Micro-Abstract
In this study we evaluated, to our knowledge, the largest blood biomarker panel ever reported. Baseline interleukin-1b and neutrophil count and early-treatment cytokeratin-19 antigen predicted lung cancer radiotherapy response. Baseline angioprotein-1 and hepatocyte growth factor (HGF) significantly correlated with the gross tumor volume. Changes in vascular cell adhesion molecule 1 (VCAM-1) correlated with proliferation imaging, highlighting for the first time a potential role of blood biomarkers as imaging surrogates.
Keywords: Circulating, Functional, Prognostic, Thoracic, Treatment
Abstract
Introduction
There is an unmet need to develop noninvasive biomarkers to stratify patients in drug-radiotherapy trials. In this pilot study we investigated lung cancer radiotherapy response and toxicity blood biomarkers and correlated findings with tumor volume and proliferation imaging.
Patients and Methods
Blood samples were collected before and during (day 21) radiotherapy. Twenty-six cell-death, hypoxia, angiogenesis, inflammation, proliferation, invasion, and tumor-burden biomarkers were evaluated. Clinical and laboratory data were collected. Univariate analysis was performed on small-cell and non–small-cell lung cancer (NSCLC) whereas multivariate analysis focused on NSCLC.
Results
Blood samples from 78 patients were analyzed. Sixty-one (78.2%) harbored NSCLC, 48 (61.5%) received sequential chemoradiotherapy. Of tested baseline biomarkers, undetectable interleukin (IL)-1b (hazard ratio [HR], 4.02; 95% confidence interval [CI], 2.04-7.93; P < .001) was the only significant survival covariate. Of routinely collected laboratory tests, high baseline neutrophil count was a significant survival covariate (HR, 1.07; 95% CI, 1.02-1.11; P = .017). Baseline IL-1b and neutrophil count were prognostic for survival in a multivariate model. The addition of day-21 cytokeratin-19 antigen modestly improved this model's survival prediction (concordance probability, 0.75-0.78). Chemotherapy (P < .001) and baseline keratinocyte growth factor (P = .019) predicted acute esophagitis, but only chemotherapy remained significant after Bonferroni correction. Baseline angioprotein-1 and hepatocyte growth factor showed a direct correlation with tumor volume whereas changes in vascular cell adhesion molecule 1 showed significant correlations with 18F-fluorothymidine (FLT) positron emission tomography (PET).
Conclusion
Select biomarkers are prognostic after radiotherapy in this lung cancer series. The correlation between circulating biomarkers and 18F-FLT PET is shown, to our knowledge for the first time, highlighting their potential role as imaging surrogates.
Introduction
Radiotherapy plays a significant therapeutic role in localized but inoperable or locally advanced lung cancer. The efficacy of radiotherapy dose escalation, using conventional fractionation with concurrent chemotherapy, has reached a plateau in patients with non–small-cell lung cancer (NSCLC).1 In patients with small-cell lung cancer (SCLC), standard of care treatments have not changed in the past 2 decades.2 Durable tumor control is rarely achieved; most patients progress locally and/or distantly.
Over the years, a number of radiotherapy-focused clinical trials in SCLC and NSCLC were conducted.3, 4 However, lung cancer 5-year age-standardized survival remains at approximately 10% to 20% with little global variation, reinforcing the inadequacy of current therapeutic strategies.5 A paradigm shift in our therapeutic approach is required, to make a substantial effect on patient outcomes. Although tumor hypoxia, repopulation, and DNA damage repair have long been linked to radiotherapy resistance,6 there is little understanding of the molecular mechanisms of radiotherapy response and toxicity. Critically, there are no biomarkers that can be applied to tailor radiotherapy to the individual molecular characteristics of the patients' tumor and normal tissues. Instead, the current focus is on combining systemic mechanism-based therapies (eg, epidermal growth factor (EGF) receptor tyrosine kinase inhibitors/immunotherapies) with radiotherapy.7, 8 Although combination trials are promising, they are equally challenging because of the potential for acute and late severe toxicities, particularly pneumonitis and esophagitis.9 Furthermore, to date no agent has shown survival advantage when combined with chemoradiotherapy in unselected patients.3 For these reasons, there is an unmet need to develop noninvasive radiotherapy response and toxicity biomarkers to tailor radiotherapy, stratify patients according to radiosensitivity, and select patients for future combination trials. It is envisioned that this could increase the chance of developing clinical trials leading to fast drug-radiotherapy combination registration.
We published on the utility of functional imaging of proliferation (18F-fluorothymidine [FLT] positron emission tomography [PET]) to predict early radiotherapy response in NSCLC patients.10 Although results were informative, serial imaging is expensive, resource-intensive, and demanding for patients. This prospective pilot study was conducted to assess blood-based biomarkers and investigate their relationship with radiotherapy response and toxicity. The relationship between blood biomarkers and tumor volume and 18F-FLT PET was also explored. A broad cytokine, growth factor, and circulating marker panel was selected to represent potential culprit biological processes (cell death, hypoxia, angiogenesis, inflammation, proliferation, invasion, and tumor burden) likely to be involved in radiotherapy response and toxicity.
Patients and Methods
Patient Population
Patients were prospectively recruited in the Christie NHS Foundation Trust (Manchester, United Kingdom) according to an ethical committee-approved protocol (reference 09/H1011/55). Eligible participants had an Eastern Cooperative Oncology Group (ECOG) performance score of ≤ 2, histologically or cytologically confirmed NSCLC or SCLC, and scheduled to receive radical radiotherapy (with or without chemotherapy). Patients with distant metastases were excluded unless a solitary metastatic site was amenable to radical-intent therapy. Radiotherapy planning was performed using 3-D or 4-dimensional computed tomography (CT). Radiotherapy doses were 50 to 55 Gy in 20 once-daily fractions or 60 to 66 Gy in 30 to 33 once-daily fractions delivered 5 d/wk. Commonly accepted dosimetric constraints were used: percentage of the lung volume receiving ≥ 20 Gy (V20Gy) ≤ 35% and maximum spinal cord dose of 40 Gy (patients treated with 20 fractions) or 48 Gy (patients treated with ≥ 30 fractions). Patients taking part in clinical trials of investigational medicinal products concurrent with radiotherapy were excluded. Chemotherapy consisted of a platinum agent (carboplatin or cisplatin) combined with etoposide for concurrent chemoradiation or gemcitabine (squamous cell carcinoma)/pemetrexed (adenocarcinoma) for sequential chemoradiation. All patients gave informed consent.
Blood Samples
Blood samples were collected before (baseline) and during radiotherapy (day 21). A panel of 26 biomarkers of radiotherapy response (primary end point) and toxicity (secondary end point), chosen a priori, were evaluated (Table 1). Additional blood samples were collected on the day of early-treatment 18F-FLT PET in patients co-recruited to this substudy.
Table 1.
The Cytokine, Growth Factor and Circulating Marker Panel Investigated, With Respective Limits of Detection and Sample Dilution Factors
| Process | Marker | Limits of Detection |
|---|---|---|
| Cell Death/Apoptosis | M30 | 75-1000 μ/L |
| M65M65 | 125-2000 μ/L | |
| Hypoxia | CA-IX | 15.6-1000 pg/mL |
| Osteopontin | 1500-1,500,000 pg/mLa | |
| Angiogenesis | Ang-1 | 40-40,000 pg/mLb |
| Ang-2 | 2.8-2800 pg/mL | |
| FGFb | 2-2000 pg/mL | |
| IL-8 | 0.4-400 pg/mL | |
| PDGFb | 1.2-1200 pg/mL | |
| PIGF | 2-2000 pg/mL | |
| Tie-2 | 200-200,000 pg/mLb | |
| VEGFA | 5-5000 pg/mL | |
| VEGFC | 16-16,000 pg/mL | |
| VEGFR-1 | 11-11,000 pg/mL | |
| VEGFR-2 | 28-28,000 pg/mL | |
| Inflammation | E-selectin | 2400-2,400,000 pg/mLc |
| IL-1b | 0.2-200 pg/mL | |
| IL-6 | 0.2-200 pg/mL | |
| IL-10 | 0.4-400 pg/mL | |
| IL-12 | 0.6-600 pg/mL | |
| TNFα | 2.4-2400 pg/mL | |
| Tumour Burden, Proliferation, and Invasion | CYFRA 21-1 | 300-50,000 pg/mL |
| EGF | 10-10,000 pg/mLc | |
| KGF | 1-1000 pg/mL | |
| VCAM-1 | 9750-10,000,000 pg/mLc | |
| Multiple processes | HGF | 3.2-3200 pg/mL |
Abbreviations: Ang = angioprotein; CA-IX = carbonic anhydrase; CYFRA 21-1 = cytokeratin-19 antigen; EGF = epidermal growth factor; FGFb = basic fibroblast growth factor; HGF = hepatocyte growth factor; IL = interleukin; KGF = keratinocyte growth factor; M30 = cytokeratin 18 cleaved; M65 = cytokeratin 18 intact; PDGFb = platelet-derived growth factor B; PIGF = placenta growth factor; Tie-2 = tyrosine kinase 2; TNF = tumor necrosis factor; VCAM = vascular cell adhesion molecule 1; VEGF = vascular endothelial growth factor; VEGFR = vascular endothelial growth factor receptor.
