Abstract
In this study, we used Pacific Biosciences RS II long-read and Illumina HiScanSQ short-read sequencing technologies for the characterization of porcine circovirus type 1 (PCV-1) transcripts. Our aim was to identify novel RNA molecules and transcript isoforms, as well as to determine the exact 5′- and 3′-end sequences of previously described transcripts with single base-pair accuracy. We discovered a novel 3′-UTR length isoform of the Cap transcript, and a non-spliced Cap transcript variant. Additionally, our analysis has revealed a 3′-UTR isoform of Rep and two 5′-UTR isoforms of Rep’ transcripts, and a novel splice variant of the longer Rep’ transcript. We also explored two novel long transcripts, one with a previously identified splice site, and a formerly undetected mRNA of ORF3. Altogether, our methods have identified nine novel RNA molecules, doubling the size of PCV-1 transcriptome that had been known before. Additionally, our investigations revealed an intricate pattern of transcript overlapping, which might produce transcriptional interference between the transcriptional machineries of adjacent genes, and thereby may potentially play a role in the regulation of gene expression in circoviruses.
Keywords: Porcine circovirus, Transcriptome, Long-read sequencing, Isoform sequencing, Short-read sequencing, PacBio sequencing, Transcriptional interference
1. Background
Porcine circovirus type 1 (PCV-1) belongs to the Circovirus genus within the Circoviridae family. The virion was discovered by Tischer and colleagues (Tischer et al., 1974) as a contaminant particle of the porcine kidney epithelial cell line (PK-15). The virus has a 1759 nucleotide long, circular single stranded DNA molecule surrounded by a non-enveloped icosahedral capsid (LeCann et al., 1997; Tischer et al., 1982). The PCV-1 genome is one of the shortest among the DNA viruses. The single origin of DNA replication (Ori) of the virus forms a “stem-loop” structure and is bracketed by two genes, rep (ORF1) and cap (ORF2) (Fig. 1 -black arrow-rectangles). Another gene, the ORF3, situated in an antisense orientation within the coding region of the ORF1 has also been described formerly (Bratanich and Blanchetot, 2002; Chaiyakul et al., 2010; Cheung, 2003). The rep gene has been shown to encode two replication-initiation proteins, Rep and Rep’, and 9 non-coding RNAs (Rep3a, Rep3b, Rep3c1, Rep3c2, Rep3c3, Rep3c4; NS515, NS672 and NS0) (Fig. 1-white arrow-rectangles). The Rep-associated RNA molecules share common 5′- and 3′-ends, but vary in the length of their introns. The Ns-associated RNAs share common 3′-ends with the rep, but vary in their transcription start sites (TSSs) and introns. The TSS of rep has been shown to map at genomic positions 19 (Cheung, 2003) or 1754 (Mankertz and Hillenbrand, 2002) nucleotides (nts), while the polyadenylation (PA) site has been located at nt 997 on the viral genome. The 312 amino acid-long Rep protein contains motifs associated with the rolling-circle-replication of other prokaryotic and eukaryotic organisms (del Solar et al., 1998; Ilyina and Koonin, 1992; Mankertz et al., 1998). Mutation studies have shown that three motifs of Rep - the RC-I, RC-II, and RC-III - play a role in the cleavage of Ori and DNA replication, and the P-loop structure is essential for DNA replication (Steinfeldt et al., 2007). The Rep’ protein contains 168 amino acids and is produced from the spliced Rep transcript. The Rep and the Rep’ proteins differ in their C-terminals: Rep’ lacks the P-loop motif. The two Rep proteins bind the Ori of the replicating double-stranded DNA of PCV with different affinity, but they exhibit similar activity (Steinfeldt et al., 2001). They can form homo- and hetero-dimer complexes, with the latter being referred to as the Rep-complex, since both Rep and Rep’ are required for PCV-1 replication.
Fig. 1.
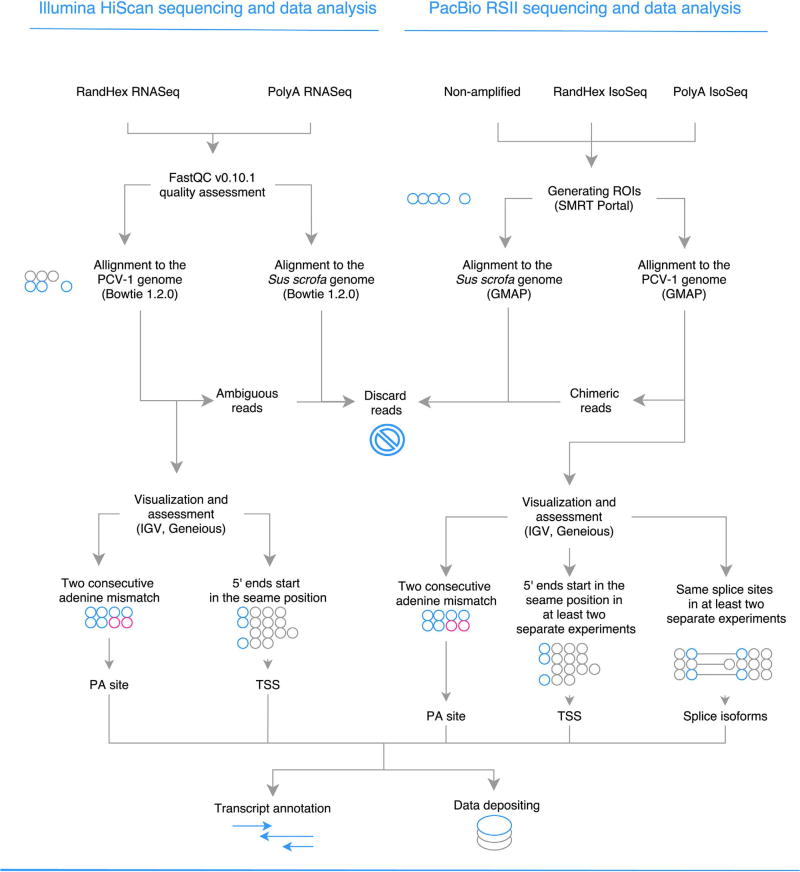
The schematic representation of the preparation of sequencing data. Circles represent nucleotides.
The cap gene codes for the capsid protein (Cap) (Cheung, 2003). The cap TSS has been localized to nts 466 (Mankertz et al., 1998) or 457 (Cheung, 2003), and the Polyadenylation sites (transcription end site, TES) of these genes are situated at 1001 (Mankertz et al., 1998) or 998 (Cheung, 2003) nts.
