ABSTRACT
Transcription of protein-encoding genes in eukaryotic cells is a dynamically coordinated process. Many of the key transcription regulators contain functionally essential intrinsically disordered regions (IDRs), the dynamic nature of which creates extra challenges to traditional biochemical analyses. Recent advances in single-molecule fluorescence imaging technology have enabled direct visualization of these rapid, complex and dynamic molecular interactions in real time.
KEYWORDS: dynamic analysis, fluorescence imaging, intrinsically disordered regions, preinitiation complex, single-molecule, transcription
Introduction
Eukaryotic RNA polymerase II (Pol II) transcription initiation requires the coordinated action of at least six general transcription factors (GTFs, i.e., TFIIA, -B, -D, -E, -F, and –H).24 These basal factors assemble into a transcription preinitiation complex (PIC) via a multi-step process that is required to initiate productive RNA synthesis. The basic properties of each factor have been extensively studied, but the timing and coordination of their interactions, and how they may behave differently in response to specific regulatory cues remains largely unknown.
A key feature of transcription factor complexes uncovered by recent studies suggests that most protein factors interact and bind to promoter DNA in a manner that is likely more dynamic than previously envisioned. One example of such transient and dynamic interactions of a protein complex with promoter DNA was seen with TFIID, an ensemble composed of the TATA binding protein (TBP) and ∼15 associated factors (TAFs). TFIID recognizes multiple DNA elements at the core promoter, including the TATA box, Initiator (Inr) and the downstream promoter elements (DPE) etc., wherein stronger promoter binding has been linked to higher transcription activity.12 However, after initial binding, TFIID has been reported to undergo a conformational isomerization to release a portion of the promoter DNA for subsequent binding by Pol II.27 The need for this isomerization arises from a steric collision between bound TFIID and sites that overlap the requisite Pol II binding at the Inr as revealed by a recent cryo-EM study.15 Given the complexity of the reaction involving >50 proteins at the PIC, such conformational changes are likely not limited to TFIID and Pol II, and might also serve as critical points of regulation targeted by activators employing novel mechanisms yet to be elucidated.
Another important but often over looked aspect of eukaryotic transcription regulation is the prevalence of so-called intrinsically disordered regions (IDRs).9 These unstructured peptide fragments were initially described as flexible linkers or missing densities in classical X-ray crystal structures of proteins. The emerging picture is that IDRs are very abundant in the eukaryotic proteome, especially amongst transcription regulators, and are thought to generally engage in heterogeneous and dynamic conformational ensembles.25 Intriguingly, IDRs constitute ∼50% of the primary sequence of eukaryotic transcription regulators, in contrast to 5% in prokaryotes.17 It had long been documented that almost all transactivation domains of eukaryotic activators are composed of polar and/or repetitive sequences characteristic of IDRs.18,2,10 Transcription activation domains are thought to interact with co-activators rather transiently (on the order of milliseconds), and it is their overall disordered nature and the presence of scattered hydrophobic residues, not a particular sequence, that seems to be critical for activity.26 How these transient weak interactions influence the kinetics of PIC assembly and lead to transcription activation remains unclear.
In this review, we will provide examples of how dynamic conformational changes controlled by an IDR play critical roles in Pol II transcription initiation, and how in vitro single-molecule imaging using a well-defined reconstituted transcription system offers a rich opportunity for mechanistic studies to address many long standing questions central to understanding transcription regulation. We note that this opportunity for detailed in vitro single molecule analysis was developed in concert with exciting advances in live cell single-molecule imaging and tracking of molecular dynamics in vivo, along with advances in high resolution electron microscopy, that were well covered in recent reviews,19,6,11,20 and thus will not be discussed here.
Chemical perturbation reveals a critical role of an IDR in controlling TFIID dynamics and transcription initiation
With structural and biochemical evidence for conformational changes occurring for TFIID during transcription initiation,13,16,27,5,15 it seemed reasonable to probe this aspect of PIC formation using small molecule modulators. There was therefore an effort to screen for chemicals that can bind and perturb the function of the TFIID complex, which led fortuitously to the isolation of an inhibitor targeting the isomerization step of TFIID29 (Fig. 1).
