Abstract
Survival of children with relapsed acute lymphoblastic leukemia is poor, and understanding mechanisms underlying resistance is essential to developing new therapy. Relapse-specific heterozygous deletions in MSH6, a crucial part of DNA mismatch repair, are frequently detected. Our aim was to determine whether MSH6 deletion results in a hypermutator phenotype associated with generation of secondary mutations involved in drug resistance, or if it leads to a failure to initiate apoptosis directly in response to chemotherapeutic agents. We knocked down MSH6 in mismatch repair proficient cell lines (697 and UOCB1) and showed significant increases in IC50s to 6-thioguanine and 6-mercaptopurine (697: 26- and 9-fold; UOCB1: 5- and 8-fold) in vitro, as well as increased resistance to 6-mercaptopurine treatment in vivo. No shift in IC50 was observed in deficient cells (Reh and RS4;11). 697 MSH6 knockdown resulted in increased DNA thioguanine nucleotide levels compared to non-targeted cells (3070 vs. 1722 fmol/μg DNA) with no difference observed in mismatch repair deficient cells. Loss of MSH6 did not give rise to microsatellite instability in cell lines or clinical samples, nor did it significantly increase mutation rate, but rather resulted in a defect in cell cycle arrest upon thiopurine exposure. MSH6 knockdown cells showed minimal activation of checkpoint regulator CHK1, γH2AX (DNA damage marker) and p53 levels upon treatment with thiopurines, consistent with intrinsic chemoresistance due to failure to recognize thioguanine nucleotide mismatching and initiate mismatch repair. Aberrant MSH6 adds to the list of alterations/mutations associated with acquired resistance to purine analogs emphasizing the importance of thiopurine therapy.
Introduction
Relapsed B-precursor acute lymphoblastic leukemia (B-ALL) is a leading cause of cancer mortality amongst children. Development of chemoresistance is a crucial factor contributing to relapse, therefore understanding the biological mechanisms underlying this resistance is imperative for discovering innovative treatment strategies.1 Recent work has begun to highlight the direct role of relapse specific/enriched genetic alterations in the emergence of clones that have gained a selective advantage under the pressure of specific chemotherapeutics, such as NT5C2, TBL1XR1, PRPS1, and CREBBP.1–5 Many of these mutations cause resistance specifically to thiopurines, which are the backbone of maintenance therapy and have proven vital for achieving cures.6 Our analysis of copy number alterations (CNAs) in diagnosis/relapse pairs revealed a relapse specific hemizygous deletion on chromosome 2p16.3 involving MSH6 in 4–10% of patients.7,8
MutS homolog 6 (MSH6) is a major component of the mismatch repair (MMR) system, which is a highly conserved biological process that recognizes and repairs errors in nascent DNA strands during replication to maintain genomic integrity. Initial recognition of replicative errors is carried out by protein heterodimers consisting of either MSH6 and MSH2 (hMutSα), or MSH3 and MSH2 (hMutSβ). Upon recognition of a mismatch, hMutSa recruits MutLα (MLH1-PMS2) which engages downstream proteins and enzymes involved in DNA repair.9,10
Constitutional defects in MMR, including monoallelic mutations in Lynch syndrome and biallelic loss in constitutional mismatch repair deficiency (CMMRD), are strongly linked to carcinogenesis, where loss of MMR functionality causes increased mutability and predisposition to malignancy.11–15 Previous work has linked defects in MMR to drug resistance, including thiopurines, in various cancers.16–19 However, it is uncertain if resistance in MMR defective clones occurs through the acquisition of secondary mutations as a consequence of mutagenic therapy, or the outgrowth of clones that have intrinsic drug resistance. Our lab previously demonstrated that lower expression of MSH6 in patient samples was associated with increased ex vivo resistance to 6-mercaptopurine and prednisone,7 highlighting the clinical importance of understanding the role of this genetic alteration in B-ALL.
The mechanism of action of thiopurines is based upon the insertion of a false nucleotide, namely a thioguanine (TGN), into DNA that when thiomethylated pairs with a thymine instead of a cytosine.18 Cytotoxicity is thought to be dependent on the MMR machinery recognizing the mismatch and attempting to match the TGN on the parental strand with an appropriate base on the daughter strand.19,20 Whether the DNA damage induced by the repetitive, futile cycles of DNA excision and repair, or simply the recognition of mismatches by hMutSα is enough to initiate a signaling cascade culminating in cell cycle arrest and apoptosis is not entirely understood.
We sought to delineate whether reduced expression of MSH6 could give rise to chemoresistance in B-precursor ALL and elucidate the mechanism responsible for the resistance. Our data here support the view that reduced MSH6 directly results in an increased tolerance to incorporated TGN and subsequent mismatches through a failure to initiate MMR, thus allowing cells to proliferate and survive under thiopurine treatment both in vitro and in vivo. We demonstrate that ALL cell lines with a functional MMR trigger a CHK1-mediated cell cycle arrest in response to thiopurines that is followed by DNA damage and apoptosis. In contrast, upon reduction of MSH6, the MMR signaling cascade is not fully activated and cells do not undergo apoptosis.
Methods
Cells and reagents
The B-lineage leukemia cell lines RS4;11 (ATCC, Manassas, VA, USA), Reh (ATCC), 697 (DSMZ, Braunschweig, Germany), and UOCB1 (a kind gift from Dr. Terzah Horton at Texas Children’s Cancer Center/Baylor College of Medicine) were grown in RPMI1640 medium. HEK293T (ATCC) cells were grown in DMEM medium. All media were supplemented with 10% FBS, 1% penicillin/streptomycin under 5% CO2 at 37°C.
Drug preparation, viral preparation, immunoblotting, apoptosis assays, and cell cycle
Standard protocols were followed and have been previously described.3,21 More detailed information is provided in the Online Supplementary Appendix.
Patients’ samples
Cryopreserved pediatric B-ALL specimens were obtained from the Children’s Oncology Group (COG) ALL cell bank. All patients were treated on COG protocols for newly diagnosed ALL. All subjects provided consent for banking and future research use of these specimens in accordance with the regulations of the institutional review boards of all participating institutions.
