Abstract
The intestinal epithelium is highly proliferative and consists of crypt invaginations that house stem cells and villus projections with differentiated cells. There exists a dynamic equilibrium between proliferation, migration, differentiation and senescence that is regulated by several factors. Among these are RNA binding proteins (RBP) that bind their targets in a both context dependent and independent manner. RBP:RNA complexes act as rheostats by regulating expression of RNAs both co- and post-transcriptionally. This is important especially in response to intestinal injury, to fuel regeneration. The manner in which these RBPs function in the intestine and their interactions with other pivotal pathways in colorectal cancer may provide a framework for new insights and potential therapeutic applications.
Keywords: RNA binding proteins, LIN28, Musashi (MSI), IGF2BP/IMP (Insulin-like growth factor 2 mRNA binding proteins), MEX3A, CELF1 (CUGBP Elav-Like Family Member 1), RBM3 (RNA binding protein 3) and HUR (Hu-Antigen R), intestinal stem cells, colorectal cancer
The proliferative and dynamic intestinal epithelium
Tissue homeostasis is a consortium of fundamental physiological processes involving proliferation, differentiation, apoptosis and senescence. There is a disparity in tissues that are proliferative with rapid turnover (e.g. intestine, skin) versus those that are largely quiescent (e.g. neurons, smooth muscle cells, endothelial cells, kidney). The small intestinal epithelium has proliferative crypt cells at its base. Daughter cells migrate to the luminal surface undergoing differentiation into cells that comprise the villus compartment. Thus, there is a proliferation-differentiation gradient from the crypt compartment to the villus compartment, which comprises two key lineages: absorptive enterocytes (most of the cells) and secretory cells (Paneth, enteroendocrine, goblet) [1]. The large intestinal epithelium differs from the small intestinal epithelium in terms of differences in the Paneth cells and the nature of the surface (absence of villi) (Figure 1). The intestinal proliferation-differentiation gradient and lineage specification is regulated to a large extent through two stem cell populations: the active crypt base columnar (CBC) cells and reserve +4 cells [2, 3]. A number of genes and pathways annotate these two populations, especially as related to Wnt signaling [4], Notch signaling [5], BMP pathway [6], amongst others.
Figure 1.
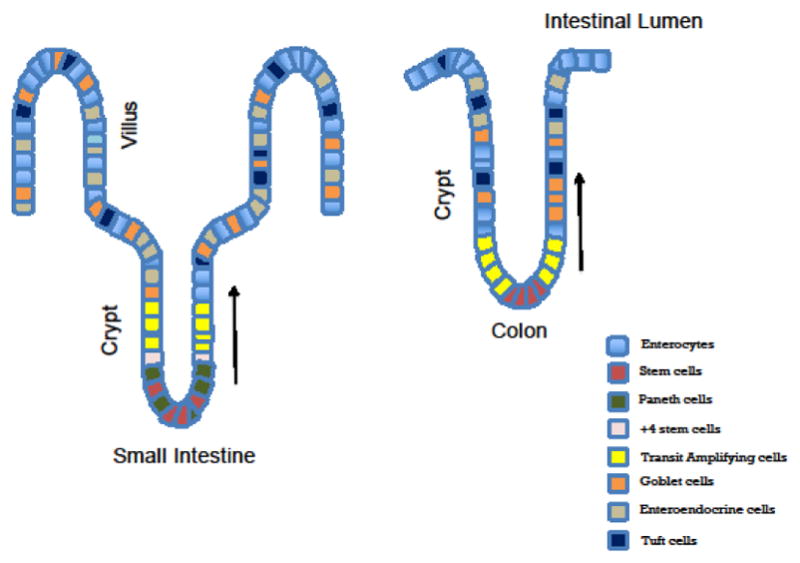
Schematic representation of the crypt-villus axis and the major intestinal cell types for small intestine and colon
The figure depicts the major cell types in the small intestine and colon. The stem cells reside at the crypt base and proliferate (transit amplifying cells) and differentiate into secretory (Paneth, enteroendocrine, goblet, tufts cells) and absorptive lineages (enterocytes). These differentiated cells migrate towards the villi (in the small intestine).
Injurious agents include infectious organisms, inflammatory conditions that trigger immune mediated responses (e.g. inflammatory bowel diseases) and subversion through activation of oncogenes and inactivation of tumor suppressor genes that drive malignant transformation (especially colorectal cancer). An emerging node of regulation of intestinal epithelial homeostasis, response to injury and malignant transformation is through RNA binding proteins (RBP) [7]. We will focus on the published cohort of RBPs in the context of intestinal homeostasis, regeneration and colorectal cancer. These include LIN28, MSI (Musashi), IGF2BP/IMP (Insulin-like growth factor 2 mRNA binding proteins), MEX3A, CELF1 (CUGBP Elav-Like Family Member 1), RBM3 (RNA binding protein 3) and HUR (Hu-Antigen R).
RNA binding proteins and their functions
Broadly speaking, RNA binding proteins (RBPs) are vital for regulation of several essential cellular processes such as RNA splicing, modifications, transport, localization, stability, degradation and translation [10]. Several RBPs are expressed ubiquitously and are evolutionarily conserved [11] to maintain their roles in basic cellular functions. Any significant change or disturbance in the RBPs regulating these essential cellular functions can lead to different diseases, including cancer [10]. RBPs function by binding to their target RNA, forming ribonucleoprotein (RNP) complexes [12] and regulating gene expression post-transcriptionally in a plethora of ways. Since RBPs can regulate already transcribed RNAs, they act in a rapid and efficient manner to alter gene expression, especially during changes in the microenvironment. A single RBP can bind to hundreds, if not thousands of targets, and a combination of several RNP interactions contribute to cellular identity and response to stimuli [13]. RBPs can help recruit translation machinery to activate translation [14]. By contrast, RBPs involved in the RNA-induced silencing complex (miRISC) result in decapping, deadenylation and translational repression of the target mRNAs [15]. They can also suppress translation and induce degradation [16]. In some cases, two RBPs can bind to the same RNA target to stabilize it, either enhancing or repressing translation [17]. RBPs can also have dichotomous functions where they can both enhance [18, 19] or repress [20, 21] tumorigenesis depending upon the cellular context. Figure 2 shows a simplistic schematic of the functional consequences of RBP binding to mRNA targets (as the RBPs discussed in this review bind mainly to mRNAs).
Figure 2.
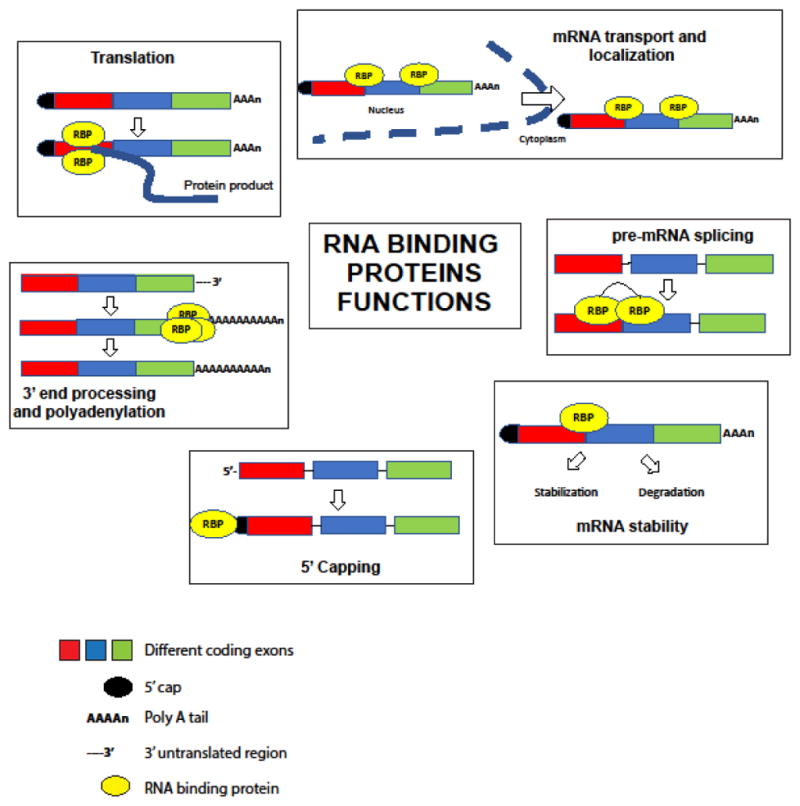
Schematic representation of the different functional roles of RBPs
The figure depicts the major functional roles of the RBPs discussed in this review. The majority of the RBPs discussed here bind to mRNAs and regulate their processing (5′ capping, 3′ end processing, splicing), stability, localization and translation. This figure does not depict the non-conventional RBPs.
