Abstract
Surviving the nutrient-poor aquatic environment for extended periods of time is important for the transmission of various water-borne pathogens, including Legionella pneumophila (Lp). Previous work concluded that the stringent response and the sigma factor RpoS are essential for the survival of Lp in water. In the present study, we investigated the role of the LetA/S two-component signal transduction system in the successful survival of Lp in water. In addition to cell size reduction in the post-exponential phase, LetS also contributes to cell size reduction when Lp is exposed to water. Importantly, absence of the sensor kinase results in a significantly lower survival as measured by CFUs in water at various temperatures and an increased sensitivity to heat shock. According to the transcriptomic analysis, LetA/S orchestrates a general transcriptomic downshift of major metabolic pathways upon exposure to water leading to better culturability, and likely survival, suggesting a potential link with the stringent response. However, the expression of the LetA/S regulated small regulatory RNAs, RsmY and RsmZ, is not changed in a relAspoT mutant, which indicates that the stringent response and the LetA/S response are two distinct regulatory systems contributing to the survival of Lp in water.
Introduction
Legionella pneumophila (Lp) is a bacterial contaminant of anthropogenic water distribution systems, where it replicates as an intracellular parasite of amoeba1–3. In the context of human infection, Lp preferentially targets alveolar macrophages causing a severe pneumonia termed Legionnaires’ disease4. The inhalation of Lp contaminated aerosols that are generated from water systems transmit the bacterium to the human lungs, where it proliferates5–7. Therefore, identifying the molecular mechanisms that Lp uses to survive in water is crucial, not only to better understand the operating system of the organism, but also for improving water systems management.
Transcriptional profiling revealed a drastic shut down of major pathways in Lp exposed to water8. Experiencing this starvation condition increases the resistance of Lp to antibiotics8, and likely other stresses. Indeed, one study exposed stationary phase Lp cultures, which are naturally more stress resistant than exponential (E) phase bacteria, to a nutrient-poor buffer9. The authors found that resistance to acid, hydrogren peroxide and antibiotic stresses acquired in the stationary phase were further enhanced by this treatment9. The current data suggests that Lp initiates unique transcriptomic and proteomic changes to adapt to and survive in water. At present, only a few genes or regulatory pathways are known to contribute to the survival of Lp in water8,10–13. The general silencing of gene expression in water seems to be orchestrated mainly by RpoS and the stringent response14.
Bacteria employ two-component systems (TCSs) to sense and respond to a variety of cues, ranging from temperature, antibiotics, quorum sensing autoinducer molecules and intermediates of the TCA cycle15,16. Upon sensing an activating environmental signal, the sensor kinase (SK) autophosphorylates a conserved histidine residue on its C-terminus. This phosphoryl group can then be shuttled to an aspartate residue on the cognate response regulator (RR), a DNA-binding protein, which will initiate the downstream transcriptional changes that allow the bacterium to adapt and respond to the aforementioned environmental stimulus17,18.
The LetA/S TCS of Lp19 is the ortholog of BarA/UvrY in Escherichia coli20 and GacS/GacA in Pseudomonas spp.21. LetS belongs to a family of tripartite sensor kinases which deviate from the traditional SK model. Similar to its well-studied counterpart BvgS in Bordetella spp., LetS architecture includes 3 major domains (transmitter (T), receiver (R) and histidine phosphotransfer (HPT) domains) that are involved in an internal phosphorelay activating its cognate response regulator LetA22,23. Upon activation, the T domain is phosphorylated by an ATP molecule. This, in turn, phosphorylates the R domain. Finally, the HPT domain receives the phosphate from R and relays it to the response regulator LetA23. The modular nature of this SK allows it to respond to multiple stimuli, where each stimulus leads to the activation of a different set of genes23–25. In Lp, LetS regulates a subset of post-exponential phase genes in response to nicotinic acid23.
The RRs orthologous to LetA activate transcription of the Csr/Rsm-type small regulatory RNAs (sRNAs)23,26–30. The CsrA/RsmA protein binds target mRNA and mainly serve to inhibit their translation29–32. Competitive binding of Csr/Rsm sRNAs to CsrA/RsmA relieves the inhibitory effects of the latter on its target mRNA, thereby promoting their translation26,30–33. Orthologs of LetA/S and their accompanying regulatory cascades are involved in the virulence phenotypes of a number of pathogens, including regulation of pathogenicity islands in Salmonella34–36, fimbriae and exopolysaccharide production in E. coli37–39, quorum sensing and production of extracellular lipase, cyanide and pyocyanin in Pseudomonas spp.21,28,33,40–42 and the ToxR virulence regulator in Vibrio cholerae43. Members of this TCS family are also involved in stress resistance, biofilm formation, the switch between glycolytic and gluconeogenic carbon sources, and iron acquisition in various bacterial species30,33,37,39,44,45. In Lp, LetA binds the promoters and positively affects transcription of three sRNAs, RsmX, RsmY and RsmZ46,47. RsmX is, however, absent in the Lp Philadelphia 1 strain and L. longbeachae47. CsrA in Lp represses post-exponential (PE) phase traits and promotes the expression of E phase genes48. RsmY/Z antagonizes CsrA and activates PE (in broth) and transmissive (in vivo) phase traits in Lp19,46,48,49. As a result, mutations within this cascade have been linked to attenuated virulence within host cells and reduced motility, as well as sensitivity to heat, oxidative and acid stress19,46,48,50.
The aim of the present study is to investigate the role of the LetA/S regulatory cascade in a relevant water model and to elucidate the transcriptome under LetS control in water. We report that LetS is responsible for a genome wide repression of metabolic pathways in response to water. Using Northern blotting, we confirm that LetS forms a regulatory cascade under the control of RpoS. Despite the dependence on RpoS we found that the cellular alarmone, ppGpp, is not the main activating signal for the sensor kinase, advocating for other environmental stimuli to be investigated.
Results
LetS is important for the culturability of Lp in water
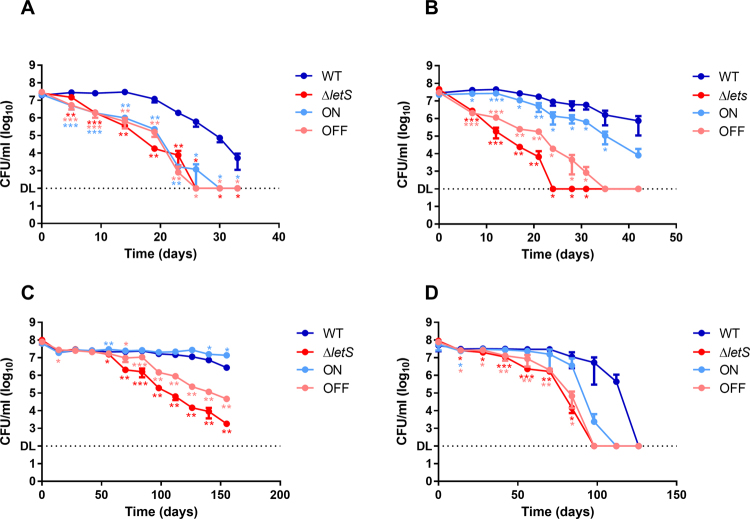
The culturability of the ∆letS mutant was compared to that of the wild-type (WT) in water at 42 °C using CFU counts (Fig. 1A). An inducible plasmid carrying the letS gene (pletS) was introduced into ∆letS to complement the mutant strain. Expression of the plasmid-borne copy was induced with 0.1 mM IPTG, herein referred to as ON, or was left uninduced, herein referred to as OFF. A significant (p < 0.005) defect was observed in the ability of ∆letS to form colonies compared to the WT starting at early time points (Fig. 1A and B). Inducing the expression of letS in the ON strain solely in water did not correct this defect (Fig. 1A); however, when induced prior to and during water exposure, ON performed markedly better than the mutant (Fig. 1B). ∆letS and OFF behaved similarly to eachother. These results implicate the LetA/S two-component system as an important tool for the adaptation and potential survival of Lp in water at 42 °C.
Figure 1.
