ABSTRACT
Master regulatory transcription factors cooperate in networks to shepherd cells through organogenesis. In the Drosophila eye, a collection of master control proteins known as the retinal determination gene network (RDGN) switches the direction and targets of its output to choreograph developmental transitions, but the molecular partners that enable such regulatory flexibility are not known. We recently showed that two RDGN members, Eyes absent (Eya) and Sine oculis (So), promote exit from the terminal cell cycle known as the second mitotic wave (SMW) to permit differentiation. A search for co-factors identified the ubiquitously expressed Combgap (Cg) as a novel transcriptional partner that impedes cell cycle exit and interferes with Eya-So activity specifically in this context. Here, we argue that Cg acts as a flexible transcriptional platform that contributes to numerous gene expression outcomes by a variety of mechanisms. For example, Cg provides repressive activities that dampen Eya-So output, but not by recruiting Polycomb chromatin-remodeling complexes as it does in other contexts. We propose that master regulators depend on both specifically expressed co-factors that assemble the combinatorial code and broadly expressed partners like Cg that recruit the diverse molecular activities needed to appropriately regulate their target enhancers.
KEYWORDS: cell cycle, Development, eye developent, gene regulation, gene regulatory network, molecular genetics, second mitotic wave, transcription, transcriptional repressor, transcription factor
Master regulatory function depends on interactions with other transcription factors
Developing animal cells progress through sequences of cellular behaviors to assemble functional adult organs. Cells that take on different fates share identical genomic information and basal gene regulatory machinery, so they specialize their developmental paths by expressing unique subsets of proteins. Transcription factors provide this discrimination by controlling which loci each cell transcribes. One uniquely powerful category of regulators, the master control transcription factors, operates in small networks that continuously adapt their transcriptional interactions to adjust the selection and regulation of downstream genes over time and drive the cellular transitions that generate the target organ [2,13,15,31]. This rewiring strategy is thought to contribute to the astonishing developmental potency of master regulatory networks [13]: it both underlies their ability to drive the sequence of cellular transitions that generate the target organ during normal development and enables their mis-expression to redirect cells through an inappropriate developmental trajectory. A classic example is that of the retinal determination gene network (RDGN) of Drosophila, which orchestrates normal retinal development and can hijack the genetic machinery of cells in the primordial wings, legs, or antennae to generate ectopic eyes [6,10,17,38,44].
Flexible regulation of cellular events by the archetypal RDGN hierarchy illustrates the importance of master control networks rewiring to propel development. Four transcription factors comprise the core network and are expressed in an overlapping sequence that drives retinogenesis. Eyeless (Ey) is first expressed early in the development of the larval eye precursor, the eye-antennal imaginal disc, where it establishes regional identity, promotes tissue growth, and suppresses differentiation [4,18]. Later, it reverses the latter regulation by initiating expression of Eyes absent (Eya) and Sine oculis (So) [18,35,36,39], which form a bipartite transcription factor that activates dachshund (dac) transcription [10,37,38]. Together, the four network members reinforce one another's expression and choreograph the first steps of retinal specification and differentiation in a domain known as the morphogenetic furrow (MF) [4,12,16,25,48,55]. Once differentiation begins, Eya-So and Dac switch their effect on ey to terminate its expression [2,23], and Eya-So directs subsequent specification and differentiation events [24,26].
Two related mysteries cloud our understanding of the RDGN: what biochemical changes reshape network activities to initiate these developmental transitions, and how does this flexibility contribute to network function during normal and ectopic development? The answers must lie in the way master control transcription factors interact with additional co-factors and how these extra-network interactions influence network relationships to orchestrate appropriate changes in target gene selection and regulation. Consistent with this idea, both negative and positive regulators can tune RD transcriptional output, but taking Eya-So as an example, only a handful of co-factors are known and none have been assigned rigorously to a specific transcriptional event that directs a developmental outcome (Table 1) [1,8,10,14,23,30,32,34,38,45,54]. Even in the best understood example, where a switch in RD transcriptional output from activating to repressive at the ey locus is required to initiate differentiation, neither the composition of the different Eya-So-Dac-containing transcriptional complexes that assemble nor how they produce activating versus repressive outputs is known [2].
Table 1.
