ABSTRACT
The processes involved in ribosome biogenesis, including synthesis of ribosomal proteins, ribosome biogenesis-related factors, and ribosomal RNAs (rRNAs), must be coordinately orchestrated in response to changes in energy supply. In animal cells, defects in ribosome biogenesis induce a nucleolar stress response through the p53-mediated pathway. Our recent finding that an essential, sugar-inducible Arabidopsis gene, APUM24, encoded a pre-rRNA processing factor allowed the relationships between rRNA biogenesis, nucleolar stress, sugar response, and growth regulation to be understood in plants. A knockdown mutant of APUM24 developed sugar-dependent phenotypes including pre-rRNA processing defects, reductions in nucleolar size, and limited promotion of leaf and root growth. Alongside the absence of plant p53 homologs and the synchronous sugar-induced expression of ribosome biogenesis-related genes, these findings suggest the following hypothesis. Sugar supply may enhance ribosome biogenesis defects, leading to p53-independent induction of nucleolar stress responses that include negative regulation of growth and development in plants.
KEYWORDS: Ribosome biogenesis, ribosomal RNA, nucleolar stress, sugar
Ribosome biogenesis, which is one of the most energy-consuming events in the cell, involves a number of essential processes, including transcription of the genes encoding pre-ribosomal RNA (pre-rRNA), ribosomal proteins (RPs), and processing factors, as well as the subsequent processing of pre-rRNAs and assembly of ribosomal components. Ribosomal gene expression and processing phases must be coordinately regulated in phase with changes in extracellular and intracellular environments, particularly with respect to energy status.1-5 Pre-rRNA processing is maturation of pre-rRNA into rRNAs, which function as scaffolds in the large and small ribosome subunits.6-8 Pre-rRNA processing begins at the nucleolus and proceeds with the help of a number of rRNA processing factors such as endo- and exonucleases and RNA helicases.6-8 Although many processing factors have been identified in yeast and animals, only a limited number of these factors have been identified in plants.3,7
Recently, we showed that APUM24, which was originally identified as the sugar-inducible gene-encoded nuclear protein NuGAP1,9 was a novel pre-rRNA processing factor.10 APUM24/NuGAP1 is a member of the Arabidopsis APUM protein family, members of which have a conserved RNA-binding domain: the pumilio domain (alternatively called the PUM-HD domain).11,12 Our analysis revealed that the APUM24 null mutant, apum24-1 (GABI_461E08), was lethal. The transmission efficiency of the paternal apum24-1 mutation was 100%, whereas that of the maternal apum24-1 mutation was much lower than 100%.10 Another recent report investigating the GABI_461E08 line (referenced as the apum24-3 mutant) also exhibited lethality caused by the apum24-1 mutation.13 Transmission efficiencies in this study agreed with those in our previous research. The study also observed no defect in male and female gametogenesis in the apum24-1 mutant, but found that fertilization was delayed and that embryogenesis stalled at the late globular stage,13 indicating that APUM24 was required for embryonic development.
Our phenotypic and molecular biological analyses with APUM24 knockdown mutants, generated either by T-DNA insertion(s) into an intron (the apum24-2 mutant (SALK_033623)) or by APUM24-targeted artificial microRNA, provided further insights into the function of APUM24.10 The knockdown mutants displayed pinned and serrated first leaves, inhibition of root elongation, and resistance to antibiotics,10 characteristics known to be frequently associated with Arabidopsis ribosome-related mutants.3,14 Furthermore, in addition to nucleolar localization of APUM24, we found that APUM24 formed complexes with RPs and rRNA processing-related factors, including a newly identified putative endonuclease (AtLAS1). AtLAS1 is probably an Arabidopsis homolog of yeast Las115 that catalyzes cleavage at a site (called the C2 site) in the Internal Transcribed Spacer 2 (ITS2) region10 (Fig. 1). In the apum24-2 mutant, over-accumulation of processing intermediates caused by incomplete digestion at the C2 site was consistently detected through monitoring of pre-rRNA processing by RT-qPCR, Northern blot, and circular RT-PCR analyses.10 Thus, APUM24 is a pre-rRNA processing factor involved in the removal of ITS2.
Figure 1.