LetS increases the culturability of Lp in water. (A and B) CFU counts of the WT, ∆letS and the induced (ON) or uninduced (OFF) ∆letS + pletS was monitored in water at 42 °C. In panel A, ON was induced only in water, while in panel B, the ON strain was induced on agar prior to water exposure and during water exposure. Panel C and D show CFU counts of the WT, ∆letS, ON and OFF at 4 °C and 25 °C respectively. The ON strain in panels C and D was induced on agar prior to water exposure and during water exposure. Strains were suspended in water at an OD600 of 0.02. ON was induced with 0.1 mM IPTG. DL, detection limit. An unpaired, one-tailed Student’s t-test was used to assess statistical significance versus the WT. *p < 0.05; **p < 0.005; ***p < 0.0005.
Culturability of the strains was also tested at 4 °C and 25 °C to determine whether the defect observed at 42 °C was temperature specific. As a general trend, strains reached the detection limit slower as the incubation temperature decreased (Fig. 1), as previously described51. Similar to Fig. 1B, the loss of letS caused a defect when Lp was exposed to water at 4 °C and 25 °C, a phenotype that was complemented in the ON strain (Fig. 1C and D). At 25 °C, ON mirrored the WT strain better during the early stages of water exposure (Fig. 1D). IPTG is reported to be a stable inducer of gene expression with minimal decay in broth; however, its long-term stability in water over the course of several months has never been investigated to our knowledge. Alternatively, the uptake of IPTG may be compromised over the long-term. Indeed, Lp repressed the majority of its genes 24 hours after water exposure8, including channels and transporters that may be involved in IPTG uptake. Taken together, these experiments suggest that the LetA/S TCS is an important regulator that allows Lp to successfully adapt to and, likely, survive the nutrient poor aquatic environment.
The effect of the letS mutation on culturability of Lp was most pronounced at 42 °C (Fig. 1). Similarly, previous reports from our group found that the deleterious effect of some mutants was observed more readily at 42 °C than at lower temperatures8,11,13. A higher metabolic rate at elevated temperatures can explain the faster decline in CFU counts in mutant strains that are less fit than the wild-type. As such, when faced with starvation in water, internal energy sources would be depleted more rapidly at warmer temperatures. Accordingly, we expected the transcriptomic and physiological responses initiated by the LetA/S cascade to be apparent at earlier time points at 42 °C. Therefore, subsequent experiments were conducted at 42 °C to facilitate the study of the regulatory effects exerted by LetS.
LetS influences morphological changes in water, pigment production and resistance to heat shock
WT, ∆letS, ON and OFF strains were exposed to water for 24 hours at 42 °C. Microscopic analysis determined that ∆letS cells were significantly (p < 0.0005) longer than WT cells after water exposure. Interestingly, the ON strain produced Lp cells that were significantly (p < 0.0005) shorter than the WT (Fig. 2A,C and E), which may be due to a higher level of letS expression. Notably, we observed that filamentous cells were more commonly found within the ∆letS and OFF populations than in the WT and ON populations. In accordance with previously published reports, cell size reduction was also observed in the post-exponential (PE) phase compared to the exponential (E) phase in the WT (Supplementary Figs S1 and S2)49. This reduction was absent in the letS and OFF strains. A strain over-expressing CsrA was similarly unable to reduce cell size upon entry into the PE phase52. Microscopic analysis highlights the contribution of LetS to cell size reduction in both the post-exponential phase and under the starvation condition of water, but not during the exponential phase of growth.
Figure 2.
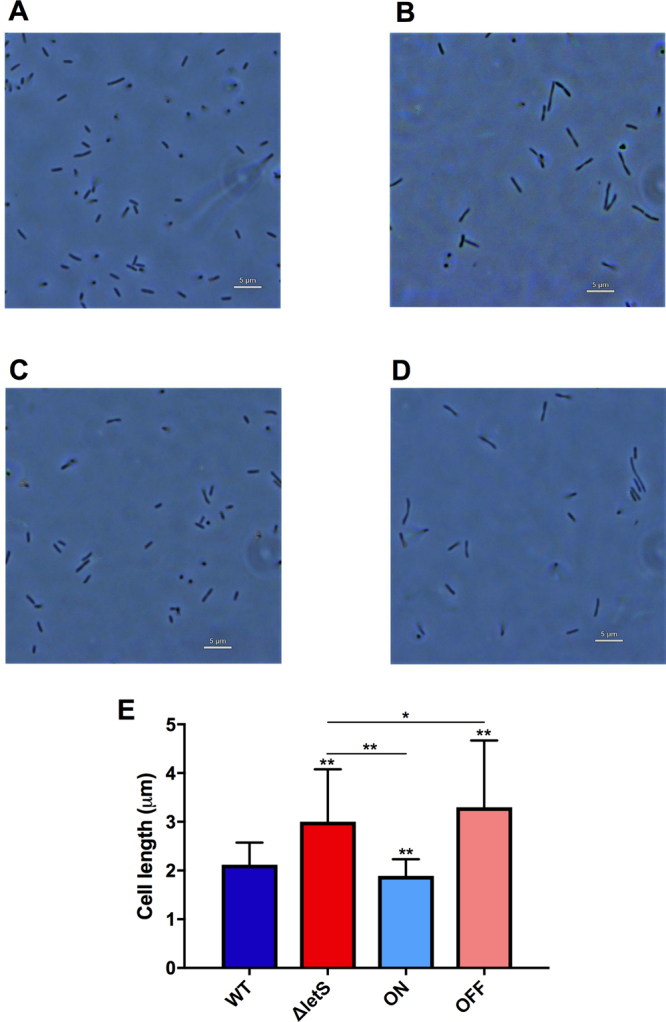
Deletion of letS affects the cell morphology of Lp in water. (A) Phase contrast microscopy was used to visualize morphological changes at 1000X magnification. A representative image of the WT (A), ∆letS (B), ON (C) and OFF (D) exposed to water for 24 hours are shown. In Panel E, Image J software was used to quantify the average length of 100 cells after exposure to water for 24 hours. The scale bar is equivalent to 5 μm. An unpaired, one-tailed Student’s t-test was used to assess statistical significance versus the WT, unless identified otherwise. *p < 0.05; **p < 0.0005.
In agreement with previously published work46,48, ∆letS was defective for pigment production during the late PE phase (Supplementary Fig. S3). It is noteworthy that pigment production was not completely abolished in the ∆letS mutant which produced visible pigmentation. Nonetheless, the amount of pigment produced by the mutant was significantly (p < 0.0005) lower than the WT, a defect that was corrected in the ON strain (Supplementary Fig. S3).
Lp frequently encounters high temperatures within man-made water distribution systems and is known to persist despite continuous heat treatments53–56. Therefore, we tested the ability of ΔletS to withstand heat shock in 55 °C water. WT CFU counts dropped by 5 orders of magnitude after 30 minutes at 55 °C, but remained above the detection limit after 60 minutes. In contrast, the letS mutant was more sensitive, as CFU counts dropped below the detection limit after a 30-minute heat shock treatment (Fig. 3). Expression of letS in ON afforded Lp an increased resistance to heat compared to the WT. The OFF strain resulted in lower heat tolerance compared to the WT and ON; however, its survival was markedly better than that of the letS mutant.
Figure 3.
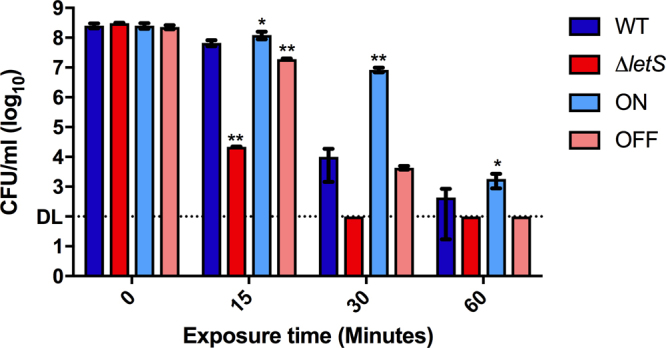
Deletion of letS affects sensitivity to heat shock. The WT, ∆letS, ON and OFF strains were suspended in water for 2 hours and subsequently exposed to a 55 °C water bath for 15, 30 or 60 minutes. CFU counts were enumerated on CYE agar before and after the heat shock treatment. DL, detection limit. An unpaired, one-tailed Student’s t-test was used to assess statistical significance versus WT. *p < 0.05; **p < 0.005.