One in 25 sample dilution (assay ranges in which the diluted samples were measured).
One in 10 sample dilution.
One in 50 sample dilution.
Blood Sample Collection, Storage, and Processing
Blood samples were collected and processed according to standard operating procedures within the Clinical and Experimental Pharmacology Group at the Cancer Research UK Manchester Institute (Manchester, United Kingdom). Blood was collected in either Monovette serum gel tubes (for processing to serum) or in Monovette Li-heparin tubes (for processing to plasma). Serum samples were left to clot for up to 120 minutes at room temperature and centrifuged at 2000g for 10 minutes. Plasma samples were stored at room temperature and were processed within 120 minutes of collection by centrifuging at 1000g for 10 minutes. Serum as well as plasma samples were transferred immediately to −80°C for storage after processing.
Blood Sample Analysis
Assay measurements were performed in the Cancer Research UK Clinical and Experimental Pharmacology Good Clinical Practice laboratories. Multiplex enzyme-linked immunosorbent assays (ELISAs; Aushon BioSystems, Boston, MA) were used in the following formats; a 6-plex containing assay to measure angioprotein (Ang)-2, basic fibroblast growth factor (FGFb), hepatocyte growth factor (HGF), platelet-derived growth factor B, vascular endothelial growth factor A, and vascular endothelial growth factor C, 2 five-plexes to measure keratinocyte growth factor (KGF), placenta growth factor, vascular endothelial growth factor receptor 1 (VEGFR-1) and VEGFR-2, and interleukin (IL)-1b, IL-6, IL-10, IL-12, and tumor necrosis factor alpha (TNFα; active trimer), a 3-plex to measure EGF, E-selectin, and vascular cell adhesion molecule 1 (VCAM-1), a 2-plex to measure Ang-1 and tyrosine kinase 2 (Tie-2), and a 1-plex to measure osteopontin. SearchLight Plus (Aushon BioSystems, Boston, MA) multiplex ELISA platform was run using the method previously described.11 Cell death (apoptosis and total cell death) was measured using cytokeratin 18 cleaved (M30) and intact (M65) ELISAs (respectively) from Peviva (now VLV Bio, Nacka, Sweden) and run as previously described.12 Carbonic anhydrase (CA-IX) was measured using a single ELISA (R&D Systems, Abingdon, United Kingdom) and cytokeratin-19 antigen (CYFRA 21-1) was measured using a single ELISA from Demeditec (Kiel, Germany); both were run according to the manufacturers' instructions. Recombinant protein quality control (QC) samples were prepared at a high and low level in kit diluent, divided into single-use aliquots and frozen at −80°C; 6 wells of each of the high and low levels of QC were added to each ELISA plate run and the results of all experiments compared to ensure consistency. The upper and lower limits of detection were taken as the highest and lowest points on the standard curve, respectively. M30, M65, and osteopontin were measured in plasma; all other proteins were measured in serum. Samples were analyzed by personnel blinded to individual patient outcome.
Data Collection
The following data were collected for all patients: clinical (pathological diagnosis, tumor, node, metastases [TNM] stage (Seventh American Joint Committee on Cancer edition13), ECOG performance score, weight, and chemotherapy schedule), demographic (age, sex, and smoking status), and routine hematology and biochemistry test results. Radiotherapy details recorded were start and end dates, dose, fractionation, gross target volume (GTV), planning target volume (PTV), radiotherapy delivery technique, lung V20Gy, and mean lung dose.
Radiotherapy-related toxicity was scored prospectively using common terminology criteria for adverse events version 4.014 during weekly on-radiotherapy and follow-up appointments (at 1, 3, 6, and 12 months post-treatment). Acute adverse events were defined as those that arise within 90 days of radiotherapy completion. Treatment response was assessed using Response Evaluation Criteria in Solid Tumors (RECIST) version 1.115 on post-treatment chest x-rays/CT scans performed at 3, 6, and 12 months as per local protocol. Progression-free survival (PFS) was defined as the time from baseline blood sample until the date of development of progressive disease according to RECIST criteria, or death (by any cause). Overall survival (OS) was defined as the time from baseline blood sample until the date of death (by any cause).
We performed 18F-FLT-PET scans at baseline (ie, before start of treatment) and 6 to 15 calendar days (median, 9 days) after start of radiotherapy in patients co-recruited to this substudy. Only a subset of patients with blood biomarkers (n = 13 baseline and n = 11 early treatment) were included in this analysis. PET data were acquired 45 to 60 minutes postinjection of a 30-second bolus of 18F-FLT (mean dose = 311 MBq; range, 254-361 MBq). Scans were reconstructed as a single frame using 3-D ordered subset expectation maximization (4 iterations, 21 subsets) into a 256 × 256 × 109, matrix with voxel sizes of 2.67 × 2.67 × 2.0 mm3 and the images were smoothed using a 4-mm Gaussian filter post reconstruction. Standardized uptake values (SUVs) were derived for the primary tumor, which was manually delineated by an oncological radiologist on the corresponding CT images. Further imaging details have previously been described.10
Statistical Methods
Data visualization methods were used to avoid multiple statistical comparisons. The significance of findings after applying the Bonferroni correction method was reported for correlations involving novel blood biomarkers. P values involving standard clinical variables were not adjusted because they have been previously identified as being significant covariates. Biomarker values were described as being below limit of quantification (BLQ) or above limit of quantification when they are not within the limit of detection (Table 1; and see Supplemental Table 1 in the online version). To visualize variability in the biomarker values within patient population, baseline biomarker data were log-transformed and subsequently each marker scaled by its mean value before generating a variance-covariance matrix. Biomarkers of interest were further explored by analyzing their distributions using histograms. The Kolmogorov–Smirnov test statistic was calculated between the distribution of values at baseline and day 21 for each biomarker.16 This test statistic represents the maximum absolute distance between 2 cumulative distributions, thus the values lie between 0 and 1; 0 implies the 2 distributions overlap whereas 1 indicates no overlap (ie, the 2 distributions are different). All statistical analyses were performed in R version 3.1.1 (https://www.r-project.org).
Gross Target Volume Correlations
The relationship between baseline blood biomarkers and GTV was explored. Correlation plots and P values are reported.
Survival Analyses
The prognostic value of baseline biomarkers and clinical, demographic, routine laboratory, and radiotherapy covariates were assessed using a univariate Cox regression analysis. To develop a multivariate baseline model, biomarkers from the univariate analysis were first ranked according to the χ2 test statistic. The highest ranking variable was designated the base model and extra variables were included in a stepwise fashion if they increased the concordance probability (CP) by a minimum of 0.01. A final prognostic model was generated by combining baseline clinical, demographic, laboratory, and radiotherapy covariates and baseline and day 21 biomarker values in a day 21 landmark Cox regression analysis. For the development of each model, P values from the likelihood ratio test and CP with standard errors were reported. Two risk groups were created from the multivariate baseline model by splitting the median risk scores. The hazard ratio (HR) of OS and PFS curves between the 2 groups is reported with 95% confidence interval (CI).
Toxicity Analysis
A toxicity data set was built by combining select baseline biomarkers (identified through data visualization; see the Statistical Methods section above) and clinical and radiotherapy covariates with Grade ≥ 3 toxicity using ordinal regression. Similar to survival analysis, a univariate analysis was performed first and variables were ranked according to the χ2 test statistic. The highest ranking variable was designated the base model and extra variables were included in a stepwise fashion with P values from the likelihood ratio test reported.
Correlations of 18F-FLT PET
The relationship between blood biomarkers and baseline/early-treatment 18F-FLT PET was explored. To avoid multiple comparisons, only biomarkers of cell death, tumor burden, proliferation, and invasion (Table 1) were investigated because they represent culprit biological biomarkers likely to be related to functional imaging of proliferation. Correlation plots and P values are reported.
Results
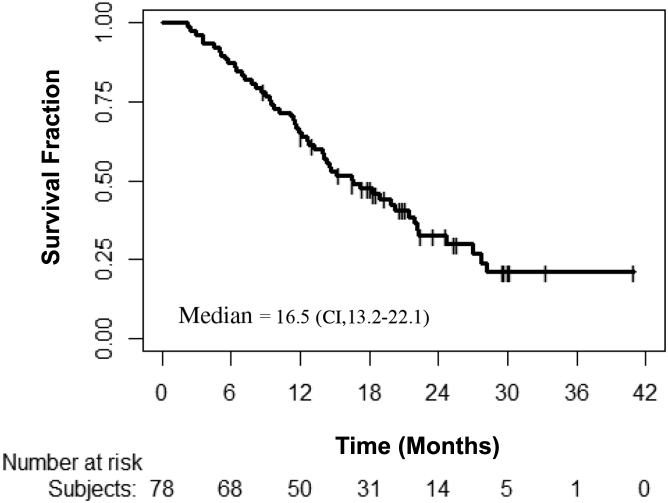
Between March 2010 and February 2012, 90 patients were registered. Eight had missing baseline biomarkers, 2 were withdrawn, 1 was subsequently recruited to a targeted drug-radiotherapy combination trial, and 1 died before the start of treatment leaving 78 analyzable patients. The median age was 68 years (range, 31-86 years). Baseline and treatment characteristics of the analyzable patients are listed in Table 2. The median OS of the entire population was 16.5 months (95% CI, 13.2-22.1; see Supplemental Figure 1 in the online version). There was a higher proportion of patients with NSCLC (78.2%) compared with SCLC. Both groups were initially combined for univariate survival and toxicity analyses but multivariate analyses was focused on NSCLC patients.
