ABSTRACT
Lineage-specific cell differentiation is a precise and coordinated biological process. To explore the roles of long noncoding RNA (lncRNA) in this process, the expression of polyA-minus RNAs was comparatively studied during the course of myocyte and adipocyte differentiation. In addition to identifying thousands of novel lncRNAs, distinct lncRNA profiles were revealed during lineage-specific differentiation, showing their active involvement in this process. This study further found that lncRNAs were organized in clusters and are co-regulated, constituting transcription open domains (TODs). In myocyte differentiation of C2C12 cells, loss-of-function screening identified three myogenic lncRNAs. Knockdown of their expression compromised not only the differentiation process, but also the essential signaling pathway. In addition to showing that lncRNAs are actively involved in cell differentiation, our results start to reveal a comprehensive signaling pathway, involving both protein and lncRNA factors.
KEYWORDS: lncRNA, Differentiation, Myogenesis, Adipogenesis
Introduction
During development of the multicellular organism, lineage-specific cell differentiation is an essential cellular process. The differentiation of tissue-specific stem cells involves increased restriction in proliferation capability, culmination in cell-cycle exit and finally terminal differentiation. Precise coordination of this process involves diverse cellular factors and their interactions [1].
Myocyte and adipocyte differentiation are two robust model systems for cell differentiation. Over the past decades, studies have focused on characterization of involved protein factors, leading to the establishment of discrete signaling pathways [2,3]. In the case of myocyte differentiation, this process is regulated mainly by master transcriptional factor MyoD, and involves multiple myogenic factors such as Myf5, myogenin, MRF4, and Mef2 [4].
More recently, emerging evidence has proposed that lncRNAs are involved in the regulation of muscle development, through a variety of molecular mechanisms occurring either in the nucleus or cytoplasm. Within the nucleus, enhancer RNAs (eRNAs) CE and DRR can induce the expression of MyoD or MyoG, which can function in cis or in trans [5]. LncRNA SRA acts as a transcription activator for MyoD, p68 and p72 [6]. LncRNAs H19 and Glt-Meg3 interact with the PRC2 complex to modulate their target genes [7–9]. While in the cytoplasm, Linc-MD1 functions as a microRNA sponge to inhibit mRNA degradation of two transcription factors MAML1 and MEF2c, mediated by miR-133 and miR-135 [10]. Short interspersed element (SINE) containing lncRNAs can bind to the UTR regions of Cdc6 and Traf6 mRNAs and promote their decay at different stages of muscle differentiation [11]. Furthermore, aberrant expression of these lncRNAs may lead to severe muscular disorders, demonstrating their physiological and pathological relevance [12–15].
Despite current progress in this field, we are still at the beginning of ‘an era of noncoding RNA’. The roles of lncRNAs in various biological processes, particularly cell differentiation, remain to be elucidated. For example, most of the characterized lncRNAs are polyadenylated RNAs due to the limitation of standard experimental design. Therefore, the present study aims to characterize polyA-minus RNA expression and regulation, specifically in the differentiation of C2C12 and 3T3-L1 cells.
Results
The expression patterns of lncRNA in myogenesis and adipogenesis
To characterize the polyA-minus lncRNAs in cell differentiation, myocyte differentiation of C2C12 cells and adipocyte differentiation of 3T3-L1 cells were comparatively investigated. C2C12 cells derived from murine satellite cells and when treated with horse serum, are capable of undergoing spontaneous myocyte differentiation [16] (Fig. S1). Mouse 3T3-L1 cell line, established in 1974, is the most used model system for in vitro adipogenesis [17]. When treated with a hormone cocktail consisting of 3-isobutyl-1-methyxanthine, dexamethasone and insulin, the cells readily differentiate into adipocytes.
To resolve the temporal regulation of lncRNA expression, several critical time-points were examined, representing the proliferating blast cells, the differentiating cells, and the lineage-specific mature cells. For convenience, the day of differentiation induction is indicated as day(0).
In myocyte differentiation, cell samples were collected at day(0), day(3) and day(5); while in adipocyte differentiation, samples were collected at day(0), day(14) and day(21). After total RNAs extraction, a reverse enrichment procedure was performed to isolate polyA-minus lncRNA components [18]. These samples were then subjected to RNA sequencing using the illuminaHiSeq™2000 platform.
For myocyte differentiation published in our earlier study [18], a total of 98.3 million reads were obtained, comprising 33.1 million reads from myoblasts, 31.7 million reads from differentiating myocytes, and 33.5 million reads from mature myocytes. Using TopHat software [19], 62.3 million reads (63%) were successfully mapped to the mouse genome (NCBI37/mm9). After filtering out rRNAs and repeated sequences, a high-confidence data set comprised of 49.4 million uniquely mapped reads was obtained. 19% of the reads were mapped to exons of Refseq genes, and the remainder were mapped to non-coding regions.
To define the boundaries of these intergenic transcripts, read-assembly was performed according to the following criteria: a maximum spacing between two neighboring reads of 150 nucleotides and a minimum of 10 mapped reads within a genomic region. This led to the identification of 856 intergenic lncRNAs in myocyte differentiation (Table S1), occupying 0.0461% of the mouse genome. Coding potentials of these sequences were examined by CPC2 program and are presented in Table S2.