Transcriptomic analysis of the LetS regulon in water
In order to identify genes affected by LetA/S in response to water, DNA microarrays were used to probe the transcriptomic differences between the WT, ∆letS, ON and OFF strains 2 hours after water exposure at 42 °C. Justification for using an early time point (2 h) were two-fold: 1) Lp is known to reduce transcription dramatically after 6 h in Fraquil8, and 2) we expect that the regulatory changes initiated by the LetA/S TCS are important for early adaptation in Lp as evidenced by the need for letS induction on agar prior to water exposure to achieve proper complementation (Fig. 1). To extract sufficient RNA for transcriptomic analysis, a high cell density was required. Therefore, the survival defect observed at low cell density (Fig. 1 – OD600 0.02) was confirmed at high cell density (Supplementary Fig. S4 - OD600 1).
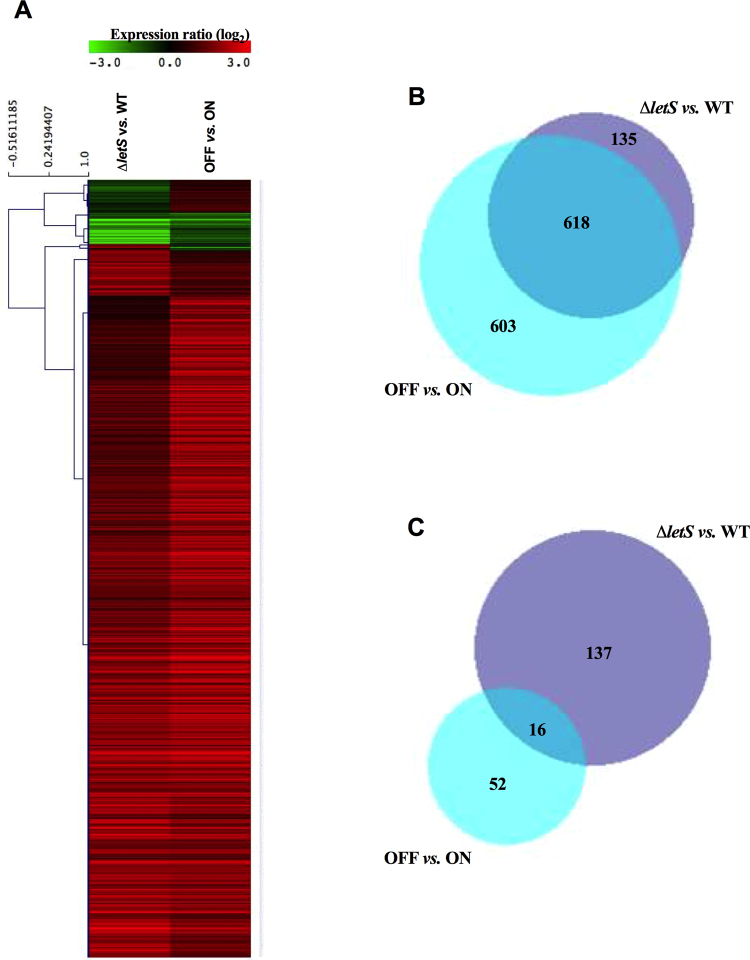
Gene expression profile of ∆letS vs. the WT was compared to that of OFF vs. ON in Fig. 4. The absence of letS led to the up-regulation of a large portion of genes (Fig. 4). Among the significantly (p < 0.05) up-regulated open reading frames (ORFs), 569 genes and 49 annotated sRNAs (a total of 618 ORFs) were shared in both ∆letS and OFF compared to the WT and ON respectively (Fig. 4B, Supplementary Table S1). On the other hand, 16 genes, including the two sRNAs RsmY and RsmZ, were significantly (p < 0.05) down-regulated in both the letS mutant and OFF (Fig. 4C, Supplementary Table S1). qPCR was performed on six randomly chosen genes; three were upregulated and three were downregulated in ΔletS compared to the WT (Fig. 5). The fold change pattern observed in the microarray analysis was mirrored in the qPCR results (Fig. 5).
Figure 4.
The absence of letS leads to ectopic up-regulation of gene expression in water. (A) A heat map showing genes differentially expressed in ∆letS compared to the WT (left), and in OFF compared to ON (right) (ratio to control value of ± 2-fold with a p < 0.05). Genes that are up-regulated in ∆letS and OFF are shown in red; genes that are down-regulated are shown in green. The number of up- (B) or down-regulated (C) genes that are shared between the ∆letS vs. WT and OFF vs. ON groups are shown in Venn diagrams.
Figure 5.
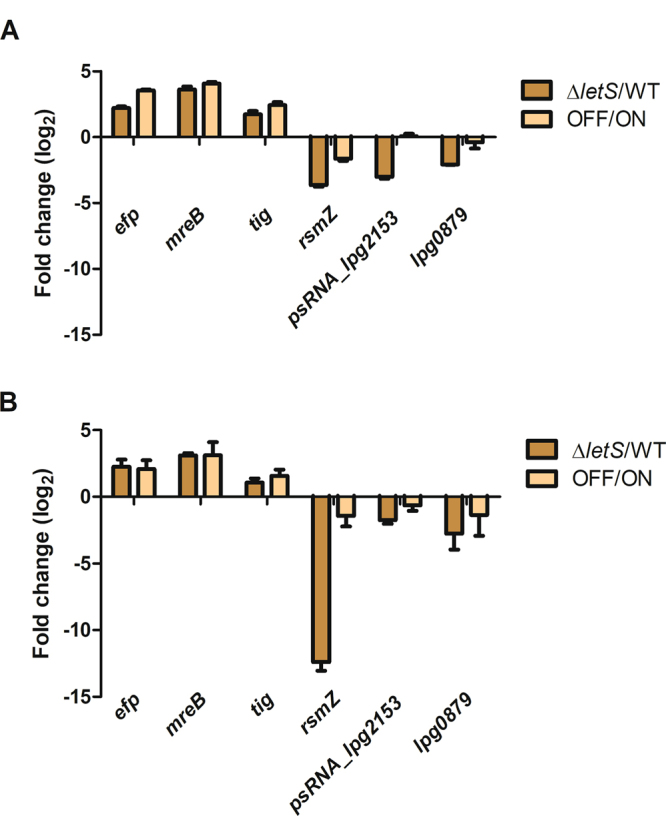
qPCR validates the DNA microarray analysis of the LetS regulon. RT-qPCR (A) was used to analyze the expression pattern of three up-regulated and three down-regulated genes in the transcriptomic analysis of WT vs. ΔletS (B). The fold change of each gene in the letS mutant (vs. the WT) and the OFF strain (vs. ON) are presented.
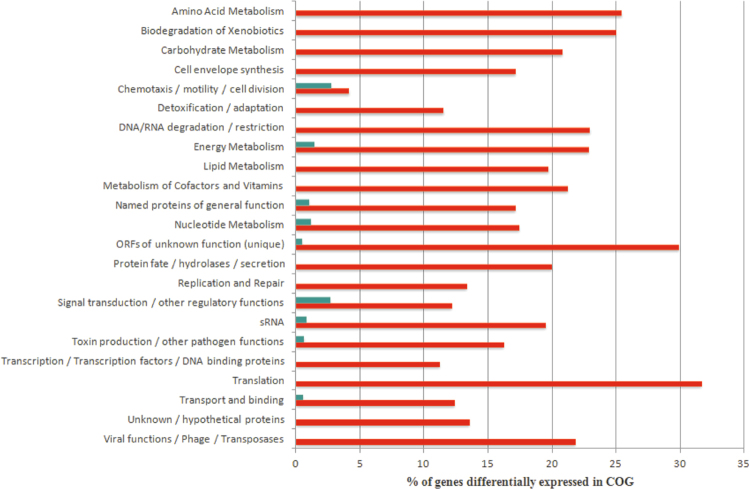
Genes that are differentially and significantly (p < 0.05) expressed in both ∆letS and OFF, relative to the WT and ON respectively, were categorized into clusters of orthologous groups (COGs) (Fig. 6). The cellular functions most affected by LetS were those associated with exponential growth or nutrient rich conditions and were mostly negatively affected by LetS in water (Fig. 6). These include “Translation”, “Amino acid metabolism”, “Lipid metabolism” and “Energy metabolism”. Notably, 30 of the 38 “Translation” genes that are normally down-regulated by LetS in water encode 30 S or 50 S ribosomal proteins (Fig. 6, Supplementary Table S1). It is also noteworthy that the rpoD transcript coding the housekeeping sigma factor was also highly up-regulated in water in the absence of letS (Table 1).