Known binding partners of Drosophila Eya and So.
| Eya Binding Partner |
Symbol |
Yeast two-hybrid |
in vitro binding |
Co-IP |
References |
| Combgap | Cg | Positive | Positive | Negative | [10] |
| Dachshund | Dac | Positive | Positive | Positive | [2-4] |
| Eyeless | Ey | Untested | Untested | Positive | [4] |
| Eyes absent | Eya | Untested | Untested | Positive | [6,8] |
| Groucho | Gro | Untested | Positive | Negative | [5,8] |
| I-κB kinase β | IKKβ | Untested | Untested | Positive | [11] |
| Optix | Optix | Negative | Untested | Positive | [1,5] |
| Relish | Rel | Untested | Untested | Positive | [11] |
| Sine oculis |
So |
Positive |
Positive |
Positive |
[1,2,4-8] |
| So Binding Partner |
Symbol |
Yeast two-hybrid |
in vitro binding |
Co-IP |
References |
| Eyeless | Ey | Untested | Positive | Untested | [9] |
| Eyes absent | Eya | Positive | Positive | Positive | [1,2,4-8] |
| Groucho | Gro | Positive | Untested | Positive | [5,8] |
| Optix binding protein | Opbp | Positive | Positive | Untested | [5] |
| Sine oculis binding protein | Sobp | Positive | Positive | Untested | [5] |
| TBP-associated factor 1 | Taf1 | Positive | Positive | Untested | [5] |
We considered yeast two-hybrid, direct in vitro binding assays, and co-immunoprecipitation experiments to be evidence of complex formation. (A) Proteins that bind Eya. (B) Proteins that bind So. 1Anderson, A. M., Weasner, B. M., Weasner, B. P. and Kumar, J. P. (2012). Dual transcriptional activities of SIX proteins define their roles in normal and ectopic eye development. Development 139, 991–1000. 2Bui, Q. T., Zimmerman, J. E., Liu, H. and Bonini, N. M. (2000). Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics 155, 709–20. 3Chen, R., Amoui, M., Zhang, Z. and Mardon, G. (1997). Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell 91, 893–903. 4Jin, M. and Mardon, G. (2016). Distinct Biochemical Activities of Eyes absent During Drosophila Eye Development. Sci. Rep. 6, 23228. 5Kenyon, K. L., Li, D. J., Clouser, C., Tran, S. and Pignoni, F. (2005). Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev. Dyn. 234, 497–504. 6Mutsuddi, M., Chaffee, B., Cassidy, J., Silver, S. J., Tootle, T. L. and Rebay, I. (2005). Using Drosophila to decipher how mutations associated with human branchio-oto-renal syndrome and optical defects compromise the protein tyrosine phosphatase and transcriptional functions of eyes absent. Genetics 170, 687–95. 7Pignoni, F., Hu, B., Zavitz, K. H., Xiao, J., Garrity, P. a and Zipursky, S. L. (1997). The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell 91, 881–91. 8Silver, S. J., Davies, E. L., Doyon, L. and Rebay, I. (2003). Functional Dissection of Eyes absent Reveals New Modes of Regulation within the Retinal Determination Gene Network. Mol. Cell. Biol. 23, 5989–5999. 9Zhang, T., Ranade, S., Cai, C. Q., Clouser, C. and Pignoni, F. (2006). Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development 133, 4881–9. 10Davis, T. L. and Rebay, I. (2017). Antagonistic regulation of the second mitotic wave by Eyes absent-Sine oculis and Combgap coordinates proliferation and specification in the Drosophila retina. Development. 11Liu, X., Sano, T., Guan, Y., Nagata, S., Hoffmann, J. A. and Fukuyama, H. (2012). Drosophila EYA Regulates the Immune Response against DNA through an Evolutionarily Conserved Threonine Phosphatase Motif. PLoS One 7, e42725.
A screen for Eya-binding proteins identifies Combgap as a novel co-factor that limits activating output from Eya-So
In an effort to identify co-factors that modulate RDGN output to effect specific developmental transitions, we performed a yeast two-hybrid screen using Eya as bait. Transcription factors that interfered with Eya-So's ability to promote gene expression were of particular interest, given that repression by this complex is poorly understood. Among the most intriguing hits was the C2-H2 zinc finger transcription factor Combgap (Cg) [14]. Initial characterization of the genetic relationship between cg and eya revealed antagonism, such that halving cg dosage strongly suppressed eya loss-of-function phenotypes at multiple developmental time points. This relationship appeared to limit transcriptional activation by Eya-So, as lowering cg levels improved misexpressed eya's ability to initiate ectopic Dac expression, and overexpressing cg attenuated Eya-So output in S2 cell-based transcription assays. Coupled with in vitro confirmation that Cg can participate in the Eya-So complex, these data established Cg as the first bona fide inhibitor of Eya-So transcriptional function.