Table 2.
Baseline and Treatment Characteristics of the Analyzable Patients
| Characteristic | Subcategory | n (%) |
|---|---|---|
| Age | <65 y | 30 (38.5) |
| ≥65 y | 48 (61.5) | |
| Sex | Male | 50 (64.1) |
| Female | 28 (35.9) | |
| Ethnicity | Caucasian | 77 (98.7) |
| Other | 1 (1.3) | |
| ECOG Performance Status | 0 | 11 (14.1) |
| 1 | 52 (66.7) | |
| 2 | 15 (19.2) | |
| Smoking Status | Never | 1 (1.3) |
| Current | 20 (25.6) | |
| Previous | 56 (71.8) | |
| No data | 1 (1.3) | |
| Weight Loss | Yes | 44 (56.4) |
| No | 34 (43.6) | |
| Histology | NSCLC | 61 (78.2) |
| Squamous cell carcinoma | 33 (42.3) | |
| Adenocarcinoma | 14 (17.9) | |
| NSCLC not otherwise specified | 9 (11.5) | |
| Undifferentiated carcinoma | 3 (3.8) | |
| Large cell carcinoma | 1 (1.3) | |
| Adenosquamous | 1 (1.3) | |
| SCLC | 16 (20.5) | |
| Mixed (SCLC and NSCLC) | 1 (1.3) | |
| Disease Status | De novo | 77 (98.7) |
| Recurrent | 1 (1.3) | |
| Stage | I | 1 (1.3) |
| IIA | 2 (2.6) | |
| IIB | 4 (5.1) | |
| IIIA | 31 (39.7) | |
| IIIB | 35 (44.9) | |
| IV (M1a) | 5 (6.4) | |
| Treatment | Radiotherapy alone | 14 (17.9) |
| Sequential chemoradiation | 48 (61.5) | |
| Concurrent chemoradiation | 16 (20.5) | |
| Radiotherapy Fractionation | 50-55 Gy | 62 (79.5) |
| 60-66 Gy | 16 (20.5) | |
| Radiotherapy Delivery | Intensity modulated radiotherapy | 13 (16.7) |
| 3-D conformal radiotherapy | 65 (83.3) |
Abbreviations: ECOG = Eastern Cooperative Oncology Group; NSCLC = non–small-cell lung cancer; SCLC = small-cell lung cancer.
Supplemental Figure 1.
Kaplan–Meier Curve for Overall Survival of the Entire Patient Population
Baseline Biomarker Analysis
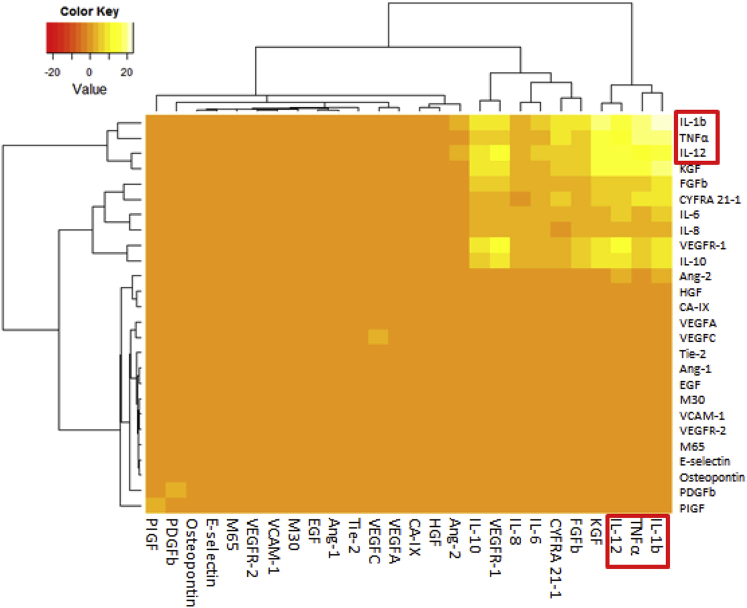
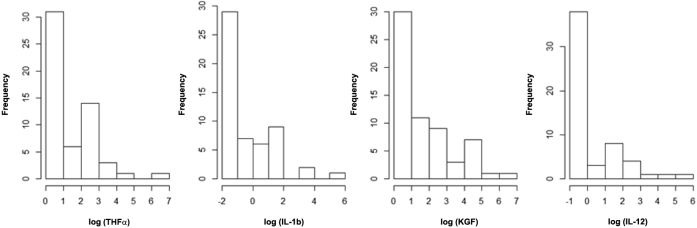
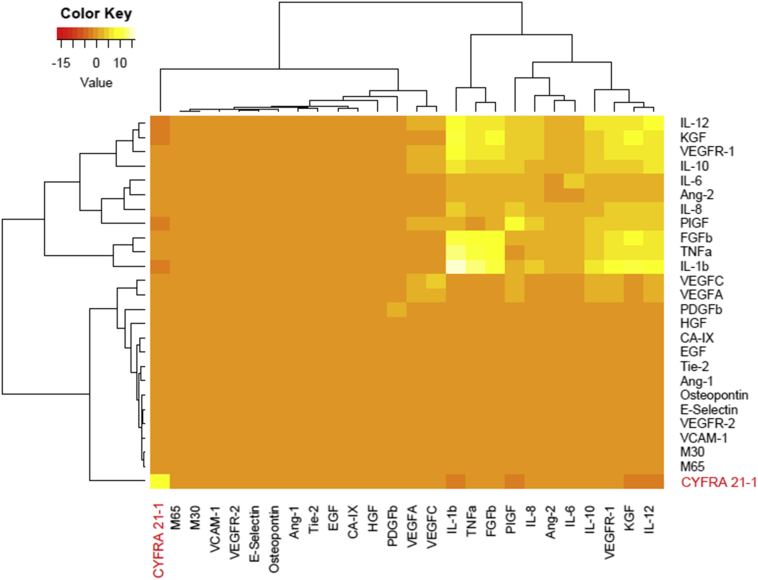
A heat map of the variance-covariance matrix can be seen in the clustergram in Supplemental Figure 2 in the online version. As shown, a subset of markers had high variance and similar covariance pattern: TNFα, IL-1b, KGF, and IL-12. The distribution of these biomarkers (see Supplemental Figure 3 in the online version) highlighted that there are 2 distinct populations, patients who have biomarker values BLQ (high frequency value of the first bar) and those who have values above (spread in frequency after the first bar; see Supplemental Table 1 in the online version). These results suggested a natural cutoff value for these biomarkers for the Cox regression analysis.
Supplemental Figure 2.
Heat Map (With Clustergram) of the Variance–Covariance Matrix After Data Were Log-Transformed and Each Marker Scaled by Its Mean Value for the Baseline Time Point
Abbreviations: Ang = angioprotein; CA-IX = carbonic anhydrase; CYFRA 21-1 = cytokeratin-19 antigen; EGF = epidermal growth factor; FGFb = basic fibroblast growth factor; HGF = hepatocyte growth factor; IL = interleukin; KGF = keratinocyte growth factor; M30 = cytokeratin 18 cleaved; M65 = cytokeratin 18 intact; PDGFb = platelet-derived growth factor B; PIGF = placenta growth factor; Tie-2 = tyrosine kinase 2; TNF = tumor necrosis factor; VCAM = vascular cell adhesion molecule; VEGF = vascular endothelial growth factor; VEGFR = vascular endothelial growth factor receptor.
Supplemental Figure 3.
