Abstract
Epidermal growth factor receptor (egfr) tyrosine kinase inhibitors (tkis) are recommended as first-line systemic therapy for patients with non-small-cell lung cancer (nsclc) having mutations in the EGFR gene. Resistance to tkis eventually occurs in all nsclc patients treated with such drugs. In patients with resistance to tkis caused by the EGFR T790M mutation, the third-generation tki osimertinib is now the standard of care. For optimal patient management, accurate EGFR T790M testing is required. A multidisciplinary working group of pathologists, laboratory medicine specialists, medical oncologists, a respirologist, and a thoracic radiologist from across Canada was convened to discuss best practices for EGFR T790M mutation testing in Canada. The group made recommendations in the areas of the testing algorithm and the pre-analytic, analytic, and post-analytic aspects of clinical testing for both tissue testing and liquid biopsy circulating tumour dna testing. The recommendations aim to improve EGFR T790M testing in Canada and to thereby improve patient care.
Keywords: Non-small-cell lung cancer, nsclc, epidermal growth factor receptor, EGFR T790M, cell-free tumour dna, ctdna
INTRODUCTION
Lung cancer is the leading cause of death from cancer worldwide, resulting in approximately 1.7 million deaths in 20151. Non-small-cell lung cancer (nsclc) accounts for most lung cancer cases2. Patients often have advanced disease at diagnosis of nsclc; of those who do not, 30%–60% will progress to advanced disease. The 5-year survival rate for nsclc patients at all stages is only 18%3.
During the last several years, a greater understanding of oncogenic driving mutations in nsclc has led to the development of new therapies. Compared with standard chemotherapy, the use of biomarker testing to guide treatment in nsclc has resulted in superior patient outcomes. For example, patients with EGFR sensitizing mutations treated with a first-line tyrosine kinase inhibitor (tki) have experienced significantly longer progression-free survival and improved quality of life4–7. Sensitizing mutations in EGFR such as L858R and exon 19 deletions cause constitutive activation of the epidermal growth factor receptor (egfr) and its downstream signalling pathways; they also decrease the affinity of the kinase for adenosine triphosphate (atp)8,9. The tkis compete with atp for binding to the kinase active site, and tki binding causes inhibition of egfr activity.
Guidelines encourage testing for EGFR mutations at the time of diagnosis of nonsquamous nsclc regardless of stage (“reflex testing”)10. Implementation of reflex testing results in better identification of patients who are eligible for first-line tkis and reduces the time to optimal systemic therapy11. However, all patients treated with a first-line tki eventually experience progression of their cancer, with a median time to progression of 8–13 months5–7.
Several mechanisms of resistance to first-line tki therapy have been identified; the most common resistance mechanism is the EGFR T790M mutation, occurring in approximately 60% of nsclcs progressing or relapsing on first-line therapy12. The EGFR T790M mutation restores the affinity of the kinase for atp to wild-type levels, and therefore at cellular atp levels, the potency of the tki is reduced. In rare cases, rather than arising through tki exposure, EGFR T790M can be found as a germline mutation that can potentially increase the risk of developing lung cancer13,14. Other resistance mechanisms include amplification of met and her2, and mutations of PIK3CA and BRAF15.
Osimertinib is a third-generation tki that is selective for both EGFR sensitizing mutations and EGFR T790M. Patients with advanced nsclc progressing on first-line tki therapy who received osimertinib in the aura3 clinical trial experienced improved response rates and longer duration of progression-free survival than did those receiving platinum–pemetrexed chemotherapy16. On the basis of those and other phase i/ii data, osimertinib was approved by Health Canada in 2016 for patients with locally advanced or metastatic EGFR T790M mutation–positive nsclc. Two kits have been approved by Health Canada for the detection of EGFR T790M in plasma and tissue biopsies17,18. Appropriately validated laboratory-developed tests can also be used for EGFR T790M mutation detection.
To appropriately target treatment after nsclc progression, all patients who progress on first-line egfr tkis should undergo testing to determine presence of the T790M mutation in the tumour19. Although tissue biopsy is generally considered the “gold standard,” tumour heterogeneity can mean that T790M status varies at different tumour sites20. Depending on the tumour sites at progression, it might be difficult to obtain a new tissue biopsy for T790M testing. As an alternative, testing for EGFR T790M in a liquid biopsy using cell-free circulating tumour dna (ctdna)21–24 from a plasma sample can also be a valid approach. The patient outcomes achieved with osimertinib therapy are similar whether T790M is identified by dna testing of a tissue biopsy or plasma ctdna24. At the time of writing, cerebrospinal fluid and urine were not well-validated sample types for EGFR T790M mutation detection.
