ABSTRACT
Telomere protects the ends of linear chromosomes. Telomere dysfunction fuels genome instability that can lead to diseases such as cancer. For over 30 years, Drosophila has fascinated the field as the only major model organism that does not rely on the conserved telomerase enzyme for end protection. Instead of short DNA repeats at chromosome ends, Drosophila has domesticated retrotransposons. In addition, telomere protection can be entirely sequence-independent under normal laboratory conditions, again dissimilar to what has been established for telomerase-maintained systems. Despite these major differences, recent studies from us and others have revealed remarkable similarities between the 2 systems. In particular, with the identification of the MTV complex as an ssDNA binding complex essential for telomere integrity in Drosophila (Zhang et al. 2016 Plos Genetics), we have now established several universal principles that are intrinsic to chromosome extremities but independent of the underlying DNA sequences or the telomerase enzyme. Telomere studies in Drosophila will continue to yield fundamental insights that are instrumental to the understanding of the evolution of telomere and telomeric functions.
KEYWORDS: CST telomere complex, drosophila telomere, epigenetic regulation, retrotransposon, ssDNA protection
Drosophila telomere and telomere protection
The very concept of “Telomere” originated from experiments performed in Drosophila. In 1938, Muller characterized chromosome rearrangements induced by X irradiation and noticed the absence of terminally deleted chromosomes, which would have arisen by losing the chromosomal materials distal to a single X-ray induced double strand break. This led him to propose that a specialized structure that he named “Telomere” serves to distinguish natural ends from broken ends of chromosomes. With this proposition, Muller also defined the capping function of telomeres.
Telomere serves a second function: to counteract incomplete end replication by providing a mechanism to elongate chromosomes. This second function is where the major difference lies between Drosophila as well as many other organisms, insects in particular, and those that uses the telomerase enzyme. In Drosophila, telomeres are elongated with retrotransposons instead, a striking discovery made over 30 y ago (reviewed in ref.1). How different is telomere maintenance between the 2 systems, and how does studying end protection in Drosophila relate to telomere maintenance in general?
An important contribution to the genetic studies of Drosophila telomere maintenance was made from mutant screens conducted over the years by the Gatti laboratory in Italy, which led to the identification of the first telomere capping mutant in Drosophila.2 Subsequent studies yielded the identities of a group of proteins called capping proteins, which are specifically enriched at chromosome ends and are essential for their stability. However, without a more detailed assignment of molecular functions to these capping proteins, earlier results seemed to further support the notion that Drosophila telomere protection might use fundamentally different mechanisms from those that are built based on the telomerase enzyme. For example, the HOAP/Cav protein, the first capping protein identified is one of the fastest evolving proteins in the genome.3 HOAP homologs based solely on sequence homology cannot be reliably identified outside of the Drosophila lineage. Indeed, all members of the capping complex subsequently identified bear signatures of fast evolution: Moi4; HipHop5; and Ver.6
In 2010, using a single telomere constructed with unique DNA sequences, we were able to reveal the distribution of the HOAP-HipHop complex at a high resolution using ChIP.5 Our results suggest that capping complex enrichment is not limited to the very end of the chromosome but encompasses a large domain of tens of kilobases from the extremity. This picture of capping protein distribution suggests that the HOAP-HipHop complex occupies primarily the dsDNA region of the telomere and is a functional equivalent to the Shelterin and related complexes in telomerase-maintained systems (reviewed in ref.7,8). Shortly after our work, Raffa et al.6 discovered that the Ver protein shares significant structural homology to ssDNA binding proteins, including those that protects telomeric ssDNA previously identified. These 2 pieces of findings form the basis for the argument that mechanisms of telomere capping, with or without the participation of the telomerase enzyme, might be more similar than the field had previously recognized. Our most recent work on the MTV complex further supports this notion.
MTV, a ssDNA binding complex essential for telomere maintenance
In telomerase-maintained systems, telomeres end as a single stranded overhang. This overhang serves essential functions not only for end protection but also for the proper function of the telomerase enzyme (reviewed in ref.9). The protection of this single strand overhang is functionally assigned to the Cdc13-Stn1-Ten1 (CST) in yeast and at least partially to its equivalent, the Ctc1-Stn1-Ten1 complex in mammals (reviewed in ref.10).
