Abstract
We aimed to identify the plasma miRNA profile of antiphospholipid syndrome (APS) patients and to investigate the potential role of specific circulating miRNAs as non-invasive disease biomarkers. Ninety APS patients and 42 healthy donors were recruited. Profiling of miRNAs by PCR-array in plasma of APS patients identified a set of miRNAs differentially expressed and collectively involved in clinical features. Logistic regression and ROC analysis identified a signature of 10 miRNA ratios as biomarkers of disease. In addition, miRNA signature was related to fetal loss, atherosclerosis, and type of thrombosis, and correlated with parameters linked to inflammation, thrombosis, and autoimmunity. Hard clustering analysis differentiated 3 clusters representing different thrombotic risk profile groups. Significant differences between groups for several miRNA ratios were found. Moreover, miRNA signature remained stable over time, demonstrated by their analysis three months after the first sample collection. Parallel analysis in two additional cohorts of patients, including thrombosis without autoimmune disease, and systemic lupus erythematosus without antiphospholipid antibodies, each displayed specific miRNA profiles that were distinct from those of APS patients. In vitro, antiphospholipid antibodies of IgG isotype promoted deregulation in selected miRNAs and their potential atherothrombotic protein targets in monocytes and endothelial cells. Taken together, differentially expressed circulating miRNAs in APS patients, modulated at least partially by antiphospholipid antibodies of IgG isotype, might have the potential to serve as novel biomarkers of disease features and to typify patients’ atherothrombotic status, thus constituting a useful tool in the management of the disease.
Introduction
Antiphospholipid syndrome (APS) is a clinical disorder characterized by the occurrence of thrombosis and/or pregnancy morbidity associated with the persistent presence of antiphospholipid antibodies (aPL), including anti-cardiolipin antibodies (aCL), anti-β2-glycoprotein 1 antibodies (anti- β2GPI) and/or lupus anticoagulant (LA). Cardiac, cerebral and vascular strokes in these patients are responsible for a significant reduction in life expectancy.1 The course of cardiovascular disease (CVD) in APS patients may rapidly change from asymptomatic to severe life-threatening manifestations that are difficult to deal with. Timely diagnosis and accurate monitoring of the course of APS are essential to improve the quality of therapy and avoid approaches based on medical empirical protocols. In the same way, like many other autoimmune diseases, APS is a heterogeneous entity, and this has a dramatic impact on diagnosis and treatment.2
Understanding of the pathophysiological mechanisms explaining how atherosclerosis and CVD are associated to APS has been greatly broadened with the application of genomic technologies.3 One emerging and important mechanism controlling gene expression is epigenetics, which regulates gene packaging and independent expression of alterations in the DNA sequence. Epigenetics, which comprises DNA methylation, histone modifications, and microRNAs (miRNAs) activity, is providing new directions linking genomics and environmental factors.4 miRNAs are small, non-coding RNAs that, depending upon base pairing to messenger RNA (mRNA), mediate mRNA cleavage, translational repression or mRNA destabilization. miRNAs are known to be involved in crucial cellular processes and their dysregulation has been described in many cell types and fluids in a broad range of diseases.5–7
In the setting of APS, a previous study by our group recognized that aPL modulate the expression of 2 miRNAs in monocytes (miR-19b and miR-20a) that control the expression of key proteins involved in the pathology of the disease, such as tissue factor (TF).8 Moreover, we recently demonstrated that both aPL and the anti-double stranded DNA antibodies (anti-dsDNA) promote specific changes in the expression of proteins related to the biogenesis of miRNAs in leukocytes of APS and systemic lupus erythematosus (SLE) patients, which are translated in the altered expression of the miRNAs profile and that of their protein targets (related to CVD) in these disorders.9
Extensive analyses have shown that miRNAs are released into the circulation where they are present in concentration levels that differ between healthy subjects and patients. Although little is known about the origin and function of such circulating miRNAs, these molecules are increasingly recognized as non-invasive and readily accessible biomarkers for risk stratification, diagnosis and prognosis of multiple forms of CVD.10
Specific profiles of circulating miRNAs are also associated to the pathophysiology of different systemic autoimmune diseases, including SLE, systemic sclerosis, and rheumatoid arthritis (RA), and some of them appear to be of diagnostic and, possibly, of prognostic value.11 To date, in the context of APS, no study has analyzed the potential role of the circulating miRNAs as biomarkers of the disease. Therefore, the present study was designed to determine the plasma miRNA specific profile of APS patients, their modulation by autoantibodies, and their potential role as non-invasive biomarkers of disease features.
Methods
Patients
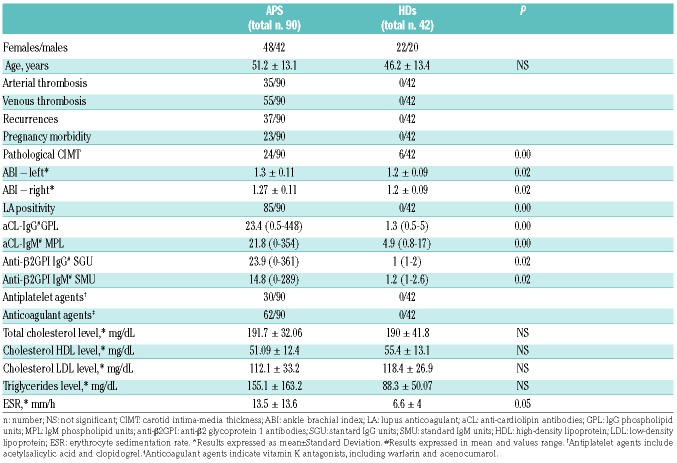
Ninety patients with primary APS and 42 healthy donors (HDs) were included in this study over a period of 24 months (see Online Supplementary Appendix). All experimental protocols were approved by the ethics committee of the Reina Sofia Hospital, Cordoba, Spain, and written informed consent was obtained. The characteristics of patients and HDs are shown in Table 1.
Table 1.
Clinical and laboratory parameters of the antiphospholipid syndrome (APS) patients and the healthy donors (HDs).
The adjusted global anti-phospholipid syndrome score (aGAPSS) was calculated for each APS patient, as previously described.12 Briefly, aGAPSS was calculated by adding the points corresponding to both the cardiovascular and thrombotic risk factors, based on a linear transformation derived from the β regression coefficient as follows: 3 for hyperlipidemia, 1 for arterial hypertension, 5 for aCL IgG/IgM, 4 for anti-β2GPI IgG/IgM, and 4 for LA.