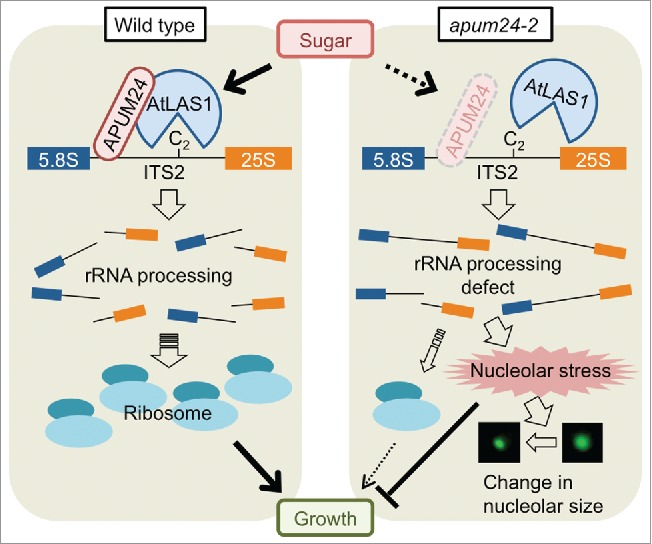
Model for the sugar-dependent nucleolar stress response in the apum24-2 mutant. In wild-type cells, sugar coordinately induces the expression of RP genes, rRNA genes, and pre-rRNA processing-related genes including APUM24 (left panel). Since APUM24 interacts with pre-rRNA at ITS2 and probably recruits AtLAS1 (the putative endonuclease involved in cleavage at the C2 site in ITS2), sugar supply results in increases in the ribosome content to promote growth in wild-type cells. However, in the apum24-2 mutant cells, pre-rRNA intermediates over-accumulate in a sugar-dependent manner because sugar supply likely enhances the disruption of coordinated expression of a set of ribosome biogenesis-related genes (right panel). This rRNA processing defect may cause nucleolar stress and inhibition of growth.
Identification of APUM24, the product of a sugar-inducible gene, as a pre-rRNA processing factor indicated connections between rRNA biogenesis, nucleolar stress (also known as ribosome stress), responses to sugar, and growth control in plants. In animal cells, nucleolar stress is triggered by defects in ribosome biogenesis and leads to cell cycle arrest, senescence, and/or apoptosis, accompanied with the disruption of the nucleolar structure.16,17,18 Two previous studies reported disruption of the nucleolar structure in two ribosome biogenesis-related mutants: the maize reas1 mutant19 and the Arabidopsis rh10 mutant.20 However, the relationships between such disruptions and physiology remained elusive in plants. We found that both over-accumulation of processing intermediates and reductions in nucleolar size occurred in the apum24-2 mutant in a sugar-dependent manner10 (Fig. 1). Furthermore, sugar-induced promotion of leaf enlargement and root growth was substantially reduced in the apum24-2 mutant.10 It remained unclear whether reductions in sugar-induced growth in the apum24-2 mutant occurred in direct response to nucleolar stress or were simply caused by reductions in the ribosome content. However, in spite of the abnormal accumulation of processing intermediates, the abundance of normal rRNAs in the apum24-2 mutant was comparable to that in the wild type.10 We therefore assumed that nucleolar stress decreased the growth rate in the presence of sugar in the apum24-2 mutant (Fig. 1). Furthermore, sugar exogenously supplied at low concentrations promotes plant growth, synchronous activation of expression of ribosome biogenesis-related genes, and an increase in mature rRNA content.5,10 We therefore assumed that the disruption of coordinated expression of a set of ribosome biogenesis-related genes in the apum24-2 mutant and subsequent nucleolar stress were enhanced by sugar (energy source) supply. Thus, we propose the hypothesis that nucleolar stress caused by defects in ribosome biogenesis is enhanced when sugar is actively supplied, leading to overt decreases in growth.