Drosophila telomeres may also end in an overhang structure. First of all, lagging strand synthesis during DNA replication likely results in a 3’ overhang after the removal of the RNA primer. In addition, retrotransposon attachment to telomeres likely requires a free single strand end to prime reverse transcription. Moreover, the Drosophila Ver protein shares substantial sequence and structural similarities with Stn1, an essential subunit of CST.6,11 Therefore, ssDNA might exist at all telomeres of linear chromosomes and a separate complex for the protection of ssDNA overhangs might be present universally. This is strongly supported by our most recent work,12 in which we identified the fifth member of the capping complex, Tea, and biochemically characterized the Moi-Tea-Ver (MTV) complex.
We identified Tea from our ongoing efforts to biochemically purify the telomere capping complex.5 Tea fulfills the basic definition of a capping protein as it localizes exclusively to telomeres and its loss leads to telomere fusion, which is lethal to the animals. Similar to other members of the larger capping complex, Tea protein has no identifiable domains and sequence-based homologs exist only in a limited number of insect species. Tea threatened to be yet another novel capping protein to which a molecular function is difficult to assign.
Tea is a large protein with close to 2000 residues. We speculated that Tea serves similar function as the Cdc13 protein from Saccharomyces cerevisiae, a member of the ssDNA-protecting CST complex that is also exceptionally large when compared with other capping proteins, although much smaller fungal Cdc13 homologs have now been identified.13 We set out to test the hypothesis that Tea, along with Ver, which is homologous to Stn1, and another small capping protein Moi, constitutes the ssDNA-binding complex in Drosophila.
To probe MTV interaction, we used the assay of yeast 2 hybrid (Y2H), which has been widely applied to study CST interactions. In our work, Ver and Moi showed robust Y2H interaction. However, Tea-Ver or Tea-Moi Y2H experiments yielded negative results. Nevertheless, a modified Y2H assay, in which all 3 proteins (or protein fragments) of the MTV complex were expressed in yeast, produced strong trimeric interaction. This suggests that Ver and Moi exists as a sub-complex, similar to the situation for yeast Stn1 and Ten1.14 In addition, the presence of both Ver and Moi is required for stable interaction with Tea at least in vitro. This mode of interaction is also supported by genetic results in which Ver-Moi interaction that has been weakened by mutations can be stabilized by the presence a wildtype Tea protein in Y2H. Moreover, these ver and moi mutations that have weakened Y2H interactions behave as hypomorphic mutations in flies.
Using insect cells, we were able to partially purify a trimeric MTV complex. Biochemically, this complex binds ssDNA but not dsDNA in a standard EMSA assay. Interestingly, ssDNA binding of MTV in vitro requires no sequence requirement, reminiscent of the in vivo situation. Strikingly, binding of MTV protects the single stranded oligoes from degradation by an exonuclease in vitro suggesting an ssDNA protective role of MTV in vivo.
Future directions for MTV biology
Much remains to be learned about MTV functions. For examples, what is the functional allocation among MTV subunits? In yeast, the Stn1-Ten1 sub-complex mainly serves a capping function and this function is normally recruited to telomeres by the Cdc13 subunit.15 Consistently, overexpression of Stn1 and Ten1 is sufficient to bypass Cdc13 and restore capping to Cdc13 mutants.16 Can overexpression of Ver and Moi restore capping function to Tea mutants? The Cdc13 protein serves as a platform to recruit the Stn1-Ten1 encoded capping function and the telomerase encoded elongation function.15 Is Tea similarly bi-functional? If Tea's primary function is to serve as a platform for recruiting various factors to the ends, one can expect that Tea exists in functional domains: perhaps an ssDNA-binding domain and various protein-protein interaction domains. Our genetic studies of capping protein localization to telomeres seemed to support that Tea might serve as a recruiting platform as neither Ver nor Moi is required for Tea's recruitment to telomeres. However, which subunit(s) of MTV is primarily responsible for ssDNA binding becomes a contentious issue as Cicconi et al.17 recently presented evidence that recombinant Ver alone can bind ssDNA. Interestingly, Tea likely participates in regulating telomeric retrotransposons since its interacting partners of Ver and Moi are essential for the localization of retrotransposon RNPs to telomeres (11, and our unpublished results). Perhaps, Tea possesses a specialized domain to interact with transposon encoded proteins and RNAs.