Figure 1.
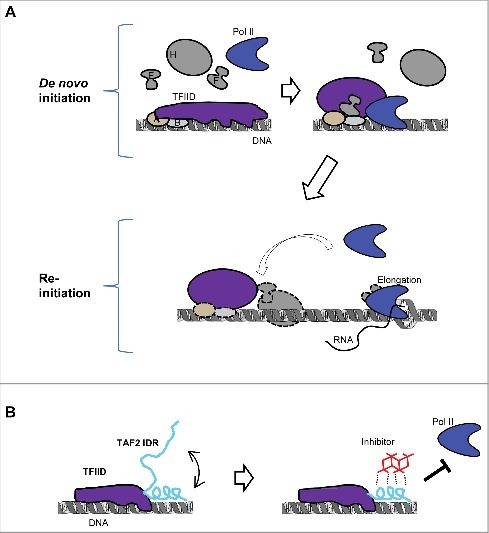
Conformational changes during transcription initiation and its modulation by an IDR. (A) Different functional states of TFIID during two modes of transcription initiation. At the beginning of de novo PIC assembly, TFIID binding covers an expanded area of the promoter, followed by an isomerization stage to release some DNA for Pol II engagement. This changed conformation is preserved after the first Pol II escapes the promoter, which might facilitate reinitiation. (B) Model of how TAF2 IDR may modulate TFIID-promoter interaction. This IDR (in cyan) may exist in multiple inter-changeable configurations, represented by a random coil and a more structured helix. The latter may directly bind DNA, thus modulates TFIID-promoter interaction. When this structured configuration of IDR is stabilized by a chemical inhibitor, release of promoter elements by TFIID is blocked, which in turn prevents Pol II engagement. IDR-mediated modulation only occurs during de novo PIC assembly, thus the inhibitor has no effect on reinitiation.
Several mechanistic insights were revealed by this novel inhibitor. First, contrary to the common expectation that the strength of TFIID-promoter binding is positively correlated with the transcription activity of promoters,12 this TFIID modulator inhibits transcription by stabilizing TFIID promoter binding. More specifically, it prevents the release of promoter DNA from TFIID after initial binding, which is required for Pol II engagement during PIC assembly (Fig. 1A). This highlights the importance of timing and coordination of multiple molecular transactions, not simply high affinity binding, for regulating a complex biochemical reaction like transcription. Second, the modulator only inhibits de novo transcription initiation, but not reinitiation, pointing to different functional states of TFIID during these two modes of transcription initiation, and how the so-called reinitiation scaffold28 might be different from de novo PIC in supporting multiple rounds of transcription efficiently (Fig. 1A). Indeed, reinitiation could represent an important mechanism for maintaining ongoing transcription in non-dividing somatic cells, but it has been very challenging to biochemically dissect such a mechanism. Third, the structural basis for the inhibitor-TFIID interaction is unusual and intriguing. The target of the inhibitor was found to be an evolutionarily conserved region at the C-terminus of the TAF2 subunit, which is mostly composed of repetitive polar residues such as lysines, histidines, aspartates, and glutamates, characteristic of low complexity (LC) IDRs. Additionally, this unusual inhibitor itself did not turn out to be a conventional organic small molecule, but rather a specific metal cluster. With a surface containing polar and periodically arrayed atoms, the metal cluster actually was a good match for the IDR target. A reasonable model hypothesizes that the IDR may assume a transient structure that allows both inhibitor and DNA to bind simultaneously, and that such a DNA/inhibitor co-binding stabilizes this transient structure (Fig. 1B). IDRs are very abundant in the human proteome and seem to be especially prevalent in regulatory pathways involved in human disease.25 However, with elusive structures and binding partners, how they function is poorly understood. Additionally, they are not suitable for conventional small-molecule targets. With a well-defined functional output, this TFIID inhibitor presents an intriguing case study to probe the function of an IDR and how a chemical moiety can interact and perturb this function.