The 234 amino acid-long Cap protein is localized within the nucleus and has an arginine-rich DNA-binding N-terminus (Cheung, 2003; Mankertz et al., 1998). The Cap contains the pat4 and the bipartite motifs, which are homologous to the nuclear localizing signal of other molecules (Shuai et al., 2008). The Cap monomers form the 16–21 nm capsid of the virion.
Intriguingly, no transcript has yet been detected from the 621 bplong ORF3 however, the expression of a highly cytotoxic pro-apoptotic protein encoded by this genomic region has been reported (Chaiyakul et al., 2010).
A large proportion of the mammalian DNA is transcribed, despite the fact that barely more than one percent of the genome codes for proteins (Katayama et al., 2005). There is an ongoing debate as to whether the majority of non-coding (nc)RNAs represent mere transcriptional noise, or if they have functions that are yet unknown (Bertone et al., 2004). The most abundant and least studied ncRNAs are the long non-coding (lnc)RNAs which are transcripts with more than 200 bp (Mattick, 2004). The lncRNAs have recently been discovered in various organisms, such as viruses, mice and humans (Mattick and Makunin, 2006; Stroop et al., 1984; Tombácz et al., 2016).
The various transcript isoforms, including the transcript end and splice variants, enhance the complexity of viral gene expression. Transcript isoforms with variations within the open reading frames (ORFs) encode different proteins with potentially different function. Variations within the non-coding parts of mRNAs or within ncRNAs may also have an effect for example on the regulation of gene expression or on the determination of the half-life of the gene products. The latest transcriptomics technologies explored a complex meshwork of transcriptional overlaps in many organisms. The transcriptional machineries have been shown to collide with each other at the overlapping regions (Wilusz et al., 2009). It has been hypothesized that transcriptional interferences form an interaction network, which might play an essential role in the regulation of global gene expression (Boldogköi, 2012).
It has been shown that type I and type II interferons modulate the infection of PCV-2 by the interferon-stimulated response element (ISRE) that regulates the expression of the viral rep gene (Gu et al., 2012). ISRE located in the promoter region of rep and other transcription factor response elements were reported in PCV-1 (Mankertz and Hillenbrand, 2002).
The PacBio RSII DNA sequencing technology, which based on single-molecule sequencing with real-time (SMRT) detection, allows the determination of the base sequences of long RNAs without PCR amplification or fragmentation, and can also be used for the analysis of kinetic properties of viral transcriptomes (Tombácz et al., 2017). Both the amplified and the non-amplified versions of isoform sequencing (Iso-Seq) are excellent tools for the identification of transcript end and splice isoforms, as well as long RNA molecules including overlapping and polycistronic transcripts. At the same time these sequencing techniques produce a low amount of systematic errors, and those that do appear can be easily corrected thanks to its high consensus accuracy (Miyamoto et al., 2014). The main advantage of Illumina sequencing is the large number of reads produced by this technology, which allows for deep sequencing coverage, which serves as a prerequisite of the identification of rare transcripts. On the other hand, the short sequencing reads can result in inefficient transcript assembly.
In this study, we aimed to identify novel transcripts and transcript isoforms and map the already described RNA molecules with single base pair precision using a multiplatform approach, which included two short-read and three long-read RNA sequencing methods, as well as a PCR technique.
2. Methods
2.1. Cell and virus
The endogenous PCV-1 was propagated in the immortalized porcine kidney cell line PK-15 (ATCC CCL-33). The cells were cultivated in Dulbecco’s modified Eagle’s medium (Gibco Invitrogen) supplemented with 5% fetal bovine serum (Gibco Invitrogen) with 80 µg/ml gentamycin (Gibco Invitrogen) at 37 °C, 5% CO2. Cells were frozen and thawed three times and centrifuged at 10,000g for 15 min.
2.2. RNA purification
2.2.1. Total RNA isolation
Total RNA isolation from the PK-15 cells was carried out using the Nucleospin RNA kit (Macherey-Nagel), according to the kit manual. Contaminating DNA was removed by on-column RNase free rDNase treatment (supplied by the kit). The isolated total RNA was further treated by TURBO DNA-free™ Kit (Life Technologies) to remove potential residual DNA contamination. RNA concentration was determined by Qubit 2.0 using the RNA BR Assay Kit (Life Technologies), and Agilent 2100 Bioanalyzer was used for assessing RNA integrity. The RNA sample was kept at −80 °C until use.
2.2.2. Isolation of polyadenylated RNAs
The PolyA+ RNA fraction was selected from total RNA with the Oligotex mRNA Mini Kit (Qiagen) following the Oligotex mRNA Spin-Column Protocol according to the manufacturer’s recommendations, and the quantity was measured by using the Qubit RNA HS Assay Kit (Life Technologies).
Library preparation and sequencing.
2.3. Illumina HiScanSQ sequencing
Two different approaches were used for sequencing on Illumina HiScanSQ system. For random hexamer primed amplification and sequencing strand-specific total RNA libraries were prepared using the Illumina ScriptSeq v2 RNA-Seq Library Preparation Kit (Epicentre, Madison, WY USA). For poly(A)-Seq, a single-end library was constructed by using oligo(VN)T20 primers with custom-anchored adapter sequence for compensating for loss in throughput compared to solely oligo(d)T primers. Quality assessment of raw read files was achieved with FastQC v0.10.1. Reads were deposited in NCBI SRA database under accession PRJNA353911.
2.4. PacBio RSII isoform sequencing
Three library preparation approaches was carried out for SMRTbell template preparation. Reads were deposited in NCBI SRA database under accession PRJNA353911.
2.4.1. Non-amplified protocol
Polyadenylated RNAs were converted to cDNAs with SuperScript Double-Stranded cDNA Synthesis Kit (Life Technologies; the included SuperScript II was changed to SuperScript III enzyme). Anchored Oligo (dT)20 primers (Life Technologies) were used for the reverse transcription reactions. Obtained cDNAs were quantified with the Qubit HS dsDNA Assay Kit (Life Technologies). SMRTbell template preparation was performed from cDNAs with PacBio DNA Template Prep Kit 1.0, based on the following protocol: Very Low (10 ng) Input 2 kb Template Preparation and Sequencing with Carrier DNA. The quality of the libraries was analyzed by Agilent 2100 Bioanalyzer. DNA polymerase binding kit XL 1.0 and v2 primers were used for polymerase binding. The polymerase-template complexes were bound magbeads using MagBead Binding Kit (Pacific Biosciences). DNA sequencing was carried out using Pacific Biosciences RSII sequencer with P5-C3 reagents. Movie lengths were 180 min (one movie was recorded for each SMRT cell).