Two additional cohorts of patients were further analyzed as disease control, including 23 patients with thrombosis in the absence of an associated autoimmune disease [12 non-pregnant women and 11 men; mean age 44 (range: 21–73 years), including patients with objectively verified thrombotic events: 14 deep venous thrombosis and 9 thrombosis in intra-cerebral vessels], and 25 SLE patients without aPLs (Online Supplementary Table S1).
For details of blood sample collection and assessment of biological parameters, B-Mode Ultrasound IMT and Ankle Brachial Index measurements see the Online Supplementary Appendix.
Isolation of miRNAs and analysis of miRNAs expression profiling
Total RNA, including the miRNA fraction, was extracted from both plasma and supernatants obtained from in vitro studies by using the QIAzol miRNeasy kit (Qiagen, Valencia, CA, USA) following the manufacturer’s instructions13 (Online Supplementary Appendix). To identify the changes that occurred in the expression levels of miRNAs in plasma from APS patients and HDs, a Human Serum & Plasma miRNA PCR-array (Qiagen) was performed (Online Supplementary Appendix) on an exploratory cohort (Online Supplementary Table S2).
Quantitative real-time PCR
A fixed volume of 3 μl of RNA solution from the 14 μl-eluate from RNA isolation of 200 μl plasma sample was used as input into the reverse transcription. Input RNA was reverse transcribed using the TaqMan miRNA Reverse Transcription kit and miRNA- specific stem-loop primers (Life Technologies, Madrid, Spain) (Online Supplementary Appendix). The expression levels of miRNAs were calculated by using 2−Ct and reciprocal ratios were performed [Ratio miR-A/miR-B = log2 (2−Ct miR-A/2−Ct miR-B)], as previously described.14–19 Reciprocal ratios analysis is an approach that bypasses the controversial issue of data normalization of miRNAs in plasma (self-normalization). Furthermore, miRNAs whose concentrations are changed because of a pathology in opposite directions can be effective in differentiating investigated populations.
Target gene prediction and integrated analysis by Ingenuity Pathway Analysis
The altered miRNAs were further analyzed to obtain information about biological functions, pathways and networks by using the web-based bioinformatics tool QIAGEN’s Ingenuity Pathway Analysis (IPA; Ingenuity Systems, http://www.INGENUITY.com). For this purpose, all differentially regulated miRNAs and fold changes were imported into IPA20 (Online Supplementary Appendix).
Details of purification of IgG, in vitro exposure of monocytes and endothelial cells to aPL antibodies, and the statistical analysis are available in the Online Supplementary Appendix.
Results
Differentially expressed miRNAs in the plasma of APS patients and HDs
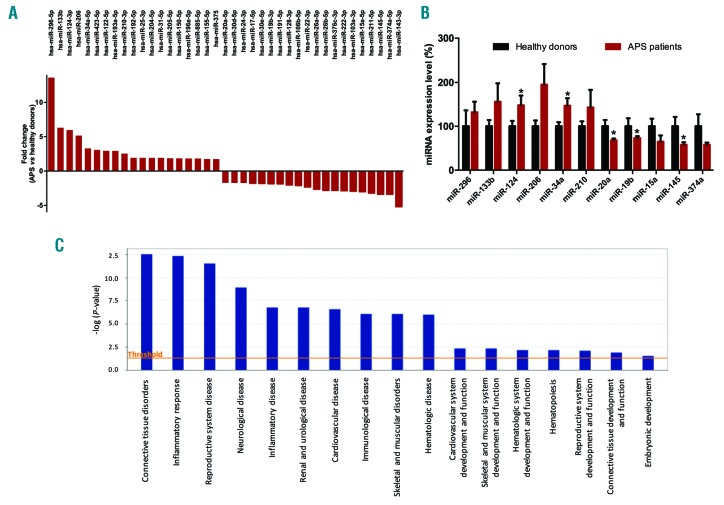
In the discovery phase (exploratory cohort), we identified 39 miRNAs that were differentially expressed between APS patients and HDs (cut off: 1.7-fold change), including 19 up-regulated and 20 down-regulated (Figure 1A). Using the IPA software, the functional analysis of the altered miRNAs in APS patients showed that a large number of them had validated and putative target mRNAs mainly involved in connective tissue disorders, inflammatory response, reproductive system disease, CVD or skeletal and muscular disorders (Figure 1C).
Figure 1.
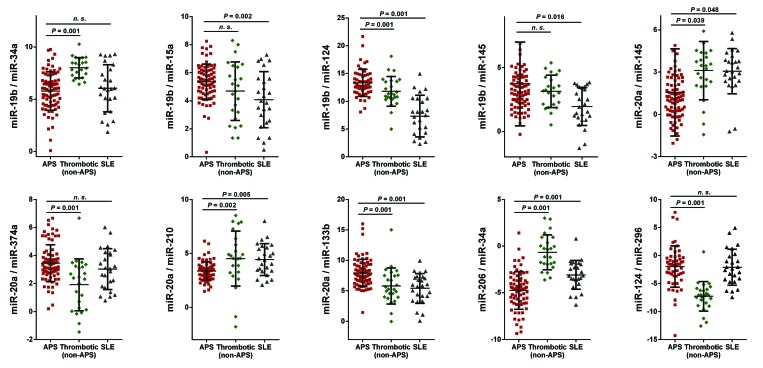
Antiphospholipid syndrome (APS) patients showed a specific circulating miRNAs profile related to clinical features of this autoimmune disorder. (A) To identify the changes that occurred in the expression levels of microRNAs (miR) in plasma from antiphospholipid syndrome versus controls, Human Serum & Plasma miRNA PCR-array (Qiagen) was performed in the study cohort. Expression levels of 19 miRNAs were found up-regulated in antiphospholipid syndrome, while 20 miRNAs were down-regulated. (B) Ingenuity Pathway Analysis (IPA) uncovered the main enriched biological functions and pathways in which these microRNAs are involved. The analysis included only the functions and pathways with average IPA score >2 [indicated as -log (P value)]. (C) Validation of selected miRNAs by RT-PCR in the whole cohort of APS patients and healthy donors. *P<0.05.