When the same procedure was performed with the sequencing data of 3T3-L1 cells, 600 intergenic lncRNAs were identified, occupying 0.0077% of the genome (Fig. S3b; Fig. 1b; Table S3 & S4).
Figure 1.
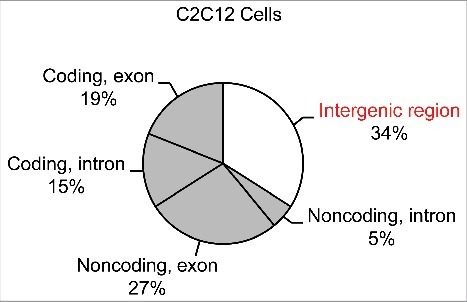
Mapping profile of the sequencing reads in myocyte differentiation of C2C12 cells.
LncRNA profiles in lineage-specific differentiation
When all of the lncRNAs were aligned to the mouse genome side by side, distinct distribution profiles were revealed (Fig. 2). For example in adipocyte differentiation, 59.3% (359) of the transcripts were mapped to a 10.3 Mbp region in chromosome 10, leading to the identification of a novel adipogenic lncRNA slincRAD as reported in our previous study (Yi 2013). However, during myocyte differentiation, the most heavily distributed chromosome is chromosome 15, followed by chromosome11, 4, 2, 1, 10, 16 and 13. Thus, 134 lncRNAs were mapped to chromosome 15 in myocyte differentiation, occupying 0.1912% of the sequence; however, only 9 adipogenesis lncRNAs, occupying 0.0017% of the sequence, were mapped to this chromosome. Interestingly, among the thousands of independent lncRNAs, only 39 (6%) lncRNAs were commonly expressed in both differentiation setting, while 94% of the lncRNAs were specifically expressed in only one of the two closely related differentiation processes. Therefore, this study has revealed novel lncRNA profiles specific to each differentiation process.
Figure 2.

Differentiation-specific profiles of lncRNAs. LncRNAs identified in myocyte and adipocyte differentiation are aligned on mouse chromosomes side by side. LncRNAs obtained from myocyte differentiation are labeled in blue, lncRNAs from adipocyte differentiation are labeled in red.
To explore the spatial distribution of the lncRNAs, sequencing reads were further analyzed by means of cumulative distribution curve and probability density analysis (Fig. S4). Given the criterion that the maximum spacing of 100 kb between two neighboring lncRNAs and no less than three lncRNAs in each cluster, 573 lncRNAs (38%) were found to be distributed in 32 gene clusters (Fig. 3 & Table S5). 28 gene clusters were identified in myocyte differentiation and 4 gene clusters were identified in adipocyte differentiation. The genomic proximity of the clustered lncRNAs and their lineage-specific co-regulation suggest that they are regulated under a common transcription program, in each differentiation processes. This led to our hypothesis that they may form transcription open domains (TODs).
Figure 3.

The clustered distribution of lncRNAs. (a) The position and the cumulative distribution curve of lncRNAs in myocyte differentiation of C2C12 (blue) and adipocyte differentiation of 3T3-L1 (red). Kolmogorov-Smirnov test was performed. *P< 0.1; **P< 0.05; ***P< 0.001. (b, c) The pairwise distance of lncRNAs of C2C12 and 3T3 cells. LncRNA clusters are shown as red blocks inside the matrix.
Regulated expression of lncRNAs
The expression of lncRNAs was examined in terms of differentiation stages. During the course of myocyte differentiation, the expression of 664 (73.5%) lncRNAs were found to be temporally regulated. Among them, consistent up-regulation of 103 lncRNAs were identified (Fig. 4a). Relative to myoblasts, 242 lncRNAs were up-regulated and 297 lncRNAs were down-regulated in the differentiating myocytes. When the cells progressed further into mature myocytes, 136 lncRNAs were up-regulated and 368 were down-regulated. Three lncRNAs overlapping the linc-MD1 locus were identified (Fig. 4c).
Figure 4.
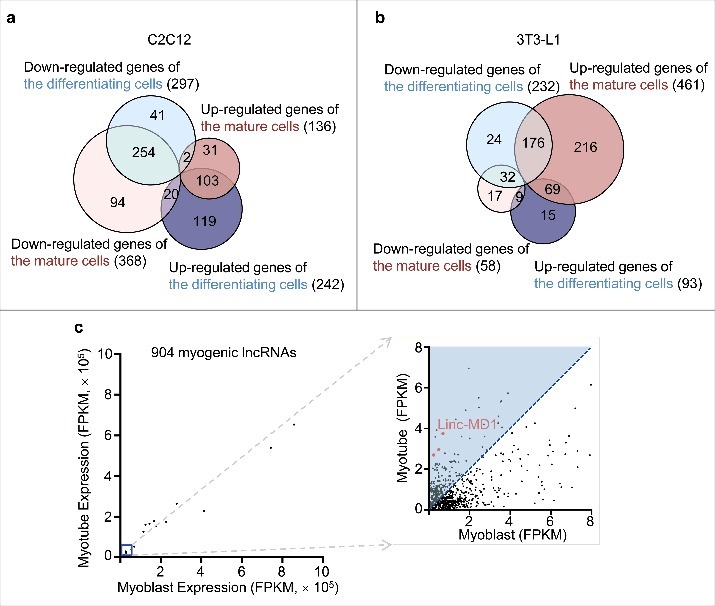
Expression regulation of lncRNAs. (a) Venn diagram analysis showing the regulation of lncRNA in the differentiation of C2C12 cells. (b) Venn diagram analysis showing the regulation of lncRNA in the differentiation of 3T3-L1 cells. (c) Expression profile of 856 myogenesis-associated lncRNAs. X axis, FPKM values of the lncRNAs in myoblasts; Y axis, FPKM values of the lncRNAs in the differentiated myocytes. Three transcripts overlapping with Linc-MD1 are indicated by red.