Histograms of the Log-Transformed Biomarker Values for (From Left to Right); Tumor Necrosis Factor (TNF)α, Interleukin (IL)-1b, Keratinocyte Growth Factor (KGF), and IL-12
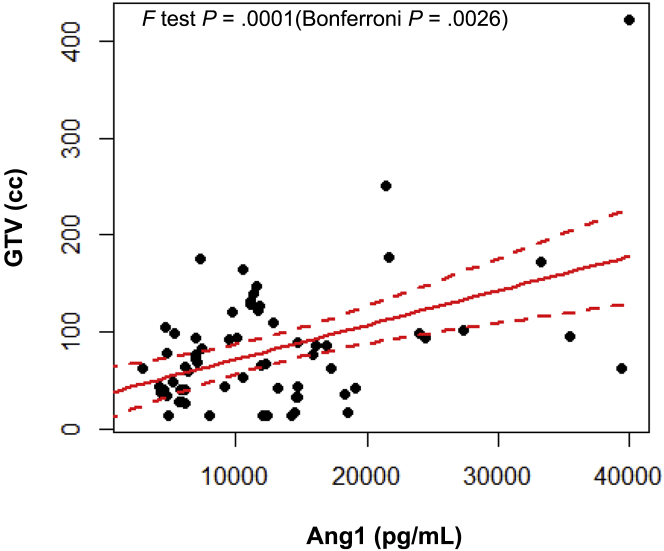
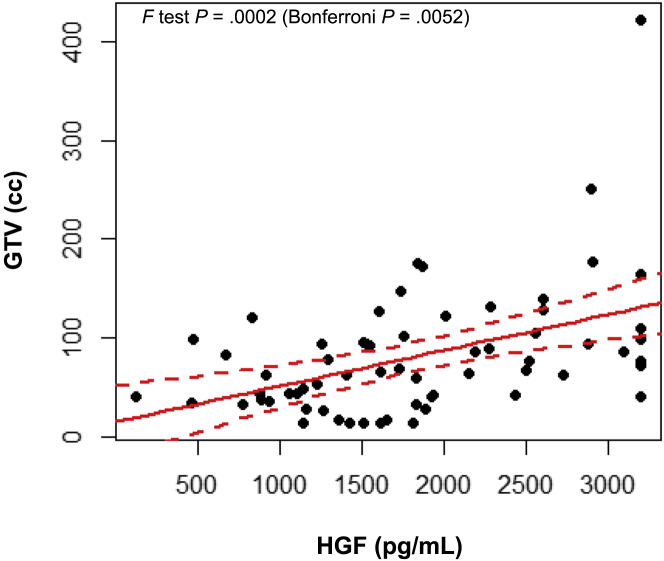
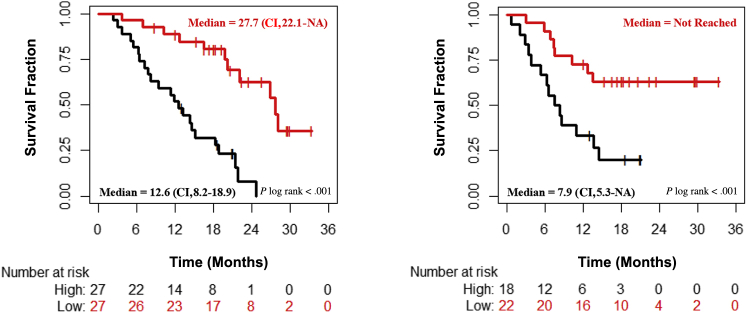
Baseline Ang-1 and HGF showed a significant positive correlation with the GTV (see Supplemental Figures 4 and 5 in the online version) even after Bonferroni correction. None of the other tested biomarkers showed any significant correlation with the GTV. Table 3 shows the correlation between biomarkers and survival after thresholding on the basis of their respective BLQ values. As shown, undetectable IL-1b and TNFα were the strongest covariates associated with poor survival, with only IL-1b remaining significant after Bonferroni correction. None of the clinical, demographic, or radiotherapy variables were prognostic (although PTV, TNM stage, and type of therapy were weakly correlated; P < .10; see Supplemental Table 2 in the online version). Of routinely collected laboratory tests, neutrophil count (but not neutrophil to lymphocyte ratio) was a significant survival covariate (the higher the neutrophil count, the worse the survival). Biomarkers taken forward into multivariate analysis were IL-1b and neutrophil count. IL-1b formed the base model because it had the highest χ2 test statistic value. The multivariate NSCLC baseline survival prediction model was a combination of IL-1b and neutrophil count. This model was then used to create 2 risk groups (low and high) by splitting the median risk score value. The difference in OS and PFS between these 2 risk groups are shown in Figure 1 and Supplemental Table 3 in the online version. The HR between low and high risk groups for OS is 0.18 (95% CI, 0.08-0.41; log-rank P < .001). For PFS, the HR between low- and high-risk groups is 0.30 (95% CI, 0.13-0.72; log-rank P = .004).
Supplemental Figure 4.
Correlation Between Baseline Angioprotein (Ang)-1 and Gross Target Volume (GTV; Solid Red Line) With 95% CI (Red Dashed Lines)
Supplemental Figure 5.
Correlation Between Baseline hepatocyte growth factor (HGF) and Gross Target Volume (GTV; Solid Red Line) With 95% CI (Red Dashed Lines)
Table 3.
Survival Concordance Probability With SE, Hazard Ratio With 95% CI, and Unadjusted P Value From LRT for the Univariate and Multivariate Analyses
| Analysis | Marker | Concordance Probability (SE) | Hazard Ratio (95% CI) | LRT P |
|---|---|---|---|---|
| Univariate Analysis (NSCLC and SCLC) | TNFα ≤ BLQ | 0.60 (0.04) | 2.27 (1.22-4.23) | .008 |
| IL-1b ≤ BLQ | 0.65 (0.04) | 4.02 (2.04-7.93) | <.001 | |
| KGF ≤ BLQ | 0.51 (0.03) | 1.16 (0.63-2.11) | .639 | |
| IL-12 ≤ BLQ | 0.56 (0.04) | 2.00 (1.05-3.82) | .030 | |
| Neutrophilsa | 0.60 (0.05) | 1.07 (1.02-1.11) | .017 | |
| Lymphocytesa | 0.48 (0.05) | 1.03 (0.97-1.09) | .410 | |
| Neutrophil to lymphocyte ratio | 0.58 (0.05) | 1.02 (0.98-1.06) | .396 | |
| Final Baseline Model (NSCLC Only) | IL-1b ≤ BLQ | 0.67 (0.05) | <.001 | |
| IL-1b ≤ BLQ + neutrophils | 0.74 (0.06) | .042 | ||
| IL-1b ≤ BLQ | 4.62 (2.11-10.14) | |||
| Neutrophilsa | 1.07 (1.01-1.14) |
Blood marker thresholds were on the basis of their respective BLQ values. Statistically significant values are shown in bold.
Abbreviations: BLQ = below limit of quantification; IL = interleukin; KGF = keratinocyte growth factor; LRT = likelihood ratio test; NSCLC = non–small-cell lung cancer; SCLC = small-cell lung cancer; TNF = tumor necrosis factor.
Continuous variable; the higher the neutrophils the worse the survival.
Figure 1.
Kaplan–Meier Curves of Overall Survival (Left Panel) and Progression-Free Survival (Right Panel) Between the High (Red) and Low (Black) Risk Groups Created Using the Multivariate Baseline Model for Non–Small-Cell Lung Cancer
Day 21 Biomarker Analysis
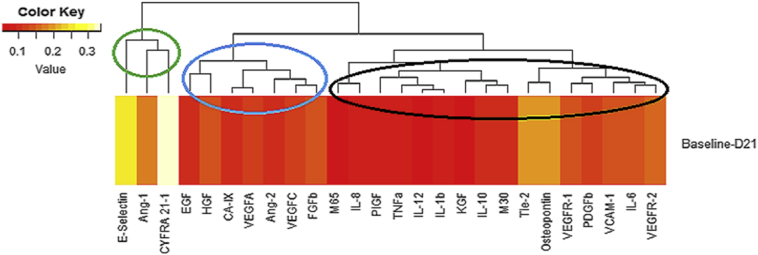
A matrix of the Kolmogorov–Smirnov test statistic values for all biomarkers can be visualized using the heat-map in Supplemental Figure 6 in the online version. This highlights 3 distinct groups of biomarkers; the group circled in green relate to biomarker distributions that change the most between baseline and day 21, the group circled in blue have modest changes, and the group circled in black show very little change. The markers circled in green and blue (E-selectin, Ang-1, CYFRA 21-1, EGF, HGF, CA-IX, VEGF-A, Ang-2, VEGFC, and FGFb) were further analyzed by creating a heat map using the same methods used for Supplemental Figure 2 in the online version. This heat map is shown in Supplemental Figure 7 in the online version. It has 1 distinct outlier, CYFRA 21-1, on the far left, suggesting relevance. Another cluster on the right side is shown. Markers that cluster together have high variance and high positive covariance pattern. Of these markers only Ang-2 and FGFb were identified in Supplemental Figure 6 in the online version. Therefore, day-21 biomarkers taken forward for further analysis were CYFRA 21-1, Ang-2, and FGFb. None of the participants had any events or were right censored before this time point.
Supplemental Figure 6.
Heat Map (With Clustergram) of the Kolmogorov–Smirnov Test Statistic Matrix Comparing Each Day-21 (D21) Value With Baseline
Abbreviations: Ang = angioprotein; CA-IX = carbonic anhydrase; CYFRA 21-1 = cytokeratin-19 antigen; EGF = epidermal growth factor; FGFb = basic fibroblast growth factor; HGF = hepatocyte growth factor; IL = interleukin; KGF = keratinocyte growth factor; M30 = cytokeratin 18 cleaved; M65 = cytokeratin 18 intact; PDGFb = platelet-derived growth factor B ; PIGF = placenta growth factor; Tie-2 = tyrosine kinase 2; TNF = tumor necrosis factor; VCAM = vascular cell adhesion molecule; VEGF = vascular endothelial growth factor; VEGFR = vascular endothelial growth factor receptor.
Supplemental Figure 7.
Heat Map (With Clustergram) of the Variance–Covariance Matrix After Data Were Log-Transformed and Each Marker Scaled by Its Mean Value for Day 21
Abbreviations: Ang = angioprotein; CA-IX = carbonic anhydrase; CYFRA 21-1 = cytokeratin-19 antigen; EGF = epidermal growth factor; FGFb = basic fibroblast growth factor; HGF = hepatocyte growth factor; IL = interleukin; KGF = keratinocyte growth factor; M30 = cytokeratin 18 cleaved; M65 = cytokeratin 18 intact; PDGFb = platelet-derived growth factor B; PIGF = placenta growth factor; Tie-2 = tyrosine kinase 2; TNF = tumor necrosis factor; VCAM = vascular cell adhesion molecule; VEGF = vascular endothelial growth factor; VEGFR = vascular endothelial growth factor receptor.