Pinpointing a specific developmental transition regulated by Cg-Eya interactions proved more challenging, as cg loss did not overtly disrupt eye formation. However, we noticed that deleting cg reduced the number of mitotic retinal precursors in a zone immediately posterior to the MF. This swath of proliferation, known as the second mitotic wave (SMW), comprises the final division of unspecified precursors in the retina and ensures that the larva generates sufficient cells with which to assemble an adult eye [3,53]. Although RDGN activity was known to regulate proliferation-differentiation transitions anterior to the MF [4,7,22,33], its role in the SMW had not been studied.
Further experiments revealed that Cg and Eya-So antagonize one another to choreograph the SMW cell cycle and prepare retinal precursors for differentiation. Knocking down eya or so caused cells that should have exited the SMW to re-enter another S phase, indicating that the Eya-So complex limits unspecified precursors to a single division after the MF. Cg and Eya-So's opposing inputs at the SMW cell cycle suggested that their antagonism calibrates the mitotic rate prior to differentiation. Consistent with this hypothesis, eya or so heterozygosity restored a nearly normal number of mitoses to cg mutant SMWs, while reducing cg dosage largely eliminated ectopic cell division when eya was knocked down. Based on these data, we proposed that Cg and Eya-So co-regulate transcription at one or more loci in the genetic circuitry surrounding the cellular decision to continue dividing or permanently exit the cell cycle.
What are Combgap's transcriptional functions?
This work not only described novel developmental roles for the Eya-So complex and Cg, but also uncovered a conspicuous paradox. Our prediction had been that Eya-So co-factors that orchestrate specific developmental transitions would be expressed in restricted patterns consistent with their function. Cg fulfilled the first expectation, in that its retinal requirement and genetic interaction with Eya and So appeared dedicated to a single cell cycle, but defied the accompanying prediction, as it is expressed ubiquitously and at uniform levels throughout the eye imaginal disc [14]. In fact, Cg is present broadly in all imaginal discs and many other tissues in the fly, and has long been known to control myriad developmental processes [9,46,47]. cg loss is not lethal until puparium formation [14,40], and amorphic and hypomorphic alleles produce pleiotropic defects throughout the larva and adult, respectively [9,14,21,40,46,47,49-51]. Together, these observations argue that Cg does not provide the spatial information that rewires Eya-So activity to promote cell cycle exit after the SMW.
If Cg does not determine specificity, then what activity might it contribute to the gene regulatory program that controls the transition from proliferation to differentiation? Coupled with its broad expression pattern, Cg's spatially restricted loss-of-function phenotype in the eye suggests that it interprets the combinatorial code, rather than supplementing it, to limit Eya-So output at target genes in cells at the SMW. Thus, while the factors that confer specificity to this regulatory circuit remain unknown, we propose that Cg recruits the repressive activity that switches Eya-So output at the SMW.
Two molecular activities could explain this role. First, work from the Gilboa laboratory determined that Cg both recruits the EcR transcription factor to target enhancers and organizes the three-dimensional configuration of these loci [21], raising the possibility that Cg could limit Eya-So output by preventing enhancer-promoter contacts that favor transcription. Second, the Kassis group showed that Cg recruits and provides DNA-binding specificity to Polycomb Group (PcG) repressive complexes [40], suggesting that Cg could help install repressive chromatin modifications that dampen activation from Eya-So.
To test the latter idea, we asked whether cg and PcG components interact synergistically to promote SMW proliferation. We first confirmed that Cg can support epigenetic repression in the developing retina by showing that deleting cg de-repressed the classic PcG target Engrailed (En) in this tissue (Fig. 1A,B) [27,28]. Lowering the dosage of PcG components in the cg mutant background did not further increase En levels, arguing that all modes of PcG-mediated en repression require Cg (Fig. 1C-G). In contrast, when we turned to SMW regulation, we found that halving the dosage of genes encoding Polycomb Repressive Complex 1 or 2 proteins strongly suppressed the cg phenotype, restoring nearly normal numbers of nuclei in S or M phase (Fig. 1H-Q). This unexpected result suggested that Cg might prevent rather than promote PcG recruitment to target genes used in SMW regulation.
Figure 1.