Bioinformatic identification and analysis of deregulated miRNAs related to the pathophysiology of APS and analysis of potential protein targets
In silico studies were performed to identify the altered miRNAs that might have as potential targets a number of genes/proteins involved in the development of clinical manifestations related to APS, such as coronary artery disease, thrombosis, abortion, and cerebrovascular dysfunction. IPA identified 11 altered miRNAs as the main regulators of proteins involved in the pathology of APS, including miRNA 34a-5p, 15a-5p, 145a-5p, 133b-3p, 124-3p, 206, 20a-5p, 19b-3p, 210-3p, 296-5p and 374a-5p. This set of 11 miRNAs included, among others, the top 5 up-regulated miRNAs and 3 out of the top 5 down-regulated miRNAs in the PCR-array. The expression levels of the 11 selected miRNAs were analyzed in all study subjects by RT-PCR (Figure 1B). MiR-124 and miR-34a were found increased in APS patients in relation to healthy donors, while miR-20a, miR-19b and miR145a were found reduced. The remaining microRNAs were also found to be altered, showing a trend to either increase or reduction as observed in the discovery phase, thus validating the data obtained by PCR-array.
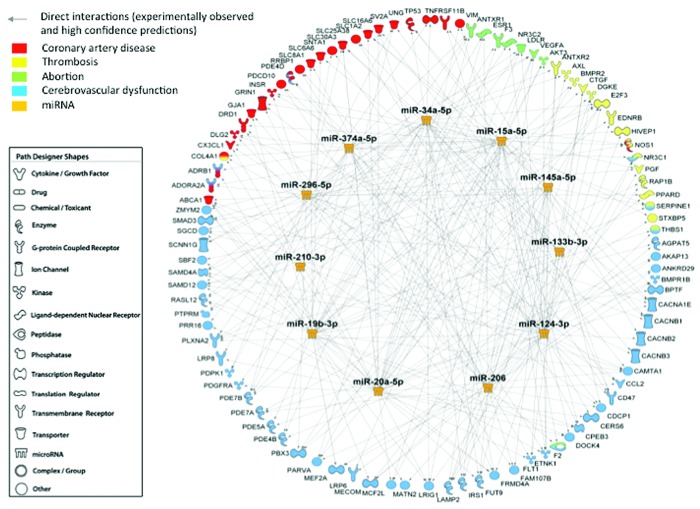
We further developed a network that defined the interaction between miRNA-mRNA targets (Figure 2). Key proteins involved in the pathophysiology of APS, and identified as potential mRNA targets of those miRNAs, were quantified in the plasma of APS patients and HDs. As previously reported,20–23 APS patients showed significantly increased plasma levels of TF, PAI-1, MCP-1, VEGF-A and VEGFR-1 (Online Supplementary Figure S1).
Figure 2.
Interaction network of microRNAs identified potential mRNA targets involved in clinical features of antiphospholipid syndrome. Using microRNA Target Filter of QIAGEN’s Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, CA, USA, www.qiagen.com/ingenuity), the software generated a network including the selected microRNAs (miRNAs or miR) and their mRNA targets, filtered by coronary artery disease, thrombosis, abortion and cerebrovascular dysfunction. Only targets experimentally observed and predicted with high confidence are shown and related by direct interactions to their specific miRNA regulators.
Circulating miRNA signature as potential biomarkers of disease in APS
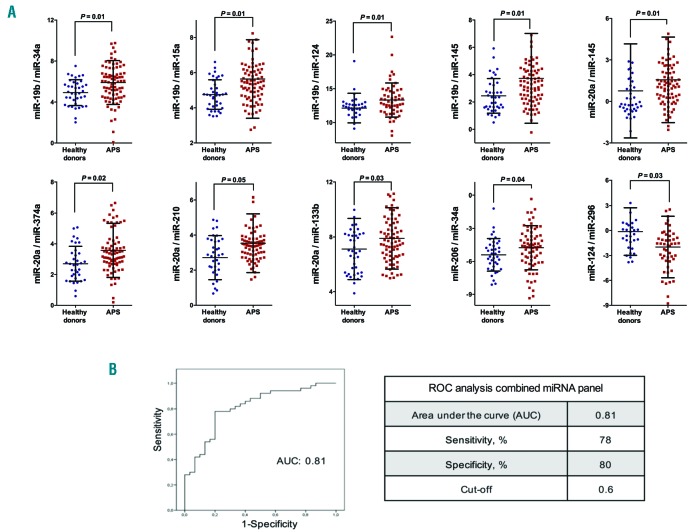
It has been shown that the combination of miRNAs improves their predictive potential to differentiate two pathological conditions.14–19 Thus, to assess the potential of specific circulating miRNAs in APS patients as biomarkers of disease features, reciprocal ratios of the miRNAs analyzed were performed by using statistical tools. By this approach, we identified 10 miRNA ratios, integrated by the 11 selected miRNAs, and differentially expressed in plasma of APS patients in comparison with HDs, including miR-19b/miR-34a, miR-19b/miR-15a, miR19b/miR- 124, miR-19b/miR-145, miR-20a/miR-145, miR-20a/miR- 374a, miR-20a/miR-210, miR-20a/miR-133b, miR- 206/miR-34a and miR-124/miR-296 (Figure 3A). To further explore the efficiency of these biomarkers to identify APS patients, a combination of the 10 miRNA ratios as a panel was carried out by using a logistic regression on the data set, as previously described.24 Thus, all miRNA-ratios were integrated into a single model or equation, which provided a single ‘score’ that allowed us to perform the ROC analysis and establish the cut off for prediction. The ROC curve for the 10 miRNA ratios signature revealed a marked accuracy, evidenced by an AUC of 0.81. At the optimal cut-off value of 0.6, the sensitivity and specificity of the combined miRNA panel for APS identification were 78% and 80%, respectively (Figure 3B).
Figure 3.
A circulating miRNA signature in antiphospholipid syndrome (APS) might have potential value as biomarkers of disease. (A) Selected microRNAs (miRNAs or miR) were analyzed in the whole cohort, including 90 APS patients and 42 healthy donors, and reciprocal ratios were performed. Beeswarm plot of each differentially expressed miR ratio is shown, along with mean, Standard Deviation, and P-value. For statistical analysis, after normality and equality of variance tests, comparisons were made by paired Student t-test or a non-parametric test (Mann-Whitney rank sum test). (B) A combination of the 10 miRNA ratios as a panel was carried out by using logistic regression on the data set. ROC curve of miRNA panel and cut off were generated based on the predicted probability (P) for each subject as a single score. The equation used in our model was: “Combined miRNA-ratio panel [Logit(p)] = − 0.64 + 0.034x(miR-19b/miR-34a) + 1.061x(miR-19b/miR-15a) + 0.248x(miR-19b/miR-124) − 1.704x(miR-19b/miR-145) + 2.34x(miR-20a/miR-145) − 0.729x(miR-20a/miR-374a) − 0.624x(miR-20a/miR-210) + 0.088x(miR- 20a/miR-133b) + 0.166x(miR-206/miR-34a) + 0.056x(mir-124/miR-296)”. The area under the curve (AUC), sensitivity and specificity are displayed, and a cut-off value with higher specificity was selected.