Figure 6.
Clusters of orthologous groups (COGs) analysis of genes affected by the absence of letS in water. Data represents the differentially expressed genes that are common to both the ∆letS vs. WT group and the OFF vs. ON group. Red bars indicate the percentage of genes upregulated in each COG, while green bars indicate the percentage of genes that are downregulated in each COG category.
Table 1.
Select genes differentially regulated in ∆letS vs. WT and OFF vs. ON.
| Lpg # | Gene Product | Gene | log2 (letS/WT)* | log2 (OFF/ON)* |
|---|---|---|---|---|
| Upregulated | ||||
| Amino Acid Metabolism | ||||
| Lpg0932 | shikimate kinase | 1.05 | 1.48 | |
| Lpg1610 | glutamate-5-kinase (gamma-glutamyl kinase) | proB | 1.72 | 1.33 |
| Lpg2278 | 4-hydroxyphenylpyruvate dioxygenase (legiolysin) oxidoreductase protein (hemolysin) | hpd | 2.12 | 1.45 |
| Lpg0890 | cystathionine beta-lyase (cystathionine gamma lyase) | metC | 1.53 | 1.40 |
| Lpg2951 | cystathionine beta synthase (cysteine synthase) | 1.62 | 1.98 | |
| Lpg0725 | serine hydroxymethyltransferase | glyA3 | 2.60 | 2.80 |
| Carbohydrate Metabolism | ||||
| Lpg2887 | phosphomannose isomerase GDP mannose pyrophosphorylase (mannose-1-phosphate guanylyltransferase/mannose-6-phosphate isomerase) | rfbA | 1.57 | 2.14 |
| Lpg0939 | 2-dehydro-3-deoxy-phosphogluconate aldolase | eda | 1.82 | 1.71 |
| Lpg0417 | 6-phosphogluconolactonase | pgl | 1.91 | 2.46 |
| Lpg0805 | phosphoenolpyruvate synthase | 2.32 | 2.85 | |
| Lpg2352 | malate dehydrogenase | mdh | 1.39 | 2.28 |
| Lpg2792 | triosephosphate isomerase (TIM) | tpiA | 2.72 | 2.66 |
| Lpg0138 | glyceraldehyde 3-phosphate dehydrogenase | gap | 1.44 | 2.40 |
| Cell Envelope Synthesis | ||||
| Lpg1753 | UDP-N-acetylmuramate:L-alanyl-gamma-D-glutamyl-meso-diaminopimelate ligase (murein peptide ligase) | mpl | 1.83 | 2.15 |
| Lpg0840 | polysialic acid capsule expression protein (carbohydrate isomerase) (KpsF/GutQ family protein) | 1.45 | 2.08 | |
| Lpg2544 | membrane-bound lytic murein transglycosylase A | mltA | 1.12 | 1.87 |
| Lpg0748 | LPS biosynthesis protein (pseudaminic acid biosynthesis and flagellin acetamidinic modification?) | 2.10 | 2.35 | |
| Lpg0811 | rod shape determining protein MreB (regulator of FtsI) | mreB | 3.07 | 2.41 |
| Motility & Cell Division | ||||
| Lpg2891 | sporulation initiation inhibitor protein Soj, chromosome partitioning protein ParA | soj | 1.38 | 1.45 |
| Lpg1553 | septum site determining protein MinC (FtsZ assembly inhibitor) | minC | 1.05 | 1.86 |
| Lpg1724 | septum site-determining protein MinD (cell division inhibitor (membrane ATPase) activates MinC) | minD | 1.24 | 1.86 |
| Detoxification/Adaptation | ||||
| Lpg0047 | chloramphenicol acetyltransferase (highly similar to antibiotic acetyltransferase) | 1.54 | 1.22 | |
| Lpg0426 | cold shock protein CspH | cspD | 1.04 | 1.71 |
| Lpg1060 | cold shock domain family protein, COG1278: cold shock proteins | 1.02 | 2.62 | |
| Lpg1971 | organic hydroperoxide resistance protein, COG1764:predicted redox protein, regulator of sulfide bond formation | 1.81 | 2.97 | |
| Lpg2967 | superoxide dismutase | sodB | 1.82 | 1.72 |
| Lpg1861 | ATP-dependent Clp protease, proteolytic subunit ClpP | clpP | 1.41 | 2.15 |
| Lpg1423 | TPR domain protein (heat shock protein) N-acetylglucosaminyl transferase | 1.93 | 1.58 | |
| DNA/RNA Degradation | ||||
| Lpg1373 | ribonuclease HII | rnhB | 1.37 | 1.11 |
| Lpg1383 | ribonuclease HI | rnhA | 1.00 | 1.73 |
| Lpg1869 | ribonuclease III (dsRNA-specific ribonuclease) (RNAse III, dsRNA) | rnc | 2.04 | 2.24 |
| Lpg0609 | alanyl tRNA synthetase | alaS | 1.16 | 2.57 |
| Lpg2004 | S-adenosylmethionine:tRNA ribosyltransferase-isomerase | queA | 1.96 | 2.09 |
| Lpg2012 | ribonuclease PH (RNAse PH) | rph | 2.33 | 2.62 |
| Energy Metabolism | ||||
| Lpg2981 | ATP synthase epsilon chain, ATP synthase F1 epsilon subunit | atpC | 2.25 | 2.74 |
| Lpg2982 | H + -transporting two-sector ATPase, ATP synthase F1 subunit beta | atpD | 1.62 | 2.12 |
| Lpg2986 | ATP synthase F0, B subunit | atpF | 1.12 | 1.75 |
| Lpg2779 | NADH dehydrogenase I, K subunit (NADH-ubiquinone oxidoreductase, chain K) | nuoK | 1.56 | 1.32 |
| Lpg2787 | NADH dehydrogenase I, C subunit (NADH-ubiquinone oxidoreductase, chain C) | nuoC | 1.99 | 2.18 |
| Icm/Dot Genes Effectors | ||||
| Lpg0483 | LegA12 | legA12 | 2.25 | 2.07 |
| Lpg2283 | small ORF (132aa) | celLp6 | 1.43 | 2.42 |
| Lpg0621 | sidA | sidA | 1.44 | 1.01 |
| Lpg0963 | ORF | 2.25 | 1.94 | |
| Lpg1110 | ORF | lem5 | 2.12 | 2.65 |
| Lpg2298 | inclusion membrane protein A | legC7/ylfA | 1.52 | 2.07 |
| Lpg2793 | LepA, interaptin | lepA | 1.24 | 2.27 |
| Lpg2999 | CG6763 gene product (eukaryotic homologs?) | legP | 1.22 | 2.14 |
| Lipid Metabolism | ||||
| Lpg0102 | 3-oxoacyl-[acyl carrier protein] synthase (beta-ketoacyl synthase) | fabF | 1.84 | 1.30 |
| Lpg1395 | 3-oxoacyl-(acyl carrier protein) reductase | fabG | 2.07 | 1.44 |
| Lpg1854 | enoyl reductase (NADH dependent enoyl ACP reductase) (enoyl [acyl carrier protein] reductase (NADH2)) | fabI | 1.26 | 1.92 |
| Lpg2228 | 3-oxoacyl (acyl carrier protein) synthase III | 1.57 | 2.03 | |
| Lpg0729 | phosphatidylglycerophosphatase A (PgpA) | pgpA | 1.73 | 2.54 |
| Lpg0920 | phosphatidylglycerophosphatase B (Pap2) | 1.34 | 2.90 | |
| Lpg1414 | glycerol kinase (probable carbohydrate kinase) | 1.88 | 1.48 | |
| Nucleotide Metabolism | ||||
| Lpg0218 | phosphoribosylaminoimidazole carboxylase, catalytic subunit PurE | purE | 1.92 | 1.69 |
| Lpg1181 | CTP synthase PyrG | pyrG | 1.52 | 2.99 |
| Lpg1411 | adenylate kinase (ATP-AMP transphosphorylase) | adK | 1.67 | 2.06 |
| Lpg1676 | phosphoribosylformylglycinamidine synthase I (FGAM synthase I) | purQ | 1.21 | 1.60 |
| Lpg1678 | phosphoribosylformylglycinamidine synthase II (FGAM synthase II) | purL2 | 1.33 | 2.10 |
| Protein Fate & Secretion | ||||
| Lpg0316 | preprotein translocase, SecE subunit | secE | 1.12 | 1.28 |
| Lpg1362 | type II protein secretion LspG (general secretion pathway protein G) | gspG | 1.60 | 1.81 |
| Lpg1463 | preprotein translocase; secretion protein SecA | secA | 2.70 | 2.49 |
| Lpg1871 | signal peptidase I (lepB-1) | lepB-1 | 1.62 | 1.56 |
| Lpg2002 | transmembrane protein YajC, preprotein translocase subunit | yajC | 1.64 | 1.62 |
| Lpg2791 | preprotein translocase, SecG subunit | secG | 2.30 | 2.94 |
| Replication & Repair | ||||
| Lpg0356 | single strand binding protein | ssb | 2.09 | 2.07 |
| Lpg0691 | DNA topoisomerase IV subunit B (DNA gyrase subunit B) | parE | 1.49 | 2.76 |
| Lpg1417 | DNA gyrase, A subunit | gyrA | 2.31 | 2.35 |
| Lpg1576 | Holliday junction DNA helicase RuvB | ruvB | 1.47 | 1.96 |