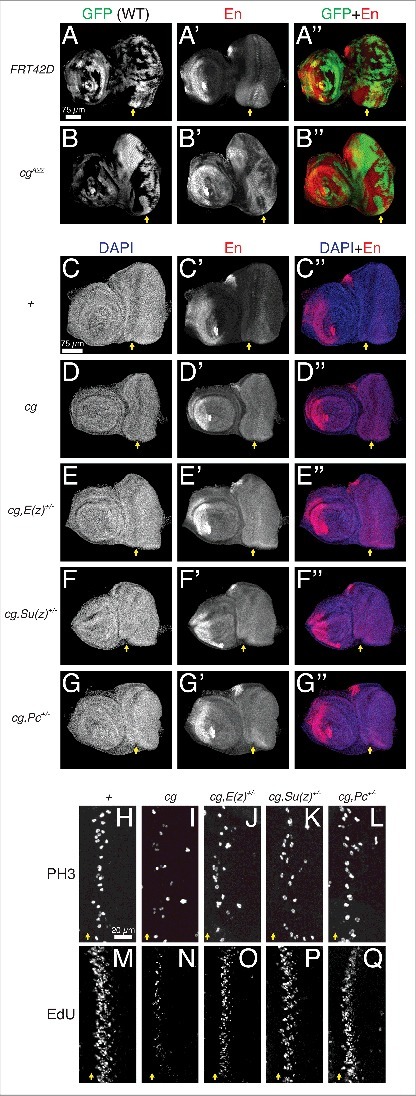
Cg may not be dedicated to repressive Polycomb group recruitment in the eye-antennal imaginal disc. All images are maximum confocal projections of late third instar eye-antennal imaginal discs oriented anterior left and dorsal up, with yellow arrows marking the MF. (A-B) Mitotic cg clones de-repress En in the antenna and retina. (C-G) Lowering the dosage of PRC1 or PRC2 components does not exacerbate En de-repression in cg null discs. (H-Q) Heterozygosity for PRC1 (Pc) or PRC2 (E(z) and Su(z)12) components suppresses the reduced mitosis and S phase entry of cg mutants. Each panel is a zoomed view centered on a representative SMW.
Given that PcG heterozygosity did not modify eya loss-of-function phenotypes in the SMW (data not shown), we speculate that the SMW target genes influenced by PcG-Cg interactions are not those regulated by Eya-So, and that at Eya-So targets, Cg repressive influence occurs via an alternate mechanism, perhaps by influencing 3D chromatin interactions. Genes that would prevent re-entry into S phase after the SMW, such as Rbf, are prime candidates for shared Cg-Eya-So regulation [14]. One model is that Cg organizes these loci into repressive configurations during the SMW, but that as cells exit this cell cycle, Eya-So directly binds Cg, disrupts Cg's interactions with proteins or DNA binding sequences that enforce the repressive 3D architecture, and installs enhancer-promoter contacts that lead to transcriptional activation. More detailed mechanistic exploration of the relationship between Cg-Eya binding, specific chromatin arrangements, and transcriptional output will elucidate how Cg and Eya-So schedule cell cycle phases during the SMW.
Considering Cg's ability to control either the chromatin environment or three-dimensional conformation of its targets in light of the idea that it selectively deploys these activities in response to the combinatorial code may explain the long-observed context specificity of its effects on gene expression. For example, Cg promotes or inhibits Cubitus interruptus expression in the two compartments of the larval wing imaginal disc [9,47] and potentiates or dampens expression of Dpp signaling targets in separate regions of the larval brain [46]. While these gene expression outcomes have not yet been assigned to direct transcriptional regulation, one model is that Cg recruits PcG complexes to loci where it represses transcription and promotes favorable chromatin conformations at genes that it activates. Cg also associates with at least one ubiquitously expressed protein complex that communicates directly with the basal transcriptional machinery [52], hinting that it may possess additional means of controlling gene expression. More broadly, we speculate that Cg interfaces with numerous transcription factors, uniquely interprets the combinatorial code at shared target enhancers, and recruits biochemical activities according to the developmental requirements of each cell.
Combgap's regulation of development is highly context-dependent
As we examined cg loss-of-function animals in more detail, we uncovered additional phenotypic complexities that support this model. First, we noted strong regionalization of the null phenotype, such that cg oppositely regulates the same cellular processes in different portions of larval tissue. In the most striking example, cg both promotes and inhibits proliferation in the eye-antennal imaginal disc. cg is required for the SMW (Fig. 2A-D) [14], but has no effect on proliferation in other portions of the developing retina. By contrast, removing cg in the adjacent presumptive head cuticle induced hyperplasia, such that this normally small patch of tissue extended basally under the eye (Fig. 2E-H) [14]. Over-proliferation likely caused this defect, as cells in this ectopic flap synthesize DNA and undergo mitosis at a higher rate than in the wild type (Fig. 2E-H). While the transcriptional targets mediating Cg's regulation of the cell cycle are not yet known, the published observation that Cg oppositely controls expression of putative targets in the larval wing and brain [9,46,47] hint that similar switching of transcriptional output, perhaps based on tissue-specific expression of transcriptional binding partners like Eya, could underlie Cg's functions in the eye-antennal disc. Alternatively, Cg may co-occupy one set of cell cycle loci with Eya-So at the SMW, but engage a different collection of targets and transcription factors in the head cuticle.
