
Materials and methods
Patients and samples
Tissue samples were collected from patients who underwent primary tumor resection from January 2017 to June 2017 in the Sir Run Run Shaw Hospital, School of Medicine, Zhejiang University. No patients received preoperative chemotherapy or radiotherapy. Diagnostic tests were performed to exclude other uterine diseases. All of the patients have given informed consent to this study. Moreover, all procedures performed in studies involving human participants have been approved by the Research Ethics Committee of Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University and conform to the provisions of the Declaration of Helsinki (as revised in Fortaleza, Brazil, October 2013).
Cell lines and transduction
The human cervical cancer cell lines were obtained from ATCC free of mycoplasma. Hela S3, SiHa and Hela cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) and C-33A cells in MEM-EBSS with 10% FBS. These cells were maintained in standard growth conditions with 5% CO2 at 37°C.
SiRNAs were synthesized by Shanghai GenePharma Co., Ltd. and transfections were performed with Lipofectamine 3000 reagent (Invitrogen, cat no. L3000015) according to the manufacturer's instructions. The lentivirus constructs expressing LINP1 shRNA and the negative control shRNA were purchased from Shanghai Genechem Co., Ltd. The siRNA and shRNA oligonucleotide sequences are listed in Table S1. For plasmid construction, the purified full length LINP1 was cloned into a pcDNA3.1 (+) vector, which was transformed into E. coli DH5α and confirmed by PCR, double endonuclease digestion and DNA sequencing. The plasmid was then transfected into SiHa cells with Lipofectamine 3000 reagent.
RNA extraction and qRT–PCR
Total RNA was extracted with TRIzol Reagent (Invitrogen, cat no. 15596-026) according to the manufacturer's instructions. Nuclear RNA isolation was performed by removing cytoplasm fraction post incubation with cytoplasmic extract (CE) buffer and followed by adding TRIzol Reagent to the nuclei enrichment pellet. cDNA was synthesized using the PrimeScript® RT reagent kit (TaKaRa, cat no. RR047A). Subsequently, PCR analysis was performed using SYBR® Premix DimerEraser kit (TaKaRa, cat no. RR091A) in a Science Light Cycle™480 System (Applied Roche). The expression of GAPDH served as the endogenous control. ΔCt was calculated by subtracting the Ct of GAPDH RNA from the Ct of LINP1 RNA, respectively. Relative LINP1 expression was quantified using the 2−ΔΔCt method after normalization for the expression of the control. The primers used for qPCR are listed in Table S1.
RNA immunoprecipitation (RIP)
RIP was conducted as previously described [22]. Cells were harvested using a scraper, washed twice with PBS and resuspended in 1 ml ice-cold RIP buffer (150 mM KCl, 25 mM Tris (pH 7.4), 5 mM EDTA, 0.5% NP-40) containing RNase and protease inhibitors. The cells were then sheared using a Dounce homogenizer on ice and the supernatant was collected by centrifugation at 15,000g for 15 minutes at 4℃. This extraction was incubated with either anti-Ku80 (Thermo, cat no. MA5-12933), anti-DNA-PKcs (Thermo, cat no. MA5-13404) antibody, or control IgG (Beyotime, cat no. A7016) at 4℃ overnight, and subsequently incubated with Protein A/G–Agarose beads (Santa Cruz Biotechnology, cat no. sc-2003) at room temperature for 2 hours. After washing with ice-cold RIP buffer three times, the RNA-protein complex bound with beads was boiled in sample buffer for western blotting or resuspended in TRIzol reagent for RNA isolation and subjected to qRT-PCR. Primers are listed in Table S1.
RNA-pulldown
RNA-pulldown was performed as previously described with some modifications [22]. Briefly, biotin-labeled LINP1 specific probes were mixed with prewashed streptavidin agarose beads (Invitrogen, cat no. 65801D) for 15 minutes at room temperature, and then these beads were added to the cell extraction and incubated overnight at 4℃. Precipitates were washed with RIP buffer three times, boiled in SDS buffer and subjected to western blotting. Sequences of biotin-labeled LINP1 probes and qPCR primers are listed in Table S1.
Protein isolation and western blotting assay
Total protein was extracted using RIPA lysis buffer (Beyotime, cat no. P0013C) containing 1% PMSF. Protein samples were separated via SDS PAGE and then transferred to polyvinyl difluoride membranes. After blocking with 5% bovine serum albumin, the membranes were incubated overnight at 4°C with primary antibody against PARP (1:1000 dilution, Epitomics, cat no. 1078-S), casepase3 (1:1000 dilution, Epitomics, cat no. 1087-1) and β-Tubulin (1:1000 dilution, Abcam, ab6046), and followed by incubation with peroxidase conjugated secondary antibodies (anti-rabbit IgG, (H+L) (Thermo, cat no. 31460) or anti-mouse IgG, (H+L) (Thermo, cat no. 31430)). The immunoreactive bands were then visualised with enhanced chemiluminescence (ECL) (Thermo, cat no. 32209).