The purpose of the present review is to provide evidence-based best-practice recommendations for EGFR T790M testing in the Canadian context. Optimizing EGFR T790M testing upon patient progression will improve patient management by ensuring accurate identification of those who can benefit from T790M-directed therapy or a third-generation egfr tki that targets T790M. Issues in EGFR T790M testing considered here include the testing algorithm; pre-analytic, analytic, and post-analytic considerations; and quality assurance.
WORKING GROUP AND METHODS
In June 2017, a Canadian multidisciplinary working group was convened to discuss the current state of EGFR T790M testing across Canada and to make recommendations for best practices into the future. Medical specialties represented were pathology and laboratory medicine, molecular genetics, medical oncology, thoracic radiology, and respirology. The initial input for the recommendations in the present report emerged from the discussion at the working group meeting; the recommendations were then refined and supplemented with information from a detailed literature review by the authors.
ISSUES CONSIDERED BY THE WORKING GROUP PARTICIPANTS
Testing Algorithm
Patients with EGFR sensitizing mutations treated with a first- or second-generation egfr tki should be tested for T790M at the time of clinical or radiographic progression, and the results should guide treatment decisions. Testing for the presence of EGFR T790M as a mechanism of resistance can be performed on either ctdna from plasma or dna from tissue.
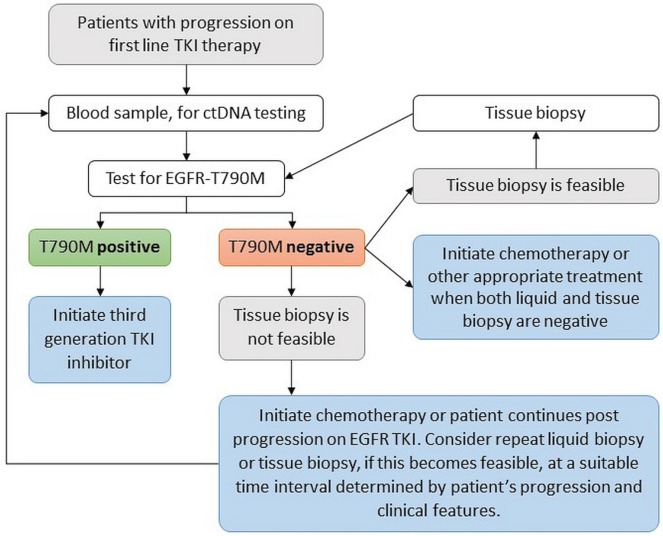
A liquid biopsy analyzes small fragments of cell-free ctdna that is shed into the blood. The test can be performed quickly and with little morbidity for the patient; it can be the first choice to assess EGFR T790M status (Figure 1). However, liquid biopsies pose challenges. They rely on the detection of the mutation in small amounts of cell-free ctdna present in a sample with a background of wild-type dna. Thus, the assay method used must have a lower limit of detection (lod) appropriate to detect a small percentage of T790M. In addition, the blood collection, processing, storage, and transport must be optimized for stability of the ctdna and minimization of genomic dna contamination.
FIGURE 1.
Assessment of EGFR T790M status. TKI = tyrosine kinase inhibitor; ctDNA = circulating tumour DNA; EGFR = epidermal growth factor receptor.
A positive result for T790M from a ctdna assay can be an indication for T790M-directed therapy; however, a negative result might reflect either the absence of T790M mutation (true negative) or a false negative resulting from a lack or a minimal amount of ctdna available to be detected by the assay. Thus, a repeat blood sample might be of benefit, although the optimal timing of a repeat blood collection is currently unknown. A negative liquid biopsy is recommended to be followed by a tissue biopsy if feasible, because of the risk of an uninformative ctdna result. For example, if the sample is negative for EGFR T790M, but also negative for the patient’s original EGFR sensitizing mutation, the results are uninformative, because they cannot confirm that ctdna from the tumour was tested24.
If tissue biopsy is not possible, consideration should be given to repeating the liquid biopsy. Even with a negative result in both tissue and liquid biopsies, EGFR T790M testing could be repeated, because T790M status is not static, and a mutation that was not detectable initially upon progression might become detectable over time, particularly if patients are maintained on egfr tki therapy beyond progression. Progression is highly individual; the optimal timing of a repeat liquid biopsy therefore depends on the extent of disease, the progression rate, and the individual clinical features of the patient. However, repeating the T790M assay in the face of a negative result yields diminishing returns, and consideration should be given to cost-effectiveness in determining how many repeat liquid or tissue biopsies to perform.
Liquid and tissue biopsy results are complementary. A positive result in either test can be an indication for using a third-generation egfr tki25, although ultimately, the choice of therapy is a clinical decision by the oncologist, individualized based on the patient’s clinical status and preference.
Although more research is required to define populations of patients that are more likely to receive positive T790M results from liquid biopsies, some evidence suggests that T790M can be detected more frequently in plasma from patients with metastatic or extrathoracic disease than from those with locally advanced disease25. Detection of the EGFR sensitizing mutations in plasma correlates with the number of metastatic sites, the number of lesions, and the sum of diameters for measurable lesions26, but no such correlation has yet been shown for T790M.