When the same analyses were performed on adipocyte differentiation, we found that 558 (93%) lncRNAs were temporally regulated (Fig. 4b). Relative to pre-adipocytes, 93 lncRNAs were up-regulated and 232 lncRNAs were down-regulated in the differentiating adipocytes. Contrastingly in the mature adipocytes, 461 lncRNAs were up-regulated and 58 lncRNAs were down-regulated.
Under further detailed examination of their regulation, we found that the early expressed lncRNAs reside mainly on chromosome 15. Moreover, the most up-regulated lncRNAs were found on chromosome 1, 8, 11 and 15 with the highest up-regulation on chromosome 15.
To corroborate their relevance to differentiation, differentiation-specific protein factors were identified and mapped to chromosomes. These included 114 myogenesis-relevant protein factors and 182 adipogenesis-relevant protein factors (Table S6 & S7). As shown in Fig. S5, these protein factors were found to reside within or close to the lncRNA clusters, lending further support to the hypothesis of TODs.
LncRNA functions in myocyte differentiation
To investigate their functions with a RNAi assay, the lncRNAs were first categorized into nine subgroups in terms of their differential regulation during the differentiation (Fig. S3c). Due to the gene silencing property of RNAi, the subgroup that was steadily up-regulated in the differentiation process was chosen for further study. Other criteria used to obtain the candidate lncRNAs are: high expression levels and proximity to critical protein factors in chromosome distribution. This led to a candidate set of 73 lncRNAs (Table S8).
For each lncRNA, its transcription orientation was first determined, using strand-specific RT-PCR (Table S9 & S10). Then, gene-specific siRNAs were transfected into C2C12 cells, on day(−1) (Table S11). On day(0), the cells were induced into myocyte differentiation, to determine lncRNAs effects on myocyte differentiation.
Five days after induction, differentiation of the cells was examined by MHC, Hoechst 33342 staining and EdU assays. The gene silencing efficacies were examined by real time PCR relative to scrambled siRNA controls (Table S11). Phenotypic assays revealed four phenotypes, in terms of the effects of siRNAs on cell proliferation and differentiation (Figs. 5, 6 & Fig. S6). For 27 lncRNAs, including myo18 and myo77, knockdown of their expression compromised both cell proliferation and differentiation, shown by the decreased cell numbers and MHC staining. For another 29 lncRNAs, including myo351, myo432, myo498 and myo586, knockdown of their expression compromised cell differentiation but not proliferation. For 10 lncRNAs, including myo40 and myo743, knockdown of their expression did not affect cell proliferation, however enhanced cell differentiation. For the remaining 7 lncRNAs, including myo90 and myo306, knockdown of their expression had no effects on either proliferation or differentiation of the cells.
Figure 5.
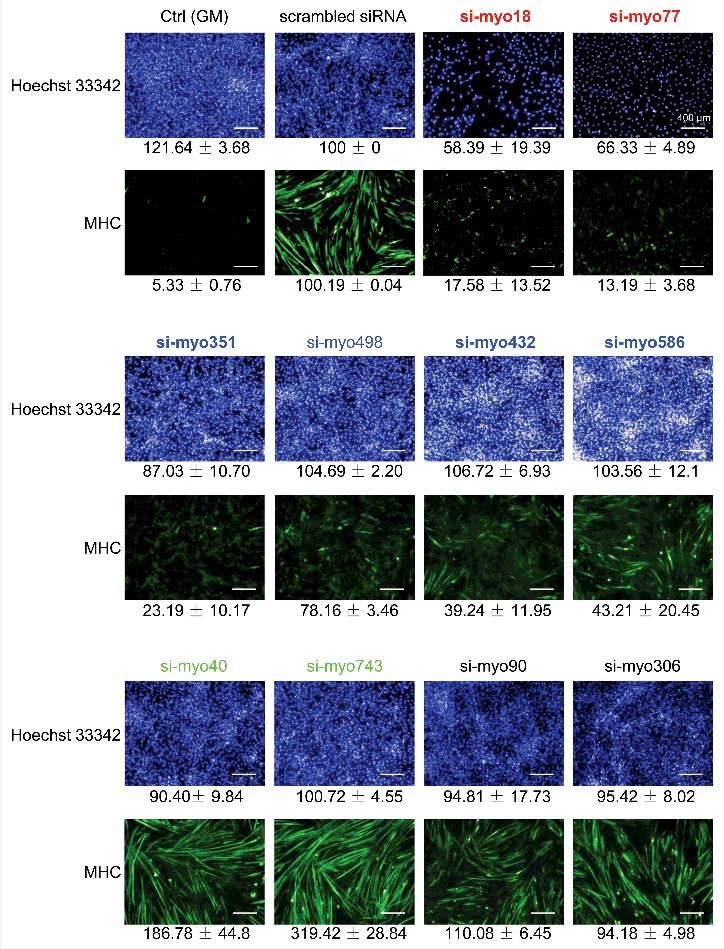
Characterization of functional lncRNAs by RNAi screening. Cultured C2C12 cells were transfected by lncRNA-specific siRNA on day(−1). Differentiation induction was performed on day(0). On day(+5), the cells were collected, examined by immunofluorescence staining of MHC and Hoechst 33342staining. Ctrl (GM), cells cultured in growth medium without siRNA treatment and induction. Scrambled siRNA, cells treated by a scrambled siRNA and then subjected to induction. lncRNA in red, down-regulation of the lncRNA led to decreased cell number and compromised MHC expression, relative to scrambled siRNA treatment. lncRNA in blue, down-regulation of the lncRNA had no effect on cell number, and led to compromised MHC expression. lncRNA in green, down-regulation of the lncRNA had no effect on cell number, however led to enhanced MHC expression. lncRNA in black, down-regulation of the lncRNA had no effect on both cell number and MHC expression.
