ABSTRACT
Histone acetylation plays a pivotal role in plant growth and development, and is regulated by the antagonistic relationship between histone acetyltransferase (HAT) and histone deacetylase (HDAC). We previously revealed that some HDAC inhibitors confer high-salinity stress tolerance in plants. In this study, we identified two HDAC inhibitors, namely Ky-9 and Ky-72, which enhanced the high-salinity stress tolerance of Arabidopsis thaliana. Ky-9 and Ky-72 are structurally similar chlamydocin analogs. However, the in vitro inhibitory activity of Ky-9 against mammalian HDAC is greater than that of Ky-72. A western blot indicated that Ky-9 and Ky-72 increased the acetylation levels of histone H3, suggesting they exhibit HDAC inhibitory activities in plants. We conducted a transcriptomic analysis to investigate how Ky-9 and Ky-72 enhance high-salinity stress tolerance. Although Ky-9 upregulated the expression of more genes than Ky-72, similar gene expression patterns were induced by both HDAC inhibitors. Additionally, the expression of high-salinity stress tolerance-related genes, such as anthocyanin-related genes and a small peptide-encoding gene, increased by Ky-9 and Ky-72. These data suggest that slight structural differences in chemical side chain between HDAC inhibitors can alter inhibitory effect on HDAC protein leading to influence gene expression, thereby enhancing high-salinity stress tolerance in different extent.
KEYWORDS: Epigenetics, high-salinity stress, histone deacetylase inhibitor, histone acetylation, transcriptome
Introduction
Histone acetylation, which is one of the most characterized epigenetic modifications, is regulated by the antagonistic activities of histone acetyltransferase (HAT) and histone deacetylase (HDAC). The HDAC family proteins normally remove the acetyl group of acetyl-lysine residues at the N-terminal tail of histones, resulting in highly condensed chromatin and repressed gene expression. Recent studies confirmed that HDAC inhibitors can enhance the high-salinity stress tolerance of plants.1-3 The effective suppression of HDAC activity by HDAC inhibitors increases the hyperacetylation of histones, ultimately resulting in the activation of transcription. Moreover, Ky-2, which is an HDAC inhibitor, reportedly induces the expression of many salt-responsive genes to decrease Na+ accumulation and increase proline and polyamine contents to enhance high-salinity stress tolerance.1 Another HDAC inhibitor, suberoylanilide hydroxamic acid, increases high-salinity stress tolerance in cassava.2 Furthermore, Ueda et al. analyzed multiple HDAC inhibitors and observed that the inhibitory activities were correlated with the histone hyperacetylation status. Moreover, treatments with Class I selective HDAC inhibitors enhance high-salinity stress tolerance in Arabidopsis thaliana. In contrast, the Class II selective inhibition is not capable of increasing high-salinity stress tolerance.3 These findings imply that the differences in the activities of HDAC inhibitors may influence which subset of genes is affected. Thus, comparisons of transcriptome data obtained following treatments using various HDAC inhibitors under high-salinity stress conditions are necessary. In this study, we observed that two novel Class I HDAC inhibitors, Cyclo (L-2-Amino-8-oxo-10-oxaundecanoyl-aminoisobutylyl-L-phenylalanyl-D-prolyl-) (i.e., Ky-9) and Cylco (L-2-Amino-8-thia-10-oxoundecanoyl-aminoisobutylyl-L-phenylalanyl-D-prolyl-) (i.e., Ky-72), enhanced high-salinity stress tolerance in A. thaliana. Both Ky-9 and Ky-72 were designed and synthesized as HDAC inhibitors based on the structure of chlamydocin.4 We confirmed the HDAC inhibitory activity of Ky-9 and Ky-72 in plants by conducting immunoblotting experiments. A transcriptomic analysis revealed that the expression levels of high-salinity stress tolerance-related genes, including anthocyanin-related genes and a small peptide-encoding gene, were upregulated by Ky-9 and Ky-72.
Results
Ky-9 and Ky-72 inhibit HDAC activities in A. thaliana
The HDAC inhibitors Ky-9 and Ky-72 are chlamydocin analogs with modified side chains4 (Fig. 1A). We completed a western blot assay to examine the A. thaliana histone acetylation levels induced by Ky-9 and Ky-72 treatments. The acetylation levels of histone H3 were higher for the plants treated with Ky-9 or Ky-72 than for the control plants, with the highest level observed for the 5 μM Ky-72 or Ky-9 treatments (Fig. 1B). These results suggested that, although Ky-9 and Ky-72 differ regarding their side chains, both inhibit HDAC in a dose-dependent manner in A. thaliana.