Structure of RNA binding proteins
The functional effects of conventional RNA binding proteins are dependent upon their binding to their target RNAs and forming ribonucleoprotein (RNP) complexes. The RNP complexes help with RNA processing, translation, export and localization. Since RBPs have multiple biological roles, their structures consist of multiple small domains. These consist of several types of RNA recognition and binding domains interspersed between catalytic domains to efficiently recognize a wide range of targets and regulate catalytic activity [22]. These catalytic domains include helicases, deaminases and RNAse III domains [23, 24]. Multiple RNA binding domains (RBDs) provide specificity to recognize and bind either long RNA sequences or sequences separated by many nucleotides or two different RNAs [22]. These can help form large complexes and regulate major signaling pathways. RBDs may comprise RNA recognition motifs (RRMs), dsRNA binding motifs (dsRBD), K-homology domain (KH), Zinc fingers, S1 domain, Piwi and PAZ (PIWI, AGO, and Zwill) domains amongst others. RRM is by far the most common and well-characterized domain and most RBPs have multiple RRMs to provide specificity. By contrast, RBPs involved in translation, such as initiation and elongation factors, bind all mRNAs and lack specificity [22]. RBPs can regulate subcellular localization of their targets due to nuclear and/or nucleolar localization signals (NLS/NoLS) or nuclear export signals (NES) depending upon their functional requirements [25, 26]. Overall, the structure of these conventional RBPs comprise multiple repeats of different RBDs with varying functional specificities and catalytic domains to regulate their target RNAs.
The target RNAs for RBPs are quite diverse. While RBPs can bind different regions of mRNAs (exonic, intronic, UTRs), there is increasing evidence of interactions with other types of RNAs, including non-coding RNAs, namely microRNAs, t-RNAs, small interfering RNAs (siRNA), telomerase RNA, small nucleolar RNAs (snoRNAs), splicesomal small nuclear RNAs (snRNA), as well as the RNA moiety of the signal recognition particle (SRP RNA or 7SL RNA). These non-coding RNAs form extensive secondary structures to associate with proteins and regulate several processes like splicing, RNA modifications, protein localization and secretion as well as chromosomal maintenance [27].
In recent years, advanced structural-analysis studies have provided evidence of complex protein–RNA interactions that do not require canonical RBDs [27]. RNA interactome capture (RIC) [28] studies have identified ‘non-conventional’ RBPs in several organisms that do not have discernible RBDs and have no known relationship to RNA biology [27]. Further studies have also shown that disordered protein regions can also facilitate protein-RNA interactions that can be specific or non-specific [29]. These unorthodox interactions can regulate RNA metabolism and different RNA processes, both co- and post- transcriptionally [29].
In this review, we will discuss only those specific RBPs that have been published in the context of intestinal homeostasis and intestinal tumorigenesis. These RBPs fall under the ‘conventional’ RBP category (the basic domain structures are summarized in Figure 3) (The role of non-coventional or non-canonical RBPs in cancer has been beautifully reviewed in [30]).
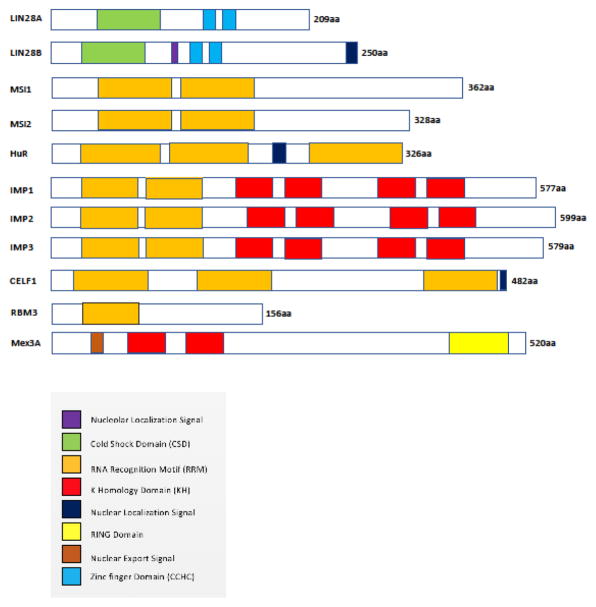
Figure 3.
Schematic representation of the structural domains of the RBPs
The figure shows a simplistic representation of the different structural domains of the RBPs discussed in this review that help them bind to their target RNAs and regulate their function. The amino acid length of the RBPs are also mentioned. The RBPs contain different types of RNA binding and catalytic domains including RRMs, KH domains, zinc finger domains and cold shock domains. Some of these RBPs also contain localization signals to help them shuttle in and out of the nucleas.
LIN28
LIN-28 was first discovered in C. elegans as a heterochronic gene that plays a vital role in developmental events [31]. LIN28 has been studied in multiple species as a promoter of pluripotency. It has been shown to be expressed highly in undifferentiated tissues and its expression is downregulated as differentiation and development progress [32]. Hence, LIN28 is evolutionarily conserved to promote pluripotency and act as a ‘gatekeeper’ of differentiation. The most well studied mechanism of LIN28B function is via its interaction with the let-7 miRNAs [33].
In mammals, there are two paralogs of LIN28; LIN28A and LIN28B that have mostly overlapping functions [34]. LIN28A and LIN28B have a cysteine cysteine histidine cysteine (CCHC) zinc finger domain and a cold shock domain [35]. LIN28B also contains an extended C terminal region with a nuclear localization signal (NLS) [36]. In mice, LIN28 proteins are expressed highly during embryonic development but their expression declines rapidly after E18.5 in the small intestine and colon correlating reciprocally with intestinal differentiation [37, 38]. In adult mice, LIN28B expression is limited to the crypt compartment [38]. This correlates with the reciprocal increase in the expression of the Let-7 microRNAs. LIN28B expression is observed in the nucleus of undifferentiated cells whereas low expression of LIN28B can be seen in the cytoplasm of differentiated intestinal cells. The constitutive knockout of either Lin28a or Lin28b causes dwarfism and a growth retardation phenotype in mice [39]. The double knockout is synthetically lethal, and the mice do not survive past E12.5. This phenotype, however, is not observed when the genes are deleted in neonatal or adult mice [39]. The intestinal epithelium specific single or double knockouts of Lin28a and Lin28b show no obvious intestinal phenotype [40]. Furthermore, these mice also do not show any difference in susceptibility to colonic tumorigenesis with dextran sodium sulphate (DSS)/azoxymethane (AOM) when compared to their wild-type littermates [40].
Several studies have shown that LIN28B is overexpressed in about 30% of colorectal tumors [41, 42]. LIN28B overexpression correlates with invasive tumor phenotype, worse survival and increased tumor recurrence in colorectal cancer (CRC) [38, 40, 43]. In mice, intestinal epithelial cell (IEC) specific Lin28b overexpression is sufficient to transform the epithelium and give rise to adenomas and adenocarcinomas between 9–12 months of age, which is accelerated by the concurrent knockout of Let7 b1/c2 with faster and greater formation of adenocarcinomas within 6 months [38, 43]. LIN28B cooperates with Wnt signaling to increase tumor formation in carcinogen-induced mouse model of colitis-associated tumorigenesis [40]. Furthermore, LIN28 overexpression increases tumor formation and decreases tumor latency in an Apc+/min model of colon cancer [40]. LIN28A, which is structurally similar to LIN28B [44], is upregulated in over 70% of CRC patients [45] and overexpression of LIN28A is functionally similar to LIN28B [40]. While silencing either LIN28 protein leads to increased apoptosis by targeting of anti-apoptotic BCL2L1 protein for degradation [46], LIN28A overexpression however, leads to increased chemosensitivity in CRC cells lines to 5FU (fluorouracil) treatment through induction of apoptosis [45]. In summary, LIN28B is critical in colorectal tumorigenesis and has been established to oncogenic effects in this context. While less studied in colorectal cancers, LIN28A has similar functions.
IGF2BPs/IMPs
The insulin-like growth factor-2 mRNA binding proteins (IGF2BPs or IMPs) belong to a conserved subfamily of RBPs. The IMPs have been studied for their roles in regulation of post-transcriptional processes such as mRNA localization, turnover, and translational control [47, 48]. In mammals, the canonical domain structure of IMPs is similar. IMP1 and IMP3 are more closely related and have 73% sequence similarity whereas IMP2 shares 56% similarity [49]. IMPs contain 2 RRMs in their N-terminal region and 4 KH domains in the C-terminal region [50]. The KH domains are the primary RBDs while the RRMs are involved in stabilization of IMP-mRNA complexes [51, 52]. The IMPs bind their targets in multiple low affinity higher-order complexes because KH domains allow recognition of only short stretches of RNA with relatively weak binding affinity [53].
Imp proteins, especially Imp1, are expressed highly during development but expression is reduced drastically after post-natal day 12 in the small and large intestine. The adult mice retain low expression of IMP1 in the crypts [54]. IMP3, an isoform of IMP1, also follows a similar pattern of expression in the intestine [55]. IMP2, by contrast, has been shown to be expressed postnatally [56] and is mainly found in Processing bodies (P bodies) in the cytoplasm [57]. Similarly, Imp1 null mice show significant growth retardation at E17.5 and more than 50% of the mice do not survive past post-natal day 3. The mice show impaired intestinal morphology and development [54]. By contrast, Imp2 null mice have no growth retardation but are highly resistant to diet induced obesity [58]. In colorectal cancer cell lines and fibroblasts, Imp2 deletion results in reduced proliferation [59].