Striking similarities between transposon-based and telomerase-based telomere maintenance
Our work and that of others continue to uncover remarkable conservations in the biologic features of chromosome ends and the protein factors that regulate their function between the Drosophila system and those involving telomerase.
For end capping, the 2 systems share the following similarities. First, the highly conserved ATM and ATR kinases and their associated factors protect telomeres in both systems (Fig. 1, and ref.18). Secondly, both systems have a dedicated complex occupying and protecting the duplex region of the telomeres (Fig. 2). Lastly, we now added that both systems have a dedicated ssDNA protecting complex (Fig. 2).
Figure 1.
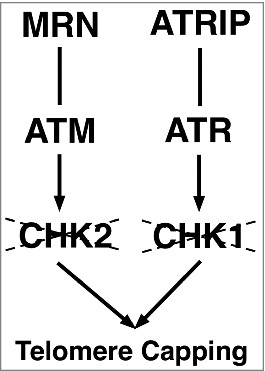
A conserved parallel pathway for telomere protection. The 2 branches are centered around 2 related proteins: the ATM and the ATR checkpoint kinases. Both kinases act with their interacting proteins to control telomere integrity: ATM with the Mre11-Rad50-Nbs (MRN) complex and ATR with ATRIP. Contrary to the pathway relationship for the response to DNA damage, the downstream kinases CHK1 and CHK2 are not part of the pathway essential for telomere protection as indicated by the strikethrough.
Figure 2.
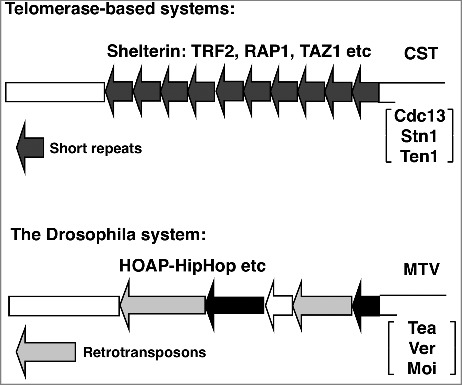
Functional similarities of capping complexes between telomerase-based and transposon-based telomere maintenance. For telomerase-maintained systems, 2 separate complexes exist that protect the dsDNA and ssDNA regions of the telomeres. For dsDNA protection, mammalian TRF2, Rap1, and yeast Taz1 and Rap1 are essential for preventing telomere fusions similar to their Drosophila counterparts of HipHop and HOAP. For ssDNA protection, the CST complex might be functionally equivalent to the newly identified MTV complex in flies.
For end elongation, several similar themes emerge. First, Drosophila telomeres are transcribed with its transcripts remain associated with chromosome ends during the cell cycle.11 This feature resembles those of TERRA recently identified in diverse telomerase-based systems (reviewed in ref.19). Secondly, by monitoring a single telomere during the somatic cell cycle, we obtained evidence suggesting that telomere elongation might also be coupled with its replication in Drosophila similarly to what has been shown in yeast and mammals.11 Thirdly, telomeres are also clustered in Drosophila cells.20
What the Drosophila system can offer
One of the advantages of studying telomere maintenance in Drosophila is that protection mechanisms in flies are independent of the DNA sequence at telomeres (21 and references therein). Similar mechanisms exist in other systems which may serve as a backup to the one controlled by telomerase. These mechanisms have been difficult to study since the telomerase-based mechanism has to be disabled first. Nevertheless, in about 15% of tumor types, cells rely on telomerase-independent mechanisms for telomere protection, which might be based on an epigenetic mechanism similar to that in Drosophila. A fundamental question for studying the determinant of telomere identity is what epigenetic markers distinguish telomeres from broken ends. The Drosophila system may arguably be the best system to gather insights for answering this question.