| Lpg1801 | RecA bacterial DNA recombination protein (recombinase A) | recA | 1.74 | 1.68 |
| Virulence Related Genes | ||||
| Lpg0704 | enhanced entry protein EnhA | enhA | 1.29 | 1.25 |
| Lpg0791 | macrophage infectivity potentiator (mip) | mip | 1.27 | 1.83 |
| Lpg2564 | LvrA | 1.69 | 1.27 | |
| Lpg0447 | LphA (DotK) (OmpA family protein) | lphA | 1.80 | 2.51 |
| Lpg0448 | IcmM (DotJ) | icmM | 1.37 | 1.75 |
| Lpg0450 | IcmK (DotH) (TraN) | icmK | 1.13 | 1.24 |
| Lpg2674 | DotD (TraH) | dotD | 1.08 | 2.12 |
| Lpg1862 | trigger factor TF (FKBP-type peptidyl prolyl cis-trans isomerase) | tig | 2.62 | 1.64 |
| Lpg2702 | stringent starvation protein A (transcription activator) | sspA | 1.42 | 1.77 |
| Transcription | ||||
| Lpg2624 | transcription elongation factor GreA | greA | 1.66 | 3.10 |
| Lpg2934 | transcription termination factor rho | 1.52 | 1.68 | |
| Lpg0232 | transcriptional regulator np20 (Fur family) (ferric uptake) | np20 | 2.26 | 2.39 |
| Lpg0542 | DNA binding protein Fis (recombinational enhancer binding protein; factor-for-inversion stimulation protein) | fis | 2.15 | 2.63 |
| Lpg1743 | Fis transcriptional activator (factor for inversion stimulation) (DNA-binding protein) | fis | 1.05 | 1.00 |
| Lpg2361 | RNA polymerase sigma 70 factor (sigma factor RpoD) | rpoD | 1.45 | 2.00 |
| Translation | ||||
| Lpg0287 | translation elongation factor P (EF-P) | efp | 1.28 | 1.60 |
| Lpg0339 | 50S ribosomal protein L14 | rplN | 1.05 | 1.58 |
| Lpg0341 | 50S ribosomal protein L5 | 1.18 | 1.74 | |
| Lpg1592 | 30S ribosomal protein S6 | rpsF | 3.92 | 3.25 |
| Lpg1711 | ribosome recycling factor (ribosome releasing factor) | frr | 1.34 | 2.09 |
| Lpg1713 | translation elongation factor Ts (EF-Ts) (ubiquitin associated domain:elongation factor Ts) | tsf | 2.51 | 2.27 |
| Lpg1714 | 30S ribosomal protein S2 | rpsB | 1.62 | 1.39 |
| Lpg2713 | translational initiation factor IF-3 | infC | 2.80 | 2.65 |
| Transport & Binding | ||||
| Lpg1277 | ABC transporter ATP binding protein (abcT3) (multidrug resistance ABC transporter) (hemolysin secreting ATP binding protein) | abcT3 | 2.80 | 2.64 |
| Lpg2245 | C4-dicarboxylate transport protein (Na+/H+dicarboxylate symporter) | dctA | 1.51 | 1.66 |
| Lpg2321 | serine transporter | sdaC | 1.79 | 2.75 |
| Lpg2475 | hydrogenase expression/formation protein (hydrogenase nickel incorporation protein HypB) | hypB | 1.81 | 1.59 |
| Lpg2476 | hydrogenase nickel incorporation protein HypA | hypA | 2.71 | 2.88 |
| Lpg2658 | ferrous iron transporter A | feoA | 1.89 | 1.79 |
| Lpg2878 | cobalt/magnesium uptake transporter | corA | 1.39 | 2.30 |
| Small Regulatory RNA | ||||
| Small regulatory RNA | lprC | 1.08 | 2.02 | |
| Small regulatory RNA | lpr0035 | 1.65 | 1.91 | |
| Small regulatory RNA | lprD | 1.67 | 2.26 | |
| Downregulated | ||||
| Small regulatory RNA | rsmZ | −14.43 | −1.32 | |
| Small regulatory RNA | rsmY | −3.69 | −4.21 | |
| Lpg1337 | flagellar protein FliS | fliS | −2.77 | −1.72 |
| Lpg1170 | pyruvate formate lyase-activating enzyme PflA | −2.32 | −1.38 | |
| Lpg0605 | nitrogen fixation protein (Fe-S cluster formation) NifU | −2.28 | −1.36 | |
| Lpg0894 | cytokinin oxidase (cytokinin dehydrogenase) | −2.92 | −1.77 | |
| Lpg2829 | SidH (myosin-like protein) Icm/Dot Effector | sidH | −2.81 | −1.04 |
| Lpg1169 | hypothetical (dioxygenase) | −1.04 | −1.05 | |
| Lpg1080 | deoxyguanosine triphosphate triphosphohydrolase (dGTP triphosphohydrolase) | −2.70 | −1.10 | |
| Lpg1925 | ORF of Uknown Function | −3.50 | −1.92 | |
| Lpg0995 | ORF of Uknown Function | −1.36 | −2.20 | |
| Lpg2458 | sensory box histidine kinase (two-component sensor histidine kinase, signal transducing histidine kinase) | −3.33 | −1.15 | |
| Lpg0879 | two component response regulator with GGDEF domain (regulatory components of sensory transduction system) | −2.92 | −1.49 | |
| Lpg0627 | type IV pilin (competence and adherence associated pilin PilA) | pilE3 | −2.86 | −1.43 |
| Lpg0628 | type IV fimbrial biogenesis PilY1-related protein | −2.26 | −1.09 | |
| Lpg1949 | Icm/Dot Effector | lem17 | −1.34 | −1.31 |
*Only significant values (P < 0.05) are shown.
The LetA/S cascade is linked to the RpoS regulatory cascade, but its activation is ppGpp-independent
Lp responds to water by shutting down major metabolic gene groups, such as replication, transcription, translation and amino acid metabolism8. In the absence of letS, Lp is unable to mount such a response, which likely results in overconsumption of internal resources. The transcriptomic response of ∆letS shows similarity to the response of an rpoS insertion mutant exposed to water; both mutants demonstrate abhorrent up-regulation of gene transcription in response to water14. Moreover, there is considerable phenotypic overlap between the activation of the stringent response and the LetA/S cascade both in vitro19,57,58 and in vivo19,59. As a result, LetS has been proposed to respond to the stringent response alarmone, ppGpp, during starvation60. We decided to investigate whether this hypothesis holds true when Lp is exposed to water. The NCBI Conserved Domain Database (CDD) was used to identify protein domains within LetS that could be responsible for environmental signal recognition. Figure 7A is a graphical representation of LetS topology as predicted by the bioinformatic tools described herein. The transmitter (T), receiver (R) and histidine phosphotransfer (HPT) domains, characteristic of tripartite sensor kinases, are depicted by blue boxes, preceded by two transmembrane domains (TMDs) in green. In addition to these domains that comply with a previous study23, we report the presence of signal sensing domains and their topology, thereby fine-tuning our knowledge of LetS structure. The N-terminus of LetS contains two conserved signal sensing domains; DUF2222 and HAMP. The former is located between residues 37–180, while the latter spans residues 185–241. The two TMDs are located between residues 15 to 34 and 182 to 204 (TMHMM), or 11 to 31 and 181 to 201 (TOPCONS). While the DUF2222 domain is found between the two helices, the N-terminus of the HAMP domain overlaps the second transmembrane (TM) helix by 19 (TMHMM) or 16 residues (TOPCONS) (Fig. 7A). Therefore, approximately 30% the HAMP domain lies within the membrane, and 70% in the cytoplasm. LetS may be activated by an extracellular (via DUF2222), intracellular (via HAMP) or by an intermembrane (via the TM region of HAMP) signal. Here, we investigated the possibility that ppGpp may act as an intracellular signal that activates the LetA/S cascade in response to water exposure as previously postulated48,61.