IMP1 plays a functional role in the RNA stability by binding and shielding several mRNAs that play critical roles in cell growth and proliferation from proteolytic degradation [60]. IMP1 also regulates cell cycle progression and migration in human CRC cells [61]. IMP1 is overexpressed in more than 80% of human CRC [62] and correlates with invasion, lymph node metastasis, and worse prognosis [18, 38, 63]. IMP1 overexpression in CRC cell lines causes a significant increase in tumor volume in xenograft models [18]. By contrast, Imp1 loss in the stroma is associated with increased tumor number in a AOM-DSS model of colonic carcinogenesis. This dichotomous role of Imp1 is seen in other instances where IMP1 stabilizes β-catenin mRNA in breast cancer cells [64] and is in turn activated by it in a feedback mechanism [65]. In others studies, IMP1 was shown to bind and stabilize beta-TRCP1, a β-catenin antagonist in CRC cell lines [66]. IMP2 gene is amplified at a higher frequency in several solid tumors. IMP2 depletion inhibits proliferation of mouse embryonic fibroblasts (MEFs) and well as several human cancer cell lines. It is also shown to stabilize oncogenic transcriptional regulator HMGA1 in MEFs [59]. IMP3 expression has been shown to correlate with worse prognosis and increased recurrence in colon cancer patients [67]. It has also been associated with low progression-free survival in small-intestinal neuroendocrine neoplasms [68] and studied as an immunohistochemical marker in small intestinal adenocarcinomas [69].
These studies imply divergent roles for IMP1, depending upon whether one considers the epithelial vs. stromal compartment.
Musashi
The Drosophila musashi gene was discovered in 1994 as a regulator of asymmetric cell division of Drosophila sensory organ precursor (SOP) cells [70]. Since then, the Musashi (Msi) proteins have been shown to be expressed in the stem cell compartments of different tissues such as brain, intestine and blood and are known to be upregulated in cancers [71–73]. They function as regulators of stem cell renewal, cell cycle progression and metabolism [72, 73]. The msi gene is evolutionarily conserved and humans have two related genes, Musashi-1 (MSI1) and Musashi-2 (MSI2) with 75% amino acid identity in structure [74]. Both MSI1 and MSI2 contain 2 N-terminal RRM RBDs. Biochemical and structural studies show that the RRM1 contributes the majority of the binding energy and specificity, while RRM2 has a more supportive role [74]. They usually bind to 3′ ends of target RNAs [75]. MSI1 also contains domains to interact with other RBPs such as PABP1 and LIN28B [76].
In the intestinal epithelium, the MSI family of proteins are expressed in the crypts in mice [73]. Their expression is observed in adult mice in both the active and reserve stem cell compartments [72, 77]. The MSI family of proteins consist of the functionally redundant MSI1 and MSI2. Ablation of Msi proteins (Msi1 or 2) in IEC, either individually or together, showed no changes in morphology, proliferation or differentiation [78]. Although, intestinal epithelial specific double knockout of Msi1 and Msi2 (Msi1ΔIEC Msi2ΔIEC) in mice does not show any overt phenotype under basal conditions [78]. However, following 12Gy radiation injury, Msi1ΔIEC Msi2ΔIEC mice show a significant impairment in the regenerative response. MSI proteins are also up-regulated during activation of reserve intestinal stem cells and are required for lineage tracing from these cells under basal conditions by enabling their S-phase entry [78]. MSI1 overexpression has been shown to induce tumorigenesis by activation of Wnt and Notch pathways in primary intestinal cells and xenograft models [79]. The overexpression of either MSI is sufficient to transform the intestinal epithelium and form tumors [72, 73, 78] via activation of the mTORC1 complex with inhibition of Pten [72, 73]. In patients with small intestinal adenocarcinomas, MSI1 is overexpressed in 71% of the tumors as compared to the normal tissue and correlated with depth of wall invasion [80]. In patients with Irritable bowel syndrome (IBS) the density of MSI1+ cells is significantly reduced and correlates with dysfunctional stem cell potential [81]. MSI1+ cells have been shown to be involved in repair of the intestinal epithelium induced by 5-FU [82].
Due to their roles in EMT, stem cell identity, and oncogenesis, the MSI proteins have increasingly been linked to therapeutic resistance in cancer treatments [83–85]. This has resulted in efforts to develop inhibitors of MSI proteins as potential therapeutic targets [86, 87].
HuR
HuR, a member of ELAV family of RBPs (reviewed extensively in [88]) consists of 2 RRM domains, a hinge region and a third RRM [89] that helps it bind to adenylate uridylate (AU) rich regions in 3′ UTRs of target RNAs involved in cell survival and tumorigenesis [90]. HuR is mainly expressed in the nucleus but can shuttle between the nucleus and cytoplasm due to the nucleo-cytoplasmic shuttling sequence present in the hinge region of the protein [91].
HuR is expressed throughout the intestinal epithelium in mice [92] [93]. Although mice with intestinal epithelial cell (IEC) knockout specific HuR show signs of mucosal atrophy, there are no changes in body weight or other abnormalities [92]. These mice show reduction in proliferating cells in the intestine and shorter crypts and villi but are otherwise healthy and reproduce normally [92] [93].
High HuR protein expression is found in both the nucleus and cytoplasm of human colon cancers [94]. While low HuR protein expression is observed in the normal colon [95], it is increased significantly in the cytoplasm of colorectal tumors [95]. Mice with intestinal specific HuR deletion (HuRΔIEC) show increased injury in a doxorubicin induced acute injury model [92]. These mice also show increased regeneration and compensatory proliferation during the peak damage phase. Furthermore HuRΔIEC mice show more than 60% attenuation in the polyposis phenotype in the Apcmin/+ mice [92]. By contrast, HuRΔIEC mice show increased protection in the AOM-DSS model of tumorigenesis [92]. In intestinal cell lines, HUR inhibition causes a significant decrease in Wnt signaling, thereby suggesting a potential role in the regulation of the Wnt pathway [93]. HuR also has tumor suppressive functions via the regulation of tumor suppressors p21 and Wnt family protein Wnt-5a [96]. HuR is known to mediate post-transcriptional regulation of its target mRNAs and is critical for neoplastic transformation and cancer development. Furthermore, HuR is activated in response to various stressors [97].
HuR is being explored as a therapeutic target and small molecule inhibitors are being developed [90, 97, 98]. One such molecule, MS-444, has been shown to inhibit HuR that in turn decreases GI tumorigenesis and the proliferation of colon cancer cells [98, 99].
Mex3A
Mex-3 protein was discovered as a translational regulator in C. elegans that helps to maintain germline totipotency. In humans, MEX3 has 4 homologous isoforms MEX3A-3D [100]. The MEX3 proteins consist of 2 KH domains at the N terminal and a RING finger module domain at the C terminal end. The KH domain helps bind target RNAs whereas the nuclear export signal (NES) helps in shutting between the nucleus and cytoplasm [100]. Recently, MEX3C has been identified as a E3 ubiquitin ligase [101], whereas a variant of MEX3D has been shown to negatively regulate the anti-apoptotic protein BCL-2 in HeLa cells [102]. MEX3B (as well as MEX3A) has been shown to be a novel component of the RNA granules called P bodies [103].
In mice, MEX3A is expressed in the crypt base and labels a slowly cycling subpopulation of Lgr5+ intestinal stem cell population that can give rise to all lineages [104]. These MEX3A- high cells appear to resist the deleterious effects of chemotherapy or irradiation and play an important role in regeneration of damaged crypts [104]. Previous studies have shown that MEX3A regulates CDX2 in human colon cancer and correlates with “stemness” [105]. Mex3a deletion in IEC does not cause any changes in reproduction and intestinal morphology in mice [104]. In Caco2 cells that can spontaneously differentiate into an enterocytic-like phenotype upon reaching confluence [106], inhibition of endogenous MEX3A using siRNA resulted in higher CDX2 expression [105]. MEX3A overexpressing Caco2 cells show increased RNA expression of stem cell markers [105].
Mex3a is overexpressed in cancers like bladder urothelial carcinoma [107] and Wilm’s tumor [108] whereas knockdown of MEX3A in human gastric cancer cells has been shown to significantly reduce cell proliferation [108] thus indicating its role in carcinogenesis and potential as a therapeutic target. This indicates the potential to study Mex3a in colorectal cancer.
CELF1
CUG binding protein 1 (CUBP1) or CELF1 is a multifunctional RBP studied primarily for its role in RNA metabolism related processes like decay, translation and splicing. CELF1 is known to bind GU rich elements in 3′UTR of target RNAs to regulate RNA stability [109]. CELF1 contains three highly conserved RRMs, two near the N terminal and one at the C terminal region. The three RRMs help recognize different motifs and form conformational changes to dictate specificity and range of binding partners [110].