Figure 2.

cg has opposite effects on the cell cycle in different regions of the eye-antennal imaginal disc. All panels are confocal projections of four optical sections centered on the apical or basal domains of late third instar eye-antennal imaginal discs. Anterior is to the left and dorsal is up. (A-D) cg promotes S and M phase entry at the SMW (white arrows). (E-H) cg limits proliferation in the presumptive head cuticle (yellow arrows).
We also discovered regional specificity in Cg's requirement for fate specification in the ocelli. Normal flies generate one medial and one lateral ocellar field in each of their eye-antennal imaginal discs, separated by presumptive interocellar cuticle [41,42]. During pupal stages, the medial fields from each imaginal disc fuse to produce a single mature ocellus, while the lateral fields form two separate ocelli, in a tissue known as the head vertex (Fig. 3A) [19,29]. During our investigation of cg's role in the retina, we noticed that animals carrying cg null mitotic clones frequently lost lateral ocelli, produced macrochaetae in their place, and had larger vertexes (Fig. 3A-D). We never observed wild type coloration of ectopic bristles when clones were marked by the yellow mutation (Fig. 3E), confirming that ectopic macrochaetae developed cell-autonomously from cg null tissue. To determine when during development cg controls lateral ocellar fate, we examined the levels of So, which marks both developing ocelli and is required for their specification [5,11,20,43,55]. Deleting cg ablated So expression in the larval lateral ocellar fields but did not affect So levels in the medial fields (Fig. 3F-H), confirming that cg is required only for lateral ocellar specification. In keeping with the theme of context specificity, cg biases the decision between ocellar and bristle fate in only one portion of the head vertex.
Figure 3.
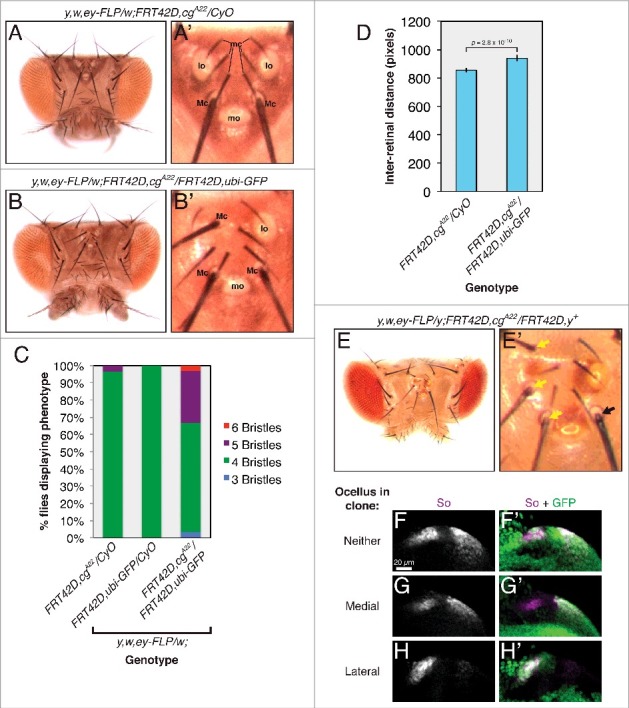
cg controls fate specification in the head vertex. Dorsal views of adult heads are oriented posterior up. For larval tissues, all images are zoomed views of the developing head vertex in late third instar eye imaginal discs, oriented anterior to the left and dorsal up. (A) The wild type arrangement of structures in the adult head vertex (zoomed view in A'). mc: microchaetae; lo: lateral ocellus; Mc: macrochaetae; mo: medial ocellus. (B) Lateral ocelli are lost and ectopic macrochaetae form in flies carrying cg clones. Note three macrochaetae, one protruding from between the microchaetae, in the zoomed view in B'. (C) The number of head vertex macrochaetae in control flies or those carrying cg clones. n > 28 for each genotype. (D) The inter-retinal distance increases in flies carrying cg clones. (E) Ectopic macrochaetae arise from cg null tissue marked by yellow (y). Zoomed view in E'. Yellow arrows mark y macrochaetae. The black arrow marks a wild type y+ bristle. (F) So expression marks the two ocellar fields in a wild type head vertex. Wild type cells are marked with GFP. (G) So expression is subtly reduced in the presumptive medial ocellus compared to the lateral field in cg null clones (marked by the absence of GFP). (H) So expression is lost in cg mutant lateral ocellar fields.