Figure 6.
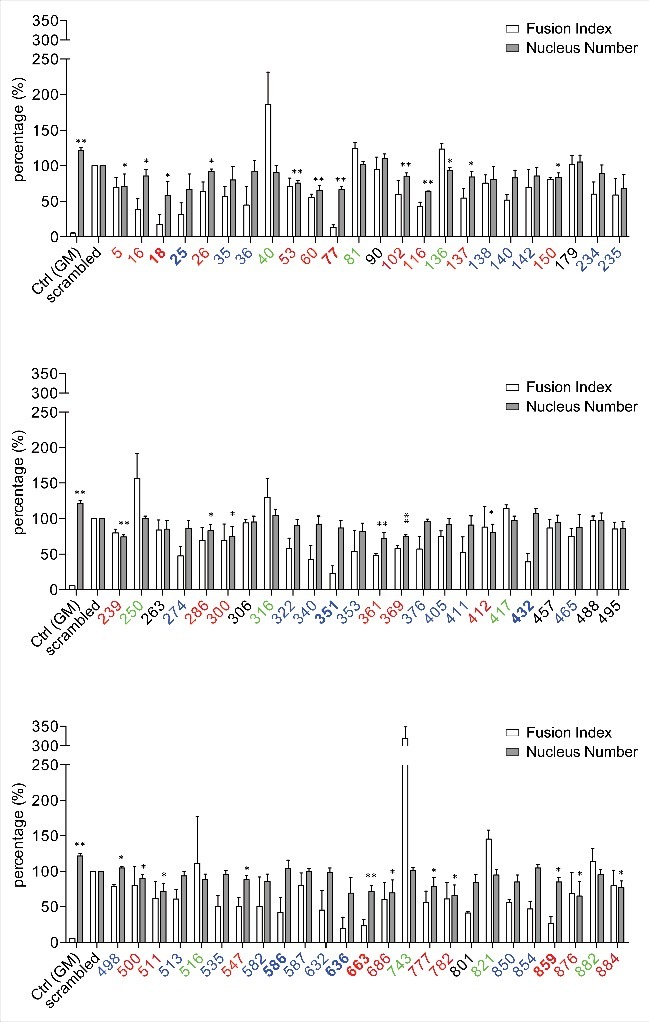
High content screening. Cultured C2C12 cells were transfected by lncRNA-specific siRNA on day(−1), induced to myocyte differentiation on day(0), and collected on day(+5) for analysis. Cell nucleus and MHC expression were detected by Hoechst 33342 and immunofluorescence staining. Data analysis was performed using a High Content Imaging System. Ctrl(GM), cells cultured in growth medium without siRNA treatment and induction. Scrambled, cells treated by a scrambled siRNA and then subjected to induction. X axis, lncRNA code; Y axis, the value of fusion index and nucleus number relative to that of scrambled siRNA-treated cells. Data are presented as mean± SD; three independent assays were performed in triplicate. Student t test was performed (*,P< 0.05; **, P < 0.01).
It has been shown that prior to terminal differentiation, myoblast cells proliferate for a few rounds to increase cell population size. Accordingly, our results clearly identified two groups of lncRNAs that function specifically in the proliferation or differentiation process. When a lncRNA functioning in cell proliferation was repressed, both proliferation and differentiation was affected. Contrastingly, when a lncRNA functioning in cell differentiation was repressed, only differentiation was affected. Interestingly, silencing of a few lncRNAs led to enhanced adipocyte differentiation.
LncRNA expression in vivo
To explore their regulation in vivo, tissue expression profiles of 9 lncRNAs were examined in mouse, including myo18, myo25, myo77, myo351, myo432, myo586, myo636, myo663 and myo859. As expected, all the lncRNAs were found to express in mouse skeletal muscles, except for myo25 (Fig. 7a). While Myo432, myo586, myo636, myo663 and myo859 were similarly expressed in all the tissues, tissue-specific expression was found for myo18, myo77 and myo351 (Fig. S7). Similar to that of MyoD, myogenin and MyHC [20], the expression of myo18 was only identified in skeletal muscle, suggesting it is a skeletal muscle-specific lncRNA. In contrast, the highest expression of myo351 was identified in skeletal and heart muscles. For myo77, the highest expression level was found in lung, followed by skeletal muscle and heart. As a reference, a previously reported myogenic lncRNA Linc-MD1 was also examined in this assay.