Flow cytometry analysis of cell apoptosis
Twenty-four hours after 6 Gy irradiation treatment, cells were harvested and washed twice with ice-cold PBS. Flow cytometry analysis for apoptosis was performed using Annexin V-APC/7-AAD apoptosis kit (MultiSciences, cat no. AP105-100) according to the manufacturer's protocol. Data were then analyzed using FlowJo vX 0.7 software.
Immunofluorescence staining
Cells were seeded on coverslips and treated with 6 Gy IR following cell adhesion. After 24 hours, the cells were fixed using 4% paraformaldehyde for 10 minutes, subsequently permeabilized with 0.5% Triton solution for 15 minutes at room temperature, and then incubated with anti-γ-H2AX antibody (Ser139) (1:500 dilution, Epitomics, cat no. 2212-1) overnight at 4°C. After washing with PBST three times, the cells were then incubated with secondary antibody for 1 h at 37°C. Cell nuclei were counterstained with 4’6-diamino-2-phenylindole (DAPI) (Beyotime, cat no. C1006) and visualized with an Axiovert 200M inverted microscope (Zeiss).
Fluorescence in situ hybridization
Formalin-fixed paraffin-embedded sections were mounted onto poly-L-lysine-coated slides and baked at 70°C overnight. RNA in situ hybridization was performed using biotin-labled probes. After deparaffinization and rehydration, the tissues were digested in pepsin. Slides were then incubated with biotinylated probes at 42°C overnight and further incubated with Streptavidin/FITC conjugates (Solarbio, cat no. SF068) at 37°C for 2 hours. The cell nuclei were counterstained with DAPI (Beyotime, cat no. C1006). Finally, the slides were viewed using an Axiovert 200M inverted microscope (Zeiss).
Cervical cancer cells were cultured on coverslips and treated with 6 Gy irradiation. Cells were fixed with 4% paraformaldehyde 30 minutes after IR, then treated with 0.1 M triethanolamine containing 0.25% aceticanhydride and subsequently permeabilized with 0.2 M HCl for 10 minutes at room temperature. After pre-hybridization, the slides were incubated with hybridization buffer containing denatured biotin-labeled probes at 60°C in a humidity incubator overnight. After stringent washes (0.1X, 2XSSC), slides were blocked with 5% BSA for 1 hour at room temperature followed by incubation with Streptavidin/FITC conjugates at 37°C for 2 hours. DAPI was used as a nuclear counterstain and the cells were finally visualized using an Axiovert 200M inverted microscope (Zeiss). Sequences of biotin-labeled LINP1 probes are listed in Table S1.
Irradiation and Colony-forming assay
Irradiation was performed using 6 MV X-rays from a linear accelerator at a dose rate of 2 Gy/min(PRIMUS-M, Siemens). Exponential phase cells were seeded in six-well plates and irradiated with single doses ranging from 0 to 6 Gy. After 10–14 days, cells were washed with cold PBS, fixed with 75% methanol and stained with 0.5% crystal violet. The colonies consisting of more than 50 cells were counted and a multitarget/single-hit model (SF = 1–[1–e–D/D0]N) was used to plot the surviving fraction curve. The radiosensitivity parameters were calculated using SPSS software based on the formula previously described [23].
Statistical analysis
SPSS 22.0 software was used for the data analysis. Data were shown as mean ± SD, and P < 0.05 indicated statistically significant.
Results
Expression levels of LINP1 in cervical cancer tissues and cell lines
To evaluate the LINP1 levels in CC, we extracted total RNA from tissues of patients and cell lines and then performed qRT-PCR assays. All of the tumor tissues from five CC patients showed higher levels of LINP1 compared to adjacent tissues, while significant difference in three of them was observed (Figure 1(a)). These data were confirmed by performing RNA fluorescence in situ hybridization (RNA-FISH) to visualize LINP1 in tissue samples (Figure 1(b)). However, LINP1 was undetectable in all the CC cell lines (SiHa, Hela and C-33A), except Hela S3 cells (Figure 1(c)). Therefore, Hela S3 cells were used to define the function of LINP1 in cervical cancers in the following studies.
Figure 1.