Univariate analysis showed that detectable on-treatment CYFRA 21-1 was the highest ranking biomarker to correlate with worse survival and remained so after Bonferroni correction (see Supplemental Table 4 in the online version). The addition of on-treatment CYFRA 21-1 to the NSCLC baseline survival prediction model modestly improved this model's survival prediction (CP, 0.75; P = .029-.78, P = .004).
Toxicity Covariates
The relationship between clinical and radiotherapy covariates and biomarkers with Grade ≥ 3 acute pneumonitis and esophagitis is shown in Table 4. Chemotherapy (P < .001) and baseline KGF (P = .019) predicted Grade ≥ 3 acute esophagitis in univariate analysis but only chemotherapy remained significant after Bonferroni correction. As shown, none of the tested variables correlated with Grade ≥ 3 acute pneumonitis.
Table 4.
Results of the Univariate Ordinal Regression Analysis of Toxicity Data
| Toxicity | Variable | LRT P |
|---|---|---|
| Grade ≥3 Acute Esophagitis | Chemotherapya | <.001 |
| IL-1b ≤ BLQ | .240 | |
| TNFα ≤ BLQ | .511 | |
| KGF ≤ BLQ | .019 | |
| IL-12 ≤ BLQ | .295 | |
| Grade ≥3 Acute Pneumonitis | Mean lung dose | .497 |
| Lung V20Gy | .745 | |
| Chemotherapya | .546 | |
| IL-1b ≤ BLQ | .824 | |
| TNFα ≤ BLQ | .529 | |
| KGF ≤ BLQ | .610 | |
| IL-12 ≤ BLQ | .445 |
Blood marker thresholds were on the basis of their respective BLQ values. Statistically significant values are shown in bold. Unadjusted P value from LRT are reported.
Abbreviations: BLQ = below limit of quantification; IL = interleukin; KGF = keratinocyte growth factor; LRT = likelihood ratio test; TNF = tumor necrosis factor; V20Gy = percentage of the lung volume receiving ≥20 Gy.
Chemotherapy was modeled by investigating concurrent versus none, concurrent versus sequential, and sequential versus none.
Correlation of 18F-FLT PET
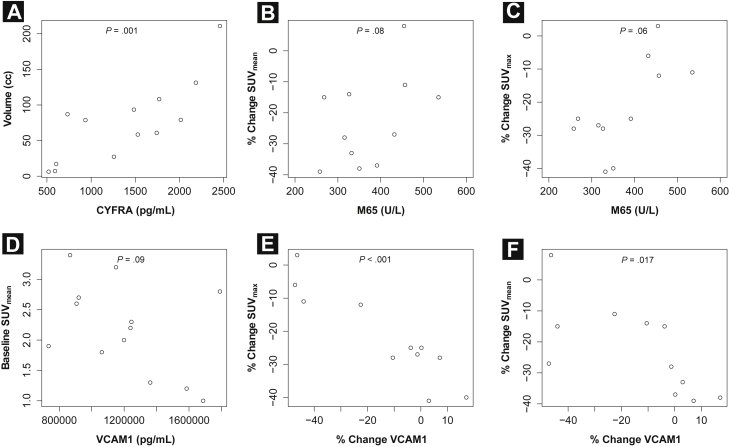
Baseline CYFRA 21-1 and EGF showed a positive correlation with the volume of the primary tumor on baseline 18FLT-PET CT (P = .001 and .011, respectively), with CYFRA 21-1 remaining significant after Bonferroni correction. There was a trend for an inverse correlation between baseline VCAM-1 and baseline mean 18FLT-PET SUV (18FLT-PET SUVmean; P = .09). Further, there was a trend for baseline M65 to predict early-treatment changes in maximum 18FLT-PET SUV (18FLT-PET SUVmax; P = .06) and SUVmean (P = .08) with lower levels associated with greater reduction in SUV values. However, none of these findings remain significant after applying Bonferroni correction. Last, early-treatment changes (baseline compared with blood sample taken on the day of 18FLT-PET) in VCAM-1 correlated inversely with early-treatment changes in 18FLT-PET SUVmax (P < .001) and 18FLT-PET SUVmean (P = .017), with only the former remaining significant after applying Bonferroni correction. These results are depicted in Figure 2. None of the other tested biomarkers showed any significant association with 18FLT-PET.
Figure 2.
Correlation Between (A) Baseline Cytokeratin-19 Antigen (CYFRA) and the Volume of the Primary Tumor on Baseline [18]fluorothymidine (18FLT)-Positron Emission Tomography (PET) Computed Tomography; (B) Baseline VCAM-1 and Baseline 18FLT-PET Mean Standardized Uptake Value (SUVmean); (C) Baseline M65 and Early-Treatment Changes in 18FLT-PET Maximum Standardized Uptake Value (SUVmax); (D) and SUVmean; (E) Early-Treatment Changes in VCAM-1 (Baseline Compared With Blood Sample Taken on Day of 18FLT-PET) and Early-Treatment Changes in 18FLT-PET SUVmax; (F) Early-Treatment Changes in VCAM-1 (Baseline Compared With Blood Sample Taken on Day of 18FLT-PET) and Early-Treatment Changes in 18FLT-PET SUVmean
Abbreviation: VCAM-1 = vascular cell adhesion molecule 1.
Discussion
This pilot study evaluated a broad cytokine, growth factor, and circulating marker panel as predictors of lung cancer radiotherapy response and toxicity. We showed that select inflammation and tumor-burden biomarkers (TNFα, IL-1b, IL-12, and CYFRA 21-1) and baseline neutrophil count were associated with patient outcomes in univariate analysis. IL-1b, IL-12, and TNFα are known proinflammatory cytokines.17, 18 IL-1b is associated with tumor proliferation, invasion, and migration and is upregulated in NSCLC patients.17 Elevated blood TNFα level has been linked with advanced/metastatic NSCLC and tumor progression, but not survival.19, 20 No published studies have investigated the effect of blood IL-12 on lung cancer patient survival. We showed that baseline undetectable IL-1b is an independent significant prognostic factor of survival in lung cancer patients treated with radiotherapy. The addition of baseline neutrophil count to IL-1b led to improvement in the CP of the final baseline prognostic model in NSCLC patients. The prognostic value of pretreatment neutrophil count was previously shown in stage IIIB-IV NSCLC patients treated with chemotherapy within a randomized trial but has not been reported for patients treated with radiotherapy.21 Neutrophils inhibit apoptosis and promote angiogenesis and metastases, thus exerting protumorigenesis effects.22 Interleukins, particularly IL-1b and IL-8, are involved in neutrophil priming and migration.23, 24 IL-1b is also involved in tumor-associated neutrophil recruitment leading to tumor growth inhibition.25 In our study, undetectable IL-1b is associated with poor prognosis which agrees with these preclinical observations, but not with a clinical study that reported that high IL-1b was independently associated with worse OS (HR, 2.24; 95% CI, 1.01-4.98; P = .047) in stage IIIB-IV NSCLC patients treated with chemotherapy.26 Possible explanations for this discrepancy include the application of different biomarker cutoffs (3.0 pg/mL vs. 0.2 pg/mL in our study) and treatments (palliative chemotherapy vs. curative-intent [chemo]-radiotherapy in our study) and the small number of patients (n = 10) who showed an IL-1b value ≥ 3.00 pg/mL in the study by Kim et al.26 We have previously reported the negative prognostic effect of high C-reactive protein, another marker of acute injury and inflammation, in the proteomics analysis of this data set.27 However, we failed to detect any prognostic value of neutrophil to lymphocyte count on OS or PFS. This is in contradiction to results synthesized from 2 meta-analyses22, 28 and a growing number of subsequent studies.29, 30 Similarly, the prognostic value of circulating osteopontin31 and M6532 was not reproduced in our study. This might be because of a number of factors, such as sample size, the lack of specificity of these biomarkers (eg, osteopontin is elevated in nononcological diseases), and variation in protocols for blood sampling, collection, storage, and analysis. Our study was conducted using a validated assay11 to reduce the possibility of results due to artifacts from inconsistent biomarker processing, storage, and analysis. The lack of correlation between clinical outcome and established tumor variables (eg, tumor stage) in our study could be explained by the predominance of patients with stage III disease (84.6%), limiting the ability to detect the prognostic capacity of this variable because of the small number of patients with early (stage I-II) or advanced (stage IV) tumor stages. The same explanation applies to tumor size.
Early-treatment blood sampling was incorporated to inform on temporal biomarker changes and their clinical significance. Day 21 was chosen because this is a clinically-relevant time point that could permit midtreatment risk stratification and adaptation in future clinical trials. We have previously shown significant reductions in 18FLT-PET SUVmax and SUVmean in the primary tumor after 5 to 11 radiotherapy fractions in NSCLC patients in the absence of tumor volume changes.10 The prognostic significance of baseline CYFRA 21-1 was established in numerous NSCLC clinical studies, with higher levels being associated with worse prognosis.33, 34, 35 Because CYFRA 21-1 is related to tumor burden,36 determination of early-treatment CYFRA 21-1 was proposed as a potential treatment response biomarker. We show that early-treatment CYFRA 21-1 is associated with worse prognosis. This is in agreement with previous studies that reported that early reduction in CYFRA 21-1 is associated with improved NSCLC treatment response.35, 37, 38, 39 Very few studies have established the prognostic effect of baseline CYFRA 21-1 in SCLC patients.40, 41 Although our study only included 16 SCLC patients (20.5%), it suggests the potential utility of this marker, when quantified early during treatment, in these patients. The significance of high circulating levels of FGFb (a known mediator of angiogenesis) on survival in cancer patients is not clear. A few studies have shown a negative prognostic effect,42, 43 but this relationship is not consistent across studies44 and might even be reversed in SCLC patients.45 In our study, detectable day-21 FGFb was correlated with improved survival in SCLC as well as NSCLC in univariate analysis, albeit of borderline significance (P = .045).