With the importance of TFIID conformational dynamics revealed by the chemical perturbation, it was natural to next attempt to directly capture this dynamics during transcription initiation, and to better understand how the inhibitor, and activators may modulate this dynamics in real time. Another related question is which subunits of TFIID still remain at the promoter after releasing promoter DNA and contribute to the reinitiation scaffold. To further understand the biological function and mechanism of chemical perturbation of the TAF2 IDR, it would be important to know which region of the promoter DNA the TAF2 IDR directly contacts during PIC assembly, what is the kinetics of the interaction, and how that is affected by inhibitor binding. Additionally, are these interactions also affected by other protein factors and the underlying promoter DNA sequence and structure? This line of investigation would require more precise analysis of the dynamics of inter-molecular interactions and intra-molecular conformational changes, which are big challenges to conventional biochemistry but may become addressable using the advanced single-molecule imaging platform described below and will likely form the basis of future studies.
Single-molecule analysis revealed unexpected promoter binding dynamics during PIC assembly
Classical biochemical analysis relies on the average of millions to billions of molecules in ensemble assays. Thus, for complex reactions involving dozens of factors occurring in multiple steps, it is a major challenge to measure dynamics of macro-molecular interactions, to reveal intermediate states of the reaction, and to understand how these transitional stages may be regulated. Recent developments in site-specific fluorescent labeling of proteins coupled with advanced optical imaging technologies have enabled direct visualization of individual molecules operating in real time during complex biochemical reactions. Such super-resolution imaging modalities have subsequently led to the development of research platforms such as Co-localization Single-Molecule Spectroscopy (CoSMoS) used for the study of bacterial transcription regulation.8
To render human Pol II transcription suitable for single-molecule biochemistry studies, the reaction was reconstituted with highly purified factors on a single-molecule imaging surface.22 This system has several critical components specifically developed for human Pol II transcription: (1) a large optical field of view and high-resolution position mapping were used in combination to image several thousand immobilized DNA molecules simultaneously, ensuring the statistical significance of the data; (2) a novel surface passivation allowing transcription from surface-immobilized DNA templates to occur as efficiently as those from DNA in solution; and (3) “fast-FISH” probes capable of detecting ∼80% of the nascent transcripts within less than a second under single-molecule imaging conditions.31 The scheme of this platform is summarized in Fig. 2A.
Figure 2.
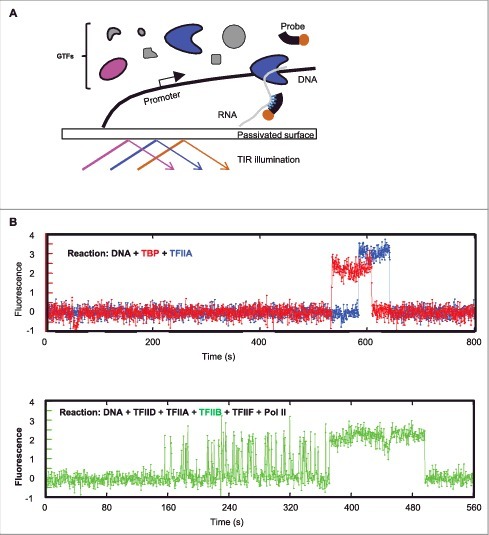
A single-molecule imaging platform for human Pol II transcription studies. (A) Scheme of platform setup. Promoter-containing DNA templates are attached to a passivated imaging surface. Fluorescently labeled (highlighted in magenta and cyan, respectively) or unlabeled (grey) GTFs may be added to the reaction. Nascent RNA transcripts can be detected by labeled hybridization probes (brown). TIR (total internal reflection) illumination minimizes fluorescent background from bulk of the reaction solution and allows the detection of single fluorescent molecules interacting with the immobilized DNA templates by high sensitivity camera (not shown). (B) Two different types of GTF-promoter binding dynamics at the single-molecule level. Top are typical fluorescent time traces of TBP (red) and TFIIA (blue) on one DNA template. The observation time of the fluorescent signals is mostly limited by photo-bleaching under the experimental conditions (1∼2 minutes). Bottom is a typical fluorescence time trace of TFIIB (green) promoter binding in the presence of others GTFs as specified (black). Note the initial TFIIB binding is very transient and repetitive. These example fluorescence traces were taken from single-molecule imaging experiments as described (30).