Stability of miRNA expression profile over time in APS
Plasma from 21 APS patients included in the study was evaluated again three months after the first blood sample collection to analyze the stability of the circulating miRNA profile. Results demonstrated that miRNA expression in the second sample collection did not change in relation to the first analysis (Online Supplementary Figure S2A). Moreover, the levels of miRNA ratios at baseline correlated significantly with the levels of these ratios three months later (Online Supplementary Figure S2B). Thus, our data support the theory that there is a specific circulating miRNA signature in APS which remains stable over time.
APS patients show a specific miRNA profile different from both non-autoimmune patients with previous thrombotic events and aPL-negative SLE patients
To assess the specificity of the miRNA signature found in APS patients, and in order to analyze whether the altered expression of the circulating miRNAs evaluated was linked to their thrombophilic status, an additional disease group including 23 patients with thrombosis in the absence of an associated autoimmune disease was evaluated. In these patients, the ratios formed by the expression levels of the 11 selected miRNAs were significantly different from those described in APS patients, except for the ratios miR-19b/miR-15a and miR-19b/miR-145 which exhibited non-significant differences (Figure 4). To evaluate if the altered expression of the miRNA signature was a sign of an autoimmune status, an additional disease group, including 25 SLE patients negative for aPL, was analyzed. In this SLE cohort, the ratios produced by the selected circulating miRNAs were significantly different from those found in APS patients, except for the ratios miR-19b/miR-34a, miR-20a/miR-374a and miR-124/miR- 296 which exhibited non-significant differences (Figure 4).
Figure 4.
Antiphospholipid syndrome (APS) patients show a specific miRNA profile distinct from both non-autoimmune patients with previous thrombotic events and aPL-negative systemic lupus erythematosus (SLE) patients. Twenty-three thrombotic non-antiphospholipid syndrome patients (non-APS) and 25 aPL-negative SLE patients were included, and the circulating microRNA (miRNA or miR) signatures of APS were compared. One-way ANOVA was used for statistical comparisons. A Bonferroni correction was applied for multiple testing. P<0.05 was considered statistically significant. Beeswarm plot of each differentially expressed miRNA ratio is shown along with mean, Standard Deviation and P-value. n.s.: no significant statistical differences.
Potential influence of standard therapy on the profile of circulating miRNAs in APS
APS patients were classified into two groups based on the treatment received, including 30 primary APS patients treated with antiplatelet agents and 62 primary APS patients treated with anticoagulant drugs. The statistical comparison between patients treated with antiplatelet and anticoagulant agents showed no significant differences in miRNA signature, except for the ratio miR- 20a/374 (Online Supplementary Figure S3).
Circulating miRNAs are associated with clinical features of APS and show potential as biomarkers for the development of atherosclerosis
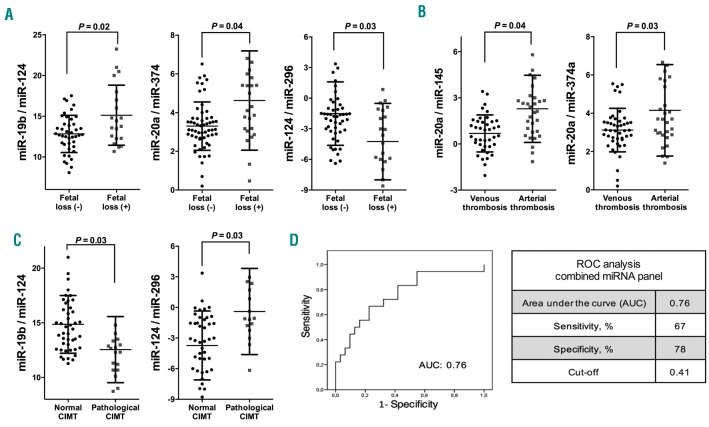
The levels of some circulating miRNA ratios that integrate the signature in APS were associated with the ocurrence of fetal losses in these patients, including elevated levels of miR-19b/miR-124 and miR-20a/miR-374, and reduced levels of miR-124/miR-296 (Figure 5A). Associations between miRNA ratios and the type of thrombosis suffered by APS patients were also identified. Thus, elevated levels of ratios miR-20a/miR-145 and miR- 20a/miR-374 were significantly associated with the ocurrence of arterial thrombosis in APS patients (Figure 5B). Furthermore, the ratios of miR-19b/miR-124 and miR- 124/miR-296 were also found to be associated with the presence of a pathological CIMT in these patients (Figure 5C). To accurately evaluate their relevance as biomarkers of early atherosclerosis, we conducted combined ROC analyses of these miRNA ratios. The combination of both circulating miRNA ratios as a panel showed an evident accuracy, with an AUC of 0.76 at a sensitivity of 67% and specificity of 78% from a cut-off value of 0.41 (Figure 5D).
Figure 5.
Circulating miRNAs are related to clinical features of antiphospholipid syndrome (APS) and show potential as biomarkers for the development of atherosclerosis. Association studies of altered circulating microRNA (miRNA or miR) ratios and the occurrence of previous fetal loss (A), the type of thrombosis suffered (B) and the presence of a pathological carotid intima-media thickness (CIMT) (C). Beeswarm plot of each miR ratio is shown, along with mean, Standard Deviation, and P-value. (D) A combination as a panel of the 2 miRNA ratios associated to the pathological CIMT was carried out by using logistic regression on the data set and receiver operator characteristics (ROC) curve analyses were performed. ROC curve of miRNA panel and cut off were generated based on the predicted probability (P) for each patient as a single score. The equation used was: “Combined miRNA-ratio panel [Logit(p)] = 0.599 − 0.133x(miR-19b/miR-124) + 0.007x(miR-124/miR- 296)”. The area under the curve (AUC), sensitivity and specificity are shown, and a cut-off value with higher specificity was selected.