Microsatellite instability analysis
Microsatellite instability (MSI) analysis was performed using MSI Analysis System, v.1.2 (Promega, Madison, WI, USA) following the manufacturer’s protocol. Detailed information is provided in the Online Supplementary Appendix.
Measurement of mutation rate
Spontaneous mutation rate was measured using a flow cytometry assay previously described by Araten et al.22 that detects the presence of numerous glycosylphosphatidylinositol-linked (GPI) membrane proteins (see Online Supplementary Appendix). Briefly, GPI(+) isolated clones from the NT and MSH6-KD cell lines were expanded either untreated or treated with 6-TG (0.040 μg/mL and 0.100 μg/mL, respectively, based on IC50 values determined for clones). Cells were then stained for GPI-dependent markers including FLAER-Alexa 488 (Pinewood), CD48, CD52, and CD59 (Serotec),23 and analyzed by flow cytometry. The mutant frequency (f) was calculated as the number of GPI(-) events divided by the total number of live events, and mutation rate (μ) was calculated as f divided by cell divisions.22
Thioguanine quantification assay
Cells were treated with 6-thioguanine (6TG) and collected every day for four days. DNA was extracted using Puregene Core Kit A (QIAGEN). DNA TGN levels were measured using liquid chromatography-tandem mass spectrometry as described previously.24
In vivo mouse model of chemoresistance
All experiments were conducted on protocols approved by the Institutional Animal Care and Use Committee and Institutional Review Board of the Children’s Hospital of Philadelphia. Briefly, 1 million UOCB1 NT GFP-CBG or MSH6 shRNA1 GFP-CBR cells were injected into NSG mice via tail vein on day 0 (total 20 mice; 10 per cell line). On day 6 leukemic burden was confirmed via bioluminescence imaging (BLI) (IVIS Spectrum imaging system, Perkin Elmer) and animals were randomized to treatment groups [PBS vehicle or Purixan (50 mg/kg) diluted in PBS]. Mice were treated on day 7 by gavage (0.2 mL/mouse). For BLI, 3 mg of luciferin was injected intraperitoneally and mice were imaged ten minutes post injection. Quantification of total flux was determined by analyzing the BLI images using Living Image Software (Perkin Elmer) (see Online Supplementary Appendix).
Statistical analysis
Statistical significance was calculated using unpaired t-test for IC50s, paired t-test for mutation rates, one-way ANOVA for cell cycle analysis, and two-way ANOVA for mutation rates with and without treatment as well as in vivo studies.
Results
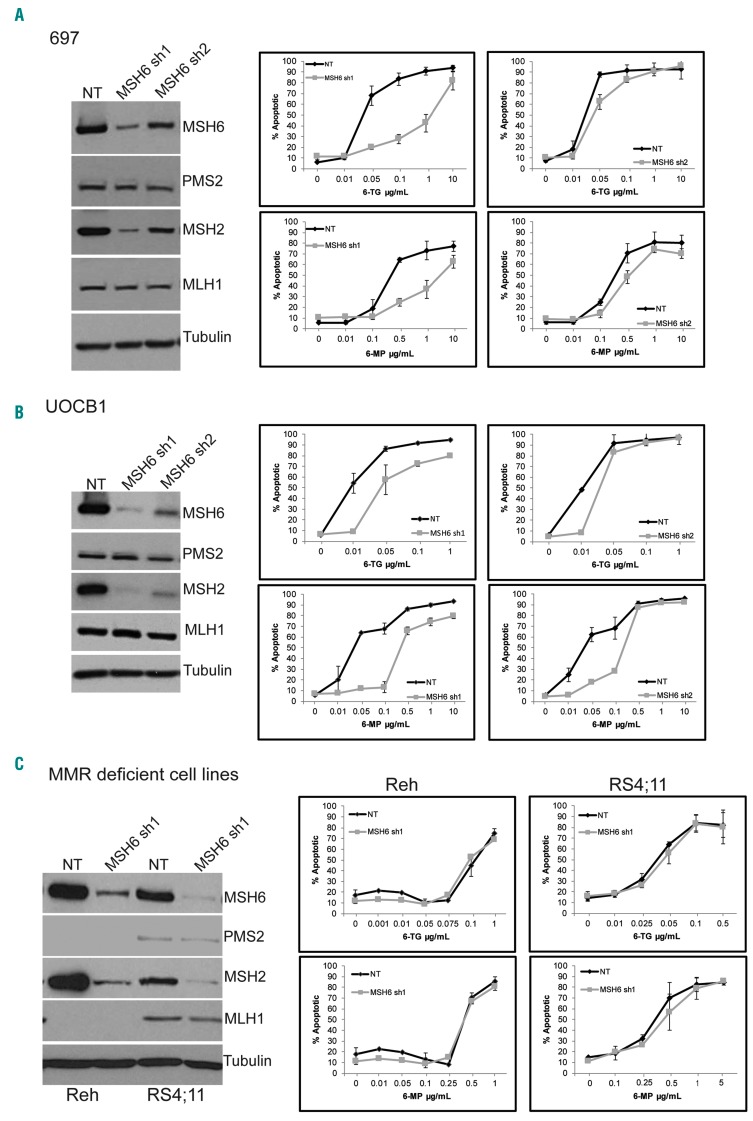
Previously we noted relapse-specific heterozygous deletions in MSH6 in 4 out of 76 patients that were near identical and deleted MSH6 only for 3 patients while one harbored a larger deletion involving more genes within the region (Online Supplementary Figure S1). To begin to elucidate the impact of MSH6 deletion on the development of relapsed disease, we knocked down expression of MSH6 using shRNA in 697 cells, a B-ALL cell line that expresses all four MMR proteins (Figure 1A) and is MMR proficient,25 and tested for changes in chemosensitivity. We observed approximately 80–90% (shRNA1) and 50–60% (shRNA2) knockdown of MSH6 expression, as well as decreased expression of MSH2, compared to non-targeting (NT) control cells (Figure 1A), which is consistent with literature on the loss of protein stability of MSH2 and MSH6 when not dimerized.17,26 Knockdown of MSH6 with both shRNA1 and shRNA2 leads to a significant decrease in apoptotic cells when treated with thiopurines for five days (Figure 1A). A 26-fold increase in IC50 with 6-TG (NT: 0.027 vs. shRNA1: 0.716 μg/mL; P=0.007) and 8.5-fold increase for 6-MP (NT: 0.340 vs. shRNA1: 2.89 μg/mL; P=0.006) was observed for shRNA1 (Online Supplementary Figure S2). A 1.7-fold (NT: 0.015 vs. shRNA2: 0.025 μg/mL; P=0.015) and a 2.6-fold (NT: 0.143 vs. shRNA2: 0.373 μg/mL; P=0.032) increase in IC50 for 6- TG and 6-MP, respectively, were observed for shRNA2 cells compared to NT cells (Online Supplementary Figure S2). However, no significant differences were observed when cells were treated with prednisolone (Pred), doxorubicin (Doxo), cytarabine (Ara-c), or methotrexate (MTX) (Online Supplementary Figure S3A). Interestingly, we found that knockdown of MSH6 also resulted in decreased sensitivity to temozolomide (TMZ), an alkylating agent used to treat glioblastomas, as reported previously (Online Supplementary Figure S3B).27,28
Figure 1.