Using such a single-molecule imaging platform, the initial steps of PIC assembly of fluorescently labeled TFIID, TFIIA and TFIIB was determined.30 By monitoring their co-localization at the promoter DNA, sequence-dependent binding of these factors showed a hierarchical assembly order of TFIID, -A, and -B, consistent with models derived from decades of ensemble biochemical studies.24 However, when the time-lapse fluorescence traces at each DNA template was examined to directly measure when each labeled protein factor binds and how long it stays, some very unexpected findings emerged (Fig. 2B). In stark contrast to the stable binding interaction expected from the traditional step-wise PIC assembly model, these single-molecule imaging assays revealed that TFIIB binding is highly transient and repetitive, lasting 1∼2 seconds on average. This transient and dynamic binding was observed in both the TFIID- or TBP-dependent reactions (TBP is the “core” part of TFIID capable of supporting basal level of transcription), suggesting that it is likely an intrinsic property of the interaction between TFIIB and the TATA-bound TBP. Interestingly, although unexpected from conventional in vitro based biochemical models, this result may explain a previous puzzling in vivo finding where TFIIB was found to be much more dynamic than TBP.3
The transient and dynamic TFIIB-promoter binding can be stabilized by TFIIF-Pol II, which, according to the step-wise PIC assembly model,24 were expected to join the PIC assembly after TFIIB. Therefore, TFIIB is not simply a factor passively siting at the promoter waiting for TFIIF and Pol II, but rather actively responding to the presence of these downstream factors (Fig. 2B). The transient nature of TFIIB-DNA interaction and dependence on TFIIF-Pol II for stable association was confirmed with in vivo single-molecule imaging as well, suggesting that this transient-to-stable transition in TFIIB binding likely reflects a stage of productive Pol II engagement during PIC assembly in live cells.30
In contrast to the single-step binding of TFIID, TBP, and TFIIA, the dynamic behavior of TFIIB offers the following advantages. First it is fast. The on-rate of TFIIB is 1∼2 orders of magnitude higher than that of the other factors tested. This may render TFIIB binding not a rate-limiting step for PIC assembly, bringing speed at the cost of regulation. On the other hand, the short residence time makes this binding unstable, which in turn might serve as a “checkpoint” that must be overcome and stabilized by regulators. One aspect of such a proposed checkpoint function could be to make sure the right downstream components, e.g. TFIIF-Pol II, are available to complete PIC assembly. Additionally, in cases where local TFIIF-Pol II availability is limited, it creates an opportunity for other factors to stabilize TFIIB binding, which in turn increases the chance of TFIIF-Pol II loading, and thus could represent an additional regulatory step. Because we only recently came to appreciate the dynamics of TFIIB binding, factors regulating this dynamics remain largely unknown and somewhat speculative. However, given the complexity of the interaction network regulating the eukaryotic transcription machinery, it is not a great leap to posit the existence of such factors. One such factor could be activators like VP16, which is known to target TFIIB.14 Therefore, this dynamic behavior, in contrast to the relatively static one-step stable binding process, might be more efficient and flexible in integrating regulatory signals.
Although largely overlooked by most conventional ensemble biochemical studies, rapid dynamic interactions actually could be a relatively common mechanism in transcription regulation. For example, the residence times of the stem cell activator Sox2 in vitro and in vivo on cognate DNA sequences was found to be only a few seconds using single-molecule imaging technologies.4 Additionally, transactivation domains of activators seem to interact with coactivators even more transiently, with residence times in the range of milliseconds or shorter, which is likely due to the intrinsically disordered nature of these transactivation domains.2 How could these rapid transient interactions lead to stable PIC assembly? The answer might lie in multiple rounds of frequent binding events. In support for this possibility, eukaryotic transcription regulatory regions usually contain clusters of activator binding sites, which might help increase the frequency of activator binding and thereby effect the local transcription factor concentrations in the nucleus.21 Also, activators tend to interact with multiple partners within coactivator complexes as well as the transcription core machinery, which might increase the frequency of activator-coactivator interactions. Thus rapid and frequent interactions, followed by transitioning to a more stable steady state in subsequent assembly steps might be a common mechanism to regulate transcription. It seems clear that deciphering such dynamic interactions would greatly benefit from emerging single-molecule imaging technologies capable of rapid direct measurements of protein-protein and protein-nucleic acid interactions both in vitro and in vivo.