2.4.2. Amplified Iso-Seq protocol using oligo(dT) primers or random hexamer primers
The following PacBio protocols were used for the cDNA production: Isoform Sequencing (Iso-Seq) using the SMARTer PCR cDNA Synthesis Kit (Clontech) and No Size Selection (for the sequencing of short transcripts) or Manual Agarose-gel Size Selection (for the analysis of long RNAs). RNAs were converted to single-stranded cDNAs with 3′ SMART® CDS Primer II A (supplied by the Clontech kit) or adapter-linked GC-rich random primers.
2.4.3. No size selection
The first-strand cDNAs were amplified by PCR, using the SMARTer PCR cDNA Synthesis Kit (Clontech) and the KAPA HiFi Enzyme (Kapa Biosystems) according to the PacBio protocol’s recommendations. 500 ng of cDNA sample was used for the SMRTbell template preparation, using the PacBio DNA Template Prep Kit 1.0.
2.4.4. Manual agarose-gel size selection
Two different PCR reactions were performed out for the different transcripts. Twelve PCR cycles and 1:45 min extension was used for the production of transcripts between 2 and 3 kb, while 15 cycles and 3 min extension was set for the longer transcripts.
DNA/Polymerase Binding Kit P6 kit was used for the library-polymerase bound reaction. Sequencing was carried out on RS II platform with DNA Sequencing Reagent 4.0 (P/N 100-356-200). Movie lengths were 240 min (one movie was recorded for each SMRT cell).
2.5. PCR analysis
RT-PCR analysis was performed for the validation of previously described transcripts (Cap, Rep, rep-associated - data not shown for the last two), putative novel splice isoforms and lncRNAs. Single stranded cDNA library was created from total RNA with SuperScript III reverse transcriptase (Life Technologies) according to the manufacturer’s instructions. Primers used in the study are listed in Table 1. The single-stranded cDNAs were amplified with PCR cycler (Veriti, Applied Biosystems) using the KAPA HiFi PCR Kit (KAPA Biosystems), according to the instructions supplied by the manufacturer. 0.8% agarose gel electrophoresis and GeneRuler 1 kb DNA Ladder (Thermo Fisher Scientific) was used for products larger than 1000 bp, and 12% acrylamide gel electrophoresis and GeneRuler Low Range DNA Ladder (Thermo Fisher Scientific) and GeneRuler 1 kb Plus DNA Ladder (Thermo Fisher Scientific) for products smaller than 1000 bp. Staining was performed with GelRed (Biotium) in both cases.
Table 1.
Primers used for RT-PCR for the confirmation of novel transcripts and transcript isoforms.
| Primer | Primer sequence | Genomic position | Orientation (strand) | Product sizes (transcript) |
|---|---|---|---|---|
| ORF1.1fw | CTTTATTCTGCTGGTCGGTT | 301–320 | − | 211 nt (Rep) |
| ORF1.1r | TCCGAGGAGGAGAAAAACAA | 110–129 | + | |
| ORF1 → ORF2.1 | TGGAAGACTGCTGGAGAACA | 887–906 | + | |
| ORF2 → ORF1.1 | GCTATGCGCTCCAAAATG | 1108–1125 | − | 239 nt (Ctr) |
| Cap2fw | CTGCTCGGCTACAGTCACCA | 432–451 | − | 175 nt (Cap), 558 nt (Cap-nsp) |
| Cap2r | CGGAGGATGTTTCCAAGATG | 1653–1672 | + | |
| ORF3fw | ACATCGAGAAAGCGAAAGGA | 282–301 | + | |
| ORF3r | CTTCCAATCACGCTGCTGCA | 516–535 | − | 254 nt (Atr) |
| Ctr’fw | GATGACGTGGCCAAGGAGGC | 1705–1724 | − | 259 nt (Ctr’) |
| Ctr’r | CCCAGGAATGGTACTCCTCA | 813–832 | + | |
| Rep”5Lfw | GAAACCGTTACAGATGGCGC | 1601–1620 | − | 83 nt (Rep”5L1), 412 nt (Rep’5L1) |
| Rep”5Lr | ATTGTTTGGTCCAGCTCAGG | 1202–1228 | + |
2.6. Data analysis and visualization
Reads from the Illumina sequencing were aligned with Bowtie 2 (Langmead and Salzberg, 2012), while reads from PacBio sequencing with GMAP mapper (Wu and Watanabe, 2005) to the host genome (Sus scrofa, assembly: Sscrofa10.2) and to the genome of PCV-1 strain Szeged (GeneBank accession: KX816645) previously sequenced and aligned by our group, and duplicated to mimic circularization (Fig. 1.). For visualization of the mapped reads we used IGV (Thorvaldsdóttir et al., 2013). Poly(A) signals were predicted using the online prediction tool PolyApred (Ahmed et al., 2009). TATA, GC and CAAT-boxes were predicted using the GPminer web application (Lee et al., 2012).
3. Results and discussion
3.1. Analysis of PCV-1 transcriptome using multi-platform techniques
In this study, two Illumina HiScanSQ RNA-Seq techniques were applied: the PA-Seq, which yielded 104,032 sequencing reads, and random hexamer-based RNA-Seq yielding 93,613 reads mapping to PCV-1 strain Szeged genome (Gene Bank accession number: KX816645). Furthermore, we used three PacBio-based long-read sequencing techniques, as follows: a random-hexamer-based isoform sequencing (Iso-Seq; yielding 101 reads) and two PA-Seq techniques, the non-amplified (SMRT; 145 reads) and the amplified (510 reads) Iso-Seq platforms. PCR was used for detecting splice variants and confirming the existence of long, whole genome spanning transcripts, detected by PacBio sequencing. The novel transcripts are illustrated by grey arrow-rectangles in Fig. 2. The criteria and evidence for the existence of novel transcripts are described in Supplementary Table 1.