Master regulators interface with complex transcriptional machinery
Returning to our goal of understanding how the RDGN integrates with the transcriptional machinery of the cell, Cg's participation in the Eya-So complex presages complexity in the biochemical basis of master regulatory function. Cg can initiate repressive or activating chromatin states, controls numerous developmental events across the fly, and appears to tune its regulation of individual cellular processes according to context, implying unusual flexibility of function for such a broadly expressed transcription factor. Consequently, adding Cg to the repertoire of proteins that bind Eya-So hints that this complex may rely on interactions with multifunctional, pervasive adaptors to communicate with the basal transcriptional and epigenetic machinery. Moving forward, the field's challenge is to assign changes in the compositions of RD transcriptional complexes to the network rewiring that initiates transitions between cellular behaviors. We expect that identifying both specifically expressed co-regulators, which comprise the combinatorial code that interprets enhancer sequences, and ubiquitous platforms for recruiting activating or repressive biochemical activities, such as Cg, will be essential to assembling a complete model of master regulatory biology.
Disclosure of potential conflicts of interest
Authors declare no conflict of interest and no competing financial interests.
Acknowledgements
We thank members of the Rebay lab for helpful discussions and Nicelio Sanchez-Luege for comments on the manuscript. This work was supported by NIH R01 EY12549 to I.R. T.L.D. was supported in part by NIH T32 HD055164.
Funding
NIH, T32 HD055164, NIH, R01 EY12549.
References
- [1].Anderson AM, Weasner BM, Weasner BP, et al.. Dual transcriptional activities of SIX proteins define their roles in normal and ectopic eye development. Development. 2012;139:991–1000. doi: 10.1242/dev.077255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Atkins M, Jiang Y, Sansores-Garcia L, et al.. Dynamic rewiring of the Drosophila retinal determination network switches its function from selector to differentiation. PLoS Genet. 2013;9:e1003731. doi: 10.1371/journal.pgen.1003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Baker NE, Yu SY. The EGF receptor defines domains of cell cycle progression and survival to regulate cell number in the developing Drosophila eye. Cell. 2001;104:699–708. doi: 10.1016/S0092-8674(01)00266-5. [DOI] [PubMed] [Google Scholar]
- [4].Bessa J, Gebelein B, Pichaud F, et al.. Combinatorial control of Drosophila eye development by eyeless, homothorax, and teashirt. Genes Dev. 2002;16:2415–27. doi: 10.1101/gad.1009002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Blanco J, Pauli T, Seimiya M, et al.. Genetic interactions of eyes absent, twin of eyeless and orthodenticle regulate sine oculis expression during ocellar development in Drosophila. Dev. Biol. 2010;344:1088–99. doi: 10.1016/j.ydbio.2010.05.494. [DOI] [PubMed] [Google Scholar]
- [6].Bonini NM, Bui QT, Gray-Board GL, et al.. The Drosophila eyes absent gene directs ectopic eye formation in a pathway conserved between flies and vertebrates. Development. 1997;124:4819–26. [DOI] [PubMed] [Google Scholar]
- [7].Brás-Pereira C, Casares F, Janody F. The retinal determination gene Dachshund restricts cell proliferation by limiting the activity of the Homothorax-Yorkie complex. Development. 2015;142:1–10. doi: 10.1242/dev.113340. [DOI] [PubMed] [Google Scholar]
- [8].Bui QT, Zimmerman JE, Liu H, et al.. Molecular analysis of Drosophila eyes absent mutants reveals features of the conserved Eya domain. Genetics. 2000;155:709–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Campbell GL, Tomlinson A. Transcriptional regulation of the Hedgehog effector CI by the zinc-finger gene combgap. Development. 2000;127:4095–103. [DOI] [PubMed] [Google Scholar]
- [10].Chen R, Amoui M, Zhang Z, et al.. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell.1997;91:893–903. doi: 10.1016/S0092-8674(00)80481-X. [DOI] [PubMed] [Google Scholar]
- [11].Cheyette BN, Green PJ, Martin K, et al.. The Drosophila sine oculis locus encodes a homeodomain-containing protein required for the development of the entire visual system. Neuron. 1994;12:977–96. doi: 10.1016/0896-6273(94)90308-5. [DOI] [PubMed] [Google Scholar]
- [12].Curtiss J, Mlodzik M. Morphogenetic furrow initiation and progression during eye development in Drosophila: the roles of decapentaplegic, hedgehog and eyes absent. Development. 2000;127:1325–1336. [DOI] [PubMed] [Google Scholar]