Challenges and opportunities for single-molecule imaging
Direct expansion of the Pol II single-molecule imaging system would include analysis of promoter binding of additional protein factors, how these dynamic interactions may be regulated by activators, and how these interactions correlate with RNA synthesis in real time. This would be quite feasible given what has been learned from TFIIB by imaging at sub-second temporal resolution and real-time transcription detection.22,31 Using various native promoters, this line of analysis would provide a more complete dynamics of transcription initiation.
Going deeper into the mechanisms behind the apparent binding and dissociation of protein factors would require acquisition of the conformational changes often associated with different functional states. Of all the conformational changes, those associated with IDRs probably are both the most interesting and most challenging. IDRs empower the assembly of dynamic structures with very low energy barriers capable of rapidly transitioning between different conformations. These rapid conformational inter-exchanges may respond to small changes in local molecular environments that could have big effects on the distribution of such complexes.7 This also makes it critical to address the dynamics of IDR driven conformational changes in the context of other components of the reaction, which is a challenge to conventional approaches.
Conformational changes can be analyzed by a strategy called Fluorescence Resonsance Energy Transfer (FRET) at the single-molecule level.23 In this assay, a pair of fluorescent dyes, a donor and a receptor, are attached to two interacting molecules or two different positions of one molecule. Within a distance of 2 ∼ 10 nm, the energy from the donor fluorophore at an excited state can be transferred to the receptor via non-radioactive dipole-dipole coupling. The efficiency of the energy transfer is inversely proportional to the sixth power of the distance between the two dyes, thus FRET is extremely sensitive to small changes in distance, and is suitable for measuring conformational changes within macromolecules. Single-molecule FRET (sm-FRET) has been successfully applied to study dynamics of intrinsically disordered proteins,1 although they have mostly been used to monitor the dynamics of IDRs themselves, in the absence of interacting partners. During single-molecule imaging of Pol II PIC assembly, sm-FRET signals have been observed between certain pairs of fluorescently labeled TFIIB and TFIIA molecules (Zhang and Tjian, unpublished data). Thus, there is no major technical barrier preventing the adaption of this approach to the Pol II transcription system. Ideally, such a system should have three independent fluorescent imaging channels, two for the FRET pair monitoring the conformational changes of the protein factors, and one for the tracking of another interacting partner, to establish a causal relationship. This would make many of the long standing questions in transcription control addressable, such as how activators work, how different molecular players coordinate to integrate regulatory signals, how the dynamics of IDR-mediated interactions affect transcriptional output, and how these dynamic interactions may be modulated by natural or artificial ligands.
Conclusion remarks
Transcription represents a critical step during gene expression. Eukaryotic cells have evolved a transcription machinery and control system that is much more complicated than is found in prokaryotic cells. The elaborate nature of the Pol II system is underscored by the large number of regulatory factors and abundance of IDRs required just to assemble the pre-initiation complex. Simple models like competitive or cooperative promoter binding, widely accepted as dominant mechanisms for prokaryotic transcription regulation, is not sufficient to explain the mechanisms driving eukaryotic transcription regulation. One key differentiating aspect may lie in the apparently redundant, weak and repetitive interactions, which have largely been overlooked by conventional biochemical analysis. Single-molecule in vitro biochemistry, in combination with advances in imaging technology to track single-molecule dynamics in vivo along with EM based high-resolution structural studies, represent new opportunities to address many of the most pressing unanswered fundamental questions in eukaryotic transcription regulation.
Funding Statement
Research in the Tjian Laboratory is supported by Howard Hughes Medical Institute and California Institute for Regenerative Medicine Research grant RB4-06016.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
Robert Tjian is an HHMI investigator.