Drosophila can also be an excellent system for the study of the evolution of telomere and telomeric functions. It is remarkable that capping proteins in Drosophila fulfill the universal function of end protection yet they seem to all evolve at a fast rate.22 What is the driving force behind this fast evolution? Could it be that telomeric retrotransposons that these proteins normally bind to drive this fast evolution? Despite having been known to the field for over 30 years, it remains largely unknown how these selfish elements functionally interact with host proteins that bind to them. But recent progress has started to shed lights onto this important aspect of telomere evolution (eg. ref.11,23,24).
Drosophila studies have also produced unexpected results that are novel to the telomere field in general. In one example, we and others showed that a paternally installed protein imprint on sperm telomeres protects them from DNA repair activities in early embryos,25-27 and a similar paternal imprint likely exists universally. The fact that the Drosophila imprint consists of the K81 protein, a sperm specific variant of HipHop, suggests that the mammalian imprint could also be a component of the duplex binding complex.
Disclosure of potential conflicts of interest
The authors declare no conflict of interest.
Funding
Research in our laboratory is supported in part by a grant from NSFC #31371364 to YSR.
References
- [1].Pardue ML, DeBaryshe PG. Drosophila telomeres: A variation on the telomerase theme. Fly (Austin) 2008; 2(3):101-10; https://doi.org/ 10.4161/fly.6393 [DOI] [PubMed] [Google Scholar]
- [2].Cenci G, Rawson RB, Belloni G, Castrillon DH, Tudor M, Petrucci R, Goldberg ML, Wasserman SA, Gatti M. UbcD1, a Drosophila ubiquitin-conjugating enzyme required for proper telomere behavior. Genes Dev 1997. April 1; 11(7):863-75; https://doi.org/ 10.1101/gad.11.7.863 [DOI] [PubMed] [Google Scholar]
- [3].Shareef MM, King C, Damaj M, Badagu R, Huang DW, Kellum R. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol Biol Cell 2001; 12(6):1671-85; PMID:11408576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Raffa GD, Siriaco G, Cugusi S, Ciapponi L, Cenci G, Wojcik E, Gatti M. The Drosophila modigliani (moi) gene encodes a HOAP-interacting protein required for telomere protection. Proc Natl Acad Sci U S A 2009. February 17; 106(7):2271-6; https://doi.org/ 10.1073/pnas.0812702106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gao G, Walser JC, Beaucher ML, Morciano P, Wesolowska N, Chen J, Rong YS. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J 2010; 29(4):819-29; https://doi.org/ 10.1038/emboj.2009.394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Raffa GD, Raimondo D, Sorino C, Cugusi S, Cenci G, Cacchione S, Gatti M, Ciapponi L. Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev 2010. August 1; 24(15):1596-601; https://doi.org/ 10.1101/gad.574810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet 2008; 42:301-34; PMID:18680434; https://doi.org/ 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- [8].Zakian VA. Telomeres: the beginnings and ends of eukaryotic chromosomes. Exp Cell Res 2012; 318(12):1456-60; https://doi.org/ 10.1016/j.yexcr.2012.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bonetti D, Martina M, Falcettoni M, Longhese MP. Telomere-end processing: mechanisms and regulation. Chromosoma 2014; 123(1):57-66. [DOI] [PubMed] [Google Scholar]