Figure 1.
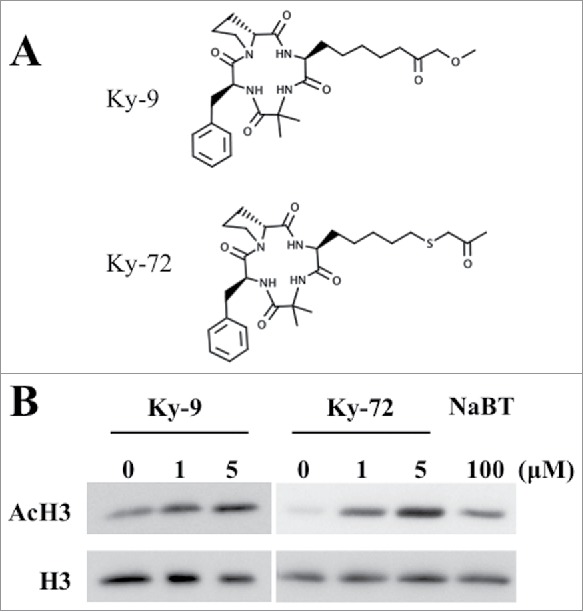
Changes in histone H3 acetylation levels during Ky-9 and Ky-72 treatments. (A) Chemical structure of Ky-9 and Ky-72. (B) Five-day-old seedlings treated with Ky-9 or Ky-72 for 6 h were analyzed. Histone acetylation levels were determined with a western blot using an anti-AcH3 antibody. The DMSO- and sodium butyrate (NaBT)-treated plants were used as negative and positive controls, respectively. The experiment was completed using three biological replicates.
Ky-9 and Ky-72 enhance high-salinity stress tolerance in A. thaliana
Previous studies confirmed that HDAC inhibitors enhance high-salinity stress tolerance in A. thaliana and cassava.1,2 In this study, we evaluated the effects of Ky-9 and Ky-72 on the high-salinity stress tolerance of A. thaliana. Wild-type plants grown in liquid culture medium were treated with 1 μM Ky-9, Ky-72, or dimethyl sulfoxide (DMSO) for 24 h, with or without a subsequent treatment with 100 mM NaCl for 4 days (Fig. 2). The plants treated with DMSO were unable to survive under high-salinity stress conditions. However, approximately 50 and 90% of the plants treated with Ky-9 and Ky-72 survived, respectively, after being exposed to highly saline conditions (Fig. 2). Our data suggested that Ky-9 and Ky-72 enhanced the tolerance of A. thaliana to high-salinity stress.
Figure 2.
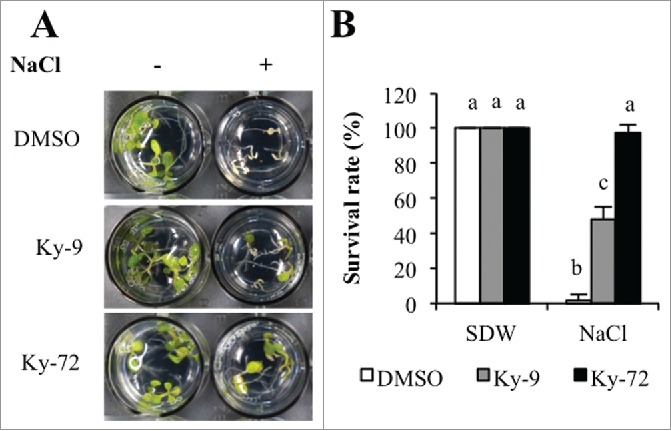
Ky-9 and Ky-72 treatments enhanced the high-salinity stress tolerance of Arabidopsis thaliana. (A) Phenotype of A. thaliana seedlings treated with 1 μM Ky-9 or Ky-72, with or without a subsequent treatment with 100 mM NaCl for 4 days. Seedlings treated with DMSO were used as negative controls. (B) Seedling survival rates under high-salinity stress conditions with or without the Ky-9 and Ky-72 treatments. The survival rate of 20 plants was calculated at 4 days after the NaCl treatment. The experiment was completed using three biological replicates. Error bars represent the mean ± standard deviation. Significance was determined according to Tukey's test (P < 0.01).