Cluster analysis
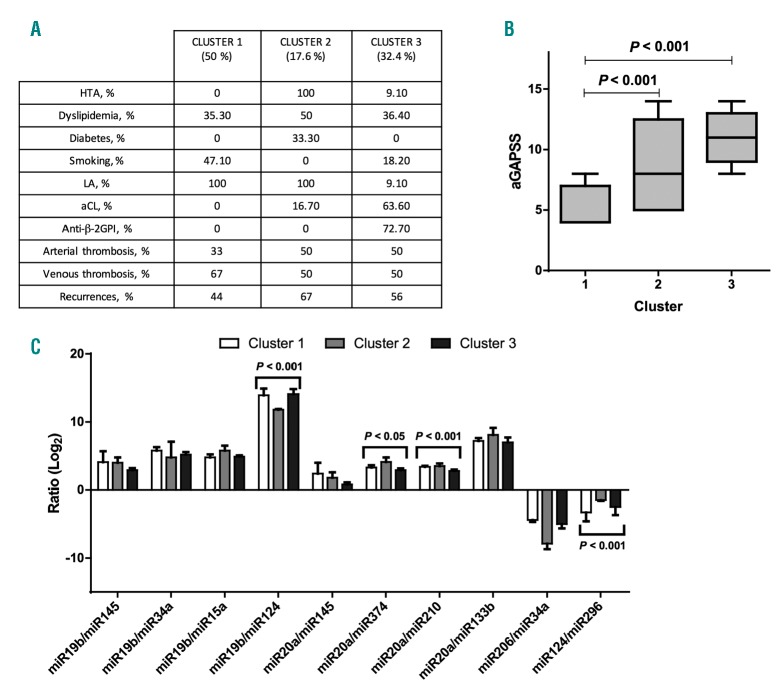
Hard clustering analysis in the APS cohort differentiated 3 clusters representing different thrombotic risk profile groups. Clinical and laboratory parameters of each cluster are resumed (Figure 6A). Briefly, cluster 1 (50% of the clustered cohort) was characterized by lower prevalence of cardiovascular risk factors and aPL multiple positivity. Conversely, cluster 1 shows a higher rate of venous thrombotic event when compared to the other clusters. Cluster 2 (17.6% of the clustered cohort) was characterized by a higher rate of cardiovascular risk factors, arterial thrombotic events, recurrences and a low prevalence of multiple aPL positivity. Cluster 3 (32.4% of the clustered cohort) was represented by a higher rate of multiple aPL positivity, arterial thrombotic events, and lower rate of cardiovascular risk factors. When evaluating different miRNA ratio expression among clusters, we found a statistically significant difference between groups for the following miRNA ratios: miR-19b/miR-124 (P<0.001, ANOVA), miR-20a/miR-374 (P<0.05, ANOVA), miR- 20a/miR-210 (P<0.001, ANOVA) and miR-124/miR-296 (P<0.05, ANOVA). miRNA ratio expression in the different clusters are summarized in Figure 6C.
Figure 6.
Specific miRNA signatures might identify subgroups of antiphospholipid syndrome (APS) patients showing different thrombotic risk profiles: cluster analysis. (A) Clinical and laboratory parameters of the 3 clusters. (B) Comparison of the adjusted global anti-phospholipid syndrome score (aGAPSS) values among the different clusters. (C) Evaluation of different microRNA (miR) ratios expression among clusters. HTA: arterial hypertension; LA: lupus anticoagulant; aCL: anti-cardiolipin IgG/IgM; anti- β2GPI: anti-β2 glycoprotein 1 IgG/IgM.
When comparing the aGAPSS values among the different clusters, we found a significant difference (P=0.008, t-test) between cluster 1 [mean aGAPSS 5.38; 1.628±Standard Deviation (SD)] and Cluster 2 (mean aGAPSS 8,67; 3.67±SD). Similarly, we found a significant difference (P<0.001, t-test) between cluster 1 and cluster 3 (mean aGAPSS 10.82; 2.316±SD). aGAPSS values stratifying for clusters are represented in Figure 6B.
Circulating miRNAs correlate with clinical and serological parameters in APS
The miRNA ratios that integrate the signature in APS were linked with clinical parameters, such as ABI, presence of elevated titers of aPL, particularly aCL and anti- β2GPI antibodies, and erythrocyte sedimentation rate (Online Supplementary Table S3). Correlation analyses with serological markers related to atherothrombosis further showed significant positive correlations with the expression levels of various miRNA ratios and with levels of TF, PAI-1, VEGF-A, VEGF-R1 and MCP-1 (Online Supplementary Table S3). Some of these correlations were also found among various miRNA ratios in plasma of APS patients.
Antiphospholipid antibodies modulate the expression of both the circulating miRNAs that integrate the signature in APS and their potential protein targets
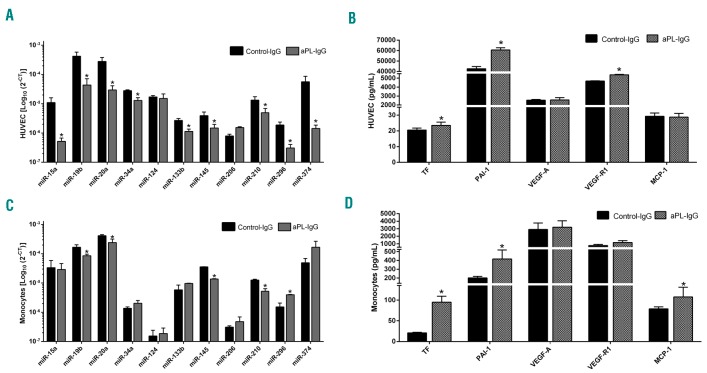
The expression of the 11 selected miRNAs was significantly altered in the supernatant of HUVECs treated with aPL-IgG in relation to those treated with a non-immune- IgG (Figure 7A), except for the miR-124 and miR-206. Accordingly, this treatment promoted in HUVECs the secretion of atherothrombotic proteins, such as TF, PAI-1 and VEGF-R1 (Figure 7B), potential targets of the miRNAs analyzed. On the other hand, the expression levels of several miRNAs were deregulated in the supernatant of monocytes treated with aPL-IgG, including miR-19b, miR- 20a, miR-145, miR-210 and miR-296 (Figure 7C). Concomitantly, aPL-IgG treatment promoted in monocytes an increase in the secretion of TF, PAI-1 and MCP-1 (Figure 7D).