Figure 7.
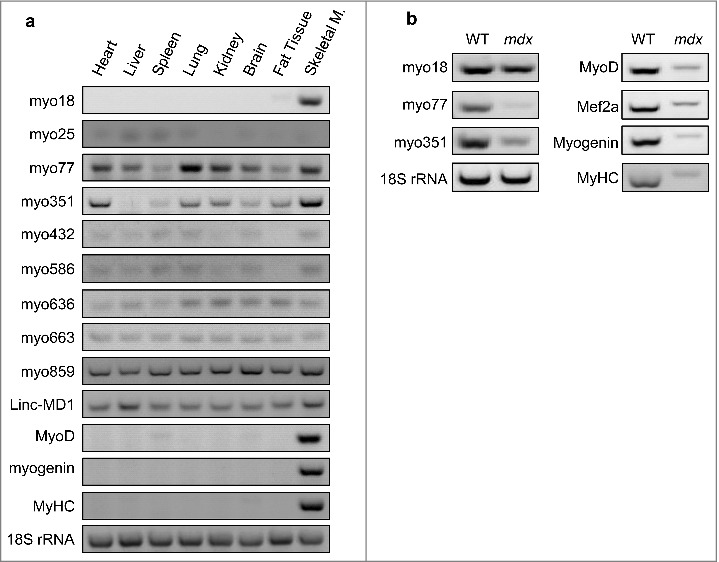
Cellular distribution of lncRNAs. (a) Using RT-PCR, the expression of lncRNAs and protein factors were determined in WT mouse tissues. (b) LncRNA expression was examined by RT-PCR in skeletal muscles, derived from WT and mdx mice.
To corroborate their in vivo activities, the expression of myo18, myo77 and myo351 were further examined with dystrophic mdx mice. Carrying a point mutation in dystrophin gene, mdx mice is a most used model system to investigate muscle degeneration and regeneration in Duchenne Muscular Dystrophy. Compared to wild-type controls, mdx mice displayed significantly decreased myo77 and myo351 levels in the muscle. However, comparable myo18 levels were observed in wild-type and mdx mice (Fig. 7b). Taken together, this led to the identification of three novel myogenic lncRNAs.
Functional mechanism of myogenic lncRNAs
To pursue their functional roles, the expression of these lncRNAs was further investigated during the course of myocyte differentiation using qRT-PCR (Table S12). While the levels of myo77 remained relatively stable throughout differentiation, myo18 and myo351 increased steadily during this process, showing an inducible property (Fig. 8a).
Figure 8.
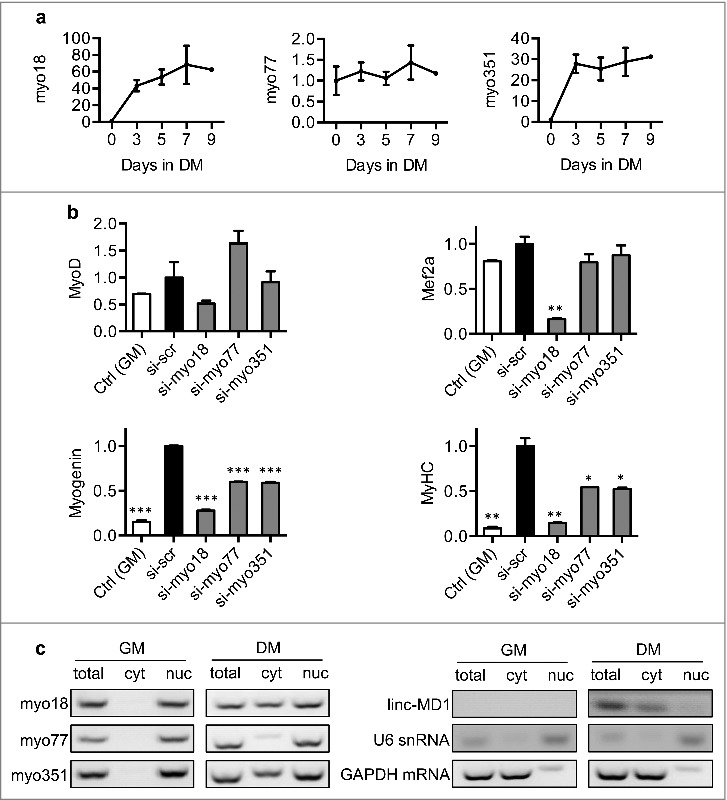
Cellular functions of lncRNAs. (a) Using qRT-PCR, the expressional profiles of myo18, myo77 and myo351 were established at specific stages in the differentiation. Expression levels are shown as fold-changes relative to that of day(0). (b) Effects on the major myogenic factors. The expression of MyoD, Mef2a, Myogenin and MHC were measured in siRNA-transfected cells. Gene-silencing activity was calculated relative to that of scrambled siRNA-treated cells. Data are presented as mean ±SD. Student t test was performed; *P< 0.05; **P< 0.01; ***P< 0.001. (c) Cellular distribution of lncRNAs. Using RT-PCR, lncRNA levels were determined in myoblasts or differentiated myocytes. Total RNA samples, nuclear (nuc) and cytoplasmic (cyt) RNA fractions were individually examined. U6 snRNA and GAPDH mRNA were include as references.