In mammalian cells, the RP-MDM2-p53 pathway plays a central role in controlling the nucleolar stress response.16,17,18 The key step in this pathway is release of the transcription factor p53 from ubiquitination and destabilization by MDM2 ubiquitin ligase.16,17,18 Because yeast lacks any p53-like protein, a p53-independent pathway(s) is proposed to mediate nucleolar stress in yeast.17 Plants also lack p53-like proteins.21,22 However, very recently, an Arabidopsis NAC protein, ANAC082, was proposed to play a role comparable to that of p53 in the nucleolar stress response, based on analysis of a loss-of-function mutation in ANAC082.23 The mutation rescued developmental abnormalities and cell proliferation defects caused by pre-rRNA processing-related mutations, such as rid2, rid3, and rh10-1, without restoring the pre-rRNA processing defect.23 NAC proteins generally function as transcription factors, and the potential of ANAC082 as a transcriptional activator was also demonstrated in budding yeast.23 Therefore, ANAC082 very likely mediates nucleolar stress and activates the nucleolar stress response through transcriptional regulation in Arabidopsis. In addition to ANAC082, another Arabidopsis NAC protein, ANAC008/SOG1, may also play a role similar to that of p53 in the DNA damage response.24,25 SOG1 contributed to the maintenance of genome stability after DNA damage by inducing cell cycle arrest, DNA repair, and programmed cell death through the regulation of downstream genes,24,25 similar to the activity of p53 in animal cells. These findings strongly suggest that some members of the NAC transcription factor family mediate the nucleolar stress and DNA damage responses in plants in a similar manner to p53 in mammalian cells. Thus, we speculate that these members may mediate negative regulation against sugar-induced growth promotion. ANAC082 target genes remain to be identified, although SOG1 was shown to directly regulate expression of SMR5 and SMR7, which encoded plant-specific cyclin-dependent kinase inhibitors.26 Comprehensive identification of the target genes of the NAC proteins involved in the nucleolar stress response would contribute to a deeper understanding of how nucleolar stress suppresses sugar-induced growth promotion in plants.
As interest in nucleolar stress in plants has grown, our understanding of the processes involved has improved, and important questions have been identified. First, how are the defects in ribosome biogenesis and nucleolar stress sensed and transduced to produce a signal that inhibits growth? Second, how does nucleolar stress repress cell proliferation and lead to cell cycle arrest in plants? Further analyses of pre-rRNA processing complexes, including processing intermediate RNAs in the apum24 knockdown mutant, would address these questions. Such analyses might also reveal new regulatory connections between seemingly antagonistic regulatory systems governing growth and development, namely, stress signaling for repression of growth and energy signaling for promotion of growth.
Funding Statement
This work was supported in part by JST (Japan Science and Technology Agency) CREST Grant Number JPMJCR15O5 and JSPS KAKENHI Grant Numbers JP25252014, JP26221103 to SY and by JSPS KAKENHI Grant Number JP15J08368 to SM.
Abbreviations
- ITS2
internal transcribed spacer 2
- rRNA
ribosomal RNA
- RP
ribosomal proteins.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–63. doi: 10.1016/S0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- 2.Lempiäinen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855–63. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Weis BL, Kovacevic J, Missbach S, Schleiff E. Plant-specific features of ribosome biogenesis. Trends Plant Sci. 2015;20:729–40. doi: 10.1016/j.tplants.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 4.Kojima H, Suzuki T, Kato T, Enomoto K, Sato S, Kato T, Tabata S, Sáez-Vasquez J, Echeverría M, Nakagawa T, et al.. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 2007;49:1053–63. doi: 10.1111/j.1365-313X.2006.03016.x. [DOI] [PubMed] [Google Scholar]
- 5.Ishida T, Maekawa S, Yanagisawa S. The pre-rRNA processing complex in Arabidopsis includes two WD40-domain-containing proteins encoded by glucose-inducible genes and plant-specific proteins. Mol Plant. 2016;9:312–5. doi: 10.1016/j.molp.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Salifou K, Ray S, Verrier L, Aguirrebengoa M, Trouche D, Panov KI, Vandromme M. The histone demethylase JMJD2A/KDM4A links ribosomal RNA transcription to nutrients and growth factors availability. Nat Commun. 2016;7:10174. doi: 10.1038/ncomms10174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henras AK, Plisson-Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip Rev RNA. 2015;6:225–42. doi: 10.1002/wrna.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673–83. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 9.Aki T, Yanagisawa S. Application of rice nuclear proteome analysis to the identification of evolutionarily conserved and glucose-responsive nuclear proteins. J Proteome Res. 2009;8:3912–24. doi: 10.1021/pr900187e. [DOI] [PubMed] [Google Scholar]
