ABSTRACT
Anthocyanins are water-soluble pigments with antioxidant activities. In plants, multiple factors can trigger the accumulation of anthocyanins, including chemicals and environmental factors. Reactive oxygen species (ROS) are common by-products produced under different biotic and abiotic conditions and cause oxidative stress when accumulated at a high level in plant cells. This in turn leads to the production of anthocyanins. However, the mechanisms of ROS-induced anthocyanin accumulation and the role of anthocyanins in the response of plants to different stresses are largely unknown. We have recently reported the cross-regulation between ROS and anthocyanin production through analyzing ten Arabidopsis mutants covering the main anthocyanin regulatory and biosynthetic genes grown under different ROS-generating stresses. Here, we describe the general phenotypic response of anthocyanin mutants under normal and ROS-generating stress conditions, showing the changing levels of anthocyanin accumulation and their sensitivity to stresses. In addition, we propose a model that describes a particular gene interaction that highlights how the cross-regulation mechanisms between ROS and anthocyanin production are essential for plant resistance to various stresses through removing excessive ROS and maintaining photosynthetic capacity.
KEYWORDS: Anthocyanin, Reactive oxygen species (ROS), ttmutants, cross-regulation, photosynthetic capacity, Arabidopsis
In Arabidopsis, anthocyanins accumulate mainly in leaves and stems with various levels of accumulation depending on different developmental and environmental factors.1-4 The production of anthocyanins is catalyzed by a series of enzymes in the anthocyanin biosynthetic pathway.5 The genes encoding these enzymes are positively regulated by three types of transcription regulators that form a protein complex: MYB/bHLH/WD40.6 Anthocyanin biosynthetic genes are generally in two groups: the early biosynthetic genes (EBG), which form precursors for anthocyanins; and late biosynthetic genes (LBG), which are down-stream genes of this pathway.7 They are regulated by different members of the three types of transcription regulators.8,9 Extensive researches on the regulation of anthocyanin production under different growth and developmental conditions have been reported.4,6,10-12 Reactive oxygen species (ROS) are one type of stress that are induced by developmental and environmental stimuli, resulting in oxidative damage to the cells. Among the enzymatic and non-enzymatic mechanisms in maintaining the homeostasis of ROS in plant cells, the accumulation of anthocyanin is reported to be important in contributing to the protection against ROS.13,14 However, the regulatory mechanisms between anthocyanin and ROS in Arabidopsis are largely unknown.
Recently, we performed a systematic study on the cross-regulation between anthocyanin and ROS by analyzing the phenotypic, physiological and molecular changes of ten anthocyanin mutants having various accumulation levels of anthocyanin under different stress conditions.15 Generally, anthocyanin deficient mutants are sensitive to ROS and ROS-generating stresses as the endogenous ROS are accumulated at a high level in those mutants.15 This is further confirmed by two facts: (1) supplementation of cyanidin to the growth medium recovers the hypersensitivity of the anthocyanin deficient mutants to ROS and (2) the radical scavenging activity of those anthocyanin deficient mutants are reduced.15 However, a gain-of-function mutant, pap1-D, which accumulates more anthocyanin than wild-type (WT) plants shows the opposite responses.15 A model for the general phenotypic responses of anthocyanin mutants under different growth conditions is shown in Fig. 1. Under normal growth conditions, anthocyanin loss-of-function mutants (tt mutants) resembles the WT in growth and development, except for the absence of anthocyanins which usually accumulate in the inflorescence, basal section of the stem and back of the leaves. The growth and development of the pap1-D mutant is also similar with WT, but with an obvious accumulation of anthocyanin throughout the whole plant (Fig. 1). Under ROS-generating stress conditions, the growth of both WT and the tt mutants are inhibited, reflected by the reduced plant size. The growth of the tt mutants are even more reduced compared to the WT, indicating that they are more sensitive to ROS-generating stresses. However, the growth of the pap1-D mutant is not significantly reduced (Fig. 1). In addition, the accumulation of anthocyanin is significantly increased in WT under stress conditions. In contrast, the obvious senescence phenotype in the tt mutants is observed with the leaves turning yellow. The whole plant of the pap1-D mutant turns a dark purple colour due to the excess accumulation of anthocyanin with reduced growth inhibition (Fig. 1). These general responses for plants under different growth conditions may vary depending on the plant material and experimental conditions.16
Figure 1.