Knockdown of MSH6 in mismatch repair (MMR) proficient cells lead to decreased sensitivity to thiopurines. (A-C, left) Western blot analysis of whole cell lysates from 697 (A), UOCB1 (B), and Reh and RS4;11 (C). (A-C, right) Apoptotic cells measured by Annexin V and 7AAD staining followed by flow cytometry after 5 days of treatment. Graphs represent 3 experiments each performed with duplicates. Bars indicate mean+Standard Deviation.
To further support the role of MSH6 in chemoresistance, we knocked down expression in UOCB1 cells, another B-ALL cell line that expresses all four MMR proteins (Figure 1B). Similar to the effect observed in 697 cells, depletion of MSH6 with either shRNA significantly reduced the induction of apoptosis upon treatment with thiopurines (Figure 1B) [fold increase in IC50 as compared to NT with 6TG: 4.8 for shRNA1 (P=0.007) and 3 for shRNA2 (P<0.001); 6MP: 8.3 for shRNA1 (P<0.001) and 9.2 shRNA2 (P<0.001)] (Online Supplementary Figure S4). Additionally, a similar impact on TMZ resistance was observed with UOCB1 MSH6 shRNA1 expressing cells compared to NT control cells, although shRNA2 did not show the same effect, possibly due to less depletion by shRNA2 (Online Supplementary Figure S3B).
To determine the specificity of the phenotype observed for MSH6 depletion versus defects in other MMR proteins, we assessed the effect of MSH6 knockdown in MMR deficient B-ALL cell lines Reh and RS4;11.25,29 Both Reh and RS4;11 have minimal to no expression of MLH1 and PMS2 (Figure 1C). Knockdown of MSH6 expression had no effect on the sensitivity of either Reh or RS4;11 to 6-TG or 6-MP (Figure 1C and Online Supplementary Figure S5).
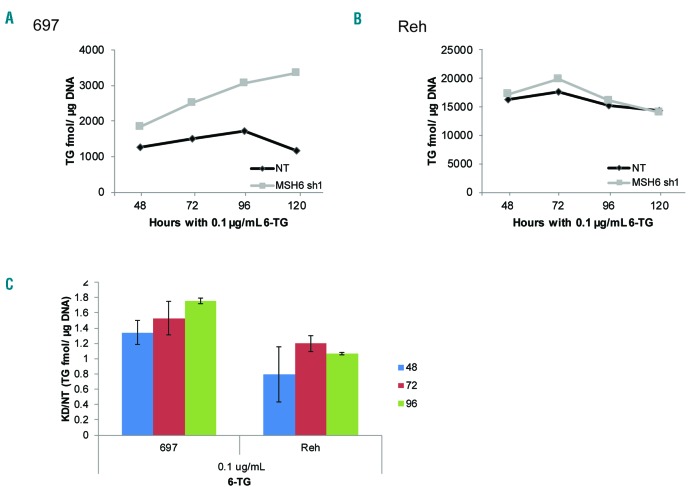
To begin to elucidate the mechanism of resistance, we measured the level of TGN incorporation into DNA upon treatment with 6-TG. 697 MSH6 shRNA1 cells accumulated more TGN/μg DNA over time than NT cells (NT 1722 and KD 3070 fmol/μg DNA) (Figure 2A and C). In contrast, no difference in TGN levels was observed in Reh cells (Figure 2B and C). Additionally, Reh cells had approximately 10-fold higher TGN levels compared to 697 cells (Figure 2A and B), highlighting the difference between MMR deficient and proficient cells in their ability to respond to and survive thiopurine exposure. Thus, MMR proficient cells with high TGN succumb to the damage and therefore display less TGN/μg DNA over time, mean- while deficient cells tolerate higher levels of TGN.
Figure 2.
Knockdown of MSH6 leads to increased tolerance of incorporated thioguanine (TGN). (A and B) TGN incorporation into DNA was measured over time after treatment with 0.1 μg/mL of 6-thioguanine (6-TG) in 697 (A) and Reh (B) cells using liquid chromatography-tandem mass spectrometry. A representative graph from 3 independent experiments is shown. (C) Combined ratio of TGN fmole/μg of DNA in MSH6 shRNA1 knockdown (KD) cells compared to non-targeting (NT) cells from 3 experiments. Bars indicate mean+Standard Deviation.