Comparative transcriptomic analysis of Ky-9 and Ky-72 treatments under high-salinity stress conditions
We conducted a microarray analysis to clarify the effects of Ky-9 and Ky-72 on high-salinity stress tolerance at the molecular level. Four-day-old plants treated with Ky-9, Ky-72, or DMSO for 24 h, with or without a subsequent treatment with 100 mM NaCl for 2 h, were examined (Fig. 3). Genes exhibiting upregulated expression according to the transcriptome data underwent a principal component analysis to investigate the global gene expression patterns induced by Ky-9 and Ky-72 treatments under normal and high-salinity stress conditions. These genes were selected because histone acetylation is associated with transcriptional activation in general. The clusters representing the treatments with and without NaCl were easily distinguishable (Fig. 3A). Moreover, the Ky-9 and Ky-72 treatment clusters were closer to each other than to the DMSO treatment cluster under control and high-salinity stress conditions. The data indicated that the NaCl treatment generally altered gene expression and the clusters of Ky-9- and Ky-72-treated plants were similar under control and high-salinity stress conditions.
Figure 3.

Upregulated gene expression profiles induced by HDAC inhibitors (Ky-9 and Ky-72) and high-salinity stress treatments. (A) Principal component analysis results for genes whose expression levels were upregulated by Ky-9, Ky-72, or DMSO treatments under control or high-salinity stress conditions. (B-E) Venn diagrams of the genes exhibiting upregulated expression in response to Ky-9, Ky-72, and DMSO treatments under control or high-salinity stress conditions. Each treatment was analyzed using 30 plants, with three biological replicates.
The Venn diagrams prepared based on the microarray data indicated that 492 and 282 genes were more highly expressed in Ky-9-treated plants than in DMSO-treated plants under control and high-salinity stress conditions, respectively (Fig. 3B). Among these genes, 91 exhibited upregulated expression under control and high-salinity stress conditions. Similarly, we detected 256 and 152 genes that were more highly expressed in Ky-72-treated plants than in DMSO-treated plants under control and high-salinity stress conditions, respectively (Fig. 3C). These genes included 67 that were detected under both control and high-salinity stress conditions. Additionally, the 492 and 256 genes whose expression levels were upregulated by Ky-9 and Ky-72, respectively, under control conditions included 169 overlapping genes (Fig. 3D). Under high-salinity stress conditions, the expression levels of 96 overlapping genes were upregulated by both Ky-9 and Ky-72 (Fig. 3E, Table S1). However, 186 and 56 genes were specifically affected by Ky-9 and Ky-72, respectively, under high-salinity stress conditions. The Ky-9 treatment generally upregulated the expression of more genes than the Ky-72 treatment. Furthermore, many of the genes whose expression was upregulated by Ky-72 also exhibited upregulated expression in response to Ky-9 (66 and 63% under control and high-salinity stress conditions, respectively).
Ky-9 and Ky-72 induce the expression of salt-responsive genes under high-salinity stress conditions
We focused on the 96 overlapping genes between the Ky-9 and Ky-72 treatments under high-salinity stress conditions. We performed the gene ontology (GO) analysis of the 96 overlapping genes (Fig. S1). This analysis revealed enrichment of genes responding to stimulus. Among the 96 genes were five salt-related genes that were identified under high-salinity stress conditions (Table 1), including At5g42800 (DFR; dihydroflavonol 4-reductase)5; At4g22880 (LDOX; leucoanthocyanidin dioxygenase), which is also known as ANS (anthocyanidin synthase)6; and At2g18660 (PNP-A; plant natriuretic peptide A).7,8 A quantitative real-time polymerase chain reaction (qRT-PCR) assay was completed to validate the expression of these candidate genes. We observed that the expression of these genes was upregulated by Ky-9 and Ky-72 treatments under control and high-salinity stress conditions (Fig. 4), suggesting that these genes might contribute to the high-salinity stress tolerance phenotype induced by Ky-9 and/or Ky-72.
Table 1.