- [10].Price CM, Boltz KA, Chaiken MF, Stewart JA, Beilstein MA, Shippen DE. Evolution of CST function in telomere maintenance. Cell Cycle 2010; 9(16):3157-65; https://doi.org/ 10.4161/cc.9.16.12547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang L, Beaucher M, Cheng Y, Rong YS. Coordination of transposon expression with DNA replication in the targeting of telomeric retrotransposons in Drosophila. EMBO J 2014; 33(10):1148-58; https://doi.org/ 10.1002/embj.201386940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhang Y, Zhang L, Tang X, Bhardwaj SR, Ji J, Rong YS. MTV, an ssDNA Protecting Complex Essential for Transposon-Based Telomere Maintenance in Drosophila. PLoS Genet 2016; 12(11):e1006435; https://doi.org/ 10.1371/journal.pgen.1006435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lue NF, Chan J. Duplication and functional specialization of the telomere- capping protein Cdc13 in Candida species. J Biol Chem 2013; 288(40):29115-23; https://doi.org/ 10.1074/jbc.M113.506519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, Lei M. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev 2009; 23(24):2900-14; https://doi.org/ 10.1101/gad.1851909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 2001; 104(3):387-96; https://doi.org/ 10.1016/S0092-8674(01)00226-4 [DOI] [PubMed] [Google Scholar]
- [16].Petreaca RC, Chiu HC, Eckelhoefer HA, Chuang C, Xu L, Nugent CI. Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat Cell Biol 2006; 8(7):748-55; https://doi.org/ 10.1038/ncb1430 [DOI] [PubMed] [Google Scholar]
- [17].Cicconi A, Micheli E, Vernì F, Jackson A, Gradilla AC, Cipressa F, Raimondo D, Bosso G, Wakefield JG, Ciapponi L, et al.. The Drosophila telomere-capping protein Verrocchio binds single-stranded DNA and protects telomeres from DNA damage response. Nucleic Acids Res 2016; 45(6):3068-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bi X, Srikanta D, Fanti L, Pimpinelli S, Badugu R, Kellum R, Rong YS. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc Natl Acad Sci U S A 2005; 102(42):15167-72; https://doi.org/ 10.1073/pnas.0504981102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cusanelli E, Chartrand P. Telomeric repeat-containing RNA TERRA: a noncoding RNA connecting telomere biology to genome integrity. Front Genet 2015; 6:143; https://doi.org/ 10.3389/fgene.2015.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Wesolowska N, Amariei FL, Rong YS. Clustering and protein dynamics of Drosophila melanogaster telomeres. Genetics 2013; 195(2):381-91; https://doi.org/ 10.1534/genetics.113.155408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Beaucher M, Zheng XF, Amariei F, Rong YS. Multiple pathways suppress telomere addition to DNA breaks in the Drosophila germline. Genetics 2012; 191(2):407-17; https://doi.org/ 10.1534/genetics.112.138818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lee YC, Leek C, Levine MT. Recurrent Innovation at Genes Required for Telomere Integrity in Drosophila. Mol Biol Evol 2017; 34(2):467-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].López-Panadès E, Gavis ER, Casacuberta E. Specific Localization of the Drosophila Telomere Transposon Proteins and RNAs, Give Insight in Their Behavior, Control and Telomere Biology in This Organism. PLoS One 2015; 10(6):e0128573; https://doi.org/ 10.1371/journal.pone.0128573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Morgunova V, Akulenko N, Radion E, Olovnikov I, Abramov Y, Olenina LV, Shpiz S, Kopytova DV, Georgieva SG, Kalmykova A. Telomeric repeat silencing in germ cells is essential for early development in Drosophila. Nucleic Acids Res 2015; 43(18):8762-73; https://doi.org/ 10.1093/nar/gkv775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dubruille R, Orsi GA, Delabaere L, Cortier E, Couble P, Marais GA, Loppin B. Specialization of a Drosophila capping protein essential for the protection of sperm telomeres. Curr Biol 2010; 20(23):2090-9; https://doi.org/ 10.1016/j.cub.2010.11.013 [DOI] [PubMed] [Google Scholar]
- [26].Gao G, Cheng Y, Wesolowska N, Rong YS. Paternal imprint essential for the inheritance of telomere identity in Drosophila. Proc Natl Acad Sci U S A 2011; 108(12):4932-7; PMID:21383184; https://doi.org/ 10.1073/pnas.1016792108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yamaki T, Yasuda GK, Wakimoto BT. The Deadbeat Paternal Effect of Uncapped Sperm Telomeres on Cell Cycle Progression and Chromosome Behavior in Drosophila melanogaster. Genetics 2016; 203(2):799-816; https://doi.org/ 10.1534/genetics.115.182436 [DOI] [PMC free article] [PubMed] [Google Scholar]