Esophageal ulceration was shown to induce KGF (an epithelial fibroblast growth factor) overexpression in the adjacent esophageal stroma in rates in a previously published preclinical study.46 Interestingly, in our study undetectable baseline KGF was associated with Grade ≥ 3 acute radiation esophagitis in univariate analysis, but did not remain significant after Bonferroni correction. Acute radiation esophagitis is relatively common in lung cancer patients treated with radiotherapy, particularly in the context of mediastinal involvement, concurrent chemotherapy, and radiotherapy dose escalation. Currently, there are no known circulating biomarkers to accurately identify patients at increased risk of developing clinically significant acute radiation esophagitis. There was no link between acute radiation pneumonitis and blood biomarkers. Dosimetric parameters (eg, lung V20Gy) are known to predict symptomatic acute, but not late radiotherapy-related lung toxicity.47 Surprisingly, there was no correlation between lung dosimetric parameters and Grade ≥ 3 acute pneumonitis in our study. This could be explained by the predominance of patients with stage III disease (84.6%) and strict adherence with dosimetric lung constraints (none of the included patients had a V20 > 35%).
We show a significant positive correlation between baseline Ang-1 and HGF with the GTV even after Bonferroni correction. In a previous study of 115 surgically resected lung adenocarcinoma patients, tumor coexpression of HGF and neuregulin 1 (NRG1; a cell adhesion molecule) occurred more frequently in tumors > 3 cm in size.48 Ang-1 is a known promoter of tumor angiogenesis, which is essential for tumor growth.49, 50
By targeting the activity of the thymidine salvage pathway, 18FLT-PET is able to image tumor proliferation.51 We have previously shown that radiotherapy induces early reduction in tumor FLT uptake (exceeding test-retest variability) in the absence of significant mean volumetric change.10 Functional tumor proliferation imaging could provide useful information for drug development; however, routine integration within clinical trials is likely to be met with difficulty. Blood biomarkers could be used to select patients for assessment with functional imaging in an attempt to decrease the unnecessary use of these expensive, resource intensive, and patient-demanding procedures.34 We have shown that baseline CYFRA 21-1 (which is related to tumor burden36) showed a positive correlation with the volume of the primary tumor on baseline 18FLT-PET CT whereas early-treatment changes in VCAM-1 were inversely correlated with changes in 18FLT-PET. VCAM-1 is a cell adhesion molecule that plays an important role in the vascular endothelium and inflammatory reaction.52 A few published preclinical studies have reported on the role of VCAM-1 in cellular proliferation, but none specifically addressed the role of VCAM-1 in tumor proliferation.53, 54 Although we acknowledge the small number of patients included in this imaging substudy, it noteworthy that the direction of the correlation between VCAM-1 and 18FLT-PET was upheld for SUVmax as well as SUVmean. For this reason, we believe these findings support future investigation of a potential role of VCAM-1 as a 18FLT-PET surrogate.
The advantages of blood biomarkers as predictors of radiotherapy response and toxicity cannot be overstated. Measurements are simple and can be repeated without subjecting patients to overly invasive tests or ionizing radiation. The improved understanding of the mechanisms of radiotherapy response and toxicity could allow radiotherapy dose individualization to achieve a balance between optimal tumor control and acceptable normal tissue toxicity. The inclusion of SCLC as well as NSCLC patients in our study resulted in a heterogeneous population. However, the distribution of included patients closely reflects the typical patient population who are offered curative-intent radiotherapy in the clinical setting.
In our study we chose to combine both groups (SCLC and NSCLC) initially for univariate survival and toxicity analyses but multivariate analysis subsequently focused on NSCLC patients only because of the small number of SCLC patients included in this study, and the inability to combine patients into 1 model because of the differences in natural history between SCLC and NSCLC patients. We acknowledge that this approach could have missed biomarkers specific for either disease. Biomarkers that show clinical outcome prediction in one study should be independently replicated in other studies to ensure validity of the generated results.55 This independent validation was not possible in our study and this is an additional study limitation.
According to our knowledge, this study evaluated the largest panel of cytokines, growth factors, and circulating markers ever reported, which represent a wide spectrum of molecularly relevant tumor and normal tissue characteristics, investigating their clinical significance in lung cancer patients. Our findings were also assessed in conjunction with routinely acquired blood tests, such as full blood count, showing the merit of this combination. Further, the longitudinal study design allowed us to highlight the additional advantage of a prognostic model on the basis of a combination of biomarkers sampled over different time points (baseline and early-treatment). According to our knowledge, this study is the first to report a relationship between blood biomarkers and functional imaging of proliferation in lung cancer patients. These preliminary results show, in principle, that this approach is worthy of further investigation in larger populations.
Conclusion
In this study, a wide panel of candidate circulating biomarkers were assessed for clinical utility in a radiotherapy-treated population. Baseline biomarkers of inflammation (IL-1b and neutrophil count) and early-treatment tumor burden (CYFRA 21-1) predict for survival in lung cancer patients treated with radiotherapy. Together with our finding of circulating biomarker correlation with functional imaging of proliferation, these results provide new candidate, minimally invasive, blood-borne biomarkers to incorporate into mechanism-based therapy-radiotherapy combination trials.
Clinical Practice Points
-
•
There is an unmet need to develop noninvasive radiotherapy response and toxicity biomarkers to tailor radiotherapy, stratify patients according to radiosensitivity, and select patients for future combination trials.
-
•
Baseline IL-1b and neutrophil count and early-treatment CYFRA 21-1 predict lung cancer radiotherapy response.
-
•
Baseline Ang-1 and HGF significantly correlated with the gross tumor volume.
-
•
Changes in VCAM-1 correlated with proliferation imaging, highlighting for the first time a potential role of blood biomarkers as less-invasive imaging surrogates.
-
•
These results provide new candidate, minimally invasive blood-borne biomarkers to incorporate into mechanism-based therapy-radiotherapy combination trials.
The funders had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the report for publication.
Disclosure
The authors have stated that they have no conflicts of interest.
Acknowledgments
This is a contribution from the Cambridge-Manchester Cancer Imaging Centre, which is funded by the Engineering and Physical Sciences Research Council and Cancer Research UK (CRUK) and the Manchester-University College London Lung Cancer Centre of Excellence, which is funded by CRUK [grant No. C8742/A18097, 2013-2018]. This work was supported by core funding to CRUK Manchester Institute (C5759/A12328), and via Manchester CRUK Centre Award (A12197). The authors gratefully acknowledge the financial support from AstraZeneca Pharmaceuticals, the Oglesby Charitable Trust, the Christie Lung Cancer Research Fund, Manchester Cancer Research Centre, and CRUK. This work was supported by the National Institute for Health Research (NIHR) Christie Clinical Research Facility. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. We thank the staff at the Christie NHS Foundation Trust and the Wolfson Molecular Imaging Centre as well as all the participants for volunteering in the study.
Footnotes
Supplemental figures and tables accompanying this article can be found in the online version at https://doi.org/10.1016/j.cllc.2017.12.002.
Supplemental Data
Supplemental Table 1.
The Number of Baseline TNFα, IL-1b, KGF, and IL-12 Values That Are BLQ or Greater Than BLQ
| Biomarker | Number BLQ | Number Greater Than BLQ |
|---|---|---|
| TNFα | 30 | 26 |
| IL-1b | 25 | 29 |
| KGF | 15 | 47 |
| IL-12 | 34 | 22 |
Abbreviations: BLQ = below limit of quantification; IL = interleukin; KGF = keratinocyte growth factor; TNF = tumor necrosis factor.
Supplemental Table 2.
Summary of the Survival Analysis of Clinical/Demographic Variables Concordance Probabilities With SE, Hazard Ratio, and P Value From the LRT Reported for All Patients (NSCLC and SCLC)
| Clinical/Demographic Variable | Concordance Probability (SE) | Hazard Ratio (95% CI) | LRT P |
|---|---|---|---|
| GTVa | 0.59 (0.05) | 1.38 (0.90-2.12) | .139 |
| PTVa | 0.60 (0.04) | 1.99 (0.98-4.05) | .054 |
| ECOG Performance Status (2 vs. 1 vs. 0) | 0.50 (0.04) | 1.05 (0.65-1.71) | .832 |
| Smoking History (Any vs. None) | 0.52 (0.03) | 1.25 (0.68-2.28) | .477 |
| TNM Stage (I/II vs. III vs. IV) | 0.56 (0.03) | 2.07 (0.96-4.45) | .056 |
| Lung V20Gya | 0.59 (0.05) | 1.53 (0.69-3.38) | .280 |
| Mean Lung Dosea | 0.45 (0.04) | 0.98 (0.52-1.85) | .946 |
| Chemotherapy (Sequential vs. Concurrent vs. None) | 0.56 (0.03) | 1.92 (0.86-4.26) | .084 |
Abbreviations: ECOG = Eastern Cooperative Oncology Group; GTV = gross target volume; LRT = likelihood ratio test; NSCLC = non–small-cell lung cancer; PTV = planning target volume; SCLC = small-cell lung cancer; TNM = tumor, node, metastases; V20Gy = percentage of the lung volume receiving ≥20 Gy.