Genes with upregulateda) expression levels in response to Ky-9 and Ky-72 treatments under high-salinity stress conditions.
| Ky-9/control under salt-stress |
Ky-72/control under salt stress |
||||||
|---|---|---|---|---|---|---|---|
| Gene name | AGI code | ratiob) | p-value | FDR | ratioc) | p-value | FDR |
| PNP-A; Plant Natriuretic Peptide A | AT2G18660 | 1.23 | 5.59E-03 | 4.32E-02 | 1.42 | 2.63E-06 | 4.32E-02 |
| OSM34; Osmotin 34 | AT4G11650 | 2.01 | 6.56E-05 | 9.42E-03 | 1.20 | 7.68E-04 | 9.42E-03 |
| DALL1; DAD1-Like Lipase 1 | AT4G16820 | 1.22 | 1.41E-02 | 7.31E-02 | 1.31 | 3.91E-04 | 7.31E-02 |
| LDOX; Leucoanthocyanidin Dioxygenase | AT4G22880 | 1.80 | 1.97E-04 | 1.17E-02 | 1.51 | 3.88E-04 | 1.17E-02 |
| DFR; Dihydroflavonol 4-Reductase | AT5G42800 | 1.84 | 2.70E-04 | 1.22E-02 | 1.37 | 1.17E-03 | 1.22E-02 |
a) Genes from the following two categories are listed:
1) log2 ratio (plants treated with Ky-9 for 24 h followed by a 2-h NaCl treatment/plants treated with DMSO for 24 h followed by a 2-h NaCl treatment) ≥ 1, FDR ≤ 0.1, t-test ≤ 0.1;
2) log2 ratio (plants treated with Ky-72 for 24 h followed by a 2-h NaCl treatment/plants treated with DMSO for 24 h followed by a 2-h NaCl treatment) ≥ 1, FDR ≤ 0.1, t-test ≤ 0.1
b) Values represent the log2 ratio (plants treated with Ky-9 for 24 h followed by a 2-h NaCl treatment/plants treated with DMSO for 24 h followed by a 2-h NaCl treatment)
c) Values represent the log2 ratio (plants treated with Ky-72 for 24 h followed by a 2-h NaCl treatment/plants treated with DMSO for 24 h followed by 2-h NaCl treatment)
Figure 4.
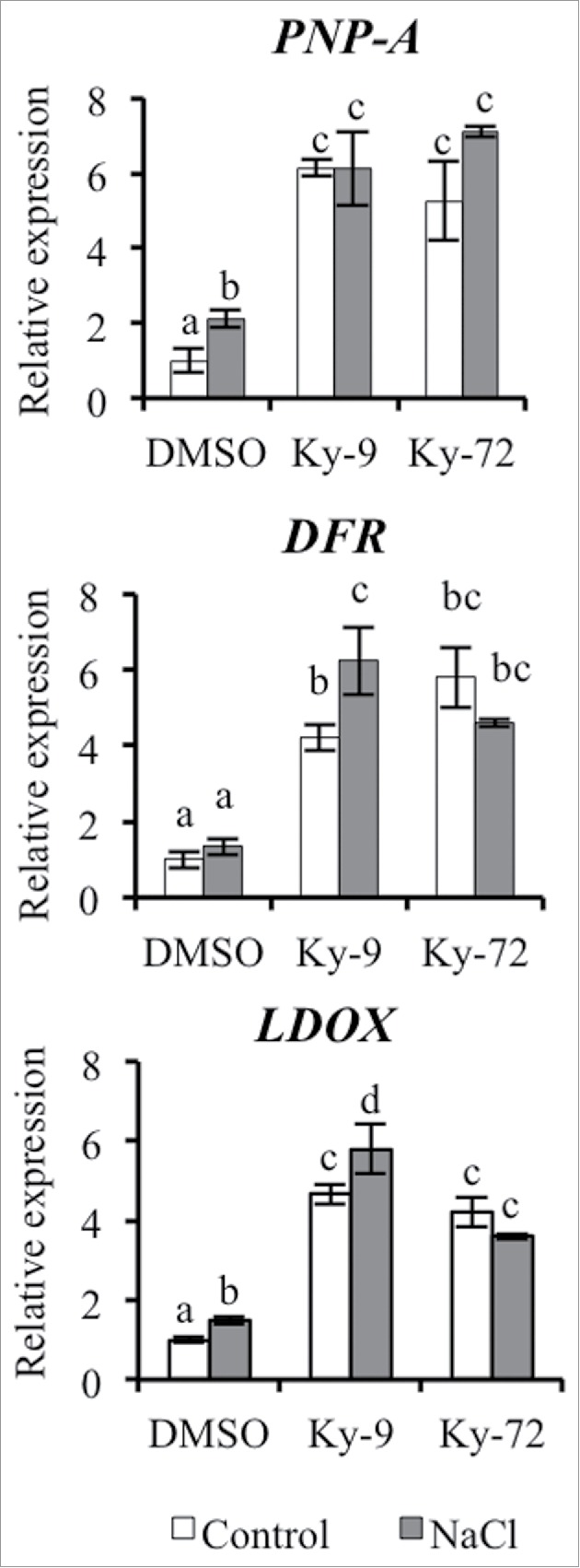
Salt-responsive genes exhibiting upregulated expression due to Ky-9 or Ky-72 treatment. Relative PNP-A, DFR, and LDOX expression levels in plants exposed to high-salinity stress conditions for 0 and 2 h in the presence or absence of Ky-9 or Ky-72. The expression levels of DMSO-treated plants were set as 1, and ACT2 was used as an internal standard. Each treatment was analyzed using 30 plants, with three biological replicates. Error bars represent the mean ± standard deviation. Significance was determined according to Tukey's t-test (P < 0.05).