If a liquid biopsy is performed first and is negative, then in addition to retesting for T790M, consideration should be given to testing for other possible resistance mechanisms in the repeat tissue biopsy (see the Introduction), if such tissue is available to the oncologist. If a patient is negative for T790M, but positive for another resistance mutation upon repeat tissue testing, then the oncologist does not have to repeat testing for the EGFR T790M mutation.
Table i summarizes the testing algorithm recommendations.
TABLE I.
Summary of testing algorithm recommendations
| ■ EGFR T790M mutation testing should be undertaken in patients with EGFR sensitizing mutation–positive NSCLC who progress on first- or second-generation EGFR TKI therapy. A positive T790M result from either liquid or tissue biopsy can be used for clinical management. |
| ■ Liquid biopsy can be performed first. Tissue biopsy can follow if the liquid biopsy is negative for T790M. |
| ■ If plasma and tissue results are both negative, testing could be repeated at a later date, because the acquisition of, and ability to detect, the T790M variant can evolve. |
NSCLC = non-small-cell lung cancer; EGFR = epidermal growth factor receptor; TKI = tyrosine kinase inhibitor.
Pre-analytic Considerations
Test Requisitions and Information Needed in Advance of Testing
When the oncologist orders a biopsy at patient progression, background information about the patient might not be readily available to the radiologist, thoracic surgeon, pathologist, and laboratory. To ensure that key information is transmitted along the testing pathway, requisitions must include relevant information about the patient such as clinical information, current disease status, and prior relevant test results. For tissue biopsy, the requisition should include either a request to biopsy a specific lesion or instructions to the radiologist to biopsy a growing lesion. Importantly, requisitions (including electronic orders) must specify that the patient has progressed on egfr tki therapy and that the test is being requested specifically for EGFR T790M mutation diagnosis. That approach allows for clear communication between the various specialties and ensures that unnecessary tests (for example, full pathology work-up for diagnosis of nsclc) are not performed, thus preserving the specimen for specific investigations.
For liquid biopsies, the patient’s original EGFR sensitizing mutation should be included on the requisition so that the laboratory can, as needed, test for the presence of that mutation as an internal control. Testing for the original sensitizing mutation confirms that the tumour is shedding ctdna at a level that is detectable by the assay used. If the original EGFR sensitizing mutation is a rare variant, the lab might not offer the associated ctdna assay.
Blood Collection
For liquid biopsies, the methods of blood collection, processing, transport, and storage can affect the mutation detection rate. Liquid biopsies depend on detecting the mutation of interest in ctdna, which generally constitutes only a small fraction of the total cell-free dna. In addition, as time elapses between blood collection and processing to plasma, genomic dna is released from the lysis of white blood cells in the sample, increasing the wild-type background on which the mutation must be detected. The result can be a decrease in the mutant allele fraction within the sample, causing it to fall below the lod of the assay.
Certain practices in blood processing, storage, and transport can optimize the detection of mutations in ctdna from blood samples. Cell-free blood collection tubes contain chemicals that prevent the lysis of white blood cells and the subsequent release of genomic dna and nucleases27–29. Two tubes of blood typically provide a sufficient quantity of plasma (8–10 mL) for ctdna analysis. Use of edta tubes is also a possibility, provided that the plasma is isolated within 6–48 hours of collection27–29. In addition, the use of two centrifugation steps, initially at low speed and then at high speed, reduces the genomic dna contamination of the isolated plasma30.
Shipping and storage temperatures are critical to optimizing mutation detection in ctdna. Blood samples shipped in cell-free blood collection tubes held below 10°C or above 40°C show increased contamination by genomic dna31. Care must be therefore be taken to ensure that the appropriate temperature range is maintained during shipping.
Tissue Collection
Several studies have shown that needle biopsy is a minimally invasive, safe, and accurate technique for tissue biopsy; it is also reliable in the assessment of progression in nsclc20,32,33. That reliability is particularly important in the setting of advanced lung cancer, because many patients have a poor performance status34.
A successful mutation analysis relies on a high enough tumour content or numbers of tumour cells and the dna yield. Fine-needle aspiration (fna) for cytology and core-needle biopsy (cnb) both have a high diagnostic yield and provide adequate tissue or numbers of tumour cells for molecular analysis35,36. As demonstrated in earlier studies, success rates for molecular testing are similar whether based on cytology (cell block) samples obtained with fna or samples obtained with cnb35,37. In fact, there is evidence to suggest that fna samples might be superior to cnb samples, possibly because of the relatively higher proportion of tumour dna present in well-obtained fna samples compared with cnb sections. Notably, however, the success rate for fna is influenced by the experience of the operator performing the biopsy and the availability of rapid on-site evaluation.