References
- [1].Brucale M, Schuler B, Samori B. Single-molecule studies of intrinsically disordered proteins. Chem Rev. 2014;114:3281-317. doi: 10.1021/cr400297g. PMID:24432838 [DOI] [PubMed] [Google Scholar]
- [2].Brzovic PS, Heikaus CC, Kisselev L, Vernon R, Herbig E, Pacheco D, Warfield L, Littlefield P, Baker D, Klevit RE, Hahn S. The acidic transcription activator Gcn4 binds the mediator subunit Gal11/Med15 using a simple protein interface forming a fuzzy complex. Mol Cell. 2011;44:942-53. doi: 10.1016/j.molcel.2011.11.008. PMID:22195967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen D, Hinkley CS, Henry RW, Huang S. TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol Biol Cell. 2002;13:276-84. doi: 10.1091/mbc.01-10-0523. PMID:11809839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Chen J, Zhang Z, Li L, Chen BC, Revyakin A, Hajj B, Legant W, Dahan M, Lionnet T, Betzig E, Tjian R, Liu Z. Single-molecule dynamics of enhanceosome assembly in embryonic stem cells. Cell. 2014;156:1274-85. doi: 10.1016/j.cell.2014.01.062. PMID:24630727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cianfrocco MA, Kassavetis GA, Grob P, Fang J, Juven-Gershon T, Kadonaga JT, Nogales E. Human TFIID binds to core promoter DNA in a reorganized structural state. Cell. 2013;152:120-31. doi: 10.1016/j.cell.2012.12.005. PMID:23332750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Coleman RA, Liu Z, Darzacq X, Tjian R, Singer RH, Lionnet T. Imaging Transcription: Past, Present, and Future. Cold Spring Harb Symp Quant Biol. 2015;80:1-8. doi: 10.1101/sqb.2015.80.027201. PMID:26763984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Flock T, Weatheritt RJ, Latysheva NS, Babu MM. Controlling entropy to tune the functions of intrinsically disordered regions. Curr Opin Struct Biol. 2014;26:62-72. doi: 10.1016/j.sbi.2014.05.007. PMID:24930020 [DOI] [PubMed] [Google Scholar]
- [8].Friedman LJ, Gelles J. Multi-wavelength single-molecule fluorescence analysis of transcription mechanisms. Methods. 2015;86:27-36. doi: 10.1016/j.ymeth.2015.05.026. PMID:26032816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fuxreiter M, Tompa P, Simon I, Uversky VN, Hansen JC, Asturias FJ. Malleable machines take shape in eukaryotic transcriptional regulation. Nat Chem Biol, 2008;4:728-37. doi: 10.1038/nchembio.127. PMID:19008886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Habchi J, Tompa P, Longhi S, Uversky VN. Introducing protein intrinsic disorder. Chem Rev. 2014;114:6561-88. doi: 10.1021/cr400514h. PMID:24739139 [DOI] [PubMed] [Google Scholar]
- [11].Hantsche M, Cramer P. Conserved RNA polymerase II initiation complex structure. Curr Opin Struct Biol. 2017;47:17-22. doi: 10.1016/j.sbi.2017.03.013. PMID:28437704 [DOI] [PubMed] [Google Scholar]
- [12].Juven-gershon T, Cheng S, Kadonaga JT. Rational design of a super core promoter that enhances gene expression. Nat Methods. 2006;3:917-22. doi: 10.1038/nmeth937. PMID:17124735 [DOI] [PubMed] [Google Scholar]
- [13].Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418-22. doi: 10.1126/science.288.5470.1418. PMID:10827951 [DOI] [PubMed] [Google Scholar]
- [14].Lin YS, Ha I, Maldonado E, Reinberg D, Green MR. Binding of general transcription factor TFIIB to an acidic activating region. Nature. 1991;353:569-71. doi: 10.1038/353569a0. PMID:1922364 [DOI] [PubMed] [Google Scholar]
- [15].Louder RK, He Y, Lopez-Blanco JR, Fang J, Chacon P, Nogales E. Structure of promoter-bound TFIID and model of human pre-initiation complex assembly. Nature. 2016;531:604-9. doi: 10.1038/nature17394. PMID:27007846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Miller G, Hahn S. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nat Struct Mol Biol. 2006;13:603-10. doi: 10.1038/nsmb1117. PMID:16819517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol, 2006;359:1137-49. doi: 10.1016/j.jmb.2006.04.016. PMID:16697407 [DOI] [PubMed] [Google Scholar]