Discussion
In this study, we revealed that two new HDAC inhibitors, Ky-9 and Ky-72, can enhance the high-salinity stress tolerance of A. thaliana. Our results are consistent with those of earlier studies that concluded HDAC inhibitors are important for plant stress tolerance.1-3,9 Ky-9 and Ky-72 were synthesized as chlamydocin analogs4 that shared a common cyclic tetrapeptide, but had different side chains. The cyclic tetrapeptide is believed to facilitate extensive interactions between HDAC and the inhibitors.4 The side chains, may inhibit the catalytic reaction by interaction with the zinc ion and catalytic residues in the active site pocket of HDAC.4 Moreover, the in vitro inhibitory activity of Ky-9 against mammalian HDAC is greater than that of Ky-72 (Table S2). Similarly, our microarray analysis indicated that Ky-9 upregulated the expression of more genes than Ky-72 under control and high-salinity stress conditions. However, the gene expression patterns were similar between the Ky-9 and Ky-72 treatments, with the expression of some salt-responsive genes induced by both Ky-9 and Ky-72. These genes included several anthocyanin-related genes, namely DFR, LDOX/ANS, and UFGT (Fig. 3, Table 1 and Table S1). In the anthocyanin biosynthesis pathway, DFR converts the dihydroflavonols generated from phenylalanine by complex upstream reactions to leucoanthocyanidins, which are converted to anthocyanidins in reactions catalyzed by LDOX/ANS.5 The anthocyanidins are converted to anthocyanins in a reaction mediated by UFGT.5 An earlier investigation demonstrated that overexpressing AtDFR genes in Brassica napus plants results in increased high-salinity stress tolerance because of an accumulation of anthocyanins.5 Additionally, LDOX/ANS is also reportedly a salt-responsive gene, with knockout mutants exhibiting significantly decreased anthocyanin accumulation.10-12 In the current study, the expression levels of several flavonoid synthesis-related genes increased by Ky-9 and Ky-72 under high-salinity stress conditions. These genes included At5g07990 (CYP75B1), At4g25310 [2OG and Fe(II)-dependent oxygenase], At4g22870 (putative LDOX/ANS), At3g29590 (5MAT), as well as At4g14090 and At2g15490 (UDP glycosyltransferases) (Table S1). The observed upregulated expression by Ky-9 and Ky-72 suggests these genes are important for high-salinity stress tolerance. The underlying mechanism enabling flavonoids, including anthocyanins, to increase stress tolerance has not been characterized. However, the correlation between the accumulation of anthocyanins and high-salinity stress tolerance has been reported.5,13,14 Our data suggest that Ky-9 and Ky-72 induce the expression of flavonoid and anthocyanin biosynthesis-related genes to protect plants from the adverse effects of highly saline conditions.
The PNP-A encodes a small peptide which is important for maintaining ion and solute homeostasis.15,16 Analyses of the data from 1,877 microarray experiments in the NASCArrays database indicated that PNP-A is involved in both biotic and abiotic stresses.17 Moreover, PNP-A expression is also reportedly upregulated by heat, salinity, and high osmotic stress conditions.8 Another study confirmed that PNP-A levels increase in plants grown under saline conditions.7 Furthermore, the application of exogenous PNP-A to an A. thaliana suspension-cultured cells resulted in the activation of salt-responsive genes, including At4g23670 (polyketide cyclase/dehydrase and lipid transport superfamily) and At1g17880 (BTF3; basic transcription factor 3).18 A recent study concluded that AtPNP-A knockout mutants and plants overexpressing AtPNP-A are hypersensitive and tolerant to high-salinity stress conditions, respectively.19 The upregulated expression of PNP-A due to Ky-9 or Ky-72 treatments may initiate downstream cascades to enable plants to overcome the effects of high-salinity stress.