In mice, CELF1 is expressed throughout the small intestinal epithelium [111] and can be repressed by mir-503 and recruited to P bodies [112]. mRNAs are localized to these cytoplasmic RNP foci and sorted for degradation and/or translational repression. CELF1 is also known to recruit certain target mRNAs like occludin to these P bodies and partially repress their translation [111]. Although CELF1 expression is found to increase proliferation and progression of several cancers [113–115], increased CELF1 causes G1 phase growth arrest in intestinal epithelial cells. By contrast, CELF1 silencing enhances cell proliferation, with an increase in cells residing in S-phase, and elevated cell number. CELF1 silencing enhances MYC translation by releasing MYC RNA from RNP complexes [111]. HUR is found to competitively repress this CELF1-MYC interaction [111]. CELF1 is mainly studied for its role in regulation of splicing in myotonic dystrophy [116, 117]. In the context of cancer, CELF1 can act as a tumor suppressor (in liver cancer [118]), increase caspase activity/apoptosis (in hepatocellular carcinoma [119] and esophageal cancer [120]) and act as a central node in post transcriptional regulatory programs underlying EMT (in breast cancer [121]) indicating its diverse role in carcinogenesis.
RBM3
RNA-binding motif protein 3 (RBM3), a glycine rich RBP [122], is an important cold shock protein that is upregulated during environmental stimuli such as hypothermia, ischemia, and hypoxia [123]. It binds to RNAs via its RRM domain and alters the secondary structure of the RNA affecting the access of mRNA initiation factor to the ribosome subunit [124], which modulates the potential activity of kinases in tumors.
RBM3 deficient mice show no overt phenotype or growth changes and are fertile [125]. RBM3 overexpression in HCT116 and DLD1 colon cancer cells increases proliferation and engenders chemotherapy resistance. These cells also exhibit increased stem cell markers via an increase in B-CATENIN activity. Therefore, the B-CATENIN signaling pathway may be regulated through alterations in expression of RBM3 [126]. In colon cancer, RBM3 is upregulated in a stage dependent manner and its overexpression is capable of inducing oncogenic transformation [127]. RBM3 is shown to increase the stability and translation of rapidly degraded mRNAs such as cyclooxygenase 2 (COX-2), interleukin-8 (IL-8), and vascular endothelial growth factor (VEGF) [127]. Like the other RBPs, the role of RBM3 can be dichotomous in different contexts. In breast cancer, higher RBM3 expression correlates with increased disease-free survival [128]. RBM3 expression is upregulated in, and correlates with, good prognosis in several cancers, including ovarian, prostate, bladder, gastric, and colorectal cancer [129–132]. RBM3 causes cellular differentiation and apoptosis in these cancers.
Identifying the RNA targets of RBPs
RNA-binding proteins are a rapid and efficient way to alter gene expression. RBPs can bind to their target mRNAs and regulate everything from developmental transitions to response to injury or stress. These RNA-protein interactions can alter gene expression on both the post-transcription and translation levels. In recent years, high throughput assays have been developed to identify RBP binding sites and enumerate their target mRNAs. Therefore, in order to elucidate the functional dynamics of RBPs, it’s important to identify the repertoire of RNAs that stably or transiently interact with the RBPs in a context-dependent and independent manner.
Large scale, high-throughput sequencing techniques, as well as mass spectrometry, have been used to identify mRNA targets and the functional effects of protein-RNA interactions [133, 134]. The widely used method for identifying RBP binding sites and partners consist of CrossLinking the RNP complexes followed by ImmunoPrecipitation and then deep SEQuencing of the bound RNA fragments also known as CLIP-Seq [135]. Several variations of CLIP or HITS-CLIP (High Throughput Sequencing - CLIP) have been described, including PAR-CLIP (photoactivatable-ribonucleoside-enhanced CLIP) [136], iCLIP (individual nucleotide resolution CLIP) [137], eCLIP (enhanced CLIP) [138], cross-linking analysis of cDNA (CRAC) [139], Fully Automated and Standardized iCLIP (FAST-iCLIP) [140] and cross-linking, ligation, and sequencing of hybrids (CLASH) [141, 142]. The main features, as well as potential advantages and disadvantages of the techniques are described in Table 1.
Table 1.
Key features of the different CLIP techniques
| Technique | Crosslinking method | Key feature | Advantages | Disadvantages |
|---|---|---|---|---|
| HITS-CLIP[150] | UV 254nm | Induction of covalent crosslinks between protein and a directly bound (within ~ 1 Å) RNA by UV irradiation | Takes advantage of the natural photoreactivity of nucleic acid bases | Less efficient for small and micro RNAs |
| PAR-CLIP[136] | UV 365nm | 4-thio-uridine incorporation into RNA | High resolution due to T->C mutations | Expensive |
| iCLIP[137] | UV 254nm | Circularization of reverse transcribed product instead of 5′ adapter ligation | Efficiency, enables identification of the cross-linking site at nucleotide resolution | Technically difficult |
| eCLIP[151] | UV 254nm | 5′ DNA adaptor ligation to the truncated cDNA | 1,000-fold increased efficiency and shorter sample-preparation times | Expensive |
| FAST-iCLIP[140] | UV 254nm | Uses biotin-streptavidin affinity purification | Does not require CLIP grade antibody | The RBP needs to be tagged and overexpressed. |
| CRAC[139] | UV 254nm | Use of affinity resins that are independent of protein–peptide interactions | Mimics more native conditions by increasing stringency | Tagging and overexpressing RBP might cause binding changes |
| CLASH[141, 142] | UV 254nm | Affinity purification and RNA-RNA intermolecular ligation | Does not require CLIP grade antibody, helps study RNA-RNA interactions | Modification of RBP might have non-physiological effects |
| iCLAP[152] | UV 254nm | Uses biotin-streptavidin -Histidine affinity purification and circularization of reverse transcribed product | Individual nucleotide resolution | Modification of RBP might have non-physiological effects |
| irCLIP[153] | UV 254nm | Use of thermostable reverse transcriptase followed by circularization of the cDNA | Increased specificity, no radioactivity, quicker method | Expensive, technically challenging |
The targets of several of the RBPs mentioned in this paper have been discovered through these high throughput sequencing techniques (Table 2). In PAR-CLIP experiments done in HEK293 cells, LIN28A and LIN28B bound to a largely overlapping set of ~3000 mRNAs at ~9500 sites located in the 3′ untranslated region (UTR) and coding DNA sequence (CDS). The binding stabilizes target mRNAs to a certain degree and increases protein abundance mainly in cell cycle regulatory genes [35, 136]. CLIP-Seq studies done in CRC cell lines and in the mouse intestinal epithelium overexpressing LIN28B indicated an enrichment in RNAs for genes regulating metabolism, protein processing in the ER, the actin cytoskeleton, mRNA processing, and focal adhesion with most of the targets being epithelial specific or associated with the translation machinery [38].
Table 2.
CLIP studies in the RBPs of interest
| RBP | Technique Used | Cell Type | Reference |
|---|---|---|---|
| CELF1 | CLIP/SoliD Sequencing | Human Hela Cells | [154] |
| CELF1 | HITS-CLIP | mouse myoblast cell line C2C12 | [146] |
| CELF1 | CLIP-Seq | Murine skeletal and heart cells | [147] |
| CELF1 | CLIP-SEQ | Mice hindbrains | [148] |
| HUR | PAR-CLIP | Human Hela Cells | [143] |
| HUR, IMP1 | fRIP-Seq | K562 cells | [155] |
| MSI2 | CLIP-Seq | Mouse epithelium | [73] |
| MSI1/MSI2 | CLIP-Seq | Mouse epithelium | [72] |
| RBM3 | RNA-Seq/CLIP-Seq | MEFs | [144] |
| LIN28A | HITS-CLIP | hESCs (H9, HUES6), 293 cells | [156] |
| LIN28A | CLIP-Seq, MS | BL21 Rosetta cell colonies | [157] |
| LIN28A | CLIP-Seq, Ribosome footprinting | Mouse embryonic stem cell A3-1 | [158] |
| LIN28 | HITS-CLIP | C. elegans | [159] |
| LIN28B | iDo-PAR-CLIP | Flp-In 293 T-REx cells | [160] |
| LIN28A, LIN28B | PAR-CLIP | HEK293 cells | [35] |
| LIN28B | CLIP-Seq, RNA Seq | LoVo cells, DLD1 cells, Mouse intestine | [38] |
| IMP1 | eCLIP | iPSCs | [138] |
| IMPs | PAR-CLIP | HEK293 | [136] |
Similarly, CLIP-Seq analysis for both endogenous and overexpressed MSI1 and MSI2 proteins in the intestine reveal that MSI1 and MSI2 drive common gene expression programs and interact with common target transcripts [72, 73, 78]. As high as 72% of gene expression changes resulting from Msi1 induction also occurred upon Msi2 induction [72, 73, 78]. The pathways found to be upregulated by gene ontology (GO) and pathway analysis were genes involved in ribosome biogenesis, signal transduction, and ErbB signaling, whereas oxidative phosphorylation and mitochondrial activity genes were downregulated [72, 80].
eCLIP studies have shown that there is substantial overlap between IMP1 and IMP2 binding but not between IMP1 and IMP3 [138]. During development and cancer, all IMP isoforms are highly expressed and might share redundant regulatory roles. IMP1 binds to and regulates genes associated with cell cycle, cell and focal adhesion and cellular integrity [138]. In HEK293 cells, separate studies with overexpression and depletion of IMP isoforms followed by PAR-CLIP showed a significant overlap of target transcripts that were mainly stabilized by IMP proteins [136].