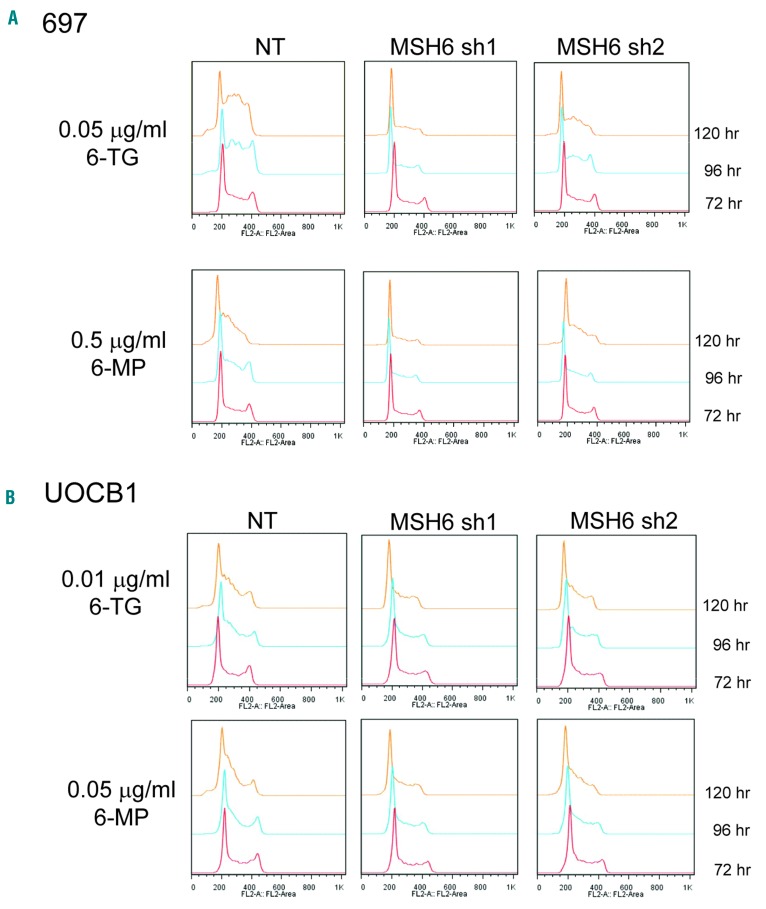
We next tested whether or not a change occurs in cell cycle progression upon treatment. 697 NT cells slowed their growth and had a significantly higher proportion of cells in S phase and less cells in G1 beginning at 96 hours (h) (6-TG, P=0.014; 6-MP, P=0.051) and progressing through 120 h of thiopurine treatment (6-TG, P=0.013; 6- MP, P=0.001) compared to the MSH6 shRNA lines by oneway ANOVA (Figure 3A and Online Supplementary Figure S6A). MSH6 shRNA1 cells had only a modest decrease in growth with no clear S phase arrest (6-TG, P=0.011, and 6-MP, P=0.001, for percent of cells in S phase compared to NT at 120 h using Tukey’s multiple comparison test), even at higher concentrations of 6-TG (Figure 3A and Online Supplementary Figure S6A and B). MSH6 shRNA2 cells had a more moderate accumulation of cells in S phase and drop of cells in G1 (Figure 3B and Online Supplementary Figure S6A), which is consistent with the modest levels of knockdown and apoptosis. Similar trends were observed with UOCB1 cells (6-TG, P=0.31; and 6-MP, P=0.34 at 120 h) (Figure 3B). This more moderate effect observed with the UOCB1 cells is consistent with the degree of impact MSH6 knockdown had on chemoresistance compared to the 697 cells. Neither NT nor MSH6 shRNA1 Reh cells showed alterations in cell cycle upon exposure (Online Supplementary Figure S6B).
Figure 3.
Thiopurine treatment resulted in an S phase arrest, which was abrogated upon knockdown of MSH6. 697 (A) and UOCB1 (B) NT and MSH6 shRNA1 and 2 expressing cells were treated with indicated drug for 5 days. Cells were fixed with 70% ethanol, treated with RNAse, and then stained with propidium iodide. DNA content was analyzed by flow cytometry. Representative images from 3 individual experiments are shown. A one-way ANOVA was performed to determine statistical significance of the increase in % of cells in S phase at each time point.
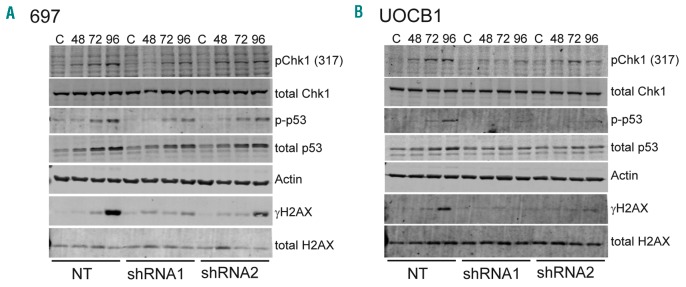
To gain a more complete understanding of the mechanism leading to apoptosis following TGN incorporation, we analyzed downstream pathways in 697 NT and MSH6 shRNA1 cells after treatment with 6-TG. Based on the observed S phase arrest and previous research demonstrating activation of the ataxia telangiectasia and Rad3-related (ATR)-Chk1 pathway downstream of MMR,30,31 we first assessed the level of activation of Chk1 by probing for phosphorylation of serine 317 (pChk1). 697 NT cells had a low level of pChk1 at 48 h with a significant increase through 96 h of exposure. 697 MSH6 shRNA1 cells had minimal to low levels of pChk1 at 72 h with minimal increase over time (Figure 4A). The 697 cells expressing shRNA2 had a similar pattern but slightly lower levels of pChk1 compared to NT cells, which is consistent with the cell cycle data. We next assessed the level of phosphorylated H2AX (γH2AX), a marker of DNA damage that is phosphorylated downstream of the ATR/ATM pathways following drug treatment,32 as well as levels of the apoptosis marker p53. There was a very modest level of γH2AX starting at 72 h that increased to a higher level at 96 h after treatment in 697 NT cells compared to very modest levels in the MSH6 shRNA1-2 cells (Figure 4A), suggesting that the functional MMR system in the NT cells was attempting to repair the DNA, leading to nicks. Additionally, the levels of phosphorylated and total p53 were higher in NT cells at 72 and 96 h compared to MSH6 shRNA1-2 cells (Figure 4A) and the shRNA2 cells had higher levels than the shRNA1 cells. Similar results were found in UOCB1 cells (Figure 4B).