Our microarray data indicated that the expression of other salt-responsive genes was upregulated by HDAC inhibitors under high-salinity stress conditions (Fig. 3, Table 1). These genes included At4g11650 (OSM34; Osmotin 34) and At4g16820 (DALL; Defective in Anther Dehiscence 1-like Lipase 1) (Table 1). The OSM34 gene reportedly encodes an osmoprotectant, and its expression was upregulated by salinity stress.20,21 In transgenic tobacco plants, the increased production of osmotin leads to the accumulation of proline, which enhances osmotic stress tolerance.22 An earlier analysis of Affymetrix microarray data revealed that DALL1 expression is upregulated in roots in response to salt stress.23 Thus, the upregulated expression of DALL1 due to HDAC inhibitors might be necessary for high-salinity stress tolerance. These results suggest that the application of exogenous Ky-9 and Ky-72 regulates the expression of salt-responsive genes to enable A. thaliana plants to overcome the detrimental effects of salt stress.
Another chlamydocin analog with HDAC inhibitor activities, namely Ky-2, has been observed to enhance high-salinity stress tolerance in A. thaliana.1 Ky-2 enhances high-salinity stress tolerance by inducing the expression of SOS1 and SOS3, resulting in decreased intracellular Na+ contents, and increasing the accumulation of proline and polyamines.1 However, microarray data indicated that SOS1 and SOS3 expression levels were not upregulated by Ky-9 and Ky-72. A qRT-PCR assay confirmed that SOS1 expression was not upregulated by Ky-9 and Ky-72 (Fig. S2). These observations imply that the Class I HDAC inhibitors have distinct targets and mechanisms responsible for enhancing high-salinity stress tolerance. Differences in the HDAC inhibitory activity among Ky-2, Ky-9, and Ky-72 might affect the number of genes targeted by each HDAC inhibitor. Moreover, the in vitro HDAC inhibitory activity of Ky-2 was greater than that of Ky-9 or Ky-72 (Table S2). Consistent with this finding was the fact that Ky-2 upregulated the expression of more genes than Ky-9 or Ky-72 (Fig. S3). Furthermore, Ky-72 appears to enhance high-salinity stress tolerance more than Ky-9. These observations imply that Ky-72 does not affect genes that suppress high-salinity stress tolerance because the HDAC inhibitory activity of Ky-72 is relatively low. In future, new HDAC inhibitors that specifically activate genes related to high-salinity stress tolerance may be developed by changing the side chain structure.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana (ecotype Columbia-0) seeds were sterilized and sown in half-strength Murashige and Skoog (MS) liquid medium supplemented with 1% sucrose and 0.1% agar. The plants were grown under previously described conditions.1 Four-day-old plants were treated with 1 μM Ky-9, Ky-72, or DMSO (Wako, Japan) for 24 h, with or without a subsequent treatment with 100 mM NaCl (Wako, Japan). The DMSO-treated plants were used as controls. The survival rate of 20 plants was calculated 4 days after the NaCl treatment. The experiment was conducted using three biological replicates.
Protein extraction and immunoblot analysis
Seeds were sown in half-strength MS medium for 5 days, after which they were treated with 1 μM or 5 μM Ky-9 or Ky-72 working solution or 100 μM sodium butyrate (NaBT) or DMSO for 6 h. The DMSO- and NaBT-treated samples were used as the negative and positive controls, respectively. The subsequent protein extraction and western blot analysis were completed as previously described.2
RNA extraction
Total RNA was extracted from 5-day-old A. thaliana seedlings treated with 1 μM Ky-9, Ky-72, or DMSO for 24 h, with or without a subsequent 2-h treatment with 100 mM NaCl. Seedlings treated with DMSO were used as the negative controls. The RNA was extracted from 30 plants per treatment (with three biological replicates) using the RNeasy® Plant Mini Kit (QIAGEN) as previously described.24 The quality of the extracted RNA was evaluated using a Bioanalyzer system (Agilent).