Using PAR-CLIP, another group identified highly conserved HUR binding sites enriched for HUR binding motifs and mainly located in 3′ untranslated regions. Furthermore, the presence of some binding sites in the intronic regions suggests HuR’s role in mRNA processing. Upon HuR knockdown, both mRNA expression and protein synthesis of thousands of target genes were downregulated, thereby suggesting a role in RNA stability and translation [143].
The increased density of RBM3 binding sites (seen via PAR-CLIP) near polyadenylation sites, especially those regulating genes that show strong circadian oscillations, has indicated the role of RBM3 in circadian gene expression [144]. In addition, CLIP-Seq analysis of alternate polyadenylation (APA) sites has elucidated the role of RBM3 in response to thermal stimuli [145]. Finally, several large-scale studies have been carried out for CELF1. HITS-CLIP has shown its preferential binding to the 3′ UTR and its role in destabilization of target mRNAs, specifically myogenic differentiation factors and RNA-binding proteins [146]. Another study has shown the role of CELF1 in stabilization and localization of developmentally regulated genes in skeletal muscle and heart cells [147]. In mice hindbrains, CELF1 binding sites were enriched in UG repeats [148] and bound to intronic and 3′UTR regions validating its role in splicing.
The CLIP studies show that all these RBPs have a huge number of targets that might overlap. Although these RBPs bind mainly to mRNAs, they can bind them at different regions and regulate a multitude of functions. Since the RBP function depends on which target RNAs are present in the microenvironment, the RBPs can function differently in different cellular contexts. In recent years, ribosome profiling studies [149] are being investigated as a means to study genome-wide translational effects of RBP overexpression or deletion in different model systems. This will help us gain a better insight into the functional effects of RBPs at the translational or protein level.
Concluding Remarks and Future Directions
The intestinal epithelium illustrates a proliferation-differentiation gradient with a rapid renewal and turnover of cells. This dynamic equilibrium can be disturbed during inflammation or injury that result from cellular stresses mediated by infectious organisms, radiation, and autoimmune diseases. These trigger a rapid protective and regenerative response that is regulated by several factors. Prolonged inflammation together with genetic alterations can result in malignant transformation. RBPs including LIN28, MSI, IMP1, MEX3A, CELF1, RBM3 and HUR-constitute a new set of regulatory proteins that play an important role in intestinal homeostasis, adaptation to injury and participation in malignant transformation. The effect of overexpression and deletion of these RBPs on intestinal development, homeostasis, response to injury and carcinogenesis is increasingly being studied both in vitro and in mouse models.
In addition to the phenotypic effects, the understanding of RBP-RNA interactions is critical. This involves identification of target RNAs, the interaction mechanism and the effect it has on the RNA metabolism. This is done initially through high-throughput approaches but mandates functional validation in model systems and in tissues. Furthermore, in RBPs that target mRNAs, the effect on translation needs to be further investigated using emerging techniques like ribosome profiling. The role of these RBPs in regulating key signaling processes and malignancies has led to their emergence as targets for therapeutics with potential implications in colorectal cancer.
Box 1. The intestinal cell types.
The intestinal epithelium consists of invaginations of crypts and protrusions villi that increase its surface manifold. The crypt base is the niche for two types of stem cells; the actively dividing crypt base columnar stem cells (CBCs) that are radiosensitive and the quiescent +4 stem cells that are radioresistant [2, 3]. All intestinal lineages (absorptive and secretory) arise from the stem cells and migrate upwards towards the villi except for Paneth cells that migrate towards the base [8]. The secretory cells consist of Paneth cells (secreting anti-microbial peptides and maintaining the stem cell niche), goblet cells (secrete mucus), enteroendocrine cells (secreting hormones) and tuft cells (secrete cytokines). The absorptive enterocytes make up the majority of the epithelium and absorb micronutrients, water and electrolytes. The transit amplifying cells are immediate progeny o stem cells. The colon epithelium is similar to the small intestinal epithelium but lacks villi and serves to absorb remaining water and provide a barrier against micro-organisms [9]. (Figure 1)
Highlights.
Intestinal epithelial cells harbor proliferation-differentiation gradient spanning crypts to villi. This is regulated by two stem cell populations.
RNA binding proteins (RBPs) provide a nexus of regulation of intestinal epithelial homeostasis, adaptation to injury and contribution to malignant transformation
These specific RBPs that have been reported in the published literature in the context of intestinal epithelial biology and colorectal cancer include: LIN28, Musashi (MSI), IGF2BP/IMP (Insulin-like growth factor 2 mRNA binding proteins), MEX3A, CELF1 (CUGBP Elav-Like Family Member 1), RBM3 (RNA binding protein 3) and HUR (Hu-Antigen R).
These specific RBPs play important roles in intestinal regeneration following injury.
These specific RBPs are overexpressed in human colorectal cancer and overexpression of some of them have been shown to be sufficient to transform the intestinal epithelium in mouse models.
Outstanding questions.
More than one RBP can bind to a transcript and regulate its function. The intestinal epithelium expresses a range of RBPs that bind common transcripts. This necessitates the need to develop high-throughput techniques to study combinatorial effects of RBPs in the intestine. Are there distinct functional effects of RBP-binding to a particular region of the RNA transcript (eg. Binding to the 3′UTR vs the coding region)?
RBPs have a range of functional effects that can regulate both the transcriptome and the translatome. What is the effect of RBP binding, not only at the RNA level, but also on protein translation and proteins? This will aid functional annotation of the RBPs.
How can RBPs be targeted therapeutically in colorectal cancer? The intestinal epithelium is highly proliferative and dynamic and the changes in microenvironment are rapid and complex. Since the function of RBPs is dependent upon which target RNAs are expressed in the microenvironment, it is necessary to study them in model systems that mimic stress and injury conditions.
Clinician’s Corner.
RNA binding proteins (RBPs) represent a newly appreciated family that serve as regulatory networks of intestinal epithelial homeostasis, adaptation to injury to enable regeneration, and contributions to malignant transformation, the latter as evident in colorectal cancer.
Many RBPs have conserved structural domains through which a repertoire of RNAs is targeted.
These RBP:RNA complexes enable functional diversity in cellular processes and a rapid response to cellular stress.
The protective role of some of these RBPs in response to injury can help inform strategies for chemotherapy and radiation therapy in colorectal cancer.
The aberrant expression and function of certain RBPs in colorectal cancer might provide the impetus for the development of inhibitors of these RBPs.
Glossary
- Paneth cells
Secretory cells in the intestinal epithelium that secrete antimicrobial peptides and proteins and help maintain the stem cell niche
- Enteroendocrine cells
Secretory cells in the intestinal epithelium that secrete gastrointestinal hormones and peptides
- Goblet cells
Mucus secreting intestinal epithelial cells
- Enterocytes
Absorptive cells in the intestinal villi that aid in digestion and transport of molecules
- Crypt base columnar cells
The radiosensitive, actively dividing stem cell population in the intestinal crypt base
- Reserve +4 cells
The radio-resistant, quiescent stem cell population in the intestinal crypt base. It is activated during injury
- Processing bodies/P bodies
The distinct foci in the cytoplasm of eukaryotic cells consisting of RNA-protein complexes that help in mRNA turnover
- Crosslinking Immunoprecipitation and Sequencing/CLIP-Seq
Technique used for genome-wide profiling of protein-RNA interactions as well as RNA modifications.