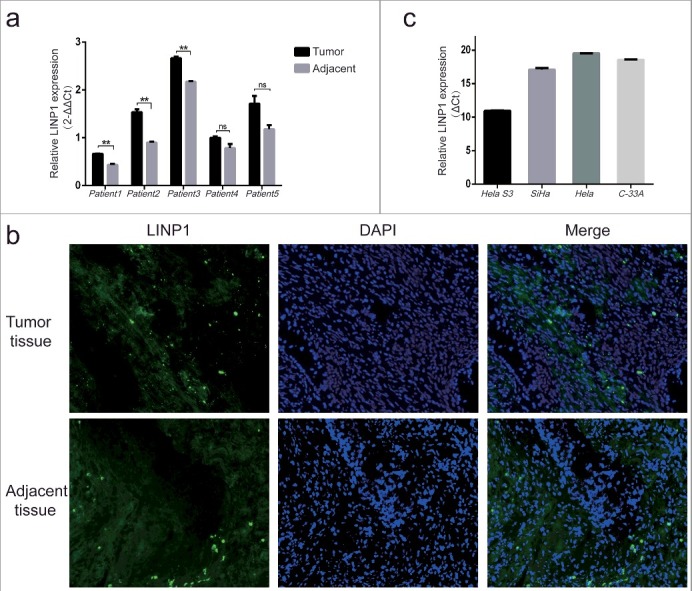
Expression of LINP1 in cervical cancer tissues and cell lines a. QRT-PCR assays showing levels of LINP1 in tumor tissues and adjacent tissues from CC patients. b. Visualization of LINP1 in CC tissues using RNA-FISH analysis. c. QRT-PCR assays showing LINP1 expression levels in different CC cell lines. The expression of GAPDH served as the endogenous control.
LINP1 associates with NHEJ pathway proteins Ku80 and DNA-PKcs in cervical cancer
To identify the function of LINP1 in CC, we performed RNA-immunoprecipitation (RIP) assays with Ku80- or DNA-PKcs-specific antibodies (Figure 2(a–c)). Compared with the IgG-bound complexes, a significantly higher fold enrichment of LINP1 RNA was detected in the RNA-protein complexes immunoprecipitated with Ku80- (Figure 2(b)) and DNA-PKcs-specific (Figure 2(c)) antibodies. To further confirm the interaction between endogenous LINP1 and these two proteins, RNA-pulldown assays with specific biotin-labled oligonucleotide probes were performed and western blotting was then used to detect the Ku80 and DNA-PKcs in the protein complexes pulled down by LINP1 probes (Figure 2(d, e)). While neither Ku80 nor DNA-PKcs was detected in proteins associated with beads alone, we observed strong signals of Ku80 and DNA-PKcs in complexes pulled down with LINP1 specific probes (Figure 2(e)). These data confirmed that LINP1 is associated with NHEJ pathway proteins Ku80 and DNA-PKcs in CC cell lines.
Figure 2.
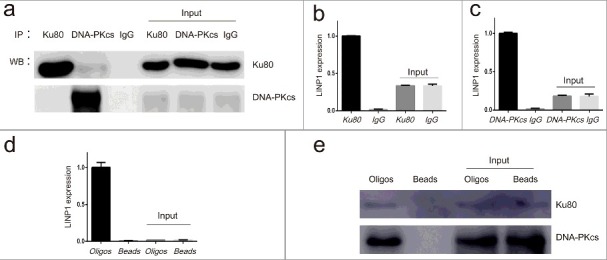
LINP1 associates with NHEJ pathway proteins Ku80 and DNA-PKcs in Hela S3 cells a. Immunoprecipitation efficiency of Ku80 and DNA-PKcs specific antibodies detected by western blots. b and c. Results from RNA-IP. QRT-PCR assays showing the relative level of LINP1 in the complex immunoprecipitated by Ku80- and DNA-PKcs-specific antibodies. d. QRT-PCR assays showing the enrichment of LINP1 with specific biotinylated oligos. e. Western blots showing the Ku80 and DNA-PKcs protein levels in the complexes pulled down by LINP1.
Changes in LINP1 subcellular localization and expression levels after IR treatment
To further characterize and quantify LINP1 expression in cells, we tracked its subcellular localization before and after IR treatment using RNA-FISH technology. The results showed that LINP1 distributed mainly in the cytoplasm of normal cells, and translocated to the nucleus in response to IR treatment (Figure 3(a)). We next measured the dynamic levels of nuclear LINP1 in different time points after irradiation. The expression level of LINP1 in the nucleus increased after irradiation, peaked in 40 minutes, and then decreased gradually (Figure 3(b)). In addition, LINP1 expression in the whole cells was increased 24 hours after IR and was significantly suppressed by EGFR inhibitor icotinib hydrochloride pretreatment (Figure 3(c)). Collectively, these results confirm the idea that LINP1 plays an important role in DSBs repair as it translocated to the nucleus within 30 minutes after irradiation and was upregulated at a later time. Furthermore, LINP1 expression may be regulated by EGFR pathway in CC cells.
Figure 3.

Changes in LINP1 subcellular localization and expression levels after IR treatment a. RNA-FISH was performed using LINP1 specific biotinylated oligos to monitor the subcellular localization patterns of LINP1 in Hela S3 cells 30 minutes after irradiated with 6 Gy X-rays. b. Levels of LINP1 in the nuclear fraction measured by qRT-PCR at different time points after 6 Gy IR treatment. c. Expression of LINP1 in the whole cells 24 hours after different treatment. IH, icotinib hydrochloride; IR, ionizing radiation. Error bars, s.d. *P < 0.05 by two-tailed Student's t test; n = 3 technical replicates.