We next examined their effects on the expression of the major myogenic factors, including MyoD, Mef2a, Myogenin and MHC. Compared to the untreated C2C12 cells, silencing the expression of myo18 led to down-regulation of all the factors while the knockdown of myo77 and myo351 only affected the expression of myogenin and MHC (Fig. 8b).
As LncRNAs function through distinct molecular mechanisms occurring either in the nucleus or cytoplasm, understanding their cellular distribution may shed light on their functional roles [21–23]. Accordingly, subcellular localization of myo18, myo77 and myo351 were examined in myoblasts and differentiated myocytes. In myoblasts, these lncRNAs were found to localize mainly within the nucleus. However, when differentiation proceeded toward the terminal stage, myo18 and myo351 were found to translocate from the nucleus to the cytoplasm. In contrast, myo77 remained within the nucleus throughout the differentiation process (Fig. 8c).
Additionally, the expression and cellular location of Linc-MD1 were also resolved. Our study showed that: Linc-MD1 was expressed in the differentiated myocytes but not the myoblasts. Also a nucleus-specific localization was revealed for Linc-MD1 transcripts (Fig. 8c).
Discussion
Higher-order chromatin structure emerges as an important issue in DNA replication and gene expression. While early studies have revealed discrete chromosome territories in interphase nuclei, recent studies have reported that each chromosome is comprised of multiple distinct topological associated domains (TADs) [24]. With an average size of one million base pairs, TADs are stable in diverse cell types and evolutionarily conserved in relevant species. Thus, they are regarded as the basic units of DNA replication and chromosome organization [25]. Interestingly, functionally relevant genes such as cytochrome genes, olfactory receptors, and protocadherin genes, are found to be organized into individual TADs, implying that they are transcriptionally co-regulated in particular cellular processes [26].
Despite this progress, another essential issue regarding the functional units of chromosome structure remains to be resolved. Accordingly, the intriguing findings in the present study are that: (1) lineage-specific lncRNAs are distributed in clusters, within distinct genomic domains; (2) these gene clusters are transcriptionally co-regulated in cell differentiation, and thus named TODs; (3) distinct TODs are involved in lineage-specific differentiation.
Among the thousands of intergenic lncRNAs identified in myocyte and adipocyte differentiation, only 39 common lncRNAs were characterized. The other 94% lncRNAs were found to express only in one of the two differentiation processes. Thus, for the first time, our study presents lineage-specific profiles of non-polyadenylated lncRNAs. This will help to establish a comprehensive regulation network involving both protein factors and lncRNAs.
Furthermore in myocyte differentiation, our study revealed two functional categories of lncRNAs. Knockdown of their expression led to proliferation or differentiation-specific phenotypes. In addition to these findings, three myogenic lincRNAs were characterized. Inhibiting their expression interfered with not only the differentiation process, but also the myogenic signaling pathway. In support of this finding, abnormal expression of these lncRNAs was also characterized in mdx mice.
In the present study, distinct lncRNA profiles were characterized in lineage-specific cell differentiation, showing that they are actively involved in these processes. Interestingly, some of them are organized in clusters and co-regulated during differentiation. Further examination of myocyte differentiation identified three myogenic lncRNAs. Knockdown of their expression compromised not only the differentiation process but also the essential signaling pathway.
Materials and methods
Ethics statement
Animals were maintained in the Center of Experimental Animals at Peking University. All procedures involving animals were performed in accordance with protocols approved by the Committee for Animal Research of Peking University.
Oligonucleotides
DNA oligonucleotides were purchased from Invitrogen (Beijing, China). RNA oligonucleotides were from RiboBio (Guangzhou, China).
Myocyte differentiation
At a subconfluent density, C2C12 myoblasts were maintained in growth medium (GM) consisting of DMEM supplemented with 10% fetal bovine serum (Gibco) and 1% penicillin/streptomycin (Gibco). To induce myogenic differentiation, the cells of ∼80% confluence were switched to differentiation medium (DM) consisting of DMEM supplemented with 2% horse serum (Invitrogen) and 1% penicillin/streptomycin, at day(0). Then, the culture medium was replaced by fresh differentiation medium daily. Myoblasts were collected at day (0), differentiating myocytes were collected at day(+3), and matured myocytes were collected at day(+5).
Adipocyte differentiation and oil red O staining
3T3-L1 pre-adipocytes were grown at a subconfluent density, in DMEM supplemented with 10% bovine serum. Two days after the cells reached confluence on day(0), they were induced to differentiate by changing the culture medium to DMEM supplemented with 0.5 mM 3-isobutyl-1-methyxanthine (Sigma), 1 μM dexamethasone (Sigma) and 167 nM insulin (Sigma). At the end of day(+1), culture medium was replaced with DMEM supplemented only with 167 nM insulin. At the end of day(+3), insulin was withdrawn and the cells were allowed to grow in DMEM throughout the rest of the differentiation. Preadipocytes were collected at day(−1), the differentiating adipocytes were collected at day(+13), and mature adipocytes were collected at day(+20).