- RNA binding proteins/RBP
The proteins that bind different types of RNAs and regulate their function in one way or another
- RNA binding domains/RBD
Structural motifs present in RBPs that help them bind to RNA
- Ribosome profiling
A high throughput technique that provides a global snapshot of actively translating RNAs in the genome by sequencing RNA sites protected by ribosomes. This technique can be used to identify translated mRNA regions, observe protein folding patterns, and measure the amount of specific proteins that are synthesized.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–60. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 2.Yan KS, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109(2):466–71. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327(5965):542–5. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gregorieff A, et al. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129(2):626–38. doi: 10.1016/j.gastro.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 5.van der Flier LG, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136(5):903–12. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 6.Kosinski C, et al. Gene expression patterns of human colon tops and basal crypts and BMP antagonists as intestinal stem cell niche factors. Proc Natl Acad Sci U S A. 2007;104(39):15418–23. doi: 10.1073/pnas.0707210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson-Rolf A, et al. Stem Cells in Repair of Gastrointestinal Epithelia. Physiology (Bethesda) 2017;32(4):278–289. doi: 10.1152/physiol.00005.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154(2):274–84. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 9.de Santa Barbara P, et al. Development and differentiation of the intestinal epithelium. Cell Mol Life Sci. 2003;60(7):1322–32. doi: 10.1007/s00018-003-2289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang ZL, et al. Comprehensive Genomic Characterization of RNA-Binding Proteins across Human Cancers. Cell Rep. 2018;22(1):286–298. doi: 10.1016/j.celrep.2017.12.035. [DOI] [PubMed] [Google Scholar]
- 11.Gerstberger S, et al. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15(12):829–45. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreyfuss G, et al. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3(3):195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 13.Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci. 2000;25(8):381–8. doi: 10.1016/s0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 14.Michlewski G, et al. The splicing factor SF2/ASF regulates translation initiation by enhancing phosphorylation of 4E-BP1. Mol Cell. 2008;30(2):179–89. doi: 10.1016/j.molcel.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 15.Fabian MR, et al. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–79. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 16.Kim HH, et al. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009;23(15):1743–8. doi: 10.1101/gad.1812509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sureban SM, et al. Functional antagonism between RNA binding proteins HuR and CUGBP2 determines the fate of COX-2 mRNA translation. Gastroenterology. 2007;132(3):1055–65. doi: 10.1053/j.gastro.2006.12.031. [DOI] [PubMed] [Google Scholar]
- 18.Hamilton KE, et al. IMP1 promotes tumor growth, dissemination and a tumor-initiating cell phenotype in colorectal cancer cell xenografts. Carcinogenesis. 2013;34(11):2647–54. doi: 10.1093/carcin/bgt217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutschner T, et al. Insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1) is an important protumorigenic factor in hepatocellular carcinoma. Hepatology. 2014;59(5):1900–11. doi: 10.1002/hep.26997. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton KE, et al. Loss of Stromal IMP1 Promotes a Tumorigenic Microenvironment in the Colon. Mol Cancer Res. 2015;13(11):1478–86. doi: 10.1158/1541-7786.MCR-15-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G, et al. IMP1 suppresses breast tumor growth and metastasis through the regulation of its target mRNAs. Oncotarget. 2016;7(13):15690–702. doi: 10.18632/oncotarget.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lunde BM, et al. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8(6):479–90. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem. 2010;79:321–49. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125(5):887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 25.Cassola A, et al. RNA recognition motifs involved in nuclear import of RNA-binding proteins. RNA Biol. 2010;7(3):339–44. doi: 10.4161/rna.7.3.12087. [DOI] [PubMed] [Google Scholar]
- 26.Rosenblum JS, et al. Nuclear import and the evolution of a multifunctional RNA-binding protein. J Cell Biol. 1998;143(4):887–99. doi: 10.1083/jcb.143.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hentze MW, et al. A brave new world of RNA-binding proteins. Nat Rev Mol Cell Biol. 2018 doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 28.Castello A, et al. System-wide identification of RNA-binding proteins by interactome capture. Nat Protoc. 2013;8(3):491–500. doi: 10.1038/nprot.2013.020. [DOI] [PubMed] [Google Scholar]
- 29.Jarvelin AI, et al. The new (dis)order in RNA regulation. Cell Commun Signal. 2016;14:9. doi: 10.1186/s12964-016-0132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore S, et al. Expanding horizons: new roles for non-canonical RNA-binding proteins in cancer. Curr Opin Genet Dev. 2017;48:112–120. doi: 10.1016/j.gde.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226(4673):409–16. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 32.Tsialikas J, Romer-Seibert J. LIN28: roles and regulation in development and beyond. Development. 2015;142(14):2397–404. doi: 10.1242/dev.117580. [DOI] [PubMed] [Google Scholar]
- 33.Balzeau J, et al. The LIN28/let-7 Pathway in Cancer. Front Genet. 2017;8:31. doi: 10.3389/fgene.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140(4):445–9. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Hafner M, et al. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA. 2013;19(5):613–26. doi: 10.1261/rna.036491.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piskounova E, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147(5):1066–79. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19(8):877–90. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 38.Madison BB, et al. LIN28B promotes growth and tumorigenesis of the intestinal epithelium via Let-7. Genes Dev. 2013;27(20):2233–45. doi: 10.1101/gad.224659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinoda G, et al. Fetal deficiency of lin28 programs life-long aberrations in growth and glucose metabolism. Stem Cells. 2013;31(8):1563–73. doi: 10.1002/stem.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tu HC, et al. LIN28 cooperates with WNT signaling to drive invasive intestinal and colorectal adenocarcinoma in mice and humans. Genes Dev. 2015;29(10):1074–86. doi: 10.1101/gad.256693.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.King CE, et al. LIN28B promotes colon cancer progression and metastasis. Cancer Res. 2011;71(12):4260–8. doi: 10.1158/0008-5472.CAN-10-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.King CE, et al. LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and -independent mechanisms. Oncogene. 2011;30(40):4185–93. doi: 10.1038/onc.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madison BB, et al. Let-7 Represses Carcinogenesis and a Stem Cell Phenotype in the Intestine via Regulation of Hmga2. PLoS Genet. 2015;11(8):e1005408. doi: 10.1371/journal.pgen.1005408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang T, et al. Comparison of the expression and function of Lin28A and Lin28B in colon cancer. Oncotarget. 2016;7(48):79605–79616. doi: 10.18632/oncotarget.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang T, et al. Lin28A enhances chemosensitivity of colon cancer cells to 5-FU by promoting apoptosis in a let-7 independent manner. Tumour Biol. 2016;37(6):7657–65. doi: 10.1007/s13277-015-4559-8. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, et al. Silencing Lin28 promotes apoptosis in colorectal cancer cells by upregulating let7c targeting of antiapoptotic BCL2L1. Mol Med Rep. 2018 doi: 10.3892/mmr.2018.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nielsen FC, et al. A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand J Clin Lab Invest Suppl. 2001;234:93–9. [PubMed] [Google Scholar]
- 48.Yaniv K, Yisraeli JK. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene. 2002;287(1–2):49–54. doi: 10.1016/s0378-1119(01)00866-6. [DOI] [PubMed] [Google Scholar]
- 49.Bell JL, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70(15):2657–75. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielsen J, et al. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19(2):1262–70. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen J, et al. Sequential dimerization of human zipcode-binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability. Nucleic Acids Res. 2004;32(14):4368–76. doi: 10.1093/nar/gkh754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wachter K, et al. Subcellular localization and RNP formation of IGF2BPs (IGF2 mRNA-binding proteins) is modulated by distinct RNA-binding domains. Biol Chem. 2013;394(8):1077–90. doi: 10.1515/hsz-2013-0111. [DOI] [PubMed] [Google Scholar]
- 53.Chao JA, et al. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev. 2010;24(2):148–58. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hansen TV, et al. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24(10):4448–64. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mori H, et al. Expression of mouse igf2 mRNA-binding protein 3 and its implications for the developing central nervous system. J Neurosci Res. 2001;64(2):132–43. doi: 10.1002/jnr.1060. [DOI] [PubMed] [Google Scholar]
- 56.Dai N, et al. mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry. Genes Dev. 2011;25(11):1159–72. doi: 10.1101/gad.2042311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lui J, et al. Granules harboring translationally active mRNAs provide a platform for P-body formation following stress. Cell Rep. 2014;9(3):944–54. doi: 10.1016/j.celrep.2014.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dai N, et al. IGF2BP2/IMP2-Deficient mice resist obesity through enhanced translation of Ucp1 mRNA and Other mRNAs encoding mitochondrial proteins. Cell Metab. 2015;21(4):609–21. doi: 10.1016/j.cmet.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dai N, et al. IGF2 mRNA binding protein-2 is a tumor promoter that drives cancer proliferation through its client mRNAs IGF2 and HMGA1. Elife. 2017:6. doi: 10.7554/eLife.27155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noubissi FK, et al. CRD-BP mediates stabilization of betaTrCP1 and c-myc mRNA in response to beta-catenin signalling. Nature. 2006;441(7095):898–901. doi: 10.1038/nature04839. [DOI] [PubMed] [Google Scholar]