Figure 7.
Antiphospholipid antibodies modulate the expression of both the circulating miRNAs that integrate the signature in antiphospholipid syndrome (APS) and their putative protein targets. Human umbilical vein endothelial cells (HUVECs) were treated with antiphospholipid antibodies and secreted selected microRNAs (miRNAs) (A) and putative target protein (B) levels were determined in the supernatant. Monocytes were also treated with antiphospholipid antibodies and secreted selected miRNAs (C) and putative target proteins (D) levels were evaluated in the supernatant of culture. Differences were analyzed by Student t-test. Values are the means and Standard Error of Mean of 4 independent experiments performed in triplicate. P<0.05 was considered statistically significant. TF: tissue factor; PAI-1: plasminogen activator inhibitor-1; VEGF-A: vascular endothelial growth factor A; VEGF-R1: VEGF-Receptor-1; MCP-1: monocyte chemotactic protein.
Discussion
The present study identifies, for the first time, a specific signature of circulating miRNAs in APS patients that might serve as potential biomarkers of clinical features of this autoimmune disorder. Moreover, this signature could represent a useful tool to typify and stratify patients based on their thrombotic status and cardiovascular risk profile (Online Supplementary Figure S3).
Circulating miRNAs were firstly described in peripheral blood as promising specific biomarkers for a wide range of diseases, such as cancer and other inflammatory pathologies.25,26 Thereafter, several studies revealed the altered expression of numerous miRNAs in plasma, blood cells, and tissues of systemic autoimmune conditions, such as RA and SLE, which were directly associated to disease activity, making them potential useful biomarkers for clinical features and follow up.9,26–29 However, to date, the specific profile of circulating miRNAs in APS patients has not been evaluated. In the present study, the profiling of miRNAs by PCR-array in plasma of APS patients has helped to identify a set of miRNAs differentially expressed and collectively associated to clinical features of the disease, such as inflammatory response, reproductive system disease, and CVD, among others. Using logistic regression, we further developed a model that identified 10 miRNA ratios, differentially expressed, that showed great potential as biomarkers of disease of APS patients.
Recent studies support the evidence that an miRNAs signature has a higher diagnostic value than individual miRNAs.14–19 The use of ratios is a feasible approach that overcomes the controversial question of normalizing plasma levels of miRNAs, given the lack of a reliable normalizer for circulating miRNAs. In addition, the establishment of these ratios allows the identification of a combination of expression profiles closer to reality in vivo in patients, where the interactions between miRNAs and their specific potential targets never occur in a unique or individualized way. In fact, it is likely that, in some cases, various miRNAs, whose concentrations are shifted in opposite directions in a particular pathology, contribute together and specifically to certain clinical profiles.
The signatures of circulating miRNAs identified in APS patients integrated miRNAs previously described to be altered in other autoimmune and CVD. Thus, miR-19b and miR-20a have been shown to be essential modulators of TF expression in APS and SLE patients,8 so that reduced expression of such miRNAs contributes to the overexpression of TF in monocytes, which is directly associated with the occurrence of thrombotic events in APS.21 On the other hand, miR-124, found altered in APS, SLE and RA patients at both cellular and plasma levels, modulates the overexpression of MCP-1, a key chemokine directly involved in CVD associated to these autoimmune conditions.30–33 Likewise, miR-133b and miR-145 have been identified as the most promising biomarkers of the pathogenesis of CVD. Both miRNAs participate in the differentiation of vascular smooth muscle cells. In addition, miR- 133b regulates angiogenesis and endothelial function, while miR-145 participates in the stabilization of atheromatous plaque.34 The miR-34a is highly expressed in endothelial cells, and elevated circulating levels of this miRNA have been associated to myocardial infarction.35 Moreover, the main target of miR-34a is VEGF-A, a key inflammatory protein involved in numerous cardiovascular and autoimmune pathologies, including APS.23,36 In the same way, miR-374 has been described as regulator of maintenance of vascular integrity.37 The remaining miRNAs members of the signature, including miR-296, miR-210, miR-206 and miRNA-15, have been found altered in severe pre-eclampsia, one of the leading causes of maternal mortality and neonatal morbidity worldwide.38–40 Thus, all the processes regulated by these miRNAs seem to orchestrate distinct aspects of APS pathogenesis.
To assess the specificity of the circulating miRNA signature in APS we evaluated the miRNA profile in an additional cohort of patients characterized by the presence of previous thrombotic events in the absence of an associated autoimmune disease. The miRNAs analysis revealed a differential pattern of expression between these two cohorts. Those results substantiate previous studies that evidenced the presence of a distinct miRNA profile in monocytes and neutrophils of thrombotic non-autoimmune patients compared to APS patients.9 This could reflect a differential mode of regulation and activity of miRNAs in thrombotic patients compared to APS patients, on which the role of autoantibodies might be crucial. Moreover, the analysis of a parallel autoimmune population (SLE patients) negative for aPL, also identified an miRNA signature distinct from that of APS, thus underlying the potential role of aPLs as regulators of thrombosis- related miRNAs in APS, and pointing to the presence of a specific miRNA profile relative to the pathogenesis of each disease.
Antiphospholipid syndrome patients recruited in this study were mainly treated with anticoagulant and/or antiplatelet agents. All of them have been shown to influence miRNAs expression, an epigenetic process that might help to delineate the mechanisms underlying their effects.9,41,42 Thus, we evaluated the potential effect of these treatments on the circulating miRNA expression profile. No significant differences were observed in our cohort of APS patients between those who received antiplatelet and those treated with anticoagulant agents, suggesting that the prothrombotic status induced by effects of aPLs, and the consequently deregulated miRNAs, were not differentially modulated among these drugs.