- [13].Davis TL, Rebay I. Master regulators in development: Views from the Drosophila retinal determination and mammalian pluripotency gene networks. Dev. Biol. 2017a;421:93–107. doi: 10.1016/j.ydbio.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Davis TL, Rebay I. Antagonistic regulation of the second mitotic wave by Eyes absent-Sine oculis and Combgap coordinates proliferation and specification in the Drosophila retina. Development. 2017b;144:2640–2651. doi: 10.1242/dev.147231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Desplan C. Eye development: Governed by a dictator or a junta? Cell. 1997;91:861–864. doi: 10.1016/S0092-8674(00)80475-4. [DOI] [PubMed] [Google Scholar]
- [16].Escudero LM, Freeman M. Mechanism of G1 arrest in the Drosophila eye imaginal disc. BMC Dev. Biol. 2007;13:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Halder G, Callaerts P, Gehring WJ. Ectopic of Induction Eyes by Targeted Expression of the eyeless in Drosophila Gene. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- [18].Halder G, Callaerts P, Flister S, et al.. Eyeless initiates the expression of both sine oculis and eyes absent during Drosophila compound eye development. Development. 1998;125:2181–91. [DOI] [PubMed] [Google Scholar]
- [19].Haynie JL, Bryant PJ. Development of the eye-antenna imaginal disc and morphogenesis of the adult head in Drosophila melanogaster. J. Exp. Zool. 1986;237:293–308. doi: 10.1002/jez.1402370302. [DOI] [PubMed] [Google Scholar]
- [20].Heitzler P, Coulson D, Saenz-Robles MT, et al.. Genetic and cytogenetic analysis of the 43A-E region containing the segment polarity gene costa and the cellular polarity genes prickle and spiny-legs in Drosophila melanogaster. Genetics. 1993;135:105 LP-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hitrik A, Popliker M, Gancz D, et al.. Combgap Promotes Ovarian Niche Development and Chromatin Association of EcR-Binding Regions in BR-C. PLoS Genet. 2016;12:e1006330. doi: 10.1371/journal.pgen.1006330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jemc J, Rebay I. Identification of transcriptional targets of the dual-function transcription factor/phosphatase eyes absent. Dev. Biol. 2007;310:416–29. doi: 10.1016/j.ydbio.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Jin M, Mardon G. Distinct Biochemical Activities of Eyes absent During Drosophila Eye Development. Sci. Rep. 2016;6:23228. doi: 10.1038/srep23228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jin M, Eblimit A, Pulikkathara M, et al.. Conditional knockout of retinal determination genes in differentiating cells in Drosophila. FEBS J. 2016;283:2754–2766. doi: 10.1111/febs.13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kango-Singh M, Singh A, Sun YH. Eyeless collaborates with Hedgehog and Decapentaplegic signaling in Drosophila eye induction. Dev. Biol. 2003;256:48–60. doi: 10.1016/S0012-1606(02)00123-9. [DOI] [PubMed] [Google Scholar]
- [26].Karandikar UC, Jin M, Jusiak B, et al.. Drosophila eyes absent is required for normal cone and pigment cell development. PLoS One. 2014;9:e102143. doi: 10.1371/journal.pone.0102143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kassis JA. Unusual properties of regulatory DNA from the Drosophila engrailed gene: Three “pairing-sensitive” sites within a 1.6-kb region. Genetics. 1994;136:1025–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kassis JA, Brown JL. Polycomb Group Response Elements in Drosophila and Vertebrates. Advances in Genetics. 2013;81:83–118. doi: 10.1016/B978-0-12-407677-8.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Kenyon KL, Ranade SS, Curtiss J, et al.. Coordinating proliferation and tissue specification to promote regional identity in the Drosophila head. Dev. Cell. 2003;5:403–14. doi: 10.1016/S1534-5807(03)00243-0. [DOI] [PubMed] [Google Scholar]
- [30].Kenyon KL, Li DJ, Clouser C, et al.. Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev. Dyn. 2005;234:497–504. doi: 10.1002/dvdy.20442. [DOI] [PubMed] [Google Scholar]
- [31].Kumar JP. The molecular circuitry governing retinal determination. Biochim. Biophys. Acta. 2009;1789, 306–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu X, Sano T, Guan Y, et al.. Drosophila EYA Regulates the Immune Response against DNA through an Evolutionarily Conserved Threonine Phosphatase Motif. PLoS One. 2012;7:e42725. doi: 10.1371/journal.pone.0042725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lopes CS, Casares F. Eye Selector Logic for a Coordinated Cell Cycle Exit. PLoS Genet. 2015;11:e1004981. doi: 10.1371/journal.pgen.1004981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mutsuddi M, Chaffee B, Cassidy J, et al.. Using Drosophila to decipher how mutations associated with human branchio-oto-renal syndrome and optical defects compromise the protein tyrosine phosphatase and transcriptional functions of eyes absent. Genetics. 2005;170:687–95. doi: 10.1534/genetics.104.039156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Niimi T, Seimiya M, Kloter U, et al.. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999;126:2253–60. [DOI] [PubMed] [Google Scholar]