Log-transformed.
Supplemental Table 3.
One-Year OS and PFS Fraction for Low- and High-Risk Groups
| Survival | Risk Group | Surviving Fraction at 1-Year (95% CI) |
|---|---|---|
| OS | Low | 0.82 (0.69-0.96) |
| High | 0.45 (0.31-0.66) | |
| PFS | Low | 0.73 (0.56-0.94) |
| High | 0.33 (0.17-0.64) |
Abbreviations: OS = overall survival; PFS = progression-free survival.
Supplemental Table 4.
Survival Concordance Probability With SE, Hazard Ratio With 95% CI, and Associated P Value From the LRT for Univariate and Multivariate Analysis Combining Baseline and Day-21 Blood Biomarkers
| Analysis | Marker | Concordance Probability (SE) | Hazard Ratio (95% CI) | LRT P |
|---|---|---|---|---|
| Univariate Analysis Day 21 (NSCLC and SCLC) | CYFRA 21-1a | 0.63 (0.05) | 2.09 (1.41-3.09) | <.001 |
| Ang-2 | 0.57 (0.05) | 0.77 (0.58-1.03) | .090 | |
| FGFba | 0.59 (0.05) | 0.78 (0.60-1.00) | .045 | |
| Final Overall Model (NSCLC Only) | IL-1b ≤ BLQ (baseline) | 0.68 (0.05) | <.001 | |
| IL-1b ≤ BLQ (baseline) + neutrophils (baseline) | 0.75 (0.06) | .029 | ||
| IL-1b ≤ BLQ (baseline) + neutrophils (baseline) + CYFRA 21-1 (day 21)a | 0.78 (0.06) | .004 | ||
| IL-1b ≤ BLQ (baseline) | 3.42 (1.38-8.51) | |||
| Neutrophils (baseline) | 1.08 (1.02-1.14) | |||
| CYFRA 21-1 (day 21)a | 2.07 (1.27-3.38) |
Blood marker thresholds were on the basis of their respective BLQ values. Statistically significant values are shown in bold.
Abbreviations: Ang = angioprotein; BLQ = below limit of quantification; CYFRA 21-1 = cytokeratin-19 antigen; FGFb = basic fibroblast growth factor; IL = interleukin; LRT = likelihood ratio test; NSCLC = non–small-cell lung cancer; SCLC = small-cell lung cancer.
Log-transformed.
References
- 1.Bradley J.D., Paulus R., Komaki R. Standard-dose versus high-dose conformal radiotherapy with concurrent and consolidation carboplatin plus paclitaxel with or without cetuximab for patients with stage IIIA or IIIB non–small-cell lung cancer (RTOG 0617): a randomised, two-by-two factorial phase 3 study. Lancet Oncol. 2015;16:187–199. doi: 10.1016/S1470-2045(14)71207-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turrisi A.T., 3rd, Kim K., Blum R. Twice-daily compared with once-daily thoracic radiotherapy in limited small-cell lung cancer treated concurrently with cisplatin and etoposide. N Engl J Med. 1999;340:265–271. doi: 10.1056/NEJM199901283400403. [DOI] [PubMed] [Google Scholar]
- 3.Christodoulou M., Bayman N., McCloskey P., Rowbottom C., Faivre-Finn C. New radiotherapy approaches in locally advanced non–small-cell lung cancer. Eur J Cancer. 2014;50:525–534. doi: 10.1016/j.ejca.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 4.Woolf D.K., Slotman B.J., Faivre-Finn C. The current role of radiotherapy in the treatment of small-cell lung cancer. Clin Oncol. 2016;28:712–719. doi: 10.1016/j.clon.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Allemani C., Weir H.K., Carreira H. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2) Lancet. 2015;385:977–1010. doi: 10.1016/S0140-6736(14)62038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boss M.K., Bristow R., Dewhirst M.W. Linking the history of radiation biology to the hallmarks of cancer. Radiat Res. 2014;181:561–577. doi: 10.1667/RR13675.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuneo K.C., Nyati M.K., Ray D., Lawrence T.S. EGFR targeted therapies and radiation: optimizing efficacy by appropriate drug scheduling and patient selection. Pharmacol Ther. 2015;154:67–77. doi: 10.1016/j.pharmthera.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naghavi A.O., Johnstone P.A., Kim S. Clinical trials exploring the benefit of immunotherapy and radiation in cancer treatment: a review of the past and a look into the future. Curr Probl Cancer. 2016;40:38–67. doi: 10.1016/j.currproblcancer.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 9.Chan C., Lang S., Rowbottom C., Guckenberger M., Faivre-Finn C. Intensity-modulated radiotherapy for lung cancer: current status and future developments. J Thorac Oncol. 2014;9:1598–1608. doi: 10.1097/JTO.0000000000000346. [DOI] [PubMed] [Google Scholar]
- 10.Trigonis I., Koh P.K., Taylor B. Early reduction in tumour [18F]fluorothymidine (FLT) uptake in patients with non–small-cell lung cancer (NSCLC) treated with radiotherapy alone. Eur J Nucl Med Mol Imaging. 2014;41:682–693. doi: 10.1007/s00259-013-2632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Backen A.C., Cummings J., Mitchell C., Jayson G., Ward T.H., Dive C. “Fit-for-purpose” validation of SearchLight multiplex ELISAs of angiogenesis for clinical trial use. J Immunol Methods. 2009;342:106–114. doi: 10.1016/j.jim.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Greystoke A., Cummings J., Ward T. Optimisation of circulating biomarkers of cell death for routine clinical use. Ann Oncol. 2008;19:990–995. doi: 10.1093/annonc/mdn014. [DOI] [PubMed] [Google Scholar]
- 13.Edge S., American Joint Committee on Cancer. American Cancer Society . Springer; New York: 2010. AJCC Cancer Staging Handbook: From the AJCC Cancer Staging Manual. [Google Scholar]
- 14.US Department of Health and Human Services . National Institutes of Health, National Cancer Institute; Bethedsa, MD: 2009. Common Terminology Criteria for Adverse Events (CTCAE) Version 4.0. [Google Scholar]
- 15.Eisenhauer E., Therasse P., Bogaerts J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 16.DeGroot M.H., Schervish M.J. Addison-Wesley; Harlow, Essex, England: 2012. Probability and Statistics. [Google Scholar]
- 17.Wang L., Zhang L.F., Wu J. IL-1beta-mediated repression of microRNA-101 is crucial for inflammation-promoted lung tumorigenesis. Cancer Res. 2014;74:4720–4730. doi: 10.1158/0008-5472.CAN-14-0960. [DOI] [PubMed] [Google Scholar]
- 18.Zijlmans H.J., Fleuren G.J., Baelde H.J., Eilers P.H., Kenter G.G., Gorter A. Role of tumor-derived proinflammatory cytokines GM-CSF, TNF-alpha, and IL-12 in the migration and differentiation of antigen-presenting cells in cervical carcinoma. Cancer. 2007;109:556–565. doi: 10.1002/cncr.22428. [DOI] [PubMed] [Google Scholar]
- 19.De Vita F., Orditura M., Auriemma A., Infusino S., Catalano G. Serum concentrations of proinflammatory cytokines in advanced non–small-cell lung cancer patients. J Exp Clin Cancer Res. 1998;17:413–417. [PubMed] [Google Scholar]
- 20.Derin D., Soydinc H.O., Guney N. Serum levels of apoptosis biomarkers, survivin and TNF-alpha in non–small-cell lung cancer. Lung Cancer. 2008;59:240–245. doi: 10.1016/j.lungcan.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Teramukai S., Kitano T., Kishida Y. Pretreatment neutrophil count as an independent prognostic factor in advanced non–small-cell lung cancer: an analysis of Japan Multinational Trial Organisation LC00-03. Eur J Cancer. 2009;45:1950–1958. doi: 10.1016/j.ejca.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 22.Gu X.B., Tian T., Tian X.J., Zhang X.J. Prognostic significance of neutrophil-to-lymphocyte ratio in non–small-cell lung cancer: a meta-analysis. Sci Rep. 2015;5:12493. doi: 10.1038/srep12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oliveira S.H., Canetti C., Ribeiro R.A., Cunha F.Q. Neutrophil migration induced by IL-1beta depends upon LTB4 released by macrophages and upon TNF-alpha and IL-1beta released by mast cells. Inflammation. 2008;31:36–46. doi: 10.1007/s10753-007-9047-x. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki O., Goto T., Yoshino T., Nakamura S., Maeda H. The role of phosphodiesterase 4B in IL-8/LTB4-induced human neutrophil chemotaxis evaluated with a phosphodiesterase 4B inhibitor. Acta Pharm. 2015;65:191–197. doi: 10.1515/acph-2015-0016. [DOI] [PubMed] [Google Scholar]