Figure 7.
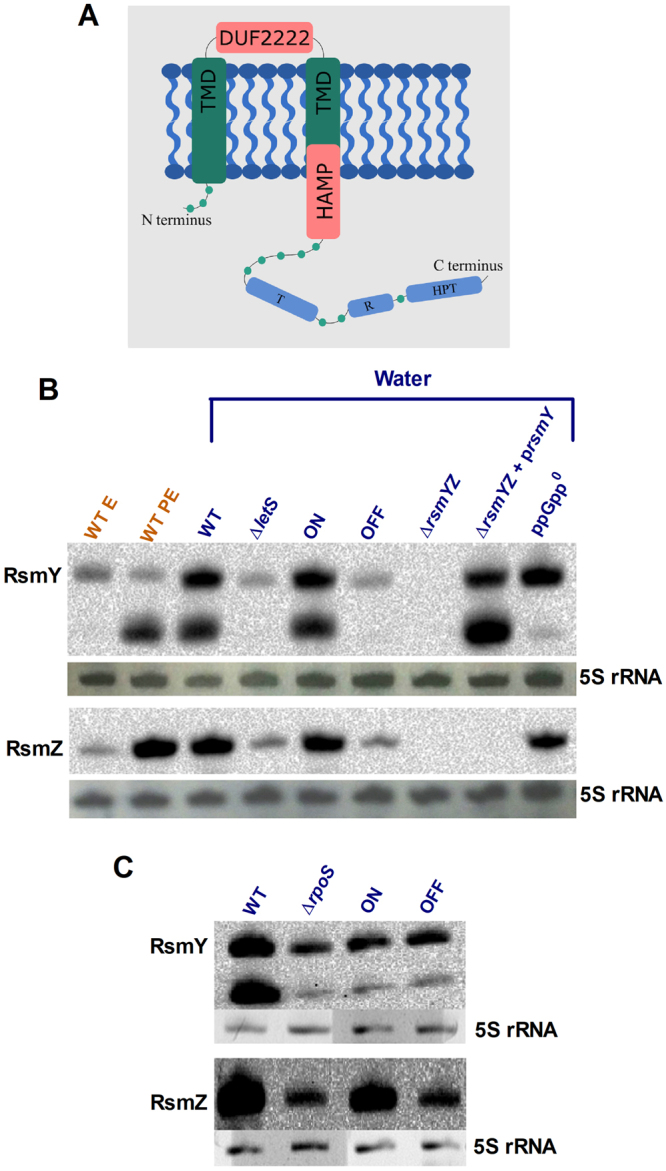
LetS topology and the impact of the stringent response elements, RpoS and ppGpp, on RsmY/Z expression. (A) Topology of the LetS protein was determined using the NCBI CDD Web server, as well as TMHMM v.2.0 and TOPCON software. Transmembrane domains (TMD) are represented by green boxes. Two putative signal sensing domains (pink boxes) are also predicted; a DUF2222 domain is located between the two TMD, and a HAMP domain overlaps the C-terminus of the second TMD. The transmitter (T), receiver (R) and phosphotransfer (HPT) domains that are involved in signal transduction are represented by light blue boxes. (B) The sRNAs RsmY (top) and RsmZ (bottom) under LetS control were probed to determine their levels in water and the influence of ppGpp on their expression. The first two lanes represent the WT strain grown in rich broth to the exponential (E) or the post-exponential (PE) phase. The remaining wells represent the respective strains exposed to water for 2 hours. (C) Impact of RpoS on the expression of RsmY/Z. The sRNAs RsmY (top) and RsmZ (bottom) under LetS control were probed to determine their levels in water. The WT (JR32), rpoS mutant, the induced (ON) or uninduced (OFF) ∆rpoS + prpoS strains were exposed to water for 2 hours. 1 μg of RNA was loaded into each well. Acrylamide gels were stained with ethidium bromide to visualize the 5 S rRNA loading control (shown beneath the respective blots). See Supplementary Figs S5 and S6 for complete gel and blot images.
The small RNAs (sRNAs) RsmY and RsmZ were used as reporters for the activation of LetS using Northern blotting. A ∆relAspoT strain (ppGpp°), incapable of producing ppGpp14, was used to determine the effect of ppGpp on RsmY/Z production. The probe for RsmY detected two bands (Fig. 7B). Similar banding patterns are reported for RsmZ in Pseudomonas fluorescens and RsmY in P. aeruginosa62–64. The top band (~110 nucleotides) was considered the active sRNA as per previous reports46,65. Corroborating previous studies65,66, the levels of RsmZ increase when cells enter the PE phase (Fig. 7B – bottom panel). This increase was, however, absent in RsmY (Fig. 6 – top panel). We postulate that the increase in RsmY may occur later during the PE phase. Nevertheless, both sRNAs were strongly expressed in water. ∆letS and OFF expressed both RsmY/Z at basal levels, while ON recovered WT level expression of RsmY/Z in water. The basal levels of RsmY/Z observed in the letS mutant is likely due to strong promoters preceding the sRNA coding regions, as the E phase WT cells exhibited similar bands (Fig. 7B). The expression of RsmY/Z was abolished in ∆rsmYZ; RsmY expression was recovered to WT levels when rsmY is expressed in trans. The absence of the alternative sigma factor, RpoS also resulted in a significant decrease in RsmY/Z expression (Fig. 7C), corroborating previous findings that the LetA/S cascade is RpoS-dependent65. Importantly, the ppGpp null mutant had WT levels of the RsmY transcript and a slightly lower level of RsmZ compared to the WT, suggesting that the alarmone is dispensable for LetS activation in water and expression of the effector sRNAs.
Discussion
Legionella pneumophila (Lp) is a resident of natural and man-made water systems, and uses aerosols generated by the latter to infect human hosts5. Lp leads a biphasic lifestyle, alternating between the replicative phase and transmissive phase during infection, and between the exponential (E) and post-exponential (PE) phase in broth culture58,67. The former stages are characterized by nutrient abundance. In contrast, the hallmark of the transmissive/PE phase is nutrient deprivation causing morphological changes, a transcriptomic shift, stress resistance and virulence phenotypes58,61,68. The LetA/S two-component system is a key tool for differentiation of Lp in response to starvation, both in artificial medium and inside the host46,50,66,69,70. The established role of LetA/S in adaptation to low nutrient conditions led us to test its contribution to surviving starvation experienced in water.
The absence of letS did not affect the growth rate of the mutant on solid medium. Pyomelanin production, which is linked to iron acquisition, was impaired in the mutant (Supplementary Fig. S3)19,46,48. However, this characteristic may be irrelevant when Lp is exposed to water, as genes related to pigmentation and iron acquisition (lpg2278/hpd/lly, feoA, lpg0232, lpg0124, lpg0746, lpg0467, np20) were repressed by the TCS (Table 1)71–73. The absence of letS also caused a slightly elongated cellular morphology in water compared to the WT (Fig. 2).
Importantly, an intact LetA/S system is required for culturability of Lp in water (Fig. 1). We corroborate previous studies showing that the LetA/S cascade exerts its regulatory activity via the two small RNAs, RsmY/Z during PE phase in broth (Fig. 7B)46. Defects previously associated with the LetA/S system occur under the starvation conditions found during the PE and transmissive phases19,50,69. Our data show that LetA/S is also activated in water, leading to RsmY and RsmZ expression (Fig. 7B). Interestingly, the transcriptomic changes initiated by the LetA/S cascade occur and are needed rapidly upon water exposure (Fig. 1).