The oncologist should specify to the respirologist, radiologist, or thoracic surgeon performing the procedure the target lesion to be biopsied. Enlarging lesions should be targeted if feasible, because tumour growth might indicate resistance to the current therapy. The EGFR T790M mutation has been found at a variety of tumour sites, including lung, lymph nodes, bone, liver, and brain20. One study reported that the prevalence of the T790M mutation was higher in specimens of lung, pleura, and lymph nodes than in distant sites, although T790M mutations were still found in 44% of specimens from distant sites overall32. Even when the T790M mutation is part of the genetic landscape of the cancer in a particular patient, it might not be found in all tumour sites. Intratumoural heterogeneity for the T790M mutation is common: In one study, 43% of the patients who had a T790M mutation at one site did not have the mutation at a second site20.
Biopsies of bone should be avoided if possible, because the resulting yield of tumour cells is generally lower. If a bone biopsy is required, soft-tissue components of the lesion should be targeted, and to preserve the integrity of the dna for molecular analysis, it is important that samples not be acid-decalcified. Fresh or formalin-fixed biopsies of bone metastases treated using a surface decalcification procedure can be used for EGFR T790M testing.
All tissue biopsy samples should be reviewed by a pathologist to determine sample suitability in terms of tumour content and viability. Given that one potential mechanism of resistance to egfr tkis is transformation to small-cell lung cancer, the tissue biopsy sample should be assessed before molecular analysis to confirm that small-cell transformation has not occurred.
The recommended tumour content varies depending on the assay method used and the lower lod of the method. Generally, 1%–5% cancer cells are required for droplet digital polymerase chain reaction (pcr) assays, 10% for real-time pcr assays, and 20%–30% for next-generation sequencing (ngs) assays. Validation of those numbers for each laboratory’s specific assay is required. Good communication between the members of the multidisciplinary team is needed to ensure that the oncologist and pathologist know the tissue requirements for the threshold of the assay that will be used.
Table ii summarizes the pre-analytic recommendations.
TABLE II.
Summary of pre-analytic recommendations
| ■ For liquid biopsies, two 10 mL tubes of blood should be collected. Cell-free blood collection tubes should be used, but if EDTA tubes are used, plasma must be isolated within 6–48 hours of blood collection. |
| ■ For tissue biopsies, core-needle or cytology specimens can be used. |
| ■ If possible, tissue biopsies should target a growing lesion. |
| ■ Samples should be reviewed by a pathologist to confirm that the tumour content of the sample is sufficient for molecular analysis. |
| ■ Bone biopsies are not recommended. If bone biopsies are taken, samples should be decalcified using a surface decalcification procedure only. |
Analytic Considerations
As is the case for any mutation detection assay for clinical use, validation studies should be conducted using appropriate mandated guidelines under regional laboratory accreditation requirements38–40.
To create a dilution series of control samples for validation of ctdna assays, EGFR T790M mutant and EGFR wild-type cell lines can be used. Laboratories should consider the similarity of the control-sample material to endogenous ctdna, such as mimicking the length of ctdna (approximately 130–180 bp41,42). In addition, clinically relevant plasma dna samples (mutant and normal) should be used for validation. To facilitate collection of such samples, laboratories currently receiving plasma samples for ctdna analysis from nsclc patients are encouraged to obtain and save extra plasma from primary diagnosis samples and recurrence or progression samples.
The sensitivity and specificity for detecting the EGFR T790M mutation in plasma, using detection of the mutation in tissue biopsies as the reference technique, varies depending on the method of analysis. Droplet digital pcr and ngs methods have higher sensitivity than real-time pcr methods: the sensitivity of droplet digital pcr and ngs can be 70%–93%, whereas the sensitivity of real-time pcr methods varies from 29% to 73%. Specificity varies from 58% to 100%24,25,43–46; studies in which specificity was low likely used tissue biopsy as the reference. Because of tumour heterogeneity, a tissue biopsy from one tumour might not reflect the total mutational landscape of a patient’s cancer; however, assays detecting T790M in plasma are evaluating the presence or absence of the mutation in all the circulating dna shed by the tumours. In studies of the specificity of T790M mutation detection in plasma that found discordant results (T790M being positive in plasma and negative in tissue), analysis of the mutation in plasma by a second, more sensitive method of analysis in most cases verified the positive result24,25. In addition, some studies have shown that plasma-positive, tissue-negative patients respond to osimertinib, also suggesting that the plasma results are true positives47. All assay methods for EGFR T790M mutation detection in plasma tend to be more prone to false negatives than to false positives, which is the reason for the general recommendation that a negative result from an analysis of ctdna be followed by a tissue biopsy or a repeat ctdna test24.