Figure 4.
Thiopurine treatment leads to activation of cell cycle regulator Chk1 and DNA repair that ultimately resulted in DNA damage and cell death. Western blot analysis of whole cell lysates from 697 (A) and UOCB1 (B) non-targeting (NT), MSH6 shRNA1, and shRNA2 cells after treatment with 6-thioguanine (0.1 μg/mL, and 0.025 μg/mL, respectively). (C) Untreated cells; numbers are hours after treatment. Blots were probed for Chk1 activation, γH2AX for DNA damage, and apoptosis marker p53. Total Chk1, actin, and total H2AX were used as loading controls. Images are representative of 3 individual experiments.
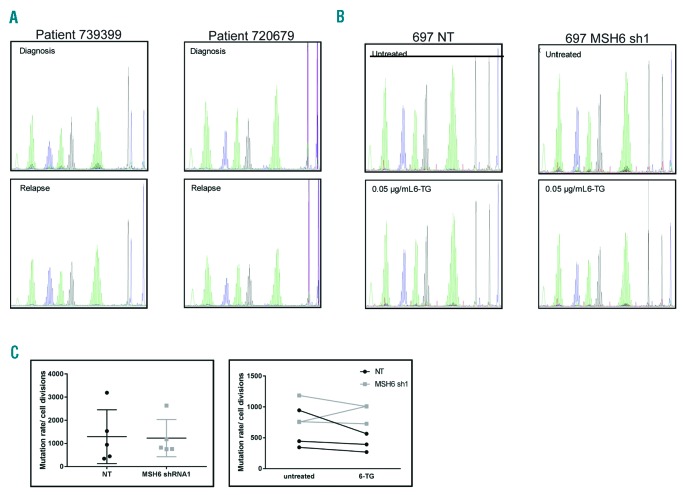
We next examined the impact of MSH6 knockdown on mutation rate by performing two assays that measure genomic instability and mutation burden. Microsatellite instability (MSI) is a marker for genomic instability and has been observed in cases where expression of MLH1 or MSH2 is lost.25,33 We investigated MSI on 2 patient sample pairs that we previously found to have deletions of MSH6 at relapse, as well as on 697 NT and MSH6 shRNA1 cells treated with 6-TG for 120 h. No MSI was observed in the patient samples comparing diagnosis to relapse or in the 697 cells comparing either untreated to 6-TG treated or NT to MSH6 shRNA1 cells (Figure 5A). These data are consistent with previous literature that found alterations in MSH6 expression alone do not lead to high MSI.34 To investigate the effect of MSH6 disruption on the rate of spontaneous mutations in PIG-A, which is required for expression of GPI, we used a flow cytometry-based assay that measures surface expression of several GPI-dependent markers (CD48, CD52, and CD59).22,35 Although there was a trend to suggest that 697 MSH6 shRNA1 cells had a slightly higher mutation rate, statistical significance was not achieved (Figure 5B). Furthermore, treatment of the clones from each cell line with 6-TG did not lead to an increased mutation rate (Figure 5B).
Figure 5.
Knockdown of MSH6 did not lead to a mutator phenotype or increased mutation rate. (A) Microsatellite instability (MSI) was measured in diagnosis/relapse pairs that had relapse specific, heterozygous MSH6 deletions and in 697 non-targeting (NT) and MSH6 shRNA1 cells left untreated or treated with 0.05 μg/mL of 6-thioguanine (6-TG) for 5 days. (B) Mutation rate in the PIC-A gene was measured in 697 NT and MSH6 shRNA1 clones that were expanded for 2–3 weeks with or without 6-TG. The cells were analyzed for loss of GPI-dependent cell surface markers, including FLAER, CD48, CD52, and CD59 using flow cytometry. (Left) Individual mutation rates/cell divisions for each clone; the line represents mean+Standard Deviation. (Right) Mutation rates/cell divisions for three clones with and without 6-TG treatment.
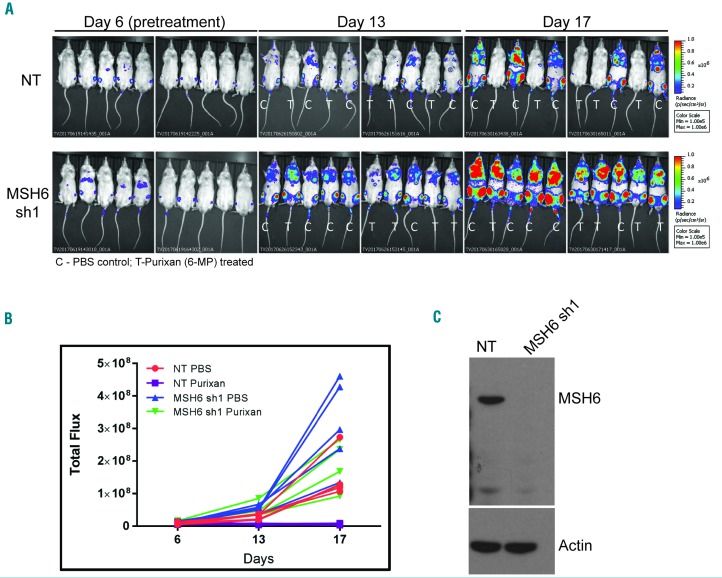
To investigate the clinical relevance of reduced MSH6 expression and drug resistance, we utilized an in vivo mouse model. We injected mice with either UOCB1 NT or UOCB1 MSH6 shRNA1 cell lines (knockdown confirmed day of injection; Figure 6C) and, following confirmation of leukemic burden on day 6, the mice were treated with PBS (control) or purixan (an oral suspension form of 6-MP). Following the 10-day course of purixan treatment, the leukemic burden was significantly diminished in the mice harboring NT cells compared to that observed in the NT PBS treated mice (P=0.0001), suggesting that these cells were unable to survive and expand under the selective pressure of the purixan (Figure 6A and B). In contrast, the MSH6 shRNA1 mice treated with purixan were not significantly different from the NT PBS group (P=0.828). Although purixan also had a statistically significant impact on MSH6 shRNA1 cells compared to MSH6 shRNA1 PBS control (P=0.0005), these cells were able to continue proliferating under the selective pressure, unlike the NT cells (Figure 6A and B). Finally, a comparison between PBS MSH6 shRNA1 and PBS control NT cells at day 17 showed that MSH6 depleted cells also had a growth advantage in vivo (P<0.0001).