Oil Red O staining was performed to monitor the progression of adipocyte differentiation. Briefly, the cells were washed three times with ice-cold PBS and fixed in 3.7% formaldehyde for 2 min, then incubated with Oil Red O reagent for 1 h at room temperature and washed with water. Oil red O reagent (0.5%) was prepared in isopropanol by mixing with water at a 3:2 ratio and filtering through a 0.45-μm filter. The stained fat droplets in the cells were visualized by light microscopy and photographed.
RNA preparation and sequencing
Total RNAs were isolated using the TRIzol reagent (Sigma-Aldrich). To isolate polyA-minus RNA components, eight micrograms of purified total RNAs were subjected to sequential depletion of rRNA, mRNA and short RNA species with an rRNA depletion kit (Jianchengda Inc., Beijing), resulting in pools of polyA-minus RNA. cDNA libraries were prepared starting from 2 µg of polyA-minus RNA, using random hexamer priming (Invitrogen). Sequencing libraries were prepared according to the single-end sample preparation protocol, and were sequenced using Illumina HiSeq™ 2000 Sequencing System by Berry Genomics Inc (Beijing, China).
Using TopHat program (v1.2.0), the sequencing reads were mapped to the mouse reference genome (NCBI37/mm9). The mapped reads were then filtered against the RepeatMask and Ensembl gene sets, leading to the identification of novel intergenic transcription regions. To evaluate the coding potential of each transcription region, a CPC (coding potential calculator) assay was performed. For each transcription region, an FPKM value was calculated to quantify its expressional abundance and variations, using cufflinks v1.0.3.
RNAi screening
C2C12 cells were seeded in 12-well dishes one day before transfection. After the cells reached ∼50% confluence, individual siRNA against lncRNA or scrambled control siRNA were transfected at a final concentration of 40 nM in growth medium, using Lipofectamine RNAimax (Invitrogen). After the cells reached ∼80% confluence, they were induced to myogenic differentiation by replacing growth medium to differentiation medium.
Immunofluorescence and high content screening assays
Prior to the assays, C2C12 cells grown in 12-well dishes were transfected with 40 nM siRNA, and cultured for 5 days in differentiation medium. Then the cells were fixed in 4% formaldehyde (Thermo Scientific) for 15 min, permeabilized in 1% Trioton X-100 (Thermo Scientific) for 15 min, blocked in 1% Albumin Bovine V (Amresco, USA) for 10 min, and incubated in monoclonal anti-myosin antibody solution (1:400; Sigma-Aldrich) for 2 h at 37°C. Then, the cells were washed with PBS and incubated with anti-mouse IgG-FITC antibody solution (1:50; Sigma-Aldrich) for 1 h at 37°C. Following this, the cells were washed again with PBS, stained the cells with Hoechst 33342 (Mac gene, China) for 20 min, and mounted. To assess myogenic differentiation, the number of Hoechst 33342-stained nuclei within the myosin positive cells (multinucleated cells, i.e., ≥2 nuclei) was determined and expressed as a percentage of the total number of nuclei analyzed per image. Immunofluorescence images were captured using Operetta High Content Imaging System (PerkinElmer, USA), and analyzed by Columbus Image Data Storage and Analysis System (PerkinElmer, USA).
Semi-quantitative RT-PCR and real-time qRT-PCR assay
Total RNA was isolated using TRIzol reagent (Sigma-Aldrich), from C2C12 cells or mouse tissues. Using PARIS™ Kit Protein and RNA Isolation System (Ambion, Grand Island), isolation of cytoplasmic and nuclear RNA was performed according to the manufacturer's instructions. cDNAs were synthesized with lncRNA-specific (Tiangen, Beijing) for determination of transcription orientation or random primer (Takara, Japan) for quantification and reverse transcriptase TIANScript M-MLV (Tiangen, Beijing, China). Semi-quantitative PCR amplification was carried out with a TC-5000PCR Thermal Cycler (TECHNE, UK), and PCR products were checked on 2% agarose gel. Real time qRT-PCR was performed with a Stratagene Mx3005P qPCR Systems (Agilent Technologies) using GoTaq® qPCR Master Mix (Promega). Relative expression levels were calculated by the comparative threshold cycle (ΔΔCT) methods, using 18s rRNA as an internal control. Primer sequences are listed in Supplementary Table.
Statistical analysis
All the experiments were repeated for at least three times. Statistical analysis was performed with GraphPad Prism (version 6) and MATLAB R2016b. Data were analyzed by 2-tailed Student's t test, and presented as means ± SD. P<0.05 was considered statistically significant.
Supplementary Material
Funding Statement
This work was supported by the National Natural Science Foundation of China (NSFC), 31571403; Beijing Natural Science Foundation, 2171001.