Microarray analysis
A microarray analysis was conducted as previously described.24 The microarray data underwent a 1-way ANOVA and were deposited in the GEO database (GEO ID: GSE108070). Each treatment was analyzed using three biological replicates. A total of 30 plants per treatment were analyzed. Genes with a log2 expression ratio ≥ 1 [t-test analysis, Benjamini–Hochberg correction (FDR) ≤ 0.1] were considered to exhibit upregulated expression.
Quantitative real-time polymerase chain reaction assay
We synthesized cDNA using the QuantiTect Reverse Transcription Kit (Qiagen) for a qRT-PCR assay, which was completed as previously described.1 We used ACT2 as a reference gene. The assay was completed using three biological replicates. A total of 30 plants per treatment were analyzed. The qRT-PCR involved the following primers: PNP-A: 5′-AGCTGCTCAAGGAAAAGCTG-3′ and 5′-TCCCGGCAGAAATCAACTAC-3′; DFR: 5′-GACGACTTATGCAACGCTCA-3′ and 5′-TCCGTCAGCTTCTTGGAACT-3′; LDOX: 5′-TGGGTCACTGCAAAATGTGT-3′ and 5′-CGGAGACTCAACACTCACCA-3′; ACT2: 5′-GATCTCCAAGGCCGAGTATGAT-3′ and 5′-CCCATTCATAAAACCCCAGC-3′.
Supplementary Material
Funding Statement
This project was financially supported by grants from RIKEN, the Japan Science and Technology Agency, the Core Research for Evolutional Science and Technology project (Grant Number JPMJCR13B4 to MS), the Ministry of Education, Culture, Sports, Science and Technology of Japan (KAKENHI Grant Number 16H01476 to MS), and the Japan Society for the Promotion of Science (KAKENHI Grant Number 16K18838 to KS).
Disclosure of potential conflicts of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be considered as a potential conflict of interest.
Acknowledgments
We thank Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
References
- 1.Sako K, Kim JM, Matsui A, Nakamura K, Tanaka M, Kobayashi M, Saito K, Nishino N, Kusano M, Taji T, et al.. Ky-2, a histone deacetylase inhibitor, enhances high-salinity stress tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2016;57:776–83. doi: 10.1093/pcp/pcv199. [DOI] [PubMed] [Google Scholar]
- 2.Patanun O, Ueda M, Itouga M, Kato Y, Utsumi Y, Matsui A, Tanaka M, Utsumi C, Sakakibara H, Yoshida M, et al.. The histone deacetylase inhibitor Suberoylanilide hydroxamic acid alleviates salinity stress in Cassava. Frontiers Plant Sci. 2017;7:2039. doi: 10.3389/fpls.2016.02039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ueda M, Matsui A, Tanaka M, Nakamura T, Abe T, Sako K, Sasaki T, Kim JM, Ito A, Nishino N, et al.. The distinct roles of class I and II RPD3-like histone deacetylases in salinity stress response. Plant Physiol. 2017;175(4):1760–73. doi: 10.1104/pp.17.01332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishino N, Jose B, Shinta R, Kato T, Komatsu Y, Yoshida M. Chlamydocin–hydroxamic acid analogues as histone deacetylase inhibitors. Bioorganic Med Chem. 2004;12:5777–84. doi: 10.1016/j.bmc.2004.08.041. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Lee WJ, Vu TT, Jeong CY, Hong SW, Lee H. High accumulation of anthocyanins via the ectopic expression of AtDFR confers significant salt stress tolerance in Brassica napus L. Plant Cell Reports 2017;36:1215–24. doi: 10.1007/s00299-017-2147-7. [DOI] [PubMed] [Google Scholar]
- 6.Bharti P, Mahajan M, Vishwakarma AK, Bhardwaj J, Yadav SK. AtROS1 overexpression provides evidence for epigenetic regulation of genes encoding enzymes of flavonoid biosynthesis and antioxidant pathways during salt stress in transgenic tobacco. J Exp Botany. 2015;66:5959–69. doi: 10.1093/jxb/erv304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rafudeen S, Gxaba G, Makgoke G, Bradley G, Pironcheva G, Raitt L, et al.. A role for plant natriuretic peptide immuno-analogues in NaCl- and drought-stress responses. Physiologia Plantarum. 2003;119:554–62. doi: 10.1046/j.1399-3054.2003.00201.x. [DOI] [Google Scholar]