Figure 6.
Knockdown of MSH6 leads to decreased sensitivity to purixan in vivo. (A) Bioluminescence imaging (BLI) of mice injected with UOCB1 non-targeting (NT) or MSH6 shRNA1 cells. Six days after injection mice were imaged and then randomized to treatment. Treatment was started on day 7 and images were taken again on days 13 and 17. C: PBS control treatment; T: purixan treatment. (B) Quantification of total flux was determined by analyzing the BLI images using Living Image software. (C) Western blot to confirm knockdown of MSH6 in cells used to inject mice. Actin was used as loading control.
Discussion
In recent years, there has been an abundance of evidence demonstrating the outgrowth of clones at relapse in ALL that are associated with unique or enriched relapse specific mutations that confer drug resistance. Some of the most common relapse specific mutations found thus far occur in NT5C2 and PRPS1 and lead to the outgrowth of thiopurine resistant clones.2,4 Our data presented here demonstrate that reduction of MSH6 in ALL also leads to decreased sensitivity to purine analogs due to a failure to initiate the apoptotic cascade directly in response to nucleotide mismatches. Even with only 50–60% reduced expression, which potentially mimics levels in patients with heterozygous loss, we demonstrate a significant decrease in sensitivity to thiopurines. Our data are consistent with the recent work of Diouf et al. who showed that lower levels of MSH2 in cell lines were associated with resistance to 6-TG and 6-MP. They found 11% of ALL samples showed decreased protein levels of MSH2 through copy number loss of genes controlling MSH2 degradation.17 Thus defects in MMR, including heterozygous deletion of MSH6, can be added to the list of genetic alterations that result in the development of resistance to purine analogs, the foundation of maintenance therapy. The variety of mutations that lead to selective outgrowth of such clones in a substantial number of patients underscores the selective pressure of thiopurines on tumor cells.
The outgrowth of MSH6 deleted/mutated clones not found at diagnosis has been observed at relapse in malignant gliomas following treatment with temozolomide,28,36,37 which produces DNA O6-methyguanine, a lesion structurally similar to 6-TG.38 Our data demonstrating decreased sensitivity of MSH6 knockdown cells to temozolomide support the hypothesis that MMR deficient clones gain an advantage under this selective pressure leading to resistant recurrences.27,39 The difference in TMZ sensitivity between the two UOCB1 shRNA knockdown cell lines could be due to the interplay between MSH6 and SETD2 protein levels since UOCB1 cells have a copy number loss of SETD2 (NA Evensen et al., 2018, unpublished data) and there is a greater reduction of MSH6 with shRNA1 compared to shRNA2. SETD2, the gene that codes for the methyltransferase responsible for the trimethylation of H3K36 that serves as the docking site for MSH6,40 is among the epigenetic regulators commonly found mutated in relapse patients.41 Ongoing studies in our lab are focused on identifying the relationship of epigenetic readers, writers, and erasers, such as SETD2, MSH6, and WHSC1 in chemoresistance.
Mechanistically, our in vitro and in vivo data support the hypothesis that the delayed cytotoxic response to thiopurines is due to the MMR system recognizing a mismatch and initiating futile, damaging DNA repair that ultimately leads to apoptosis.20,42,43 This pathway is not fully activated in cells with reduced MSH6 because the mismatch goes undetected, allowing these cells to tolerate excess TGN mismatches and, ultimately, to continue to survive and proliferate while under treatment. Our data provide evidence that, upon recognition of mismatches, NT cells slow their progression through S phase by activating Chk1 as they begin to repair their DNA. Due to the mismatch being on the daughter strand, the excision/repair process is unsuccessful, and over time nicks build up in the DNA, demonstrated by increased levels of γH2AX. Eventually, the damage becomes overwhelming and cells initiate apoptosis, as shown by increased p53. MSH6 shRNA1 cells exhibited minimal to no change in cell cycle, activation of Chk1, or increased γH2AX and p53. The moderate changes observed with the MSH6 shRNA2 cells highlight the idea that even a more modest reduction in MSH6 expression could lead to subtle changes that have a significant impact on chemoresistance. The MMR deficient Reh cells also had no alteration in their cell cycle, suggesting that recognition of mismatches by MutSa is not sufficient for full activation of this cascade, but rather damage induced by the repair, which is orchestrated by MutLa, is what initiates apoptosis. Our data demonstrating the involvement of the ATR-Chk1-H2AX signaling cascade is supported by the work of Eich et al. which demonstrated activation of this pathway upon treatment with temozolomide.32 Interestingly, understanding how MMR deficient cells respond to thiopurines in terms of TGN incorporation could prove essential given the emerging idea of measuring these parameters in patients on maintenance therapy.44
The data presented here do not support the hypothesis of increased mutation burden, genomic instability, or MSI when MSH6 is reduced. Our inability to demonstrate MSI-high, which is considered a standard method for clinical testing of MMR deficiencies in tumors,45 in MSH6 depleted cell lines and clinical samples is consistent with the lack of MSI in glioma samples with MSH6 deletions or mutations.46,47 Haploinsufficiency of MSH6 or compensation by MSH2/MSH3 may account for this observation.25,48 In addition, ALL clonal evolution from diagnosis to relapse is not associated with increased mutation burden supporting our mutation rate analysis, although Ma et al. reported a subset of hypermutated relapse cases.49 Of these, one had a bialleic mutation of PMS2, another had multiple damaging MSH6 mutations as well as an MLH1 splice site mutation, while the others harbored no MMR mutations.49 Furthermore, one case demonstrated that a heterozygous deletion of MSH6 at diagnosis was not sufficient to cause a hypermutator phenotype, but the acquisition of a second hit in the WT allele at relapse was.49 Likewise, the majority of hypermutated gliomas at relapse show defects in multiple MMR genes or loss of heterozygosity.50 Thus, our work supports a model whereby haploinsufficiency of MSH6 results in TGN tolerance and resistance directly rather than by generation of secondary mutations. However, it does not rule out the possibility that haploinsufficiency, along with other defects in the MMR pathway, may result in a mutator phenotype.