- [18].Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371-8.doi: 10.1126/science.2667136. PMID:2667136 [DOI] [PubMed] [Google Scholar]
- [19].Mueller F, Stasevich TJ, Mazza D, Mcnally JG. Quantifying transcription factor kinetics: at work or at play? Crit Rev Biochem Mol Biol. 2013;48:492-514. doi: 10.3109/10409238.2013.833891. PMID:24025032 [DOI] [PubMed] [Google Scholar]
- [20].Nogales E, Louder RK, He Y. Structural Insights into the Eukaryotic Transcription Initiation Machinery. Annu Rev Biophys. 2017;46:59-83. doi: 10.1146/annurev-biophys-070816-033751. PMID:28532216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Normanno D, Boudarene L, Dugast-Darzacq C, Chen J, Richter C, Proux F, Benichou O, Voituriez R, Darzacq X, Dahan M. Probing the target search of DNA-binding proteins in mammalian cells using TetR as model searcher. Nat Commun. 2015;6:7357. doi: 10.1038/ncomms8357. PMID:26151127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Revyakin A, Zhang Z, Coleman RA, Li Y, Inouye C, Lucas JK, Park SR, Chu S, Tjian R. Transcription initiation by human RNA polymerase II visualized at single-molecule resolution. Genes Dev. 2012;26:1691-702. doi: 10.1101/gad.194936.112. PMID:22810624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nature Methods. 2008;5:507-516. doi: 10.1038/nmeth.1208. PMID:18511918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105-78. doi: 10.1080/10409230600648736. PMID:16858867 [DOI] [PubMed] [Google Scholar]
- [25].Uversky VN. A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci. 2013;22:693-724. doi: 10.1002/pro.2261. PMID:23553817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Warfield L, Tuttle LM, Pacheco D, Klevit RE, Hahn S. A sequence-specific transcription activator motif and powerful synthetic variants that bind Mediator using a fuzzy protein interface. Proc Natl Acad Sci U S A. 2014;111:E3506-13. doi: 10.1073/pnas.1412088111. PMID:25122681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yakovchuk P, Gilman B, Goodrich JA, Kugel JF. RNA polymerase II and TAFs undergo a slow isomerization after the polymerase is recruited to promoter-bound TFIID. J Mol Biol. 2010;397:57-68. doi: 10.1016/j.jmb.2010.01.025. PMID:20083121 [DOI] [PubMed] [Google Scholar]
- [28].Zawel L, Kumar KP, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479-90. doi: 10.1101/gad.9.12.1479. PMID:7601352 [DOI] [PubMed] [Google Scholar]
- [29].Zhang Z, Boskovic Z, Hussain MM, Hu W, Inouye C, Kim HJ, Abole AK, Doud MK, Lewis TA, Koehler AN, Schreiber SL, Tjian R. Chemical perturbation of an intrinsically disordered region of TFIID distinguishes two modes of transcription initiation. Elife. 2015;4. doi: 10.7554/eLife.07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhang Z, English BP, Grimm JB, Kazane SA, Hu W, Tsai A, Inouye C, You C, Piehler J, Schultz PG, Lavis LD, Revyakin A, Tjian R. Rapid dynamics of general transcription factor TFIIB binding during preinitiation complex assembly revealed by single-molecule analysis. Genes Dev. 2016;30:2106-2118. doi: 10.1101/gad.285395.116. PMID:27798851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang Z, Revyakin A, Grimm JB, Lavis LD, Tjian R. Single-molecule tracking of the transcription cycle by sub-second RNA detection. Elife. 2014;3:e01775. doi: 10.7554/eLife.01775. PMID:24473079 [DOI] [PMC free article] [PubMed] [Google Scholar]