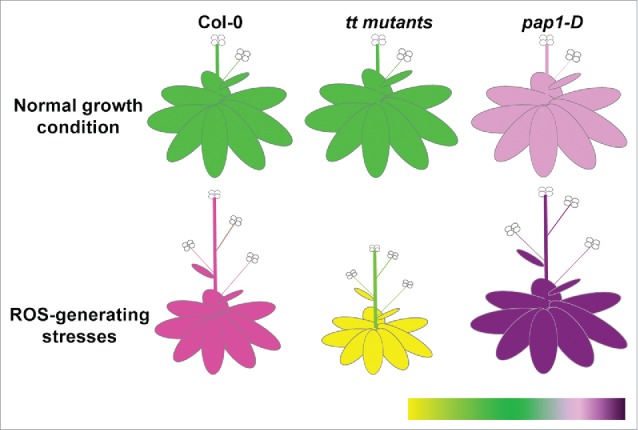
General phenotypic responses of anthocyanin mutants under different growth conditions. The color heatmap on the right-bottom corner represents the color change of the plant under different growth conditions. Green colour indicates the optimal growth status of the plants. Yellow colour indicates the sensitivity of plants to stress. Different intensity of purple indicates various levels of anthocyanin accumulation. The relative plant size reflects the relative sensitivity of plants to different growth condition.
In Arabidopsis, PAP, TT8 and TTG1 are the representative genes for the three components forming the protein complex that regulates the expression of anthocyanin biosynthetic genes. We found that ROS induce the production of anthocyanin by up-regulating the LBGs, such as TT3 and TT18, due to the up-regulation of PAP and TT8.15 Among all the anthocyanin mutants we used in the study, the response of the tt8-6 mutant is particularly interesting. Under normal growth conditions, no anthocyanin accumulation was observed phenotypically in the tt8-6 mutant. In contrast, under ROS-generating stresses, tt8-6 had a significant increase in the accumulation of anthocyanin.15 Therefore, we propose another model for the regulation of anthocyanins production in the tt8 mutant under different growth conditions as well as the physiological function of anthocyanin in protecting plants against ROS (Fig. 2). Under normal growth conditions, production of anthocyanins in Arabidopsis is maintained at a low level in most tissues. However, in the tt8 mutant, the protein complex regulating the expression of TT3 and TT18 is not as active due to the loss of function of the TT8 gene, resulting in the lack of anthocyanin production in the tt8-6 mutant. Under ROS-generating stress conditions, the expression levels of PAP and TT8 in the WT plants are up-regulated, resulting in the up-regulation of TT3 and TT18 and therefore leading to the significant induction of anthocyanins (Fig. 2).15 However, in the tt8-6 mutant, the expression of GL3 (an paralogue gene of TT8) and TT3 are still up-regulated by stress,15 suggesting that GL3 can complement the loss-of-function of TT8 in the tt8 mutant to form an alternative protein complex with PAP and TTG1 leading to anthocyanin production (Fig. 2). Therefore, in a feedback loop, anthocyanins are produced due to ROS stress and in turn protect plants from growth inhibition and cell death by scavenging excess ROS (Fig. 2). This is supported by the reduced radical scavenging activities of the anthocyanin deficient mutants and the increased level seen in the PAP1-D mutant.15 It should be noted that this is not due to changes in the expression levels of the genes encoding the main ROS scavenging enzymes.15 Meanwhile, anthocyanins also function in preventing chlorophyll degradation and in maintaining photosynthetic capacity (Fig. 2), as shown by the expression patterns of the chlorophyll and photosynthetic related genes in the mutants.15
Figure 2.
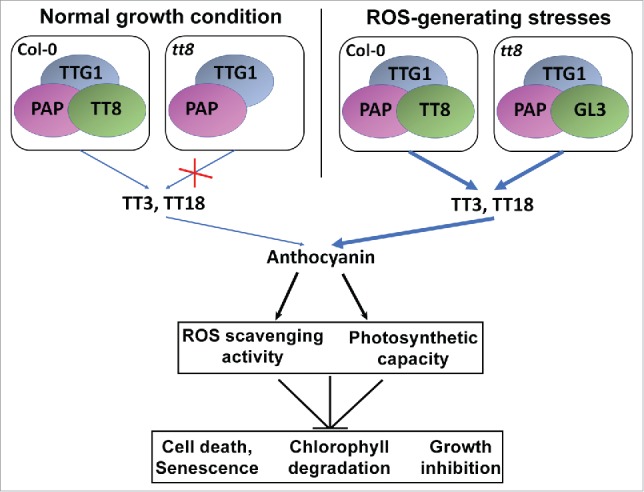
Model for the regulation of anthocyanin production in the tt8 mutant under different growth conditions and the physiological functions of anthocyanin in protecting plants against ROS. Expression of GL3 complements the loss-of-function of TT8 in the tt8 mutant only under stress conditions. The pointed arrows indicate positive regulation and blunt end arrows indicate negative regulation. The thickness of the blue arrow indicates the extent of the regulation.