Overall, it has become increasingly evident that the genetic and epigenetic landscape of cancer cells is vital to the overall effectiveness of treatment. These studies illustrate yet another example of a mutation/deletion found at relapse that directly influences the response to a therapeutic agent that is currently heavily relied on. Continuous efforts to elucidate the potential functions and mechanisms of genes found mutated at relapse will help lead us to novel treatment strategies.
Supplementary Material
Acknowledgments
We gratefully acknowledge the Children’s Oncology Group (COG) Specimen Bank for samples. Support for flow cytometry was provided by NYU School of Medicine’s Cytometry and Cell Sorting Laboratory, which is supported in part by grant P30CA016087 from the NIH/NCI, and the CHOP Flow Cytometry Core. We acknowledge the VA-Mertid award 1I01BX-000670, which helped support this work.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/5/830
Funding
This work was supported by the Leukemia and Lymphoma Society SCOR grant: 7010-14 (WLC, JY, DT, SPH), the US National Institutes of Health (NIH) funded grant RO1 CA140729 (WLC), and the Perlmutter Cancer Center Support Grant: P30 DA016087.
References
- 1.Pui CH, Evans WE. Treatment of acute lymphoblastic leukemia. N Engl J Med. 2006;354(2):166–178. [DOI] [PubMed] [Google Scholar]
- 2.Meyer JA, Wang J, Hogan LE, et al. Relapse- specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet. 2013;45(3):290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jones CL, Bhatla T, Blum R, et al. Loss of TBL1XR1 disrupts glucocorticoid receptor recruitment to chromatin and results in glucocorticoid resistance in a B-lymphoblastic leukemia model. J Biol Chem. 2014; 289(30):20502–20515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li B, Li H, Bai Y, et al. Negative feedback- defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL. Nat Med. 2015;21(6):563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mullighan CG, Zhang J, Kasper LH, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011; 471(7337):235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nielsen SN, Grell K, Nersting J, et al. DNA- thioguanine nucleotide concentration and relapse-free survival during maintenance therapy of childhood acute lymphoblastic leukaemia (NOPHO ALL2008): a prospective substudy of a phase 3 trial. Lancet Oncol. 2017;18(4):515–524. [DOI] [PubMed] [Google Scholar]
- 7.Yang JJ, Bhojwani D, Yang W, et al. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood. 2008;112(10):4178–4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hogan LE, Meyer JA, Yang J, et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood. 2011;118(19):5218–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18(1):85–98. [DOI] [PubMed] [Google Scholar]
- 10.Edelbrock MA, Kaliyaperumal S, Williams KJ. Structural, molecular and cellular functions of MSH2 and MSH6 during DNA mismatch repair, damage signaling and other noncanonical activities. Mutat Res. 2013;743–744: 53–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunlop MG, Farrington SM, Carothers AD, et al. Cancer risk associated with germline DNA mismatch repair gene mutations. Hum Mol Genet. 1997;6(1):105–110. [DOI] [PubMed] [Google Scholar]
- 12.Tiwari AK, Roy HK, Lynch HT. Lynch syndrome in the 21st century: clinical perspectives. QJM. 2016;109(3):151–158. [DOI] [PubMed] [Google Scholar]
- 13.Goecke T, Schulmann K, Engel C, et al. Genotype-phenotype comparison of German MLH1 and MSH2 mutation carriers clinically affected with Lynch syndrome: a report by the German HNPCC Consortium. J Clin Oncol. 2006; 24(26):4285–4292. [DOI] [PubMed] [Google Scholar]
- 14.Ripperger T, Schlegelberger B. Acute lymphoblastic leukemia and lymphoma in the context of constitutional mismatch repair deficiency syndrome. Eur J Med Genet. 2016;59(3):133–142. [DOI] [PubMed] [Google Scholar]
- 15.Ripperger T, Beger C, Rahner N, et al. Constitutional mismatch repair deficiency and childhood leukemia/lymphoma–report on a novel biallelic MSH6 mutation. Haematologica. 2010;95(5):841–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fink D, Aebi S, Howell SB. The role of DNA mismatch repair in drug resistance. Clin Cancer Res. 1998;4(1):1–6. [PubMed] [Google Scholar]
- 17.Diouf B, Cheng Q, Krynetskaia NF, et al. Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nat Med. 2011;17(10):1298–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swann PF, Waters TR, Moulton DC, et al. Role of postreplicative DNA mismatch repair in the cytotoxic action of thioguanine. Science. 1996;273(5278):1109–1111. [DOI] [PubMed] [Google Scholar]
- 19.Waters TR, Swann PF. Cytotoxic mechanism of 6-thioguanine: hMutSalpha, the human mismatch binding heterodimer, binds to DNA containing S6-methylthioguanine. Biochemistry. 1997;36(9):2501–2506. [DOI] [PubMed] [Google Scholar]
- 20.Karran P, Attard N. Thiopurines in current medical practice: molecular mechanisms and contributions to therapy-related cancer. Nat Rev Cancer. 2008;8(1):24–36. [DOI] [PubMed] [Google Scholar]
- 21.Morrison DJ, Hogan LE, Condos G, et al. Endogenous knockdown of survivin improves chemotherapeutic response in ALL models. Leukemia. 2012;26(2):271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araten DJ, Golde DW, Zhang RH, et al. A quantitative measurement of the human somatic mutation rate. Cancer Res. 2005;65(18):8111–8117. [DOI] [PubMed] [Google Scholar]