Abbreviations
- DM
differentiation medium
- FPKM
Reads Per Kilobase of per Million mapped reads
- GM
growth medium
- lncRNA
long noncoding RNA
- lincRNA
long intergenic noncoding RNAs
- MHC
myosin heavy chain
- siRNA
small interference RNA.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
This work was supported by the grants from the National Natural Science Foundation of China (31571403), Beijing Natural Science Foundation (2171001). The authors thank Dr. Yong Zhang (Institute of Basic Medical Sciences, Peking Union Medical College, Beijing) for providing C2C12 cells.
Author contributions
Q.Y. carried out most of the experiments and helped to draft the manuscript. L.Z. performed the experiments of real-time qRT-PCR. Y.L. carried out the bioinformatics analysis. D.L. and C.F carried out the immunofluorescence. F.Y., L.Z. and L.Z participated in the design of study. Q.D. convinced of the study, participated in the design and drafted the manuscript. All the authors read and approved the final manuscript.
References
- [1].Zhu L, Skoultchi AI. Coordinating cell proliferation and differentiation. CurrOpin Genet Dev. 2001;11:91–97. doi: 10.1016/S0959-437X(00)00162-3 [DOI] [PubMed] [Google Scholar]
- [2].Yokoyama S, Asahara H. The myogenic transcriptional network. Cell Mol Life Sci. 2011;68:1843–1849. doi: 10.1007/s00018-011-0629-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Prokesch A, Hackl H, Hakim-Weber R, et al.. Novel Insights into Adipogenesis from Omics Data. Curr Med Chem. 2009;16:2952–2964. doi: 10.2174/092986709788803132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Berkes CA, Tapscott SJ. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol. 2005;16:585–595. doi: 10.1016/j.semcdb.2005.07.006 [DOI] [PubMed] [Google Scholar]
- [5].Mousavi K, Zare H, Dell'orso S, et al.. eRNAs Promote Transcription by Establishing Chromatin Accessibility at Defined Genomic Loci. Molecular Cell. 2013;51:606–617. doi: 10.1016/j.molcel.2013.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Caretti G, Schiltz RL, Dilworth FJ, et al.. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Developmental Cell. 2006;11:547–560. doi: 10.1016/j.devcel.2006.08.003 [DOI] [PubMed] [Google Scholar]
- [7].Kallen AN, Zhou XB, Xu J, et al.. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell. 2013;52:101–112. doi: 10.1016/j.molcel.2013.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Schuster-Gossler K, Bilinski P, Sado T, et al.. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev Dynam. 1998;212:214–228. doi: 10.1002/(SICI)1097-0177(199806)212:2%3c214::AID-AJA6%3e3.0.CO;2-K [DOI] [PubMed] [Google Scholar]
- [9].Zhou YL, Cheunsuchon P, Nakayama Y, et al.. Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development. 2010;137:2643–2652. doi: 10.1242/dev.045724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cesana M, Cacchiarelli D, Legnini I, et al.. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wang J, Gong C, Maquat LE. Control of myogenesis by rodent SINE-containing lncRNAs. Genes Dev. 2013;27:793–804. doi: 10.1101/gad.212639.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Neguembor MV, Jothi M, Gabellini D. Long noncoding RNAs, emerging players in muscle differentiation and disease. Skelet Muscle. 2014;4:8. doi: 10.1186/2044-5040-4-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ballarino M, Morlando M, Fatica A, et al.. Non-coding RNAs in muscle differentiation and musculoskeletal disease. J Clin Invest. 2016;126:2021–2030. doi: 10.1172/JCI84419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Nie M, Deng ZL, Liu J, et al.. Noncoding RNAs, Emerging Regulators of Skeletal Muscle Development and Diseases. Biomed Res Int. 2015;2015:676575. doi: 10.1155/2015/676575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Simionescu-Bankston A, Kumar A. Noncoding RNAs in the regulation of skeletal muscle biology in health and disease. Journal of Molecular Medicine. 2016;94:853–866. doi: 10.1007/s00109-016-1443-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Yaffe D,Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0 [DOI] [PubMed] [Google Scholar]
- [17].Guo X, Liao K. Analysis of gene expression profile during 3T3-L1 preadipocyte differentiation. Gene. 2000;251:45–53. doi: 10.1016/S0378-1119(00)00192-X [DOI] [PubMed] [Google Scholar]
- [18].Yi F, Yang F, Liu X, et al.. RNA-seq identified a super-long intergenic transcript functioning in adipogenesis. RNA Biol. 2013;10:991–1001. doi: 10.4161/rna.24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Olson EN. Regulation of muscle transcription by the MyoD family. The heart of the matter. Circ Res. 1993;72:1–6. doi: 10.1161/01.RES.72.1.1 [DOI] [PubMed] [Google Scholar]
- [21].Tye CE, Gordon JA, Martin-Buley LA, et al.. Could lncRNAs be the missing links in control of mesenchymal stem cell differentiation? J Cell Physiol. 2015;230:526–534. doi: 10.1002/jcp.24834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36:25–64. doi: 10.1210/er.2014-1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15:7–21. doi: 10.1038/nrg3606 [DOI] [PubMed] [Google Scholar]
- [24].Dixon JR, et al.. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dekker J, Heard E. Structural and functional diversity of Topologically Associating Domains. FEBS letters. 2015;589:2877–2884. doi: 10.1016/j.febslet.2015.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nora EP, Lajoie BR, Schulz EG, et al.. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