Production of anthocyanin and the regulation mechanisms are different under various growth and developmental conditions. The incorporation of other signaling pathways such as hormone signaling makes the regulation of anthocyanin production even more complex. For example, DELLA and JAZ proteins are reported to promote anthocyanin production, indicating the involvement of GA and JA signaling in regulating anthocyanin production, respectively3,10,17 Our models revealed a general mechanism in the cross-regulation between anthocyanin and ROS, with further research necessary to delineate the interactions of other regulators with the components of the MYB/bHLH/WD40 complex, the detailed regulatory mechanisms of anthocyanin production under specific developmental stage or growth stimulus and the construction of a regulatory network for anthocyanin production.
Diclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–22. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoch WA, Zeldin EL, McCown BH. Physiological significance of anthocyanins during autumnal leaf senescence. Tree Physiol. 2001;21:1–8. doi: 10.1093/treephys/21.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Li Y, Van den Ende W, Rolland F. Sucrose induction of anthocyanin biosynthesis is mediated by DELLA. Mol Plant. 2014;7:570–2. doi: 10.1093/mp/sst161. [DOI] [PubMed] [Google Scholar]
- 4.Li S, Wang W, Gao J, Yin K, Wang R, Wang C, Petersen M, Mundy J, Qiu JL. MYB75 phosphorylation by MPK4 is required for light-induced anthocyanin accumulation in Arabidopsis. Plant Cell. 2016;28:2866–83. doi: 10.1105/tpc.16.00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lepiniec L, Debeaujon I, Routaboul JM, Baudry A, Pourcel L, Nesi N, et al.. Genetics and biochemistry of seed flavonoids. Annu Rev Plant Biol. 2006;57:405–30. doi: 10.1146/annurev.arplant.57.032905.105252. [DOI] [PubMed] [Google Scholar]
- 6.Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–85. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Butelli E, Martin C. Engineering anthocyanin biosynthesis in plants. Curr Opin Plant Biol. 2014;19:81–90. doi: 10.1016/j.pbi.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–27. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 9.Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–77. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Y, Tan H, Ma Z, Huang J. DELLA proteins promote anthocyanin biosynthesis via sequestering MYBL2 and JAZ suppressors of the MYB/bHLH/WD40 complex in Arabidopsis thaliana. Mol Plant. 2016;9:711–21. doi: 10.1016/j.molp.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21:3567–84. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian H, Qi T, Li Y, Wang C, Ren C, Song S, Huang H. Regulation of the WD-repeat/bHLH/MYB complex by gibberellin and jasmonate. Plant Signaling & Behavior. 2016;11:e1204061. doi: 10.1080/15592324.2016.1204061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, De Stefano R, Robine M, Butelli E, Bulling K, Hill L, Rejzek M, Martin C, Schoonbeek HJ. Different reactive oxygen species scavenging properties of flavonoids determine their abilities to extend the shelf life of tomato. Plant Physiol. 2015;169:1568–83. doi: 10.1104/pp.15.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, et al.. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77:367–79. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Z, Mahmood K, Rothstein SJ. ROS induces anthocyanin production via late biosynthetic genes and anthocyanin deficiency confers the hypersensitivity to ROS-generating stresses in Arabidopsis. Plant Cell Physiol. 2017;58:1364–77. doi: 10.1093/pcp/pcx073. [DOI] [PubMed] [Google Scholar]
- 16.Misyura M, Colasanti J, Rothstein SJ. Physiological and genetic analysis of Arabidopsis thaliana anthocyanin biosynthesis mutants under chronic adverse environmental conditions. J Exp Bot. 2013;64:229–40. doi: 10.1093/jxb/ers328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D, et al.. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell. 2011;23:1795–814. doi: 10.1105/tpc.111.083261. [DOI] [PMC free article] [PubMed] [Google Scholar]