- 23.Araten DJ, Sanders KJ, Anscher D, Zamechek L, Hunger SP, Ibrahim S. Leukemic blasts with the paroxysmal nocturnal hemoglobinuria phenotype in children with acute lymphoblastic leukemia. Am J Pathol. 2012;181(5):1862–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobsen JH, Schmiegelow K, Nersting J. Liquid chromatography-tandem mass spectrometry quantification of 6-thioguanine in DNA using endogenous guanine as internal standard. J Chromatogr B Analyt Technol Biomed Life Sci. 2012;881–882: 115–118. [DOI] [PubMed] [Google Scholar]
- 25.Gu L, Cline-Brown B, Zhang F, Qiu L, Li GM. Mismatch repair deficiency in hematological malignancies with microsatellite instability. Oncogene. 2002;21(37):5758–5764. [DOI] [PubMed] [Google Scholar]
- 26.Marra G, Iaccarino I, Lettieri T, Roscilli G, Delmastro P, Jiricny J. Mismatch repair deficiency associated with overexpression of the MSH3 gene. Proc Natl Acad Sci USA. 1998;95(15):8568–8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McFaline-Figueroa JL, Braun CJ, Stanciu M, et al. Minor Changes in Expression of the Mismatch Repair Protein MSH2 Exert a Major Impact on Glioblastoma Response to Temozolomide. Cancer Res. 2015; 75(15):3127–3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie C, Sheng H, Zhang N, Li S, Wei X, Zheng X. Association of MSH6 mutation with glioma susceptibility, drug resistance and progression. Mol Clin Oncol. 2016; 5(2):236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matheson EC, Hall AG. Assessment of mismatch repair function in leukaemic cell lines and blasts from children with acute lymphoblastic leukaemia. Carcinogenesis. 2003;24(1):31–38. [DOI] [PubMed] [Google Scholar]
- 30.Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol Cell. 2006;22(4):501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan T, Desai AB, Jacobberger JW, Sramkoski RM, Loh T, Kinsella TJ. CHK1 and CHK2 are differentially involved in mismatch repair-mediated 6-thioguanine- induced cell cycle checkpoint responses. Mol Cancer Ther. 2004;3(9):1147–1157. [PubMed] [Google Scholar]
- 32.Eich M, Roos WP, Nikolova T, Kaina B. Contribution of ATM and ATR to the resistance of glioblastoma and malignant melanoma cells to the methylating anti-cancer drug temozolomide. Mol Cancer Ther. 2013;12(11):2529–2540. [DOI] [PubMed] [Google Scholar]
- 33.Verma L, Kane MF, Brassett C, et al. Mononucleotide microsatellite instability and germline MSH6 mutation analysis in early onset colorectal cancer. J Med Genet. 1999;36(9):678–682. [PMC free article] [PubMed] [Google Scholar]
- 34.Bodo S, Colas C, Buhard O, et al. Diagnosis of Constitutional Mismatch Repair- Deficiency Syndrome Based on Microsatellite Instability and Lymphocyte Tolerance to Methylating Agents. Gastroenterology. 2015;149(4):1017–1029.e1013. [DOI] [PubMed] [Google Scholar]
- 35.Araten DJ, Krejci O, Ditata K, et al. The rate of spontaneous mutations in human myeloid cells. Mutat Res. 2013;749(1-2):49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cahill DP, Codd PJ, Batchelor TT, Curry WT, Louis DN. MSH6 inactivation and emergent temozolomide resistance in human glioblastomas. Clin Neurosurg. 2008;55:165–171. [PubMed] [Google Scholar]
- 37.Cahill DP, Levine KK, Betensky RA, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007; 13(7):2038–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Stevens MF, Laughton CA, Madhusudan S, Bradshaw TD. Acquired resistance to temozolomide in glioma cell lines: molecular mechanisms and potential translational applications. Oncology. 2010; 78(2):103–114. [DOI] [PubMed] [Google Scholar]
- 39.Hunter C, Smith R, Cahill DP, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006; 66(8):3987–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li F, Mao G, Tong D, et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell. 2013;153(3):590–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mar BG, Bullinger LB, McLean KM, et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun. 2014;5:3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan T, Berry SE, Desai AB, Kinsella TJ. DNA mismatch repair (MMR) mediates 6- thioguanine genotoxicity by introducing single-strand breaks to signal a G2-M arrest in MMR-proficient RKO cells. Clin Cancer Res. 2003;9(6):2327–2334. [PubMed] [Google Scholar]
- 43.Tidd DM, Paterson AR. Distinction between inhibition of purine nucleotide synthesis and the delayed cytotoxic reaction of 6-mercaptopurine. Cancer Res. 1974;34(4):733–737. [PubMed] [Google Scholar]
- 44.Nielsen SN, Grell K, Nersting J, Frandsen TL, Hjalgrim LL, Schmiegelow K. Measures of 6-mercaptopurine and methotrexate maintenance therapy intensity in childhood acute lymphoblastic leukemia. Cancer Chemother Pharmacol. 2016; 78(5):983–994. [DOI] [PubMed] [Google Scholar]
- 45.Buza N, Ziai J, Hui P. Mismatch repair deficiency testing in clinical practice. Expert Rev Mol Diagn. 2016;16(5):591–604. [DOI] [PubMed] [Google Scholar]
- 46.Yip S, Miao J, Cahill DP, et al. MSH6 mutations arise in glioblastomas during temozolomide therapy and mediate temozolomide resistance. Clin Cancer Res. 2009;15(14):4622–4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maxwell JA, Johnson SP, McLendon RE, et al. Mismatch repair deficiency does not mediate clinical resistance to temozolomide in malignant glioma. Clin Cancer Res. 2008;14(15):4859–4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Umar A, Risinger JI, Glaab WE, Tindall KR, Barrett JC, Kunkel TA. Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics. 1998;148(4):1637–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma X, Edmonson M, Yergeau D, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukaemia. Nat Commun. 2015;6:6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Thuijl HF, Mazor T, Johnson BE, et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 2015;129(4):597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.