Because tissue and liquid biopsies can, for reasons discussed earlier, both yield relatively small amounts of tumour dna, and because not all dna analyzed will contain the T790M mutation, the method used for mutation analysis must be sensitive enough to detect low levels of the T790M mutation on a background of T790 wild-type dna. Although measuring the lod in terms of copies per microlitre is a more standard approach in lod determinations, the variant allele fraction (percentage of mutant alleles per total alleles at that locus) is commonly used in clinical practice. The lod should be defined at a given amount of input dna and should, at a minimum, be between 0.2% and 0.5% (that is, 0.2%–0.5% variant allele on a background of 99.8%–99.5% wild-type allele). One study compared the ability of 3 different platforms to reliably detect the T790M mutation in a reference set of samples with variant allele fractions of 5%, 1%, and 0.1%, based on 15 ng input ctdna48. In that study, digital pcr was able to reliably detect the 0.1% variant allelic fraction using 5 dye-positive droplets, ngs was able to detect the 0.2% variant allele fraction using 20 variant reads (minimum of 5 variant reads in each direction), and quantitative pcr was able to reliably detect a 1% variant allele fraction. Using ngs, T790M was found in patient samples at variant allele frequencies of 0.3%–21.0%48.
If the patient has a common EGFR sensitizing mutation, such as L858R or exon 19 deletion, the sensitizing mutation should, as needed, be tested as an internal control to confirm that ctdna from the tumour can be detected in the dna extracted from peripheral blood. In patients who have developed resistance during egfr tki therapy, the sensitizing mutation would be expected to be present at a higher frequency than would T790M, although the detection of individual mutations by the lab’s assays can vary in efficiency. Laboratories should determine, using standardized controls and patient samples, the threshold of detection of the sensitizing mutations relative to the detectable variant allele fraction of T790M.
Table iii summarizes the analytic recommendations.
TABLE III.
Summary of analytic recommendations
| ■ Laboratories can use the method of analysis that best fits their lab, provided that the lower limit of detection is 0.2%–0.5%. |
| ■ Assays should be appropriately validated and performed in labs accredited to perform clinical testing. |
| ■ EGFR sensitizing mutations should, when possible, be tested as an internal control to ensure that ctDNA is detectable in the sample. |
ctDNA = circulating tumour DNA.
Post-analytic
Reporting Elements
The laboratory report should state whether the sample was positive, negative, inconclusive, or failed. (Inconclusive results are obtained when the controls do not give the expected results and therefore the sample cannot conclusively be called positive or negative.) It should state which variants were tested and the lod. For quantitative methods, the report should include the variant allele fraction (or copies per microlitre) of T790M and any other mutations tested. When pathology and molecular testing are performed at the same centre, the pathology and molecular reports should be linked in the laboratory information system. If T790M has not previously been analyzed for a patient, and if it is found at a fraction consistent with an inherited mutation, the report should indicate that its presence could reflect a germline mutation and that referral to a medical geneticist is recommended.
For liquid biopsies, when only T790M is tested and is negative (for example, when confirmation of a rare EGFR sensitizing mutation is not possible), the report should include the possibility that the “undetectable” result might reflect an insufficient amount of circulating tumour dna. If the patient’s EGFR sensitizing mutation is tested and is positive, and if T790M is negative, the report should include the caveat that there is a rare possibility that the T790M mutation is present in the tumour, but cannot be detected by the ctdna test. If the EGFR sensitizing mutation and T790M tests are both negative, the test is then inconclusive or uninterpretable, and a repeat ctdna assay or tissue biopsy should be considered. For tissue biopsies, the report should include the microscopically estimated cellularity of the sample, the tumour content, and any other pre-analytic issues such as necrosis that might affect the result.
Turnaround Time
Given that the need for treatment is urgent, the turnaround time for results to the oncologist must be fast enough to facilitate treatment decisions. Ideally, the turnaround time from receipt of the sample at the laboratory should be within 10 working days. The time required for shipping the sample, if it is to be sent to a reference lab, should be taken into consideration by the oncologist.
Quality Assurance
As is the case for any molecular assay, robust standard operating procedures, reagent qualification, and a quality assurance framework should be in place. All laboratory standards typically required by regional laboratory accreditation bodies for quality control and quality assurance of the assays should be used. Laboratories should participate in external proficiency testing programs, or acceptable alternatives as allowed by laboratory accreditation, for EGFR T790M assays.
CONCLUSIONS
Accurate identification of EGFR T790M status in patients with advanced nsclc is important for guiding patient care and the appropriate use of osimertinib. The field of nsclc testing is evolving rapidly with the recent advent of mutation-specific therapies and liquid biopsy testing. Laboratories desiring to incorporate this testing into their menu might need guidance about how to do so. Currently, no global guidelines have emerged specifically for EGFR T790M testing, although Australian recommendations are available49. Recently, new guidelines for using molecular testing to select lung cancer patients for tki therapy were published50; they include recommendations about testing for various biomarkers in lung cancer patients. Those recommendations accord with ours, in that they recommend a new biopsy to test for T790M status when patients progress on tki therapy, and that ctdna assays, if sufficiently sensitive, can be used for that purpose. Our review further provides a detailed overview of T790M testing from the testing algorithm to the pre-analytic, analytic, and post-analytic aspects of testing.