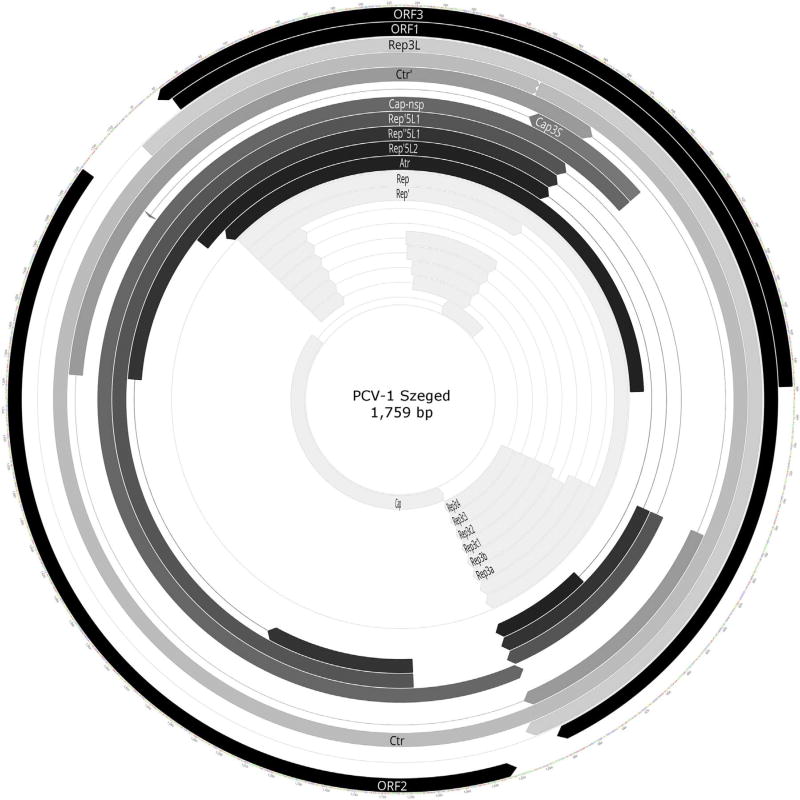
Fig. 2.
Location of already known and novel transcripts on the PCV-1 genome, created with Geneious (http://www.geneious.com/). Arrow-rectangles in black: ORFs; arrow-rectangles in white: already known transcripts; arrow-rectangles in grey: novel splice isoforms, length variants and long, complex transcripts, lines represent introns.
3.2. Determination of the 5′ and 3′-ends of already described PCV-1 transcripts with base-pair precision
Two main transcripts (Cap and Rep) and ten transcript isoforms (Rep’, Rep3a, Rep3b, Rep3c-1, Rep3c-2, Rep3c-3, Rep3c-4, NS462, NS642, and NS0) have been previously detected using Northern blot analysis. The UTRs of these RNA molecules have been identified with 5′ and 3′ rapid amplification of cDNA ends (RACE) (Cheung, 2003; Mankertz et al., 1998). In this work, we reanalyzed the 5′- and 3′-ends of the Cap and Rep RNAs with the amplified Iso-Seq method of PacBio sequencing (Table 2), which is capable of determining full-length transcripts, including the accurate TSSs and TESs. The TSS of the Cap transcript was mapped to nt 468 genomic position instead of 457, while the 3′ end is at nt 988 instead of 998, both reported by Cheung (Cheung, 2003). The Rep transcript and all of its previously detected isoforms have also been mapped in this work, and as a result, we obtained that the TSS is located at nt 6 genomic position on the viral genome instead of position 19, while the 3′ end at nt 996 instead of 997, both reported by Cheung (Cheung, 2003). Theoretically, the differences in the transcript length may indicate real genetic variation between the two PCV-1 strains or alternatively, they can be the result of the inaccuracy of the applied RACE technique.
Table 2.
Annotation of PCV-1 transcripts with base-pair precision, and the predicted location of their promoters.
| Transcript | GC box | CAT box | TATA box | TSS | 5′ variation | PA Signal | TES | 3′ variation |
|---|---|---|---|---|---|---|---|---|
| Cap | 560–565 | 527–532 | – | 468 | 471–464 | 1015–1020 | 988 | – |
| 1021–1026 | ||||||||
| 1025–1030 | ||||||||
| Cap3S | 560–565 | 527–532 | – | 468 | – | 1184–1189 | – | 1148–1175 |
| Cap-nsp | 560–565 | 527–532 | – | 468 | – | 1015–1020 | 988 | – |
| 1021–1026 | ||||||||
| 1025–1030 | ||||||||
| Rep | 1676–1681 | – | 1724–1729 | 6 | – | 972–977 | 996 | 993–1006 |
| Rep3L | 1676–1681 | – | 1724–1729 | 6 | – | 972–977 | 1006 | – |
| Rep’ | 1676–1681 | – | 1724–1729 | 6 | – | 972–977 | 996 | – |
| Rep’5L1 | – | – | 1067–1072 | 1095 | – | 972–977 | 998 | – |
| Rep”5L1 | – | – | 1067–1072 | 1095 | – | 972–977 | 998 | – |
| Rep’5L2 | – | – | 1724–1729 | 1734 | – | 972–977 | 998 | – |
| Rep3a | – | – | – | 6 | – | 972–977 | 998 | – |
| Rep3b | – | – | – | 6 | – | 972–977 | 998 | – |
| Rep3c1 | – | – | – | 6 | – | 972–977 | 998 | – |
| Rep3c2 | – | – | – | 6 | – | 972–977 | 998 | – |
| Rep3c3 | – | – | – | 6 | – | 972–977 | 998 | – |
| Rep3c4 | – | – | – | 6 | – | 972–977 | 998 | – |
| Atr | 779–784 | 737–742 | 686–691 | *658 | – | – | 1757 | – |
| Ctr | – | – | 315–320 | 345 | – | 972–977 | 998 | – |
| Ctr’ | – | – | 315–320 | 345 | – | 972–977 | 998 | – |
GC, CAT and TATA boxes were predicted in silico with GPMiner, polyA-signals were predicted in silico with PolyApred. 5′- and 3′-UTR positions were determined using sequencing data visualized with IGV. The putative 5′ end of Atr was marked with *.