- 61.Boyerinas B, et al. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68(8):2587–91. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 62.Ross J, et al. Overexpression of an mRNA-binding protein in human colorectal cancer. Oncogene. 2001;20(45):6544–50. doi: 10.1038/sj.onc.1204838. [DOI] [PubMed] [Google Scholar]
- 63.Dimitriadis E, et al. Expression of oncofetal RNA-binding protein CRD-BP/IMP1 predicts clinical outcome in colon cancer. Int J Cancer. 2007;121(3):486–94. doi: 10.1002/ijc.22716. [DOI] [PubMed] [Google Scholar]
- 64.Gu W, et al. Feedback regulation between zipcode binding protein 1 and beta-catenin mRNAs in breast cancer cells. Mol Cell Biol. 2008;28(16):4963–74. doi: 10.1128/MCB.00266-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gu W, et al. Blocking beta-catenin binding to the ZBP1 promoter represses ZBP1 expression, leading to increased proliferation and migration of metastatic breast-cancer cells. J Cell Sci. 2009;122(Pt 11):1895–905. doi: 10.1242/jcs.045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elcheva I, et al. CRD-BP protects the coding region of betaTrCP1 mRNA from miR-183-mediated degradation. Mol Cell. 2009;35(2):240–6. doi: 10.1016/j.molcel.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li D, et al. IMP3 is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Ann Surg Oncol. 2009;16(12):3499–506. doi: 10.1245/s10434-009-0648-5. [DOI] [PubMed] [Google Scholar]
- 68.Massironi S, et al. IMP3 expression in small-intestine neuroendocrine neoplasms: a new predictor of recurrence. Endocrine. 2017;58(2):360–367. doi: 10.1007/s12020-017-1249-x. [DOI] [PubMed] [Google Scholar]
- 69.Daikuhara S, et al. Insulin-Like Growth Factor II mRNA-Binding Protein 3 (IMP3) as a Useful Immunohistochemical Marker for the Diagnosis of Adenocarcinoma of Small Intestine. Acta Histochem Cytochem. 2015;48(6):193–204. doi: 10.1267/ahc.15021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakamura M, et al. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13(1):67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 71.Park SM, et al. Musashi-2 controls cell fate, lineage bias, and TGF-beta signaling in HSCs. J Exp Med. 2014;211(1):71–87. doi: 10.1084/jem.20130736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li N, et al. The Msi Family of RNA-Binding Proteins Function Redundantly as Intestinal Oncoproteins. Cell Rep. 2015;13(11):2440–2455. doi: 10.1016/j.celrep.2015.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang S, et al. Transformation of the intestinal epithelium by the MSI2 RNA-binding protein. Nat Commun. 2015;6:6517. doi: 10.1038/ncomms7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sakakibara S, et al. Rna-binding protein Musashi2: developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J Neurosci. 2001;21(20):8091–107. doi: 10.1523/JNEUROSCI.21-20-08091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sakakibara S, et al. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev Biol. 1996;176(2):230–42. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- 76.Kudinov AE, et al. Musashi RNA-Binding Proteins as Cancer Drivers and Novel Therapeutic Targets. Clin Cancer Res. 2017;23(9):2143–2153. doi: 10.1158/1078-0432.CCR-16-2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li N, et al. Single-cell analysis of proxy reporter allele-marked epithelial cells establishes intestinal stem cell hierarchy. Stem Cell Reports. 2014;3(5):876–91. doi: 10.1016/j.stemcr.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yousefi M, et al. Msi RNA-binding proteins control reserve intestinal stem cell quiescence. J Cell Biol. 2016;215(3):401–413. doi: 10.1083/jcb.201604119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rezza A, et al. The overexpression of the putative gut stem cell marker Musashi-1 induces tumorigenesis through Wnt and Notch activation. J Cell Sci. 2010;123(Pt 19):3256–65. doi: 10.1242/jcs.065284. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y, et al. Correlation of Musashi-1, Lgr5, and pEGFR expressions in human small intestinal adenocarcinomas. Tumour Biol. 2015;36(8):6075–82. doi: 10.1007/s13277-015-3288-3. [DOI] [PubMed] [Google Scholar]
- 81.El-Salhy M, Gilja OH. Abnormalities in ileal stem, neurogenin 3, and enteroendocrine cells in patients with irritable bowel syndrome. BMC Gastroenterol. 2017;17(1):90. doi: 10.1186/s12876-017-0643-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo Y, et al. Characterization of rhodamine 123 low staining cells and their dynamic changes during the injured-repaired progress induced by 5-FU. Pathol Res Pract. 2017;213(7):742–748. doi: 10.1016/j.prp.2017.04.008. [DOI] [PubMed] [Google Scholar]
- 83.Lee J, et al. Musashi-2 is a novel regulator of paclitaxel sensitivity in ovarian cancer cells. Int J Oncol. 2016;49(5):1945–1952. doi: 10.3892/ijo.2016.3683. [DOI] [PubMed] [Google Scholar]
- 84.Han Y, et al. Musashi-2 Silencing Exerts Potent Activity against Acute Myeloid Leukemia and Enhances Chemosensitivity to Daunorubicin. PLoS One. 2015;10(8):e0136484. doi: 10.1371/journal.pone.0136484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Araujo PR, et al. Musashi1 Impacts Radio-Resistance in Glioblastoma by Controlling DNA-Protein Kinase Catalytic Subunit. Am J Pathol. 2016;186(9):2271–8. doi: 10.1016/j.ajpath.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clingman CC, et al. Allosteric inhibition of a stem cell RNA-binding protein by an intermediary metabolite. Elife. 2014:3. doi: 10.7554/eLife.02848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lan L, et al. Natural product (−)-gossypol inhibits colon cancer cell growth by targeting RNA-binding protein Musashi-1. Mol Oncol. 2015;9(7):1406–20. doi: 10.1016/j.molonc.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65(20):3168–81. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58(2):266–77. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brody JR, Gonye GE. HuR’s role in gemcitabine efficacy: an exception or opportunity? Wiley Interdiscip Rev RNA. 2011;2(3):435–44. doi: 10.1002/wrna.62. [DOI] [PubMed] [Google Scholar]
- 91.Wang W, et al. AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1: involvement in the nuclear import of RNA-binding protein HuR. J Biol Chem. 2004;279(46):48376–88. doi: 10.1074/jbc.M409014200. [DOI] [PubMed] [Google Scholar]
- 92.Giammanco A, et al. Intestinal epithelial HuR modulates distinct pathways of proliferation and apoptosis and attenuates small intestinal and colonic tumor development. Cancer Res. 2014;74(18):5322–35. doi: 10.1158/0008-5472.CAN-14-0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu L, et al. RNA-binding protein HuR promotes growth of small intestinal mucosa by activating the Wnt signaling pathway. Mol Biol Cell. 2014;25(21):3308–18. doi: 10.1091/mbc.E14-03-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Denkert C, et al. Expression of the ELAV-like protein HuR in human colon cancer: association with tumor stage and cyclooxygenase-2. Mod Pathol. 2006;19(9):1261–9. doi: 10.1038/modpathol.3800645. [DOI] [PubMed] [Google Scholar]
- 95.Young LE, et al. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology. 2009;136(5):1669–79. doi: 10.1053/j.gastro.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leandersson K, et al. Wnt-5a mRNA translation is suppressed by the Elav-like protein HuR in human breast epithelial cells. Nucleic Acids Res. 2006;34(14):3988–99. doi: 10.1093/nar/gkl571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brody JR, Dixon DA. Wiley Interdiscip Rev RNA. 2018. Complex HuR function in pancreatic cancer cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blanco FF, et al. Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis. Oncotarget. 2016;7(45):74043–74058. doi: 10.18632/oncotarget.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lang M, et al. HuR Small-Molecule Inhibitor Elicits Differential Effects in Adenomatosis Polyposis and Colorectal Carcinogenesis. Cancer Res. 2017;77(9):2424–2438. doi: 10.1158/0008-5472.CAN-15-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Buchet-Poyau K, et al. Identification and characterization of human Mex-3 proteins, a novel family of evolutionarily conserved RNA-binding proteins differentially localized to processing bodies. Nucleic Acids Res. 2007;35(4):1289–300. doi: 10.1093/nar/gkm016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cano F, et al. The RNA-binding E3 ubiquitin ligase MEX-3C links ubiquitination with MHC-I mRNA degradation. EMBO J. 2012;31(17):3596–606. doi: 10.1038/emboj.2012.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Donnini M, et al. Identification of TINO: a new evolutionarily conserved BCL-2 AU-rich element RNA-binding protein. J Biol Chem. 2004;279(19):20154–66. doi: 10.1074/jbc.M314071200. [DOI] [PubMed] [Google Scholar]
- 103.Courchet J, et al. Interaction with 14-3-3 adaptors regulates the sorting of hMex-3B RNA-binding protein to distinct classes of RNA granules. J Biol Chem. 2008;283(46):32131–42. doi: 10.1074/jbc.M802927200. [DOI] [PubMed] [Google Scholar]
- 104.Barriga FM, et al. Mex3a Marks a Slowly Dividing Subpopulation of Lgr5+ Intestinal Stem Cells. Cell Stem Cell. 2017;20(6):801–816e7. doi: 10.1016/j.stem.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pereira B, et al. CDX2 regulation by the RNA-binding protein MEX3A: impact on intestinal differentiation and stemness. Nucleic Acids Res. 2013;41(7):3986–99. doi: 10.1093/nar/gkt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pinto M, et al. Enterocyte-Like Differentiation and Polarization of the Human-Colon Carcinoma Cell-Line Caco-2 in Culture. Biology of the Cell. 1983;47(3):323–330. [Google Scholar]
- 107.Shi JW, Huang Y. Mex3a expression and survival analysis of bladder urothelial carcinoma. Oncotarget. 2017;8(33):54764–54774. doi: 10.18632/oncotarget.18399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jiang H, et al. Knockdown of hMex-3A by small RNA interference suppresses cell proliferation and migration in human gastric cancer cells. Mol Med Rep. 2012;6(3):575–80. doi: 10.3892/mmr.2012.943. [DOI] [PubMed] [Google Scholar]