Liquid biopsy testing can provide valuable information about a patient’s T790M mutation status; however, the sensitivity and lower lod vary between the testing methods. Assay validation should unequivocally demonstrate mutant allele detection at a low mutant allele fraction. A negative result from either a tissue or a liquid biopsy does not conclusively indicate lack of a T790M variant; repeat testing should therefore be considered regardless of the initial method used. A multidisciplinary approach to T790M testing and reporting is important to ensure optimal communication from the oncologist through to the laboratory and back to the oncologist—from requisition to report.
ACKNOWLEDGMENTS
We thank the participants of the National Working Group meeting for their contributions. In addition to the authors, the participants included Shantanu Banerji (Department of Medical Oncology, CancerCare Manitoba, and Department of Internal Medicine, Rady Faculty of Health Sciences, University of Manitoba, Winnipeg, MB), Quincy Chu (Cross Cancer Institute and University of Alberta, Edmonton, AB), Iyare Izevbaye (Division of Molecular Pathology, Laboratory Medicine and Pathology, University of Alberta Hospital, and Alberta Health Services, Edmonton, AB), Leon van Kempen (McGill University and OPTILAB–McGill University Health Centre, Montreal, QC, and University Medical Center Groningen, Department of Pathology, Laboratory for Molecular Pathology, Groningen, Netherlands), Stephen Lam (BC Cancer, Vancouver, BC), and Danh Tran-Thanh (Centre hospitalier de l’Université de Montréal, Department of Pathology, Montreal, QC).
AstraZeneca Canada supported the working group meeting. Medical writing services, funded by AstraZeneca, were provided by Philippa Bridge-Cook phd, Senior Medical Writer from Precision Rx-Dx Inc.
CONFLICT OF INTEREST DISCLOSURES
We have read and understood Current Oncology’s policy on disclosing conflicts of interest, and we declare the following interests: PKC has received honoraria from AstraZeneca, Hoffmann–La Roche, Bristol–Myers Squibb, Boehringer Ingelheim, Novartis, Eli Lilly, Pfizer, and Merck, and research grants from Hoffmann–La Roche and Boehringer Ingelheim. MST, TS, SKR, and AK have received grants and honoraria from AstraZeneca. CAS, BM, and AS have received honoraria from AstraZeneca. CAS has received honoraria from Pfizer, Boehringer Ingelheim, and Hoffman–La Roche.
REFERENCES
- 1.World Health Organization (WHO) Cancer: fact sheet [Web page] Geneva, Switzerland: WHO; 2018. [Available at http://www.who.int/mediacentre/factsheets/fs297/en/; cited 1 March 2018] [Google Scholar]
- 2.Canadian Cancer Statistics Advisory Committee . Canadian Cancer Statistics 2017. Toronto, ON: Canadian Cancer Society; 2017. [Google Scholar]
- 3.Coleman MP, Forman D, Bryant H, et al. Cancer survival in Australia, Canada, Denmark, Norway, Sweden, and the U.K., 1995–2007 (the International Cancer Benchmarking Partnership): an analysis of population-based cancer registry data. Lancet. 2011;377:127–38. doi: 10.1016/S0140-6736(10)62231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Janne PA, Wang X, Socinski MA, et al. Randomized phase ii trial of erlotinib alone or with carboplatin and paclitaxel in patients who were never or light former smokers with advanced lung adenocarcinoma: calgb 30406 trial. J Clin Oncol. 2012;30:2063–9. doi: 10.1200/JCO.2011.40.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–8. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 6.Mitsudomi T, Morita S, Yatabe Y, et al. on behalf of the West Japan Oncology Group Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (wjtog3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–8. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin–paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 8.Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res. 2015;4:36–54. doi: 10.3978/j.issn.2218-6751.2014.05.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in egfr kinase causes drug resistance by increasing the affinity for atp. Proc Natl Acad Sci U S A. 2008;105:2070–5. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for egfr and Alk tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. Arch Pathol Lab Med. 2013;137:828–60. doi: 10.5858/arpa.2012-0720-OA. [Erratum in: Arch Pathol Lab Med 2013;137:1174] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheema PK, Menjak IB, Winterton-Perks Z, et al. Impact of reflex EGFR/ALK testing on time to treatment of patients with advanced nonsquamous non-small-cell lung cancer. J Oncol Pract. 2017;13:e130–8. doi: 10.1200/JOP.2016.014019. [DOI] [PubMed] [Google Scholar]