3.3. Minor variations at the 5′- and 3′-UTRs of PCV-1 detected by amplified Iso-Seq method
We found that the Cap transcript varies in length at its TSS with a variation ranging from nts 464–471. Our in silico analysis (using GPMiner predicted a TBP binding site (at genomic position 686–691 nts), CAAT box (at 527–532 nts) and GC box (at 560–565 nts) upstream of the TSS of Cap. These sites are within the range (556–480) reported by Mankertz and Hillenbrand (Mankertz and Hillenbrand, 2002). We found a 5% variation in the 3′-end of Cap transcripts ranging from 989 to 995 nts. A TATA box (between 1724–1729 nt) and a GC box (between 1676–1681 nt) were predicted in silico upstream of the Rep TSS, which fall in the range found by Mankertz and Hillenbrand (Mankertz and Hillenbrand, 2002) for the Cap and Rep promoters. Rep’ exhibits a 3′-end variation between nts 993–1006, but there is no variation whatsoever at its 5′-end.
3.4. Long-read sequencing identified novel transcript end and splice isoforms of PCV-1 transcripts
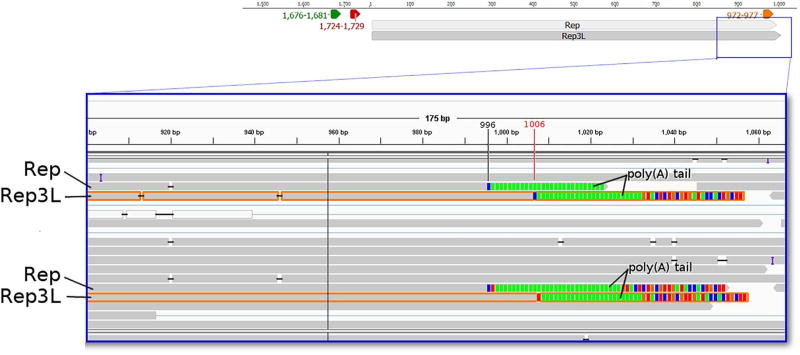
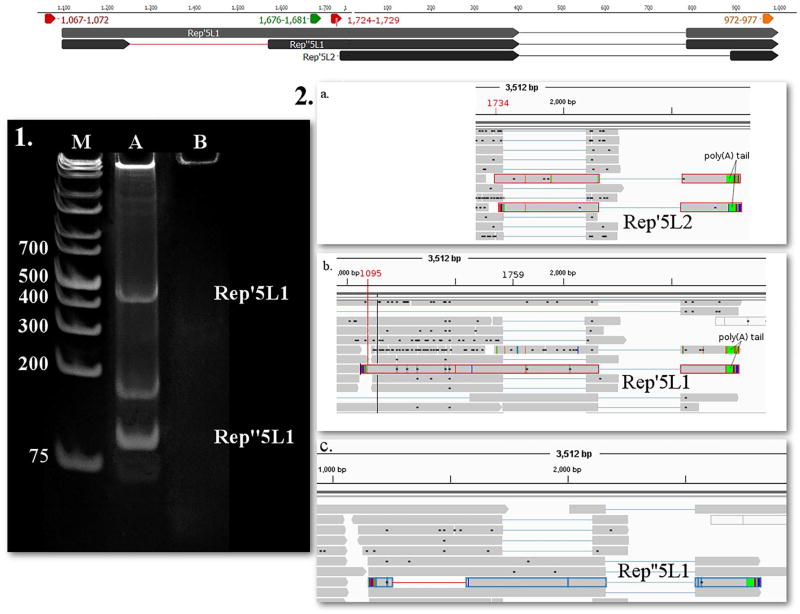
In this study, we identified four novel length variants. The TSS of Cap3S is the same as that of the Cap, but with a shorter and varying TESs. The PA signal of Cap3S is predicted to be localized within 1184–1189 bp and the TESs ranges within 1148- 1175 nts. The Cap3S transcript is terminated within the ORF, and therefore lacks an in-frame stop codon. Additionally, we detected a non-spliced version of the Rep transcript, which we termed Rep3L. Similarly to Rep, the TSS of Rep3L maps at nt 6 on the viral genome, but it has a distinct TES: the 3′-UTR of this transcript is ten bases longer than that of Rep, but the two RNA molecules utilize the same PA signal (Fig. 3). Our in silico analysis identified the ISRE in the promoter region of PCV-1 strain Szeged and other promoter response elements (Supplementary Table 2). We also identified longer variants of the spliced isoform of Rep, and named them Rep′5L1 and Rep′5L2. The TSS of Rep′5L1 maps at nt 1095 (Fig. 4a and b) while the TSS of Rep5′L2 maps at nt 1734. Rep′5L1 and is predicted to be controlled by a putative upstream TATA box being located at nts 1067–1072 (the precise splice donor and acceptor sites are listed in Supplementary Table 3).
Fig. 3.
PacBio RSII Iso-Seq read of Rep3L (highlighted with orange) yielded from the oligo(dT) primed reaction, and visualized with IGV. Green arrow-rectangle: GC box; Yellow arrow-rectangle: TATA box; Red arrow-rectangle: Poly(A) signal. The longer 3′ end of Rep3L can be observed. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Fig. 4.
1. 12% acrylamide gel electrophoresis of Rep’5L1 and Rep”5L1, and 2. PacBio RSII IsoSeq reads of (b.) Rep’5L1 (c.) Rep”5L1 and (a.) Rep’5L2 yielded from the oligo(dT) primed reaction, and visualized with IGV. Lanes were loaded with cDNA products from RT-PCR and with GeneRuler 1 kb Plus DNA Ladder (lane M) (Thermo Fisher Scientific). Staining was performed with GelRed (Biotium). On lane A three bands appear, the upper one representing Rep’5L1 (product size 412 nts) while the lower one being Rep”5L1 (product size 83 nts). The middle band could be a splice variant not detected by our sequencing. Lane B was loaded with no-RT control. Yellow arrow-rectangle: TATA box; Red arrow-rectangle: Poly(A) signal, lines represent introns. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
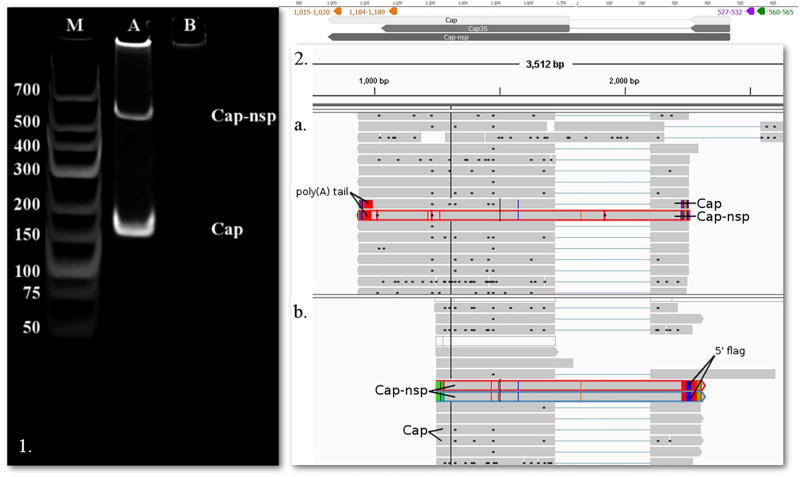
We succeeded in detecting all of the formerly reported splice variants of the Rep-associated transcripts (Cheung, 2003; Mankertz and Hillenbrand, 2002) by using the non-amplified and the two amplified Iso-Seq protocols of PacBio sequencing, as well as the Illumina HiSeq platform. We were able to detect the full-length transcripts of spliced Rep’, Rep3c2, Rep3c3 and Rep3c4 using various combinations of PacBio techniques. Our analyses also revealed a non-spliced Cap transcript variant, which was termed Cap-nsp (Fig. 5), and a splice variant of Rep′5L1, termed Rep”5L1 (Fig. 4c). Cap-nsp has the same TSS and TES as its spliced isoform. We detected this transcript with the PacBio platform by using the amplified Iso-Seq approaches.