LetS is a tripartite sensor kinase whose ortholog, BvgS, in Bordetella is known to respond to multiple environmental stimuli, activating specific regulons22,74. Accordingly, differences between the LetS regulon of Lp in water and that in the PE phase were expected46,50,75. While both are characterized by the absence of nutrients, the latter accumulates waste products likely causing additional stress. Indeed, Lp can survive for several weeks in Fraquil, but dies after only a few days in broth11. Over time, WT Lp exposed to water progressively shuts down transcription relative to exponential growth8. In stark contrast, an overwhelming majority of differentially expressed genes are up-regulated (97%) in the absence of letS (Fig. 4). In PE phase, approximately equal numbers of genes are positively and negatively influenced by the LetA/S cascade46. In water, LetS almost exclusively represses genes encoding functions that are unnecessary for survival, such as translation and metabolism of amino acids, lipids and carbohydrates (Fig. 6).
Over-expression of replicative phase genes in the absence of LetS is likely due to its downstream effect on CsrA. CsrA affects mRNA expression by: 1) blocking the ribosome binding site, 2) increasing degradation by recruiting RNase E, or 3) enhancing the stability and, thereby, expression of the target mRNA76. In ∆letS, binding of CsrA to target mRNAs is not relieved by RsmY/Z, which will favor the continued expression of replicative genes, while repressing transmissive genes48. A recent study revealed that CsrA directly regulates 479 transcripts involved in amino acid metabolism, carbon metabolism, virulence, flagella expression and iron acquisition77. For example, the binding of CsrA to the gap mRNA increases transcription by preventing rho-dependent termination77. As a result, gap expression increased in the absence of LetS in water (Supplementary Table S1). Moreover, two out of three Fis transcriptional regulators (Lpg0542/Fis1 and Lpg1743/Fis2), that are stabilized by CsrA77, are repressed by LetS in water. Strikingly, 30 ribosomal genes were highly up-regulated in the absence of letS (Supplementary Table S1). In contrast, only five ribosomal genes are reported to bind CsrA directly77, suggesting indirect regulation by CsrA. Another notable regulator repressed by LetS in water is the housekeeping sigma factor, RpoD. Increasing the amount of RpoD in the letS mutant strain could interfere with the binding of other sigma factors to the core RNA polymerase, a mechanism termed sigma factor competition78. Regulation of RpoD by LetA/S or by CsrA has not been reported in Lp46,48,77. As such, it is unclear whether the effect of CsrA on rpoD in water is direct or indirect. The general down-regulation of gene expression upon LetS activation in water is likely mediated by relieving both the direct and indirect effects of CsrA binding to target mRNAs.
The most noticeable difference between the transcriptome of the letS mutant in PE phase and in water is the marked absence of gene up-regulation. Only 16 genes were significantly (p < 0.05) induced by LetS in water, including RsmY/Z that are directly regulated by LetA (Table 1). In contrast, over 300 genes were significantly up-regulated in the PE phase when letA or letS were absent46. This is presumably because water is a less stressful condition than PE phase, during which metabolic by-products accumulate. As such, virulence regulators (RpoS, LetE and LqsR) were not affected by LetS in water (Supplementary Table S1). While LetA/S controls flagella genes in broth cultures19,23,46,69,79, we report that only one flagella-associated gene was differentially expressed in ∆letS in water (Supplementary Table S1). At 25 °C, flagella genes are maximally induced at 6 hours after exposure to water8. At 42 °C, it is possible that their induction was not captured at the 2-hour time point. Furthermore, flagellar gene expression is tightly regulated in Lp, both cooperatively and independently by several high-profile regulatory entities including RpoS, RpoN, FleQ, LetS and LqsR46,79–81.
PE phase heat shock (HS) resistance is presumed to be conferred by genes under LetS control48,50. Accordingly, ∆letS was also heat sensitive relative to the WT after water exposure (Fig. 3); however, similar to flagella gene expression, HS genes were unaffected by LetS at 42 °C. The HS response is initiated within the first few minutes of exposure to this stress, whose transcriptomic effects subside quickly thereafter82,83. It is, therefore, possible that the HS-related transcriptomic changes were not detectable by the microarray analysis, if, indeed, they were activated at 42 °C. Alternatively, the general sensitivity of the letA/S mutants to various stresses, including heat shock, (Fig. 3)48,50 may be a result of cell structure and therefore, a by-product of the cell’s inability to adapt to the respective environments.
Upon sensing starvation, bacteria deploy the stringent response (SR) network governed by the cellular alarmone ppGpp, a key signal in growth phase differentiation60,84. SR is characterized by a rapid downshift in the synthesis of stable RNAs like rRNAs and tRNAs85. For the most part, ɤ-proteobacteria synthesize ppGpp using RelA, a synthase, and SpoT, a dual-acting hydrolase with weak synthase activity60,85. ppGpp positively affects cellular levels of RpoS, an alternative sigma factor60,86. Mutation of RpoS leads to the over-expression of replicative genes in water14, similar to the letS mutant (Fig. 5). Given these transcriptomic similarities, the LetA/S system was proposed to be integrated into the SR during water exposure, which suggests that the same activating signal initiates the LetS cascade.
Northern blotting was used to probe for the direct downstream targets of LetA/S, RsmY and RsmZ (Fig. 7B). Both sRNAs were LetS-dependent and highly expressed upon water exposure (Fig. 7B). The ppGpp° strain (∆relA∆spoT) that is unable to survive in water14 was used to determine the effect of the alarmone on RsmY/Z production. We show that the ppGpp° strain did not change RsmY/Z expression considerably (Fig. 7B). Therefore, the transcriptomic changes initiated by the SR seem largely independent of those mediated by LetS, and the survival defect of ppGpp° is not due to an impaired LetS response and vice-versa. We also confirm that RsmY/Z expression is RpoS-dependent in water (Fig. 7C)65. ppGpp may contribute to RpoS-mediated RNAP binding to letS or rsmY/Z, but its effect is likely minimal (Fig. 7B). Notwithstanding its effect on transcription, the LetA/S-RsmY/Z-CsrA cascade and the SR seem to be parallel responses contributing to survival under nutrient-deprived conditions. The data presented here suggests the following model. ppGpp and the LetA/S cascade regulate similar regulons in parallel and possibly in different time frames, each promoting adaptation to and survival in water independently. A ppGpp° mutant has a survival defect that is apparent earlier than that of the letS mutant in water14, suggesting that the SR is more important than LetS and that its effect is required immediately upon exposure to water. RpoS which is positively affected by ppGpp, positively influences the letS transcript and thereby, levels of RsmY/Z (Fig. 7C). However, whether RpoS increases the level of letS in response to ppGpp in water, or whether basal RpoS levels maintain constant letS expression within the cell is not yet know.
It is likely that LetS responds to a variety of stresses fine-tuning the transcriptomic response to the challenge at hand23. In broth, LetA/S was required to increase flagella expression in response to nicotinic acid, and the expression of several virulence traits in response to free fatty acids75,87. It is unclear whether these are direct signals and whether they also represent activation signals in water. Recently, the Lp quorum sensing molecule, Legionella autoinducer-1 (LAI-1) was shown to increase RsmY/Z levels in broth88; however, it is unlikely to be a viable signal for the activation of LetS in water, because of the bacterium’s low metabolic activity, lowering LAI-1 production under this condition. As such, the environmental signal initiating the LetS/A cascade in water is yet to be determined. This study does not exclude the possibility that ppGpp is one of multiple stimuli sensed by LetS in water; its weak contribution to LetS activation may be masked by other, stronger signals.
In conclusion, we report that the LetA/S-RsmY/Z-CsrA regulatory cascade is essential for the culturability of Lp in water. In contrast to the transcriptome in broth cultures, LetS almost exclusively acts to repress genes related to growth; with RsmY/Z being the prominent exceptions. While there is overlap between the regulons and crosstalk between members of the SR and the LetA/S TCS, activation of the two systems seems to be independent of each other in water.