Knockdown of LINP1 increased IR-induced cell apoptosis and delayed repair of DNA double-strand breaks in cervical cancer cells after radiation
To study the impact of LINP1 on radiotherapy efficiency, we knocked down LINP1 using small interfering RNA (siRNA) in Hela S3 cells (Figure 4(a)) and generated cells stably expressing control or LINP1 shRNA by performing short hairpin RNA (shRNA) lentiviral transduction (Figure 4(b)). Compared with cells transduced with control siRNA, the expression levels of activated caspase3 and inactivated PARP, two markers of apoptosis, were significantly increased in LINP1-knockdown cells (Figure 4(c)). The influence of LINP1 knockdown on cell apoptosis was then measured via flow cytometry (Figure 4(d, e)). In control cells, the apoptotic rate was 3.67±0.15% before IR and increased only 0.96% after a 6 Gy irradiation. Significant differences were observed in the LINP1 knockdown groups as the apoptotic rates were 5.51±0.35% and 3.99±0.35% at the baseline level and increased to 10.2±0.51% and 7.54±0.02% respectively 24 hours post radiation (P < 0.05). These results indicate that LINP1 knockdown significantly increases IR-induced apoptosis in Hela S3 cells.
Figure 4.
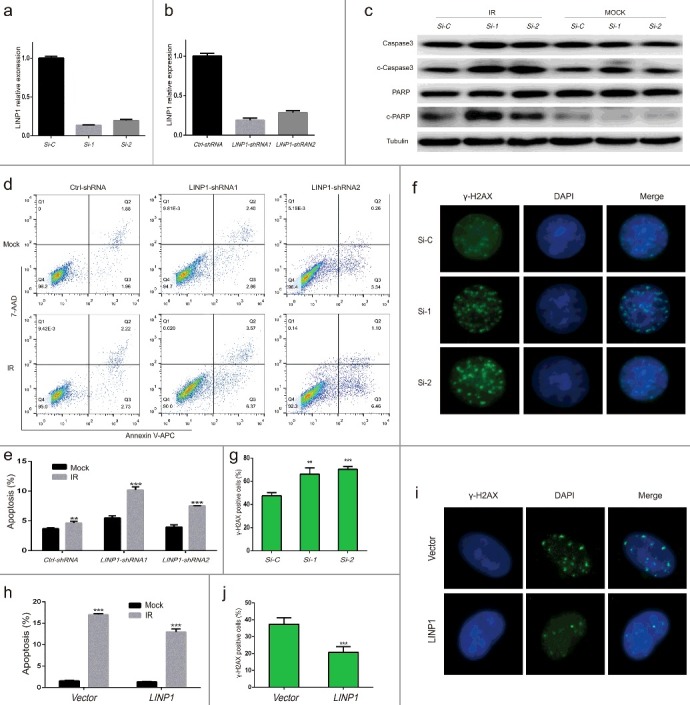
Knockdown of LINP1 increased IR-induced cell apoptosis and delayed repair of DNA double-strand breaks in cervical cancer cells treated with radiation a and b. QRT-PCR assays showing LINP1 silencing efficiency using siRNA or shRNA. c. Expression of caspase3, cleaved caspase3, PARP and cleaved PARP detected by western blotting in Hela S3 cells expressing LINP1 or control siRNAs 24 hours after treated with or without 6 Gy irradiation. d. Apoptosis was detected by flow cytometry in Hela S3 cells expressing control or LINP1-specific shRNAs 24 hours after treated with 6 Gy IR. e. The apoptotic rate was calculated as the percentage of Annexin V-APC-positive cells. f. Immunofluorescence stain visualizing IR-induced γ-H2AX foci in LINP1 silencing and control Hela S3 cells 24 hours after treatment. g. Quantification of the number of γ-H2AX foci positive cells expressing LINP1 or control siRNAs 24 hours after irradiation. h. The apoptotic rate of SiHa cells expressing control vector or LINP1 24 hours after treated with or without 6 Gy irradiation. i. Immunofluorescence stain detecting α-H2AX foci in LINP1-overexpressing and control SiHa cells 24 hours after 6 Gy IR treatment. j. Quantification of the number of γ-H2AX foci positive cells expressing control vector or LINP1 24 hours after exposure to irradiation. Cells with 5 or more γ-H2AX foci were counted as unrepaired cells or γ-H2AX foci positive cells. Si-C, cells expressing Ctrl-siRNA; Si-1, cells expressing LINP1-siRNA1; Si-2, cells expressing LINP1-siRNA2. PcDNA3.1 (+) plasmid transfected SiHa cell lines were used as controls; LINP1 indicates LINP1-overexpressing SiHa cells. Error bars, s.d. *P < 0.05 by two-tailed Student's t test; n = 3 independent cell cultures.