Fig. 5.
The Cap and Cap-nsp transcripts. 1. 12% acrylamide gel electrophoresis of Cap and Cap-nsp. The lanes were loaded with cDNA products from RT-PCR and with GeneRuler Low Range DNA Ladder (lane M) (Thermo Fisher Scientific). Staining was performed with GelRed (Biotium). On lane A two bands appear, the upper one representing Cap-nsp (product size 558 nts), while the lower one being Cap (product size 175 nts). Lane B was loaded with no-RT control. 2. PacBio RSII IsoSeq reads of Cap and Cap-nsp yielded from the oligo(d)T (a.) and random hexamer (b.) primed reaction, and visualized with IGV. Green arrow-rectangle: GC box; Purple arrow-rectangle: CAT box; Red arrow-rectangle: Poly(A) signal, lines represent introns. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Rep”5L1 has identical TSS, TES and splice junction between 404 and 786 as Rep’5L1, but it has a new splice donor site at nt 1253 and a new splice acceptor site at nt 1569 with the GT/AG consensus. Both splice sites are localized within the 5′ UTR of Rep”5L1.The existence of the two novel splice variants was confirmed by PCR.
Primers were designed for the non-spliced version of Cap (Cap-nsp) between the genomic positions 1653 to 451. The PCR amplification yielded two products, one being 558 nts long, which detected the Cap-nsp transcript and the other being 175 nts long, detecting the Cap transcript (Fig. 5.1).
For the new splice site of Rep’5L1 primers were designed to position 1209–1620 producing a 83 nts and a 412 nts product (Fig. 4.1), the first detects Rep”5L1, while the second detects the unspliced Rep’5L1. The splice junctions of Rep3a, Rep3b, Rep3c1, Rep3c2, Rep3c3 and Rep3c4 were detected using the random hexamer-based RNA-Seq protocol of Illumina sequencing (reads were deposited in NCBI SRA database under accession PRJNA353911).
3.5. Complex PCV-1 transcripts explored by long-read sequencing
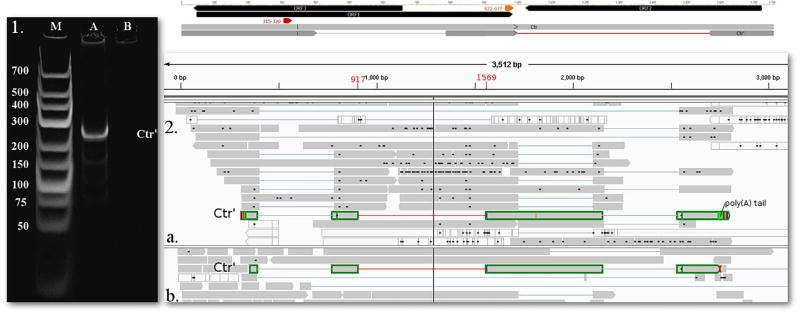
In this study, we identified two novel complex transcripts each containing oppositely-oriented genes. A 2412 nt-long RNA molecule named complex transcript (Ctr) and a 994 nt-long RNA molecule named Ctr’, both with a TSS at nt 345 on the genome, and a controlling TATA box predicted in silico to map to nts 315–320. The TES of these transcripts is located at nt 998. Ctr’ and contains three introns, two of which coincide with the intron of Rep’ and a new one with a splice donor site at nt 917 and acceptor site at nt 1569 and a GT/AG consensus at the splice junctions (Fig. 6). The new intron is localized within the 5′ UTR of the Ctr’.
Fig. 6.
1. 12% acrylamide gel electrophoresis and 2. sequencing reads of the novel intron region in transcript Ctr. 1. The lanes were loaded with cDNA products from RT-PCR and with GeneRuler Low Range DNA Ladder (lane M) (Thermo Fisher Scientific). Staining was performed with GelRed (Biotium). On lane A one pronounced band appears using primers Ctr’fw and Ctr’r, with a product size of 259 nts. b. PacBio RSII IsoSeq oligo(d)T primed (a.) (read highlighted with green) and SMRT (b.) (read highlighted with green) reads, visualized with IGV. Yellow arrow-rectangle: TATA box, Red arrow-rectangle: Poly(A) signal, lines represent introns. The novel intron of the transcript is represented by a red line between 917 and 1569 nts. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. The ORF3 transcript
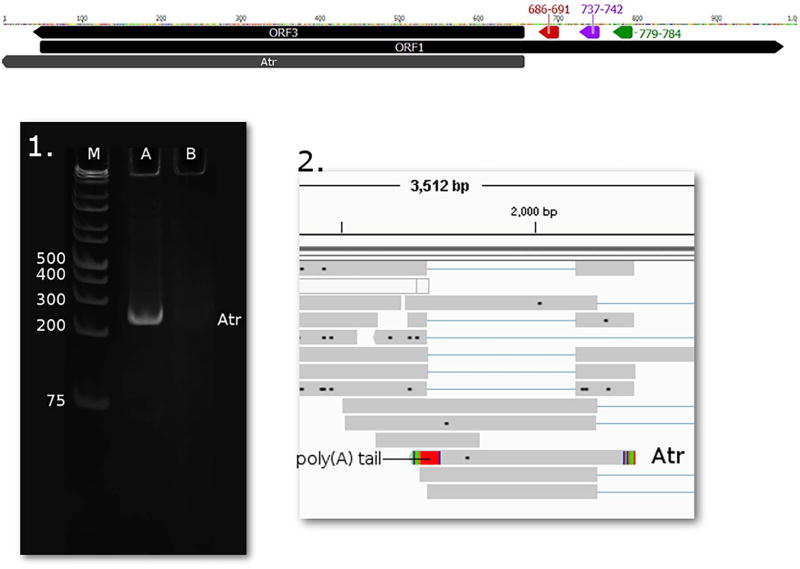
Although a previous study hypothesized the existence of a transcript encoding ORF3, and reported the detection of an apoptosis-associated protein (Chaiyakul et al., 2010) produced from this genomic region, the mRNA has not yet been detected. We were able detect this low-abundance transcript with a 5′ end at 471 nts and with a 3′ end, which was located at 1757 nts and named it apoptosis-associated transcript (Atr). Since the start of ORF3 is at genomic position 658, we hypothesize the TSS of Atr to be at this position (Fig. 7). An upstream promoter was predicted with TATA-box at position 686–691, CAT-box at 737–742 and GC-box at 779–784.