In order to understand the clinical relevance of the altered circulating miRNA signature, association and correlation studies were perfomed. Altered expression of various miRNA ratios was associated with the presence of previous fetal losses. In line with these findings, several studies have shown that the misregulation of circulating placental miRNAs in maternal blood might lead to pregnancy complications, thus acting as non-invasive diagnostic and prognostic biomarkers for pregnancy monitoring.42–44 Association studies further established a significantly increased expression of 2 miRNA ratios in APS patients that had suffered arterial thrombosis in comparison with those who experienced venous thrombotic events. Interestingly, both miRNA ratios were integrated by the miR-20a, previously reported to be the main regulator of TF, whose expression levels have been found to be related to the development of arterial thrombosis in the setting of APS.8,45 Finally, we identified 2 miRNA ratios as clinical relevant biomarkers related to early atherosclerosis development in APS patients, which were integrated by the miR-19b and miR-124, both of them critical players in the expression of proteins related to inflammation and thrombosis in APS and SLE.8,9
Correlation studies revealed that the altered circulating miRNA signature in APS is linked to parameters related to increased risk of peripheral artery disease such as ABI. Moreover, correlations between circulating miRNA levels and numerous altered parameters related to inflammation and thrombosis were also identified. These correlations support the relationship observed in the in silico study between the selected miRNAs and potential target proteins involved in various clinical features of APS. The influence of the autoimmunity in the circulating profile of miRNAs in APS was also revealed by the significant correlation between high titers of aPL-IgG and the altered expression of several miRNAs integrating the signature. These relationships further sustain those previously identified among the altered profile of miRNAs in APS and SLE at cellular level and the autoimmune and inflammatory profile of both autoimmune conditions.9 Therefore, our data suggest that the altered plasma profile of miRNAs is an important mechanisin that might contribute to the regulation of the pro-atherothrombotic status of APS patients, on which aPL seem to play a key role.
Our in vitro studies further confirmed this hypothesis, demonstrating that aPL-IgG antibodies promoted a significant deregulation in the expression levels of both the selected miRNAs and their potential protein targets in the supernatant of cultured monocytes and HUVECs, the main drivers of the CVD in the setting of APS. These results also confirm and complement previous studies which showed the in vitro effects of aPL-IgG in the induction of prothrombotic/inflammatory mediators3,31 and the modulation of specific cellular miRNAs involved in their modulation.8,9 Nevertheless, although our data show specific effects of aPL-IgG on the secretion of several circulating microRNAs related to CVD, the contribution of other components of the vascular and immune system to the altered profile of circulating miRNAs still has to be defined. In addition, since we did not perform a complete plasma human microarray analysis, we cannot exclude the complementary role of other circulating miRNAs in the physiopathology of APS.
Interestingly, our analysis supports a clinical role for the use of miRNA ratios when stratifying patients for their thrombotic risk. While studying the miRNA expression profile has widened the understanding of APS pathogenesis,9 its clinical utility is still a question of debate. Our data support the view that specific miRNA signatures could identify subgroups of APS patients showing different clinical profiles (in terms of site of thrombosis and risk of recurrences), potentially paving the way for their use as useful biomarkers that will increase the specificity and sensitivity of thrombotic risk assessment.
Taken together, our data suggest that differentially expressed miRNAs in the plasma of APS patients, modulated at least partially by aPL-IgG antibodies, might have the potential to serve as novel biomarkers of disease features and could help typify the atherothrombotic status of patients, thus constituting a useful tool in the management of this disease.
Supplementary Material
Acknowledgments
We thank all patients for their participation in this study.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/5/908
Funding
This study was supported by grants from the Junta de Andalucia (CTS-7940), the Instituto de Salud Carlos III (ref. n. PI15/01333), Cofinanciado por el Fondo Europeo de Desarrollo Regional de la Unión Europea ‘Una manera de hacer Europa’, Spain, and the Spanish Inflammatory and Rheumatic Diseases Network (RIER), Instituto de Salud Carlos III (RD16/0012/0015). CL-P was supported by a contract from the Spanish Junta de Andalucía. YJ-G was supported by a contract from the University of Cordoba (Co-financing of the Research Plan of the University of Cordoba and the Operating Program of the European Regional Development Funds -ERDF- for Andalusia).
References
- 1.Young SP, Kapoor SR, Viant MR, et al. The impact of inflammation on metabolomic profiles in patients with arthritis. Arthritis Rheum. 2013;65(8):2015–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palisi A, Grimaldi M, Sabatini P, et al. A serum nuclear magnetic resonance-based metabolomic signature of antiphospholipid syndrome. J Pharm Biomed Anal. 2017;133:90–95. [DOI] [PubMed] [Google Scholar]
- 3.Perez-Sanchez C, Barbarroja N, Messineo S, et al. Gene profiling reveals specific molecular pathways in the pathogenesis of atherosclerosis and cardiovascular disease in antiphospholipid syndrome, systemic lupus erythematosus and antiphospholipid syndrome with lupus. Ann Rheum Dis. 2015;74(7):1441–1449. [DOI] [PubMed] [Google Scholar]
- 4.López-Pedrera C, Pérez-Sánchez C, Ramos-Casals M, Santos-Gonzalez M, Rodriguez-Ariza A, Cuadrado MJ. Cardiovascular risk in systemic autoimmune diseases: epigenetic mechanisms of immune regulatory functions. Clin Dev Immunol. 2012;2012:9746–9748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67(1):129–139. [DOI] [PubMed] [Google Scholar]
- 6.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. [DOI] [PubMed] [Google Scholar]
- 7.Alevizos I, Illei GG. MicroRNAs as biomarkers in rheumatic diseases. Nat Rev Rheumatol. 2010;6(7):391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teruel R, Pérez-Sánchez C, Corral J, et al. Identification of miRNAs as potential modulators of tissue factor expression in patients with systemic lupus erythematosus and antiphospholipid syndrome. J Thromb Haemost. 2011;9(10):1985–1992. [DOI] [PubMed] [Google Scholar]