- 109.Vlasova-St Louis I, Bohjanen PR. Coordinate regulation of mRNA decay networks by GU-rich elements and CELF1. Curr Opin Genet Dev. 2011;21(4):444–51. doi: 10.1016/j.gde.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Teplova M, et al. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure. 2010;18(10):1364–77. doi: 10.1016/j.str.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liu L, et al. Competition between RNA-binding proteins CELF1 and HuR modulates MYC translation and intestinal epithelium renewal. Mol Biol Cell. 2015;26(10):1797–810. doi: 10.1091/mbc.E14-11-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cui YH, et al. miR-503 represses CUG-binding protein 1 translation by recruiting CUGBP1 mRNA to processing bodies. Mol Biol Cell. 2012;23(1):151–62. doi: 10.1091/mbc.E11-05-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xia L, et al. CELF1 is Up-Regulated in Glioma and Promotes Glioma Cell Proliferation by Suppression of CDKN1B. Int J Biol Sci. 2015;11(11):1314–24. doi: 10.7150/ijbs.11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cifdaloz M, et al. Systems analysis identifies melanoma-enriched pro-oncogenic networks controlled by the RNA binding protein CELF1. Nat Commun. 2017;8(1):2249. doi: 10.1038/s41467-017-02353-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.House RP, et al. RNA-binding protein CELF1 promotes tumor growth and alters gene expression in oral squamous cell carcinoma. Oncotarget. 2015;6(41):43620–34. doi: 10.18632/oncotarget.6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ho TH, et al. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14(11):1539–47. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 117.Zhang L, et al. The RNA-binding protein CUGBP1 regulates stability of tumor necrosis factor mRNA in muscle cells: implications for myotonic dystrophy. J Biol Chem. 2008;283(33):22457–63. doi: 10.1074/jbc.M802803200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lewis K, et al. RNA Binding Protein CUGBP1 Inhibits Liver Cancer in a Phosphorylation-Dependent Manner. Mol Cell Biol. 2017;37(16) doi: 10.1128/MCB.00128-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim JH, et al. Inhibition of CUG-binding protein 1 and activation of caspases are critically involved in piperazine derivative BK10007S induced apoptosis in hepatocellular carcinoma cells. PLoS One. 2017;12(10):e0186490. doi: 10.1371/journal.pone.0186490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chang ET, et al. The RNA-binding protein CUG-BP1 increases survivin expression in oesophageal cancer cells through enhanced mRNA stability. Biochem J. 2012;446(1):113–23. doi: 10.1042/BJ20120112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chaudhury A, et al. CELF1 is a central node in post-transcriptional regulatory programmes underlying EMT. Nat Commun. 2016;7:13362. doi: 10.1038/ncomms13362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kishore S, et al. Deciphering the role of RNA-binding proteins in the post-transcriptional control of gene expression. Brief Funct Genomics. 2010;9(5–6):391–404. doi: 10.1093/bfgp/elq028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang H, et al. Posttranscriptional Regulation of Intestinal Epithelial Tight Junction Barrier by RNA-binding Proteins and microRNAs. Tissue Barriers. 2014;2(1):e28320. doi: 10.4161/tisb.28320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chappell SA, Mauro VP. The internal ribosome entry site (IRES) contained within the RNA-binding motif protein 3 (Rbm3) mRNA is composed of functionally distinct elements. J Biol Chem. 2003;278(36):33793–800. doi: 10.1074/jbc.M303495200. [DOI] [PubMed] [Google Scholar]
- 125.Matsuda A, et al. Generation of mice deficient in RNA-binding motif protein 3 (RBM3) and characterization of its role in innate immune responses and cell growth. Biochem Biophys Res Commun. 2011;411(1):7–13. doi: 10.1016/j.bbrc.2011.06.038. [DOI] [PubMed] [Google Scholar]
- 126.Venugopal A, et al. RNA binding protein RBM3 increases beta-catenin signaling to increase stem cell characteristics in colorectal cancer cells. Mol Carcinog. 2016;55(11):1503–1516. doi: 10.1002/mc.22404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sureban SM, et al. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene. 2008;27(33):4544–56. doi: 10.1038/onc.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jogi A, et al. Nuclear expression of the RNA-binding protein RBM3 is associated with an improved clinical outcome in breast cancer. Mod Pathol. 2009;22(12):1564–74. doi: 10.1038/modpathol.2009.124. [DOI] [PubMed] [Google Scholar]
- 129.Jang HH, et al. Expression of RNA-binding Motif Protein 3 (RBM3) and Cold-inducible RNA-binding protein (CIRP) Is Associated with Improved Clinical Outcome in Patients with Colon Cancer. Anticancer Res. 2017;37(4):1779–1785. doi: 10.21873/anticanres.11511. [DOI] [PubMed] [Google Scholar]
- 130.Grupp K, et al. High RNA-binding motif protein 3 expression is an independent prognostic marker in operated prostate cancer and tightly linked to ERG activation and PTEN deletions. Eur J Cancer. 2014;50(4):852–61. doi: 10.1016/j.ejca.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 131.Melling N, et al. Loss of RNA-binding motif protein 3 expression is associated with right-sided localization and poor prognosis in colorectal cancer. Histopathology. 2016;68(2):191–8. doi: 10.1111/his.12726. [DOI] [PubMed] [Google Scholar]
- 132.Ye F, et al. High RNA-Binding Motif Protein 3 (RBM3) Expression is Independently Associated with Prolonged Overall Survival in Intestinal-Type Gastric Cancer. Med Sci Monit. 2017;23:6033–6041. doi: 10.12659/MSM.905314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Konig J, et al. Protein-RNA interactions: new genomic technologies and perspectives. Nat Rev Genet. 2012;13(2):77–83. doi: 10.1038/nrg3141. [DOI] [PubMed] [Google Scholar]
- 134.Gerstberger S, et al. Learning the language of post-transcriptional gene regulation. Genome Biol. 2013;14(8):130. doi: 10.1186/gb-2013-14-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ule J, et al. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–5. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 136.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–41. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Konig J, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17(7):909–15. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Conway AE, et al. Enhanced CLIP Uncovers IMP Protein-RNA Targets in Human Pluripotent Stem Cells Important for Cell Adhesion and Survival. Cell Rep. 2016;15(3):666–679. doi: 10.1016/j.celrep.2016.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Granneman S, et al. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci U S A. 2009;106(24):9613–8. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Flynn RA, et al. Dissecting noncoding and pathogen RNA-protein interactomes. RNA. 2015;21(1):135–43. doi: 10.1261/rna.047803.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kudla G, et al. Cross-linking, ligation, and sequencing of hybrids reveals RNA-RNA interactions in yeast. Proc Natl Acad Sci U S A. 2011;108(24):10010–5. doi: 10.1073/pnas.1017386108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Helwak A, et al. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153(3):654–65. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lebedeva S, et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43(3):340–52. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 144.Liu Y, et al. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci Rep. 2013;3:2054. doi: 10.1038/srep02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hu W, et al. Microarray meta-analysis of RNA-binding protein functions in alternative polyadenylation. PLoS One. 2014;9(3):e90774. doi: 10.1371/journal.pone.0090774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Masuda A, et al. CUGBP1 and MBNL1 preferentially bind to 3′ UTRs and facilitate mRNA decay. Sci Rep. 2012;2:209. doi: 10.1038/srep00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang ET, et al. Antagonistic regulation of mRNA expression and splicing by CELF and MBNL proteins. Genome Res. 2015;25(6):858–71. doi: 10.1101/gr.184390.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Daughters RS, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5(8):e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.McGlincy NJ, Ingolia NT. Transcriptome-wide measurement of translation by ribosome profiling. Methods. 2017;126:112–129. doi: 10.1016/j.ymeth.2017.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chi SW, et al. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Van Nostrand EL, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP) Nat Methods. 2016;13(6):508–14. doi: 10.1038/nmeth.3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wang Z, et al. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010;8(10):e1000530. doi: 10.1371/journal.pbio.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zarnegar BJ, et al. irCLIP platform for efficient characterization of protein-RNA interactions. Nat Methods. 2016;13(6):489–92. doi: 10.1038/nmeth.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Le Tonqueze O, et al. Identification of CELF1 RNA targets by CLIP-seq in human HeLa cells. Genom Data. 2016;8:97–103. doi: 10.1016/j.gdata.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.DGH, et al. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016;17:28. doi: 10.1186/s13059-016-0878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wilbert ML, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48(2):195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ransey E, et al. Comparative analysis of LIN28-RNA binding sites identified at single nucleotide resolution. RNA Biol. 2017;14(12):1756–1765. doi: 10.1080/15476286.2017.1356566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Cho J, et al. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151(4):765–77. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 159.Stefani G, et al. A novel mechanism of LIN-28 regulation of let-7 microRNA expression revealed by in vivo HITS-CLIP in C. elegans. RNA. 2015;21(5):985–96. doi: 10.1261/rna.045542.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Graf R, et al. Identification of LIN28B-bound mRNAs reveals features of target recognition and regulation. RNA Biol. 2013;10(7):1146–59. doi: 10.4161/rna.25194. [DOI] [PMC free article] [PubMed] [Google Scholar]