Fig. 7.
1. 12% acrylamide gel electrophoresis and 2. sequencing reads of the novel Atr transcript. 1. The lanes were loaded with cDNA products from RT-PCR and with GeneRuler 1 kb Plus DNA Ladder (lane M) (Thermo Fisher Scientific). Staining was performed with GelRed (Biotium). On lane A one pronounced band appears using primers ORF3Fw and ORF3r, with a product size of 254 nts. 2. PacBio RSII IsoSeq oligo(d)T primed read of Atr, visualized with IGV. Green arrow-rectangle: GC box; Purple arrow-rectangle: CAT box; Yellow arrow-rectangle: TATA box. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.7. Long-read sequencing reveals a complex meshwork of transcriptional overlaps in PCV-1
We detected novel transcriptional overlaps and also confirmed the existence of an already published one. We found that only 5% of the Cap transcripts are terminated at the genomic location reported earlier (Mankertz and Hillenbrand, 2002), and approximately 95% of the Cap transcript is longer with 10 bp, the TES of these transcripts is located at nt 988. In contrast to the short isoform, the 10 bp longer Cap transcript overlaps with Rep transcript in a convergent (tail-to-tail) manner. The same phenomenon was observed in Rep3L, Rep’5L1, Rep’5L2 overlapping Cap and Cap-nsp. Intriguingly, these transcripts also overlap each other in a head-to-head manner on the circular PCV-1 genome. Ctr and Ctr’ overlaps with Rep and rep-associated transcripts in tandem (tail-to-head), while with Cap in a convergent manner through spanning the whole genome (Fig. 2). The transcriptional overlaps may indicate a novel layer of genetic regulation (Boldogköi, 2012; Prescott and Proudfoot, 2002) acting through the collisions of the transcriptional apparatuses.
4. Conclusion
In this work, we applied a multiplatform approach to study the transcriptome of porcine circovirus type 1. Our investigations revealed a complex meshwork of overlapping RNA molecules. We identified seven transcript isoforms, including two novel TSSs (Rep’5L1, Rep’5L2) and two TESs (Cap3S, Rep3L) variants, as well as two novel splice sites (in Rep”5L1 and Ctr’), a previously known splice site in a novel transcript (Ctr’) and in two length variants (Rep’5L1 and Rep”5L1), and a novel non-spliced transcript variant (Cap-nsp). Additionally, we discovered the transcript for the putative ORF3 (Atr) and a genome spanning long transcript (Ctr). Most of the newly discovered polycistronic RNA molecules are expressed at low levels, which may explain why they had previously gone undetected.
Supplementary Material
Acknowledgments
We would like to thank Marianna Ábrahám (University of Szeged) for technical assistance.
Funding
This study was supported by the following: Centers of Excellence in Genomic Science (CEGS) - National Institutes of Health (NIH): 1P50HG007735-01 - MS; TÁMOP-Social Renewal Operational Programme: TÁMOP-4.2.6-14/1 - ZBo; Bolyai János Scholarship of the Hungarian Academy of Sciences: 2015-18-DT, and Swiss-Hungarian Cooperation Programme (https://www.palyazat.gov.hu/swiss_contribution): SH/7/2/8 - ZBo. The cost of publication was covered by the University of Szeged.
Abbreviations
- PCV-1
porcine circovirus type 1
- ORF3
open reading frame 3
- TES
transcription end site
- TSS
transcription start site
- Ori
origin of replication
- PA site
polyadenylation site
- PA signal
polyadenylation signal
- AP2
activating protein 2 response element
- AP3
activating protein 3 response element
- AP4
activating protein 4 response element
- SP1
specificity protein 1 response element
- USF/MLTF
up-stream stimulating/major later transcription factor response element
- ISRE
interferon-stimulated response element
Footnotes
Competing interests
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
The datasets generated and/or analyzed during the current study are available in the NCBI SRA database under accession PRJNA353911, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA353911/.
Authors' contributions
Zsolt Boldogkői conceived and designed the experiments; Norbert Moldován, Zsolt Csabai, Dóra Tombácz and Zsolt Boldogkői performed the experiments; Norbert Moldován, Zsolt Balázs, Dóra Tombácz, Attila Szűcs, Zsolt Csabai, and Zsolt Boldogkői analyzed the data; Zsolt Boldogkői and Michael Snyder contributed reagents/materials/analysis tools; Norbert Moldován and Zsolt Boldogkői wrote the paper.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.virusres.2017.05.010.