- 12.Yu HA, Arcila ME, Rekhtman N, et al. Analysis of tumor specimens at the time of acquired resistance to egfr-tki therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–7. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–16. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 14.Oxnard GR, Miller VA, Robson ME, et al. Screening for germline EGFR T790M mutations through lung cancer genotyping. J Thorac Oncol. 2012;7:1049–52. doi: 10.1097/JTO.0b013e318250ed9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Re M, Vasile E, Falcone A, Danesi R, Petrini I. Molecular analysis of cell-free circulating dna for the diagnosis of somatic mutations associated with resistance to tyrosine kinase inhibitors in non-small-cell lung cancer. Expert Rev Mol Diagn. 2014;14:453–68. doi: 10.1586/14737159.2014.908120. [DOI] [PubMed] [Google Scholar]
- 16.Mok TS, Wu YL, Ahn MJ, et al. on behalf of the aura3 investigators Osimertinib or platinum–pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376:629–40. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vallee A, LeLoup AG, Denis MG. Efficiency of the Therascreen rgq pcr kit for the detection of EGFR mutations in non–small cell lung carcinomas. Clin Chim Acta. 2014;429:8–11. doi: 10.1016/j.cca.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 18.Weber B, Meldgaard P, Hager H, et al. Detection of EGFR mutations in plasma and biopsies from non–small cell lung cancer patients by allele-specific pcr assays. BMC Cancer. 2014;14:294. doi: 10.1186/1471-2407-14-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanna N, Johnson D, Temin S, et al. Systemic therapy for stage iv non-small-cell lung cancer: American Society of Clinical Oncology clinical practice guideline update. J Clin Oncol. 2017;35:3484–515. doi: 10.1200/JCO.2017.74.6065. [DOI] [PubMed] [Google Scholar]
- 20.Arcila ME, Oxnard GR, Nafa K, et al. Rebiopsy of lung cancer patients with acquired resistance to egfr inhibitors and enhanced detection of the T790M mutation using a locked nucleic acid–based assay. Clin Cancer Res. 2011;17:1169–80. doi: 10.1158/1078-0432.CCR-10-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gortais H, Daniel C, Bidard FC, Jeannot E, Callens C, Cabel L. T790M EGFR mutation detection in cerebrospinal fluid and response to osimertinib in a lung cancer patient with meningeal carcinomatosis. J Thorac Oncol. 2017;12:e138–9. doi: 10.1016/j.jtho.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 22.Rong J, Chunhua M, Yuan L, et al. Detected EGFR mutation in cerebrospinal fluid of lung adenocarcinoma patients with meningeal metastasis. Open Med (Wars) 2016;11:93–6. doi: 10.1515/med-2016-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Huang J, Ji D, et al. Utility of urinary circulating tumor dna for EGFR mutation detection in different stages of non–small cell lung cancer patients. Clin Transl Oncol. 2017;19:1283–91. doi: 10.1007/s12094-017-1669-3. [DOI] [PubMed] [Google Scholar]
- 24.Oxnard GR, Thress KS, Alden RS, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:3375–82. doi: 10.1200/JCO.2016.66.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thress KS, Brant R, Carr TH, et al. EGFR mutation detection in ctdna from nsclc patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90:509–15. doi: 10.1016/j.lungcan.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Zhu YJ, Zhang HB, Liu YH, et al. Quantitative cell-free circulating EGFR mutation concentration is correlated with tumor burden in advanced nsclc patients. Lung Cancer. 2017;109:124–7. doi: 10.1016/j.lungcan.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 27.Kang Q, Henry NL, Paoletti C, et al. Comparative analysis of circulating tumor dna stability in k3edta, Streck, and Cell-Save blood collection tubes. Clin Biochem. 2016;49:1354–60. doi: 10.1016/j.clinbiochem.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 28.Sherwood JL, Corcoran C, Brown H, Sharpe AD, Musilova M, Kohlmann A. Optimised pre-analytical methods improve KRAS mutation detection in circulating tumour dna (ctdna) from patients with non-small cell lung cancer (nsclc) PLoS One. 2016;11:e0150197. doi: 10.1371/journal.pone.0150197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothwell DG, Smith N, Morris D, et al. Genetic profiling of tumours using both circulating free dna and circulating tumour cells isolated from the same preserved whole blood sample. Mol Oncol. 2016;10:566–74. doi: 10.1016/j.molonc.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swinkels DW, Wiegerinck E, Steegers EA, de Kok JB. Effects of blood-processing protocols on cell-free dna quantification in plasma. Clin Chem. 2003;49:525–6. doi: 10.1373/49.3.525. [DOI] [PubMed] [Google Scholar]