We next analyzed the effects of LINP1 knockdown on the DNA damage initiation and resolution in cells treated with IR using γ-H2AX foci formation assays. Cells with 5 or more γ-H2AX foci were counted as unrepaired cells or γ-H2AX foci positive cells. LINP1 knockdown significantly increased the number of γ-H2AX foci positive cells 24 hours post IR (Figure 4(f, g)), suggesting that LINP1 promoted the DNA damage repairs and reduced radiotherapy efficiency. To further confirm the impact of LINP1 on apoptosis and repair of DSBs, we performed over-expression experiments in the SiHa cells. Twenty four hours following irradiation, the apoptotic rates were 17.01±0.31% in cells expressing control vector and 12.97±0.91% in LINP1-overexpressing cells (Figure 4(h)), which indicated that LINP1 overexpression suppressed IR-induced apoptosis in SiHa cells. Moreover, the number of γ-H2AX foci positive cells was significantly decreased by LINP1 overexpression (Figure 4(i, j)). In summary, our findings demonstrated that LINP1 could determine the fate of CC cells exposure to radiation by suppressing cell apoptosis and facilitating DNA damage repair.
Knockdown of LINP1 enhances radiation sensitivity of cervical cancer cells
The effects of LINP1 knockdown on radiosensitivity of CC cells were measured by clonogenic survival assay (Figure 5). LINP1-specific shRNAs or control shRNAs were introduced into Hela S3 cells. These cells were then treated with different doses of IR and cell survival was assessed two weeks after the IR treatments. The results showed that the survival of LINP1-knockdown cells was significantly decreased compared to cells expressing control shRNAs. We next calculated the parameters of a multitarget/single-hit model based on clonogenic survival assay (Table 1). Their D0 values were 1.88 and 1.98 versus 2.71 and the sensitizing enhancement ratio (SER) were calculated as 1.44 and 1.37 respectively. Therefore, these data suggested that LINP1 knockdown can enhance radiation sensitivity of cervical cancer cells.
Figure 5.
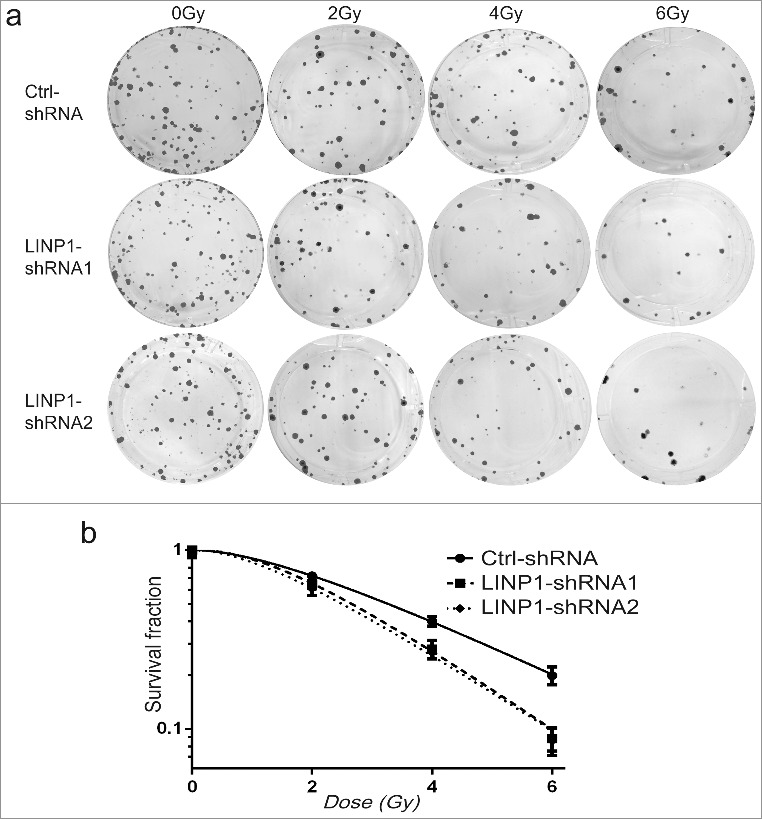
Knockdown of LINP1 enhanced radiation sensitivity of cervical cancer cells a. Survival of control or LINP1-knockdown Hela S3 cells in response to IR treatments. Cells expressing control or LINP1 specific shRNAs were treated with different doses of IR, and the survival after treatment was measured with colony formation assays. b. Survival curve of Hela S3 cells expressing control or LINP1 specific shRNAs. Error bars, s.d.; n = 3 independent cell cultures.
Table 1.
The main parameters of a multitarget model based on colony formation assays.
| N | D0 | Dq | SF2 | SER | |
|---|---|---|---|---|---|
| Ctrl-shRNA | 1.951 | 2.71 | 1.81 | 0.72 | |
| LINP1-shRNA1 | 2.484 | 1.88 | 1.71 | 0.65 | 1.44 |
| LINP1-shRNA2 | 2.088 | 1.98 | 1.45 | 0.61 | 1.37 |
N, extrapolation number; D0, mean lethal dose; Dq, quasi-threshold dose; SF2, surviving fraction at 2 Gy; SER, sensitivity enhancement ratio.