- 25.Chen L.C., Wang L.J., Tsang N.M. Tumour inflammasome-derived IL-1beta recruits neutrophils and improves local recurrence-free survival in EBV-induced nasopharyngeal carcinoma. EMBO Mol Med. 2012;4:1276–1293. doi: 10.1002/emmm.201201569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J.W., Koh Y., Kim D.W. Clinical implications of VEGF, TGF-beta1, and IL-1beta in patients with advanced non–small-cell lung cancer. Cancer Res Treat. 2013;45:325–333. doi: 10.4143/crt.2013.45.4.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walker M.J., Zhou C., Backen A. Discovery and validation of predictive biomarkers of survival for non–small-cell lung cancer patients undergoing radical radiotherapy: two proteins with predictive value. EBioMedicine. 2015;2:839–848. doi: 10.1016/j.ebiom.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin Y., Wang J., Wang X. Prognostic value of the neutrophil to lymphocyte ratio in lung cancer: a meta-analysis. Clinics. 2015;70:524–530. doi: 10.6061/clinics/2015(07)10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi Y., Horio H., Hato T. Prognostic significance of preoperative neutrophil-lymphocyte ratios in patients with stage I non–small-cell lung cancer after complete resection. Ann Surg Oncol. 2015;22(suppl 3):S1324–S1331. doi: 10.1245/s10434-015-4735-5. [DOI] [PubMed] [Google Scholar]
- 30.Zhang H., Zhang L., Zhu K. Prognostic significance of combination of preoperative platelet count and neutrophil-lymphocyte ratio (COP-NLR) in patients with non–small-cell lung cancer: based on a large cohort study. PLoS One. 2015;10:e0126496. doi: 10.1371/journal.pone.0126496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mack P.C., Redman M.W., Chansky K. Lower osteopontin plasma levels are associated with superior outcomes in advanced non–small-cell lung cancer patients receiving platinum-based chemotherapy: SWOG study S0003. J Clin Oncol. 2008;26:4771–4776. doi: 10.1200/JCO.2008.17.0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oven Ustaalioglu B., Bilici A., Ercan S. Serum M30 and M65 values in patients with advanced stage non–small-cell lung cancer compared with controls. Clin Transl Oncol. 2012;14:356–361. doi: 10.1007/s12094-012-0808-0. [DOI] [PubMed] [Google Scholar]
- 33.Brechot J.M., Chevret S., Nataf J. Diagnostic and prognostic value of Cyfra 21-1 compared with other tumour markers in patients with non–small-cell lung cancer: a prospective study of 116 patients. Eur J Cancer. 1997;33:385–391. [PubMed] [Google Scholar]
- 34.Carvalho S., Troost E.G., Bons J., Menheere P., Lambin P., Oberije C. Prognostic value of blood-biomarkers related to hypoxia, inflammation, immune response and tumour load in non–small-cell lung cancer - a survival model with external validation. Radiother Oncol. 2016;119:487–494. doi: 10.1016/j.radonc.2016.04.024. [DOI] [PubMed] [Google Scholar]
- 35.Vollmer R.T., Govindan R., Graziano S.L. Serum CYFRA 21-1 in advanced stage non–small-cell lung cancer: an early measure of response. Clin Cancer Res. 2003;9:1728–1733. [PubMed] [Google Scholar]
- 36.Dogan I., Karyagar S., Karyagar S.S., Kahraman C., Alver A. Relationship between pretreatment levels of serum Cyfra 21.1, CEA and PET metabolic parameters in NSCLC. Ann Nucl Med. 2014;28:829–835. doi: 10.1007/s12149-014-0877-y. [DOI] [PubMed] [Google Scholar]
- 37.Edelman M.J., Hodgson L., Rosenblatt P.Y. CYFRA 21-1 as a prognostic and predictive marker in advanced non–small-cell lung cancer in a prospective trial: CALGB 150304. J Thorac Oncol. 2012;7:649–654. doi: 10.1097/JTO.0b013e31824a8db0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holdenrieder S., von Pawel J., Dankelmann E. Nucleosomes and CYFRA 21-1 indicate tumor response after one cycle of chemotherapy in recurrent non–small-cell lung cancer. Lung Cancer. 2009;63:128–135. doi: 10.1016/j.lungcan.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Xu Y., Xu L., Qiu M. Prognostic value of serum cytokeratin 19 fragments (Cyfra 21-1) in patients with non–small-cell lung cancer. Sci Rep. 2015;5:9444. doi: 10.1038/srep09444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boher J.M., Pujol J.L., Grenier J., Daures J.P. Markov model and markers of small-cell lung cancer: assessing the influence of reversible serum NSE, CYFRA 21-1 and TPS levels on prognosis. Br J Cancer. 1999;79:1419–1427. doi: 10.1038/sj.bjc.6690227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giovanella L., Piantanida R., Ceriani L. Immunoassay of neuron-specific enolase (NSE) and serum fragments of cytokeratin 19 (CYFRA 21.1) as tumor markers in small-cell lung cancer: clinical evaluation and biological hypothesis. Int J Biol Markers. 1997;12:22–26. doi: 10.1177/172460089701200105. [DOI] [PubMed] [Google Scholar]
- 42.Brattstrom D., Bergqvist M., Hesselius P., Larsson A., Wagenius G., Brodin O. Serum VEGF and bFGF adds prognostic information in patients with normal platelet counts when sampled before, during and after treatment for locally advanced non–small-cell lung cancer. Lung Cancer. 2004;43:55–62. doi: 10.1016/j.lungcan.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 43.Bremnes R.M., Camps C., Sirera R. Angiogenesis in non–small-cell lung cancer: the prognostic impact of neoangiogenesis and the cytokines VEGF and bFGF in tumours and blood. Lung Cancer. 2006;51:143–158. doi: 10.1016/j.lungcan.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 44.Isa S., Kawaguchi T., Teramukai S. Serum osteopontin levels are highly prognostic for survival in advanced non–small-cell lung cancer: results from JMTO LC 0004. J Thorac Oncol. 2009;4:1104–1110. doi: 10.1097/JTO.0b013e3181ae2844. [DOI] [PubMed] [Google Scholar]
- 45.Ueno K., Inoue Y., Kawaguchi T., Hosoe S., Kawahara M. Increased serum levels of basic fibroblast growth factor in lung cancer patients: relevance to response of therapy and prognosis. Lung Cancer. 2001;31:213–219. doi: 10.1016/s0169-5002(00)00187-2. [DOI] [PubMed] [Google Scholar]
- 46.Baatar D., Kawanaka H., Szabo I.L. Esophageal ulceration activates keratinocyte growth factor and its receptor in rats: implications for ulcer healing. Gastroenterology. 2002;122:458–468. doi: 10.1053/gast.2002.31004. [DOI] [PubMed] [Google Scholar]
- 47.Marks L.B., Bentzen S.M., Deasy J.O. Radiation dose-volume effects in the lung. Int J Radiat Oncol Biol Phys. 2010;76:S70–S76. doi: 10.1016/j.ijrobp.2009.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan B., Wang R., Huang Y., Garfield D., Zhang J., Chen H. HGF and NRG1 protein expression are not poor prognostic markers in surgically resected lung adenocarcinoma. Onco Targets Ther. 2015;8:1185–1191. doi: 10.2147/OTT.S78116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Currie M.J., Gunningham S.P., Han C. Angiopoietin-1 is inversely related to thymidine phosphorylase expression in human breast cancer, indicating a role in vascular remodeling. Clin Cancer Res. 2001;7:918–927. [PubMed] [Google Scholar]
- 50.Harfouche R., Echavarria R., Rabbani S.A., Arakelian A., Hussein M.A., Hussain S.N. Estradiol-dependent regulation of angiopoietin expression in breast cancer cells. J Steroid Biochem Mol Biol. 2011;123:17–24. doi: 10.1016/j.jsbmb.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 51.McKinley E.T., Ayers G.D., Smith R.A. Limits of [18F]-FLT PET as a biomarker of proliferation in oncology. PLoS One. 2013;8:e58938. doi: 10.1371/journal.pone.0058938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tichet M., Prod'Homme V., Fenouille N., Ambrosetti D. Tumour-derived SPARC drives vascular permeability and extravasation through endothelial VCAM1 signalling to promote metastasis. Nat Commun. 2015;6:6993. doi: 10.1038/ncomms7993. [DOI] [PubMed] [Google Scholar]
- 53.Lee H.M., Kim H.J., Won K.J. Soluble form of vascular cell adhesion molecule 1 induces migration and proliferation of vascular smooth muscle cells. J Vasc Res. 2008;45:259–268. doi: 10.1159/000112941. [DOI] [PubMed] [Google Scholar]
- 54.Lukacs N.W., Strieter R.M., Evanoff H.L., Burdick M.D., Kunkel S.L. VCAM-1 influences lymphocyte proliferation and cytokine production during mixed lymphocyte responses. Cell Immunol. 1994;154:88–98. doi: 10.1006/cimm.1994.1059. [DOI] [PubMed] [Google Scholar]
- 55.Chau C.H., Rixe O., McLeod H., Figg W.D. Validation of analytical methods for biomarkers employed in drug development. Clin Cancer Res. 2008;14:5967–5976. doi: 10.1158/1078-0432.CCR-07-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]