References
- Ahmed F, Kumar M, Raghava GPS. Prediction of polyadenylation signals in human DNA sequences using nucleotide frequencies. In Silico Biol. 2009;9:135–148. [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, Gerstein M, Snyder M. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. http://dx.doi.org/10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Boldogköi Z. Transcriptional interference networks coordinate the expression of functionally related genes clustered in the same genomic loci. Front. Genet. 2012;3:122. doi: 10.3389/fgene.2012.00122. http://dx.doi.org/10.3389/fgene.2012.00122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratanich AC, Blanchetot A. PCV2 replicase transcripts in infected porcine kidney (PK15) cells. Virus Genes. 2002;25:323–328. doi: 10.1023/a:1020940411267. [DOI] [PubMed] [Google Scholar]
- Chaiyakul M, Hsu K, Dardari R, Marshall F, Czub M. Cytotoxicity of ORF3 proteins from a nonpathogenic and a pathogenic porcine circovirus. J. Virol. 2010;84:11440–11447. doi: 10.1128/JVI.01030-10. http://dx.doi.org/10.1128/JVI.01030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AK. Comparative analysis of the transcriptional patterns of pathogenic and nonpathogenic porcine circoviruses. Virology. 2003;310:41–49. doi: 10.1016/s0042-6822(03)00096-5. [DOI] [PubMed] [Google Scholar]
- del Solar G, Giraldo R, Ruiz-Echevarría MJ, Espinosa M, Díaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Zhang Y, Lian X, Sun H, Wang J, Liu W, Meng G, Li P, Zhu D, Jin Y, Cao R. Functional analysis of the interferon-stimulated response element of porcine circovirus type 2 and its role during viral replication in vitro and in vivo. Virol. J. 2012;9:152. doi: 10.1186/1743-422X-9-152. http://dx.doi.org/10.1186/1743-422X-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, Suzuki H, Carninci P, Hayashizaki Y, Wells C, Frith M, Ravasi T, Pang KC, Hallinan J, Mattick J, Hume DA, Lipovich L, Batalov S, Engström PG, Mizuno Y, Faghihi MA, Sandelin A, Chalk AM, Mottagui-Tabar S, Liang Z, Lenhard B, Wahlestedt C. RIKEN Genome Exploration Research Group, Genome Science Group (Genome Network Project Core Group), FANTOM Consortium. Antisense transcription in the mammalian transcriptome. Science (80-.) 2005;309:1564–1566. doi: 10.1126/science.1112009. http://dx.doi.org/10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. http://dx.doi.org/10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCann P, Albina E, Madec F, Cariolet R, Jestin A. Piglet wasting disease. Vet. Rec. 1997;141:660. [PubMed] [Google Scholar]
- Lee T-Y, Chang W-C, Hsu JB-K, Chang T-H, Shien D-M. GPMiner: an integrated system for mining combinatorial cis-regulatory elements in mammalian gene group. BMC Genomics. 2012;13(Suppl. 1):S3. doi: 10.1186/1471-2164-13-S1-S3. http://dx.doi.org/10.1186/1471-2164-13-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankertz A, Hillenbrand B. Analysis of transcription of Porcine circovirus type 1. J. Gen. Virol. 2002;83:2743–2751. doi: 10.1099/0022-1317-83-11-2743. http://dx.doi.org/10.1099/0022-1317-83-11-2743. [DOI] [PubMed] [Google Scholar]
- Mankertz J, Buhk HJ, Blaess G, Mankertz A. Transcription analysis of porcine circovirus (PCV) Virus Genes. 1998;16:267–276. doi: 10.1023/a:1008022521329. [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006:R17–29. doi: 10.1093/hmg/ddl046. http://dx.doi.org/10.1093/hmg/ddl046. [DOI] [PubMed]
- Mattick JS. RNA regulation: a new genetics? Nat. Rev. Genet. 2004;5:316–323. doi: 10.1038/nrg1321. http://dx.doi.org/10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Miyamoto M, Motooka D, Gotoh K, Imai T, Yoshitake K, Goto N, Iida T, Yasunaga T, Horii T, Arakawa K, Kasahara M, Nakamura S. Performance comparison of second- and third-generation sequencers using a bacterial genome with two chromosomes. BMC Genomics. 2014;15:699. doi: 10.1186/1471-2164-15-699. http://dx.doi.org/10.1186/1471-2164-15-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proc. Natl. Acad. Sci. U. S. A. 2002;99:8796–8801. doi: 10.1073/pnas.132270899. http://dx.doi.org/10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuai J, Wei W, Jiang L, Li Xl, Chen N, Fang W. Mapping of the nuclear localization signals in open reading frame 2 protein from porcine circovirus type 1. Acta Biochim. Biophys. Sin. (Shanghai) 2008;40:71–77. doi: 10.1111/j.1745-7270.2008.00377.x. http://dx.doi.org/10.1111/j.1745-7270.2008.00377.x. [DOI] [PubMed] [Google Scholar]
- Steinfeldt T, Finsterbusch T, Mankertz A. Rep and Rep’ protein of porcine circovirus type 1 bind to the origin of replication in vitro. Virology. 2001;291:152–160. doi: 10.1006/viro.2001.1203. http://dx.doi.org/10.1006/viro.2001.1203. [DOI] [PubMed] [Google Scholar]
- Steinfeldt T, Finsterbusch T, Mankertz A. Functional analysis of cis- and transacting replication factors of porcine circovirus type 1. J. Virol. 2007;81:5696–5704. doi: 10.1128/JVI.02420-06. http://dx.doi.org/10.1128/JVI.02420-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroop WG, Rock DL, Fraser NW. Localization of herpes simplex virus in the trigeminal and olfactory systems of the mouse central nervous system during acute and latent infections by in situ hybridization. Lab. Invest. 1984;51:27–38. [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. http://dx.doi.org/10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer I, Rasch R, Tochtermann G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralblatt für Bakteriol. Parasitenkunde, Infekt. und Hyg. Erste Abteilung Orig. R. A Medizinische Mikrobiol. und Parasitol. 1974;226:153–167. [PubMed] [Google Scholar]
- Tischer I, Gelderblom H, Vettermann W, Koch MA. A very small porcine virus with circular single-stranded DNA. Nature. 1982;295:64–66. doi: 10.1038/295064a0. http://dx.doi.org/10.1038/295064a0. [DOI] [PubMed] [Google Scholar]
- Tombácz D, Csabai Z, Oláh P, Balázs Z, Likó I, Zsigmond L, Sharon D, Snyder M, Boldogkői Z. Full-length isoform sequencing reveals novel transcripts and substantial transcriptional overlaps in a herpesvirus. PLoS One. 2016;11:e0162868. doi: 10.1371/journal.pone.0162868. http://dx.doi.org/10.1371/journal.pone.0162868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombácz D, Balázs Z, Csabai Z, Moldován N, Szűcs A, Sharon D, Snyder M, Boldogkői Z. Characterization of the dynamic transcriptome of a herpesvirus with long-read single molecule real-Time sequencing. Sci. Rep. 2017;7:43751. doi: 10.1038/srep43751. http://dx.doi.org/10.1038/srep43751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. http://dx.doi.org/10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. http://dx.doi.org/10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.