Discussion
Cervical cancer is one of the leading female malignancies worldwide [1,24]. For patients with locally advanced cancers, a combination of chemotherapy and radiotherapy is the standard of care [25,26]. Although certain prognostic factors such as patient age, tumor grade, pelvic node status have been used to predict survival [27,28], identification of new manageable factors mediating radioresistance and revelation of the underlying mechanism remain unsolved problems.
Recent studies have reported that lncRNAs are associated with tumor initiation, progression, and treatment resistance in certain malignancies, including cervical cancer [29,30]. For instance, decreased LET expression inhibits carcinogenesis of cervical cancer [31]. TUG1 acts as an endogenous sponge binding to miR-138-5p, thereby accelerating cervical cancer malignant progression by activating SIRT1-Wnt/β-catenin signaling pathway [32]. HOTAIR promotes tumor migration and invasion by upregulating VEGF and MMP-9 expression in cervical cancer [33] and mediates redioresistance via inhibiting p21 [34]. NHEJ is the most dominant pathway of DSBs repair in mammalian cells [35], which is initiated by binding of Ku heterodimers (Ku70 and Ku80) and DNA-PKcs to form a DNA-PK holoenzymes that greatly enhances kinase activity [36,37]. LncRNA in non-homologous end joining (NHEJ) pathway 1 (LINP1), a 917-bp lncRNA that maps to chromosome 10: 6,737,382-6,739,026, was recently identified as an integral component of the synaptic complex of NHEJ that stabilizes the structure of Ku80–DNA-PKcs interaction in TNBC cells [18]. To define the role of LINP1 in cervical cancers, we first verified that both cervical cancer tissues and cell lines expresses high levels of LINP1. Next, we found that LINP1 is associated with Ku80 and DNA-PKcs, which is consistent with previous study [18]. Furthermore, using RNA-FISH analysis and nuclear RNA extraction technique followed by qRT-PCR, we observed that LINP1 translocated to the cell nucleus after exposure to irradiation. These data suggested that LINP1 is mainly in the cytoplasm and could translocate to the nucleus in the presence of DNA damage. Intriguingly, icotinib hydrochloride, an EGFR inhibitor, not only significantly suppressed LINP1 expression, but also completely abolished IR-induced upregulation of LINP1. LINP1 knockdown enhances activation of the apoptosis pathway induced by radiation. We further confirmed these results using flow cytometry by demonstrating significant differences in apoptotic rates between the LINP1 knockdown and the control cells. γ-H2AX foci formation assays showed that decreased expression levels of LINP1 delayed DSBs repair. In addition, LINP1 overexpression decreased cell apoptosis and promoted γ-H2AX foci resolution. Finally, the negative impact of LINP1 on the radiosensitivity of CC cell lines was defined via clonogenic survival assay. These data suggested that LINP1 plays a similar role in CC as it does in breast cancer and further studies to evaluate its expression and functions in different cancer types are warranted.
In summary, our study indicated that lncRNA LINP1 is involved in radiation resistance of CC by enhancing DNA damage repair efficiency through NHEJ pathway. Although further investigations are required to define the relationship between LINP1 expression levels and tumor response to radiotherapy in CC patients, our study suggests that LINP1 may serve as an important prognostic marker and a novel therapeutic target for cervical cancers.
Supplementary Material
Funding Statement
Natural Science Foundation of Zhejiang Province (CN) [grant number LY14H160016] Major Science and Technology Program of Zhejiang Province [grant number 2013C03044-6] Chinese National Natural Science Foundation [grant number 81441086, 81672976].
Disclosure of potential conflicts of interest
The authors declare that they have no conflict of interest.
Acknowledgments
This work was supported in part by grants from the Chinese National Natural Science Foundation (No. 81441086, 81672976), Natural Science Foundation of Zhejiang Province (No. LY14H160016), Major Science and Technology Program of Zhejiang Province (No. 2013C03044-6).