- 31.Sacher AG, Paweletz C, Dahlberg SE, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016;2:1014–22. doi: 10.1001/jamaoncol.2016.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oxnard GR, Arcila ME, Sima CS, et al. Acquired resistance to egfr tyrosine kinase inhibitors in EGFR-mutant lung cancer: distinct natural history of patients with tumors harboring the T790M mutation. Clin Cancer Res. 2011;17:1616–22. doi: 10.1158/1078-0432.CCR-10-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon HJ, Lee HY, Lee KS, et al. Repeat biopsy for mutational analysis of non–small cell lung cancers resistant to previous chemotherapy: adequacy and complications. Radiology. 2012;265:939–48. doi: 10.1148/radiol.12112613. [DOI] [PubMed] [Google Scholar]
- 34.Hasegawa T, Sawa T, Futamura Y, et al. Feasibility of rebiopsy in non–small cell lung cancer treated with epidermal growth factor receptor–tyrosine kinase inhibitors. Intern Med. 2015;54:1977–80. doi: 10.2169/internalmedicine.54.4394. [DOI] [PubMed] [Google Scholar]
- 35.Shiau CJ, Babwah JP, da Cunha Santos G, et al. Sample features associated with success rates in population-based EGFR mutation testing. J Thorac Oncol. 2014;9:947–56. doi: 10.1097/JTO.0000000000000196. [DOI] [PubMed] [Google Scholar]
- 36.Yao X, Gomes MM, Tsao MS, Allen CJ, Geddie W, Sekhon H. Fine-needle aspiration biopsy versus core-needle biopsy in diagnosing lung cancer: a systematic review. Curr Oncol. 2012;19:e16–27. doi: 10.3747/co.19.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pang B, Dettmer M, Ong CW, et al. The positive impact of cytological specimens for EGFR mutation testing in non– small cell lung cancer: a single South East Asian laboratory’s analysis of 670 cases. Cytopathology. 2012;23:229–36. doi: 10.1111/j.1365-2303.2012.01000.x. [DOI] [PubMed] [Google Scholar]
- 38.Clinical and Laboratory Standards Institute (CLSI) Molecular Methods for Clinical Genetics and Oncology Testing: Approved Guideline. 3rd ed. Wayne, PA: CLSI; 2012. [Google Scholar]
- 39.Clinical and Laboratory Standards Institute (CLSI) Nucleic Acid Sequencing Methods in Diagnostic Laboratory Medicine: Approved Guideline. 2nd ed. Wayne, PA: CLSI; 2014. [Google Scholar]
- 40.Jennings LJ, Arcila ME, Corless C, et al. Guidelines for validation of next-generation sequencing-based oncology panels: a joint consensus recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn. 2017;19:341–65. doi: 10.1016/j.jmoldx.2017.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jahr S, Hentze H, Englisch S, et al. dna fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–65. [PubMed] [Google Scholar]
- 42.Underhill HR, Kitzman JO, Hellwig S, et al. Fragment length of circulating tumor dna. PLoS Genet. 2016;12:e1006162. doi: 10.1371/journal.pgen.1006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reckamp KL, Melnikova VO, Karlovich C, et al. A highly sensitive and quantitative test platform for detection of nsclc EGFR mutations in urine and plasma. J Thorac Oncol. 2016;11:1690–700. doi: 10.1016/j.jtho.2016.05.035. [DOI] [PubMed] [Google Scholar]
- 44.Wu YL, Tong RZ, Zhang Y, et al. Conventional real-time pcr–based detection of T790M using tumor tissue or blood in patients with egfr tki–resistant nsclc. Onco Targets Ther. 2017;10:3307–12. doi: 10.2147/OTT.S136823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y, Shen X, Li R, et al. The detection and significance of EGFR and BRAF in cell-free dna of peripheral blood in nsclc. Oncotarget. 2017;8:49773–82. doi: 10.18632/oncotarget.17937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoshida H, Kim YH, Ozasa H, et al. EGFR T790M detection in circulating tumor dna from non–small cell lung cancer patients using the pna-lna clamp method. Anticancer Res. 2017;37:2721–5. doi: 10.21873/anticanres.11623. [DOI] [PubMed] [Google Scholar]
- 47.Wan R, Wang Z, Lee JJ, et al. Comprehensive analysis of the discordance of EGFR mutation status between tumor tissues and matched circulating tumor dna in advanced non–small cell lung cancer. J Thorac Oncol. 2017;12:1376–87. doi: 10.1016/j.jtho.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 48.Bartels S, Persing S, Hasemeier B, Schipper E, Kreipe H, Lehmann U. Molecular analysis of circulating free dna from lung cancer patients in routine laboratory practice: a cross-platform comparison of three different molecular methods for mutation detection. J Mol Diagn. 2017;19:722–32. doi: 10.1016/j.jmoldx.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 49.John T, Bowden JJ, Clarke S, et al. Australian recommendations for EGFR T790M testing in advanced non–small cell lung cancer. Asia Pac J Clin Oncol. 2017;13:296–303. doi: 10.1111/ajco.12699. [DOI] [PubMed] [Google Scholar]
- 50.Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med. 2018 doi: 10.5858/arpa.2017-0388-CP. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]