- 8.Wang YH, Gehring C, Irving HR. Plant natriuretic peptides are apoplastic and paracrine stress response molecules. Plant Cell Physiol. 2011;52:837–50. doi: 10.1093/pcp/pcr036. [DOI] [PubMed] [Google Scholar]
- 9.Kim JM, Sasaki T, Ueda M, Sako K, Seki M. Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Frontiers Plant Sci. 2015;6:114. doi: 10.3389/fpls.2015.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Oosten MJ, Sharkhuu A, Batelli G, Bressan RA, Maggio A. The Arabidopsis thaliana mutant air1 implicates SOS3 in the regulation of anthocyanins under salt stress. Plant Mol Biol. 2013;83:405–15. doi: 10.1007/s11103-013-0099-z. [DOI] [PubMed] [Google Scholar]
- 11.Abrahams S, Tanner GJ, Larkin PJ, Ashton AR. Identification and biochemical characterization of mutants in the Proanthocyanidin Pathway in Arabidopsis. Plant Physiol. 2002;130:561–76. doi: 10.1104/pp.006189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR. The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J. 2003;35:624–36. doi: 10.1046/j.1365-313X.2003.01834.x. [DOI] [PubMed] [Google Scholar]
- 13.Oh JE, Kim YH, Kim JH, Kwon YR, Lee H. Enhanced level of anthocyanin leads to increased salt tolerance in Arabidopsis PAP1-D plants upon sucrose treatment. J Korean Soc Applied Biol Chem. 2011;54:79–88. doi: 10.3839/jksabc.2011.011. [DOI] [Google Scholar]
- 14.Wahid A, Ghazanfar A. Possible involvement of some secondary metabolites in salt tolerance of sugarcane. J Plant Physiol. 2006;163:723–30. doi: 10.1016/j.jplph.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Gehring CA, Irving HR. Natriuretic peptides — a class of heterologous molecules in plants. Int J Biochem Cell Biol. 2003;35:1318–22. doi: 10.1016/S1357-2725(03)00032-3. [DOI] [PubMed] [Google Scholar]
- 16.Ludidi N, Morse M, Sayed M, Wherrett T, Shabala S, Gehring C. A recombinant plant Natriuretic peptide causes rapid and spatially differentiated K+, Na+ and H+ flux changes in Arabidopsis thaliana Roots. Plant Cell Physiol. 2004;45:1093–8. doi: 10.1093/pcp/pch113. [DOI] [PubMed] [Google Scholar]
- 17.Meier S, Bastian R, Donaldson L, Murray S, Bajic V, Gehring C. Co-expression and promoter content analyses assign a role in biotic and abiotic stress responses to plant natriuretic peptides. BMC Plant Biol. 2008;8:24. doi: 10.1186/1471-2229-8-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turek I, Marondedze C, Wheeler JI, Gehring C, Irving HR. Plant natriuretic peptides induce proteins diagnostic for an adaptive response to stress. Frontiers Plant Sci. 2014;5:661. doi: 10.3389/fpls.2014.00661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ficarra, FA, Grandellis, C, Garavaglia, BS, Gottig, N, Ottado, J. Bacterial and plant natriuretic peptides improve plant defence responses against pathogens. Mol Plant Pathol. 2018;19(4):801–811. doi: 10.1111/mpp.12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binzel ML, Hasegawa PM, Handa AK, Bressan RA. Adaptation of tobacco cells to NaCl. Plant Physiol. 1985;79:118–25. doi: 10.1104/pp.79.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karam MA, Abd-Elgawad ME, Ali RM. Differential gene expression of salt-stressed Peganum harmala L. J Genetic Engineering Biotechnol. 2016; 14:319–26. doi: 10.1016/j.jgeb.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barthakur S, Babu V, Bansa KC. Over-expression of Osmotin induces proline accumulation and confers tolerance to osmotic stress in transgenic tobacco. J Plant Biochem Biotechnol. 2001;10:31–7. doi: 10.1007/BF03263103. [DOI] [Google Scholar]
- 23.Ma S, Gong Q, Bohnert HJ. Dissecting salt stress pathways. J Exp Botany. 2006;57:1097–107. doi: 10.1093/jxb/erj098. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen AH, Matsui A, Tanaka M, Mizunashi K, Nakaminami K, Hayashi M, Iida K, Toyoda T, Nguyen DV, Seki M. Loss of Arabidopsis 5′-3′ Exoribonuclease AtXRN4 function enhances heat stress tolerance of plants subjected to severe heat stress. Plant Cell Physiol. 2015;56:1762–72. doi: 10.1093/pcp/pcv096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