References
- [1].Torre LA, Bray F, Siegel RL, et al.. Global cancer statistics, 2012. CA Cancer J Clin. 2015. March;65(2):87–108. doi: 10.3322/caac.21262. PubMed PMID: 25651787. [DOI] [PubMed] [Google Scholar]
- [2].Hong JH, Tsai CS, Wang CC, et al.. Comparison of clinical behaviors and responses to radiation between squamous cell carcinomas and adenocarcinomas/adenosquamous carcinomas of the cervix. Chang Gung Med J. 2000. July;23(7):396–404. PubMed PMID: 10974754. [PubMed] [Google Scholar]
- [3].Zhou J, Wu SG, Sun JY, et al.. Comparison of clinical outcomes of squamous cell carcinoma, adenocarcinoma, and adenosquamous carcinoma of the uterine cervix after definitive radiotherapy: a population-based analysis. J Cancer Res Clin Oncol. 2017. January;143(1):115–122. doi: 10.1007/s00432-016-2246-9. PubMed PMID: 27646608. [DOI] [PubMed] [Google Scholar]
- [4].Chen JL, Huang CY, Huang YS, et al.. Differential clinical characteristics, treatment response and prognosis of locally advanced adenocarcinoma/adenosquamous carcinoma and squamous cell carcinoma of cervix treated with definitive radiotherapy. Acta Obstet Gynecol Scand. 2014. July;93(7):661–668. doi: 10.1111/aogs.12383. PubMed PMID: 24666257. [DOI] [PubMed] [Google Scholar]
- [5].Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009. March;10(3):155–159. doi: 10.1038/nrg2521. PubMed PMID: 19188922. [DOI] [PubMed] [Google Scholar]
- [6].Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011 Apr 13;10:38. doi: 10.1186/1476-4598-10-38. PubMed PMID: 21489289; PubMed Central PMCID: PMC3098824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rinn JL, Kertesz M, Wang JK, et al.. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007 Jun 29;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. PubMed PMID: 17604720; PubMed Central PMCID: PMC2084369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011 Jul 08;146(1):119–133. doi: 10.1016/j.cell.2011.06.026. PubMed PMID: 21729784; PubMed Central PMCID: PMC3150513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Tsai MC, Manor O, Wan Y, et al.. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010 Aug 06;329(5992):689–693. doi: 10.1126/science.1192002. PubMed PMID: 20616235; PubMed Central PMCID: PMC2967777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yap KL, Li S, Munoz-Cabello AM, et al.. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010 Jun 11;38(5):662–674. doi: 10.1016/j.molcel.2010.03.021. PubMed PMID: 20541999; PubMed Central PMCID: PMC2886305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yan X, Hu Z, Feng Y, et al.. Comprehensive Genomic Characterization of Long Non-coding RNAs across Human Cancers. Cancer Cell. 2015 Oct 12;28(4):529–540. doi: . 10.1016/j.ccell.2015.09.006. PubMed PMID: 26461095; PubMed Central PMCID: PMC4777353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Iyer MK, Niknafs YS, Malik R, et al.. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015. March;47(3):199–208. doi: 10.1038/ng.3192. PubMed PMID: 25599403; PubMed Central PMCID: PMC4417758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Fan J, Xing Y, Wen X, et al.. Long non-coding RNA ROR decoys gene-specific histone methylation to promote tumorigenesis. Genome Biol. 2015 Jul 14;16:139. doi: 10.1186/s13059-015-0705-2. PubMed PMID: 26169368; PubMed Central PMCID: PMC4499915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sun H, Lin DC, Cao Q, et al.. Identification of a Novel SYK/c-MYC/MALAT1 Signaling Pathway and Its Potential Therapeutic Value in Ewing Sarcoma. Clin Cancer Res Off J Am Assoc Cancer Res. 2017 Mar 23; doi: 10.1158/1078-0432.CCR-16-2185. PubMed PMID: 28336564. [DOI] [PubMed] [Google Scholar]
- [15].Wang X, Li M, Wang Z, et al.. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015 Feb 13;290(7):3925–3935. doi: 10.1074/jbc.M114.596866. PubMed PMID: 25538231; PubMed Central PMCID: PMC4326802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Guffanti A, Iacono M, Pelucchi P, et al.. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics. 2009 Apr 20;10:163. doi: 10.1186/1471-2164-10-163. PubMed PMID: 19379481; PubMed Central PMCID: PMC2678161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Arun G, Diermeier S, Akerman M, et al.. Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Development. 2016 Jan 01;30(1):34–51. doi: 10.1101/gad.270959.115. PubMed PMID: 26701265; PubMed Central PMCID: PMC4701977.//doi.org/ 10.1101/gad.270959.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang Y, He Q, Hu Z, et al.. Long noncoding RNA LINP1 regulates repair of DNA double-strand breaks in triple-negative breast cancer. Nat Struct Mol Biol. 2016. June;23(6):522–530. doi: 10.1038/nsmb.3211. PubMed PMID: 27111890; PubMed Central PMCID: PMC4927085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Sakthianandeswaren A, Liu S, Sieber OM. Long noncoding RNA LINP1: scaffolding non-homologous end joining. Cell Death Discovery. 2016;2:16059. doi: 10.1038/cddiscovery.2016.59. PubMed PMID: 27551547; PubMed Central PMCID: PMC4979485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010 Oct 22;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. PubMed PMID: 20965415; PubMed Central PMCID: PMC2988877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Zlotorynski E. Non-coding RNA: LINP1 joins ends with triple-negative effect. Nat Rev Mol Cell Biol. 2016. June;17(6):330–331. doi: 10.1038/nrm.2016.60. PubMed PMID: 27145720. [DOI] [PubMed] [Google Scholar]