- 10.Maekawa S, Ishida T, Yanagisawa S. Reduced expression of APUM24, encoding a novel rRNA processing factor, induces sugar-dependent nucleolar stress and altered sugar responses in Arabidopsis thaliana. Plant Cell. 2018;30:209–27. doi: 10.1105/tpc.17.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francischini CW, Quaggio RB. Molecular characterization of Arabidopsis thaliana PUF proteins-binding specificity and target candidates. FEBS J. 2009;276:5456–70. doi: 10.1111/j.1742-4658.2009.07230.x. [DOI] [PubMed] [Google Scholar]
- 12.Tam PP, Barrette-Ng IH, Simon DM, Tam MW, Ang AL, Muench DG. The Puf family of RNA-binding proteins in plants: phylogeny, structural modeling, activity and subcellular localization. BMC Plant Biol. 2010;10:44. doi: 10.1186/1471-2229-10-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shanmugam T, Abbasi N, Kim HS, Kim HB, Park NI, Park GT, Oh SA, Park SK, Muench DG, Choi Y, et al.. An Arabidopsis divergent pumilio protein, APUM24, is essential for embryogenesis and required for faithful pre-rRNA processing. Plant J. 2017;92:1092–105. doi: 10.1111/tpj.13745. [DOI] [PubMed] [Google Scholar]
- 14.Horiguchi G, Van Lijsebettens M, Candela H, Micol JL, Tsukaya H. Ribosomes and translation in plant developmental control. Plant Sci. 2012;191-192:24–34. doi: 10.1016/j.plantsci.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Gasse L, Flemming D, Hurt E. Coordinated ribosomal ITS2 RNA processing by the Las1 complex integrating endonuclease, polynucleotide kinase, and exonuclease activities. Mol Cell. 2015;60:808–15. doi: 10.1016/j.molcel.2015.10.021. [DOI] [PubMed] [Google Scholar]
- 16.Chakraborty A, Uechi T, Kenmochi N. Guarding the ‘translation apparatus’: defective ribosome biogenesis and the p53 signaling pathway. Wiley Interdiscip Rev RNA. 2011;2:507–22. doi: 10.1002/wrna.73. [DOI] [PubMed] [Google Scholar]
- 17.James A, Wang Y, Raje H, Rosby R, DiMario P. Nucleolar stress with and without p53. Nucleus. 2014;5:402–26. doi: 10.4161/nucl.32235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou X, Liao WJ, Liao JM, Liao P, Lu H. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 2015;7:92–104. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi W, Zhu J, Wu Q, Wang Q, Li X, Yao D, Jin Y, Wang G, Wang G, Song R. Maize reas1 mutant stimulates ribosome use efficiency and triggers distinct transcriptional and translational responses. Plant Physiol. 2016;170:971–88. doi: 10.1104/pp.15.01722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsumura Y, Ohbayashi I, Takahashi H, Kojima S, Ishibashi N, Keta S, Nakagawa A, Hayashi R, Saéz-Vásquez J, Echeverria M, et al.. A genetic link between epigenetic repressor AS1-AS2 and a putative small subunit processome in leaf polarity establishment of Arabidopsis. Biol Open. 2016;5:942–54. doi: 10.1242/bio.019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huart AS, Hupp TR. Evolution of conformational disorder & diversity of the p53 interactome. Biodiscovery. 2013;8:5. doi: 10.7750/BioDiscovery.2013.8.5 [DOI] [Google Scholar]
- 22.Rutkowski R, Hofmann K, Gartner A. Phylogeny and function of the invertebrate p53 superfamily. Cold Spring Harb Perspect Biol. 2010;2:a001131. doi: 10.1101/cshperspect.a001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohbayashi I, Lin CY, Shinohara N, Matsumura Y, Machida Y, Horiguchi G, Tsukaya H, Sugiyama M. Evidence for a Role of ANAC082 as a Ribosomal Stress Response Mediator Leading to Growth Defects and Developmental Alterations in Arabidopsis. Plant Cell. 2017;29:2644–60. doi: 10.1105/tpc.17.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshiyama K, Conklin PA, Huefner ND, Britt AB. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc Natl Acad Sci U S A. 2009;106:12843–48. doi: 10.1073/pnas.0810304106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshiyama KO. SOG1: a master regulator of the DNA damage response in plants. Genes Genet Syst. 2016;90:209–16. doi: 10.1266/ggs.15-00011. [DOI] [PubMed] [Google Scholar]
- 26.Yi D, Alvim Kamei CL, Cools T, Vanderauwera S, Takahashi N, Okushima Y, Eekhout T, Yoshiyama KO, Larkin J, Van den Daele H, et al.. The Arabidopsis SIAMESE-RELATED cyclin-dependent kinase inhibitors SMR5 and SMR7 regulate the DNA damage checkpoint in response to reactive oxygen species. Plant Cell. 2014;26:296–309. doi: 10.1105/tpc.113.118943. [DOI] [PMC free article] [PubMed] [Google Scholar]