- 9.Pérez-Sánchez C, Aguirre MA, Ruiz-Limón P, et al. Atherothrombosis-associated microRNAs in Antiphospholipid syndrome and Systemic Lupus Erythematosus patients. Sci Rep. 2016;6:31375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110(3): 483–495. [DOI] [PubMed] [Google Scholar]
- 11.Heegaard NHH, Carlsen AL, Skovgaard K, Heegaard PMH. Circulating Extracellular microRNA in Systemic Autoimmunity. EXS. 2015;106:171–195. [DOI] [PubMed] [Google Scholar]
- 12.Sciascia S, Sanna G, Murru V, Roccatello D, Khamashta MA, Bertolaccini ML. GAPSS: The global anti-phospholipid syndrome score. Rheumatology (Oxford). 2015;54(1):134–138. [DOI] [PubMed] [Google Scholar]
- 13.Castro-Villegas C, Pérez-Sánchez C, Escudero A, et al. Circulating miRNAs as potential biomarkers of therapy effectiveness in rheumatoid arthritis patients treated with anti-TNFα. Arthritis Res Ther. 2015;17:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boeri M, Verri C, Conte D, et al. MicroRNA signatures in tissues and plasma predict develoment and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci USA. 2011;108(9):3713–3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennessey PT, Sanford T, Choudhary A, et al. Serum microRNA biomarkers for detection of non-samll cell lung cancer. PLoS One. 2012;7(2):e32307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheinerman KS, Tsivinsky VG, Abdullah L, Crawford F, Umansky SR. Plasma microRNA biomarkers for detection of mild cognitive impairment: biomarker validation study. Aging (Albany NY). 2013;5(12):925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthaei H, Wylie D, Lloyd MB, et al. miRNA biomarkers in cyst fluid augment the diagnosis and management of pancreatic cysts. Clin Cancer Res. 2012;18(17):4713–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fortunato O, Boeri M, Verri C, et al. Assessment of circulating microRNAs in plasma of lung cancer patients. Molecules. 2014;19(3):3038–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharova E, Grassi A, Marcer A, et al. A circulating miRNA assay as a first-line test for prostate cancer screening. Br J Cancer. 2016;114(12):1362–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayburd AL, Martlínez A, Sackett D, et al. Ingenuity network-assisted transcription profiling: Identification of a new pharmacologic mechanism for MK886. Clin Cancer Res. 2006;12(6):1820–1827. [DOI] [PubMed] [Google Scholar]
- 21.Cuadrado MJ, López-Pedrera C, Khamashta MA, et al. Thrombosis in primary antiphospholipid syndrome: a pivotal role for monocyte tissue factor expression. Arthritis Rheum. 1997;40(5):834–841. [DOI] [PubMed] [Google Scholar]
- 22.López-Pedrera C, Aguirre MA, Buendía P, et al. Differential expression of protease activated receptors in monocytes from patients with primary Antiphospholipid syndrome. Arthritis Rheum. 2010; 62(3):869–877. [DOI] [PubMed] [Google Scholar]
- 23.Cuadrado MJ, Buendía P, Velasco F, et al. Vascular endothelial growth factor expression in monocytes from patients with primary antiphospholipid syndrome. J Thromb Haemost. 2006;4(11):2461–2469. [DOI] [PubMed] [Google Scholar]
- 24.Xiao B, Wang Y, Li W, et al. Plasma microRNA signature as a noninvasive biomarker for acute graft-versus-host disease. Blood. 2013;122(19):3365–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood- based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006. [DOI] [PubMed] [Google Scholar]
- 27.Dai Y, Huang YS, Tang M, et al. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16: 939–946. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J, Chen J, Sen S. MicroRNA as Biomarkers and Diagnostics. J Cell Physiol. 2016;231(1):25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamachi Y, Kawano S, Takenokuchi M. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009;60(5):1294–1304. [DOI] [PubMed] [Google Scholar]
- 31.Pérez-Sánchez C, Ruiz-Limón P, Aguirre MA, et al. Mitochondrial dysfunction in antiphospholipid síndrome: implications in the pathogenesis of the disease and effects of coenzyme Q10 treatment. Blood. 2012; 119(24):5859–5870. [DOI] [PubMed] [Google Scholar]
- 32.Ruiz-Limón P, Barbarroja N, Pérez-Sánchez C, et al. Atherosclerosis and cardiovascular disease in systemic lupus erythematosus: effects of in vivo statin treatment. Ann Rheum Dis. 2015;74(7):1450–1458. [DOI] [PubMed] [Google Scholar]
- 33.Barbarroja N, Pérez-Sanchez C, Ruiz-Limon P, et al. Anticyclic citrullinated protein antibodies are implicated in the development of cardiovascular disease in rheumatoid arthritis. Arterioscler Thromb Vasc Biol. 2014;34(12):2706–2716. [DOI] [PubMed] [Google Scholar]
- 34.Navickas R, Gal D, Laucevičius A, Taparauskaitė A, Zdanytė M, Holvoet P. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc Res. 2016;111(4):322–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Y, Qi Y, Du JQ, Zhang DF. MicroRNA-34a regulates cardiac fibrosis after myocardial infarction by targeting Smad4. Expert Opin Ther Targets. 2014; 18(12):1355–1365. [DOI] [PubMed] [Google Scholar]
- 36.Li N, Wang K, Li PF. MicroRNA-34 Family and Its Role in Cardiovascular Disease. Crit Rev Eukaryot Gene Expr. 2015;25(4):293–297. [DOI] [PubMed] [Google Scholar]
- 37.Licholai S, Blaż M, Kapelak B, Sanak M. Unbiased Profile of MicroRNA Expression in Ascending Aortic Aneurysm Tissue Appoints Molecular Pathways Contributing to the Pathology. Ann Thorac Surg. 2016;102(4):1245–1252 [DOI] [PubMed] [Google Scholar]
- 38.Choi SY, Yun J, Lee OJ, et al. MicroRNA expression profiles in placenta with severe preeclampsia using a PNA-based microarray. Placenta. 2013;34(9):799–804. [DOI] [PubMed] [Google Scholar]
- 39.Sheikh AM, Small HY, Currie G, Delles C. Systematic Review of Micro-RNA Expression in Pre-Eclampsia Identifies a Number of Common Pathways Associated with the Disease. PLoS One. 2016;11(8): e0160808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akehurst C, Small HY, Sharafetdinova L, et al. Differential expression of microRNA-206 and its target genes in preeclampsia. J Hypertens. 2015;33(10):2068–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan ES, Cronstein BN. Methotrexate–how does it really work? Nat Rev Rheumatol. 2010;6(3):175–178. [DOI] [PubMed] [Google Scholar]
- 42.Tsochandaridis M, Nasca L, Toga C, Levy-Mozziconacci A. Circulating microRNAs as clinical biomarkers in the predictions of pregnancy complications. Biomed Res Int. 2015;2015:294954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Z, Moley KH, Gronowski AM. Diagnostic potential for miRNAs as biomarkers for pregnancy-specific diseases. Clinical Biochemistry. 2013;46(10–11):953–960. [DOI] [PubMed] [Google Scholar]
- 44.Wu L, Zhou H, Lin H, et al. Circulating microRNAs are elevated in plasma from severe preeclamptic pregnancies. Reproduction. 2012;143(3):389–397. [DOI] [PubMed] [Google Scholar]
- 45.Tatsumi K, Mackman N. Tissue Factor and Atherothrombosis. J Atheroscler Thromb. 2015;22(6):543–549 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.