- [22].Feng Y, Hu X, Zhang Y, et al.. Methods for the study of long noncoding RNA in cancer cell signaling. Methods Mol Biol. 2014;1165:115–143. doi: 10.1007/978-1-4939-0856-1_10. PubMed PMID: 24839023; PubMed Central PMCID: PMC4142042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cariveau MJ, Tang X, Cui XL, et al.. Characterization of an NBS1 C-terminal peptide that can inhibit ataxia telangiectasia mutated (ATM)-mediated DNA damage responses and enhance radiosensitivity. Mol Pharmacol. 2007. August;72(2):320–326. doi: 10.1124/mol.107.036681. PubMed PMID: 17507690. [DOI] [PubMed] [Google Scholar]
- [24].Vaccarella S, Lortet-Tieulent J, Plummer M, et al.. Worldwide trends in cervical cancer incidence: impact of screening against changes in disease risk factors. European Journal of Cancer . 2013. October;49(15):3262–3273. doi: 10.1016/j.ejca.2013.04.024. PubMed PMID: 23751569. [DOI] [PubMed] [Google Scholar]
- [25].Waggoner SE. Cervical cancer. Lancet. 2003 Jun 28;361(9376):2217–2225. doi: 10.1016/S0140-6736(03)13778-13776. PubMed PMID: 12842378. [DOI] [PubMed] [Google Scholar]
- [26].Rogers L, Siu SS, Luesley D, et al.. Radiotherapy and chemoradiation after surgery for early cervical cancer. Cochrane Database Syst Rev. 2012. May;16(5):CD007583. doi: 10.1002/14651858.CD007583.pub3. PubMed PMID: 22592722; PubMed Central PMCID: PMC4171000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Stehman FB, Bundy BN, DiSaia PJ, et al.. Carcinoma of the cervix treated with radiation therapy. I. A multi-variate analysis of prognostic variables in the Gynecologic Oncology Group. Cancer. 1991 Jun 01;67(11):2776–2785. PubMed PMID: 2025841. doi: . [DOI] [PubMed] [Google Scholar]
- [28].Rose PG, Java J, Whitney CW, et al.. Nomograms Predicting Progression-Free Survival, Overall Survival, and Pelvic Recurrence in Locally Advanced Cervical Cancer Developed From an Analysis of Identifiable Prognostic Factors in Patients From NRG Oncology/Gynecologic Oncology Group Randomized Trials of Chemoradiotherapy. J Clin Oncol: Off J Am Soc Clin Oncol. 2015 Jul 01;33(19):2136–2142. doi: 10.1200/JCO.2014.57.7122. PubMed PMID: 25732170; PubMed Central PMCID: PMC4477785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hosseini ES, Meryet-Figuiere M, Sabzalipoor H, et al.. Dysregulated expression of long noncoding RNAs in gynecologic cancers. Mol Cancer. 2017 Jun 21;16(1):107. doi: 10.1186/s12943-017-0671-2. PubMed PMID: 28637507; PubMed Central PMCID: PMC5480155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chi S, Shen L, Hua T, et al.. Prognostic and diagnostic significance of lncRNAs expression in cervical cancer: a systematic review and meta-analysis. Oncotarget. 2017 May 31; doi: 10.18632/oncotarget.18323. PubMed PMID: 28620127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jiang S, Wang HL, Yang J. Low expression of long non-coding RNA LET inhibits carcinogenesis of cervical cancer. Int J Clin Exp Pathol. 2015;8(1):806–811. PubMed PMID: 25755778; PubMed Central PMCID: PMC4348863. PMID:25755778 [PMC free article] [PubMed] [Google Scholar]
- [32].Zhu J, Shi H, Liu H, et al.. Long non-coding RNA TUG1 promotes cervical cancer progression by regulating the miR-138-5p-SIRT1 axis. Oncotarget. 2017 May 26; doi: 10.18632/oncotarget.18224. PubMed PMID: 28615533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Kim HJ, Lee DW, Yim GW, et al.. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015. February;46(2):521–530. doi: 10.3892/ijo.2014.2758. PubMed PMID: 25405331; PubMed Central PMCID: PMC4277242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Jing L, Yuan W, Ruofan D, et al.. HOTAIR enhanced aggressive biological behaviors and induced radio-resistance via inhibiting p21 in cervical cancer. Tumour Biol. 2015. May;36(5):3611–3619. doi: 10.1007/s13277-014-2998-2. PubMed PMID: 25547435. [DOI] [PubMed] [Google Scholar]
- [35].Ceccaldi R, Rondinelli B, D'Andrea AD. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016. January;26(1):52–64. doi: 10.1016/j.tcb.2015.07.009. PubMed PMID: 26437586; PubMed Central PMCID: PMC4862604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Bakkenist CJ, Kastan MB. Initiating cellular stress responses. Cell. 2004 Jul 09;118(1):9–17. doi: 10.1016/j.cell.2004.06.023. PubMed PMID: 15242640. [DOI] [PubMed] [Google Scholar]
- [37].Spagnolo L, Rivera-Calzada A, Pearl LH, et al.. Three-dimensional structure of the human DNA-PKcs/Ku70/Ku80 complex assembled on DNA and its implications for DNA DSB repair. Mol Cell. 2006 May 19;22(4):511–519. doi: 10.1016/j.molcel.2006.04.013. PubMed PMID: 16713581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