- [36].Ostrin EJ, Li Y, Hoffman K, et al.. Genome-wide identification of direct targets of the Drosophila retinal determination protein Eyeless. Genome Res. 2006;16:466–76. doi: 10.1101/gr.4673006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pappu KS, Ostrin EJ, Middlebrooks BW, et al.. Dual regulation and redundant function of two eye-specific enhancers of the Drosophila retinal determination gene dachshund. Development. 2005;132:2895–905. doi: 10.1242/dev.01869. [DOI] [PubMed] [Google Scholar]
- [38].Pignoni F, Hu B, Zavitz KH, et al.. The eye-specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–91. doi: 10.1016/S0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- [39].Punzo C, Seimiya M, Flister S, et al.. Differential interactions of eyeless and twin of eyeless with the sine oculis enhancer. Development. 2002;129:625–34. [DOI] [PubMed] [Google Scholar]
- [40].Ray P, De S, Mitra A, et al.. Combgap contributes to recruitment of Polycomb group proteins in Drosophila. Proc. Natl. Acad. Sci. 2016;201520926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Royet J, Finkelstein R. Pattern formation in Drosophila head development: the role of the orthodenticle homeobox gene. Development. 1995;121:3561–72. [DOI] [PubMed] [Google Scholar]
- [42].Royet J, Finkelstein R. Hedgehog, wingless and orthodenticle specify adult head development in Drosophila. Development. 1996;122:1849–1858. [DOI] [PubMed] [Google Scholar]
- [43].Serikaku MA, O'Tousa JE. sine oculis is a homeobox gene required for Drosophila visual system development. Genetics. 1994;138:1137–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Shen W, Mardon G. Ectopic eye development in Drosophila induced by directed dachshund expression. Development. 1997;124:45–52. [DOI] [PubMed] [Google Scholar]
- [45].Silver SJ, Davies EL, Doyon L. et al.. Functional Dissection of Eyes absent Reveals New Modes of Regulation within the Retinal Determination Gene Network. Mol. Cell. Biol. 2003;23:5989–5999. doi: 10.1128/MCB.23.17.5989-5999.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Song Y, Chung S, Kunes S. Combgap relays wingless signal reception to the determination of cortical cell fate in the Drosophila visual system. Mol. Cell. 2000;6:1143–54. doi: 10.1016/S1097-2765(00)00112-X. [DOI] [PubMed] [Google Scholar]
- [47].Svendsen PC, Marshall SD, Kyba M, et al.. The combgap locus encodes a zinc-finger protein that regulates cubitus interruptus during limb development in Drosophila melanogaster. Development. 2000;127:4083–4093. [DOI] [PubMed] [Google Scholar]
- [48].Tanaka-Matakatsu M, Du W. Direct control of the proneural gene atonal by retinal determination factors during Drosophila eye development. Dev. Biol. 2008;313:787–801. doi: 10.1016/j.ydbio.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Waddington CH. Growth and determination in the development of Drosophila. Nature. 1942;149:264–265. doi: 10.1038/149264a0. [DOI] [Google Scholar]
- [50].Waddington CH. The development of some “leg genes” in Drosophila. J. Genet. 1943;45:29–43. doi: 10.1007/BF02982772. [DOI] [Google Scholar]
- [51].Waddington CH. The interactions of some morphogenetic genes in Drosophila melanogaster. J. Genet. 1953;51:243–258. doi: 10.1007/BF03023296. [DOI] [Google Scholar]
- [52].Weake VM, Dyer JO, Seidel C, et al.. Post-transcription initiation function of the ubiquitous SAGA complex in tissue-specific gene activation. Genes Dev. 2011;25:1499–1509. doi: 10.1101/gad.2046211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wolff T, Ready DF.. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991;113:841–50. [DOI] [PubMed] [Google Scholar]
- [54].Zhang T, Ranade S, Cai CQ, et al.. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development. 2006;133:4881–9. doi: 10.1242/dev.02669. [DOI] [PubMed] [Google Scholar]
- [55].Zhou Q, Zhang T, Jemc JC, et al.. Onset of atonal expression in Drosophila retinal progenitors involves redundant and synergistic contributions of Ey/Pax6 and So binding sites within two distant enhancers. Dev. Biol. 2014;386:152–64. doi: 10.1016/j.ydbio.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]