Experimental Procedures
Bacterial strains and media
KS79 is a ∆comR mutant of the JR32 strain rendering it constitutively competent. JR32 is a salt-sensitive, streptomycin-resistant, restriction negative mutant of Lp strain Philadelphia 189. The increased competence of KS79 renders allelic exchange mutations through natural transformation possible. It was, therefore, used as the wild-type (WT) strain in this study. A complete list of strains used in this study can be found in Table 2. Bacterial strains stored at −80 °C in 10% glycerol were grown on CYE (ACES-buffered charcoal yeast extract) agar supplemented with 0.1 mg ml−1 α-ketoglutarate, 0.25 mg ml−1 L-cysteine and 0.4 mg ml−1 ferric pyrophosphate90. AYE broth (CYE without agar and charcoal) was used as the liquid medium90. When necessary, media were supplemented with 5 μg ml−1 chloramphenicol, 2.5 μg ml−1 kanamycin and/or 0.1 mM IPTG.
Table 2.
Strains used in this study.
| Strain Name | Relevant Genotypea | Source or Reference |
|---|---|---|
| Legionella pneumophila | ||
| JR32 | Philadelphia-1; Smr; r- m+ | 89 |
| KS79 (WT) | JR32 ∆comR | 96 |
| ∆letS (GAH338) | KS79 letS::aptII, Knr | 65 |
| ∆letS + pletS (SPF39) | ∆letS + pMMB207c Ptac-letS; Cmr | 65 |
| ∆rsmYZ (SPF41) | KS79 rsmY::aptII, rsmZ::aacC1; Knr, Gmr | This study |
| ∆rsmYZ + prsmY (SPF291) | ∆rsmYZ + pXDC39-prsmY; Knr, Gmr, Cmr | This study |
| ∆rpoS | JR32 rpoS::Tn903dGent; Gmr | 96 |
| ∆rpoS + prpoS | ∆rpoS + pMMB207c Ptac-rpoS; Cmr | 14 |
| ∆relA∆spoT (ppGpp°) | KS79 ∆relA::aacC1 spot::aptII; Gmr, Kmr | 14 |
| Plasmid Name | ||
| pBBR1MCS-5 | pBBR1MCS Gmr | 97 |
| pSF6 | DH5α, pGEMT-easy-rrnb | 98 |
| pMMB207c | RSF1010 derivative, IncQ, lacIq Cmr Ptac oriT ∆mobA | 99 |
| pXDC39 | pMMB207c ∆Ptac, ∆lacI, Cmr | Xavier Charpentier |
| prsmY | pXDC39-rsmY, Cmr | This study |
aSmr, streptomycin resistance; Cmr, chloramphenical resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance.
Deletion of rsmY and rsmZ
Construction of the ∆rsmYZ strain was performed by allelic exchange as described before65, by replacing, first, rsmY with a kanamycin resistance cassette and, then, rsmZ with a gentamicin resistance cassette. Two 1-kb fragments corresponding to the upstream and downstream of rsmY were amplified using primers rsmY-BF/rsmY-BR and rsmY-EF/rsmY-ER, respectively. A kanamycin cassette was amplified from pSF6 using primers rsmY-BRKN/rsmY-EFKN. PCR fragments were purified on gel using a gel extraction kit (Qiagen). The three fragments were ligated together by PCR using primers rsmY-BF/RsmY-ER. The resulting 3 kb fragment was purified on gel, as described above, and introduced into KS79 by natural transformation91. The recombinants were selected for kanamycin resistance and the allelic exchange confirmed by PCR. Deletion of rsmZ was performed similarly by using primers rsmZ-BF/rsmZ-BR and rsmZ-EF/rsmZ-ER to amplify the upstream and downstream fragments, and rsmZ-BRGT and rsmZ-EFGT to amplify a gentamycin cassette from pBBR1MCS-5. The resulting 3 kb fragment was introduced into the ∆rsmY strain, recombinants were selected for kanamycin and gentamicin resistance and the deletion of rsmZ was confirmed by PCR. The resulting strain was named SPF41. Northern blot was used to confirm absence of expression of both sRNA. All PCR amplifications were carried with Phusion polymerase (NEB) according to the manufacturer’s protocol. Primer sequences used in this study are found in Supplementary Table S2.
Cloning of rsmY
The rsmY gene was amplified with its own promoter from the KS79 wild-type strain using primers rsmY-F and rsmY-R (Supplementary Table S2). The pXDC39 vector and the amplicon were then digested with BamHI and HindIII (New England Biolabs) according to the manufacturer’s protocol, and purified using a MinElute purification kit (Qiagen). The digested vector and insert were ligated overnight using T4 DNA Ligase (New England Biolabs) at 16 °C. The recombinant plasmid was then transformed into E. coli DH5α (pSF86). The transformed population was incubated at 37 °C shaking for 90 minutes before plating on 25ug ml−1 chloramphenicol plates. Colonies that grew on antibiotic plates were patched and tested by PCR for insertion of the rsmY gene into the vector. The recombinant plasmid was extracted using a plasmid extraction kit (Qiagen) and introduced into the ∆rsmYZ mutant to produce the ∆rsmYZ + prsmY strain (SPF291).
Survival in water
Survival in water was tested in the artificial freshwater medium, Fraquil, as described previously8,51. Briefly, Lp strains cultured on CYE agar at 37 °C for 3 days were suspended in Fraquil at an OD600nm of 0.1 and washed three times with Fraquil. One millilitre of this bacterial suspension was mixed with 4 ml of fresh Fraquil in a 25 cm2 cell culture flasks (Sarstedt) and incubated at 4 °C, 25 °C or 42 °C. To test survival in water at high cell densities, strains were suspended in Fraquil at an OD600nm of 1 and washed three times with Fraquil. Five millilitres of this bacterial suspension was placed in a 25 cm2 cell culture flasks (Sarstedt) and incubated at 42 °C. Survival of the strains in water was monitored using CFU counts.
Microscopic analysis
Lp strains were grown on CYE agar for 3 days at 37 °C. AYE broth was inoculated with the respective strains and grown to exponential phase (OD600 0.4–0.7) or post-exponential phase (OD600 > 3) at 37 °C shaking (200 rpm). To test the effect of water, strains were suspended in Fraquil at an OD600 of 1 and incubated at 42 °C for 24 hours. 20 μl of each sample was placed on a clean microscope slide, covered with a cover slip and observed under 1000X magnification under oil immersion using digital phase contrast microscopy (Nikon Eclipse 80i). For each strain, 10 images of random microscopic fields were captured using the NIS Element Software (Nikon Instruments, Inc.). ImageJ software92 was used for quantitative analysis of cell length. 10 cells from 10 different fields of view were randomly chosen and analyzed per strain per treatment (n = 100). Multiplying cells (presence of a septum) were excluded and only individual, non-filamentous cells were used for analysis.
Pigment production
Lp strains grown on CYE for 3 days at 37 °C were inoculated into AYE broth. Strains were grown to late post-exponential phase at 37 °C shaking at 200 rpm. 10 ml of each strain was pelleted at 4500 rpm for 10 minutes. The supernatant was then removed and filtered using 0.2μm pore sized syringe filter. The optical density of the supernatant was measured at 550 nm.
Heat shock
Lp strains cultured on CYE agar at 37 °C for 3 days were suspended in Fraquil at an OD600nm of 0.1, as described above for the survival assay. One milliliter aliquots of each strain were transferred to 13 ml tubes (Sarstedt) and were allowed to acclimate to the water environment for 2 hours at room temperature. At the end of the incubation period, tubes were submerged in a 55 °C water bath. At each time point tested, three biological replicates from each strain were removed from the water bath and the CFU counts enumerated on CYE agar.
DNA microarray
The WT, ∆letS, and the complemented strain induced (ON) or not (OFF) with IPTG were grown on CYE agar for 3 days at 37 °C. Each culture was suspended in Fraquil in triplicate at an OD600 of 1. 20 ml of each strain was placed in 75 cm2 cell culture flasks and incubated at 42 °C for 2 hours. After incubation, 10 ml aliquots were pelleted for 5 minutes at 5000 g. After centrifugation, the supernatant was removed, and the cell pellets were re-suspended in 1 ml of TRIzol reagent (Invitrogen). Three biological replicates of each strain were used for the transcriptomic analysis. The three remaining replicates of each strain were preserved at −20 °C for further experimentation. RNA extractions were done according to the manufacturer’s protocol. To remove DNA contamination, extracted RNA was subsequently treated with Turbo DNase (Ambion) as per the manufacturer’s protocol. The purity and concentration of RNA were determined by UV spectrophotometry. Fifteen micrograms of RNA was labeled with aminoallyl-dUTP (Sigma) during reverse transcription (ProtoScript II, New England Biolabs) using random hexamers (Life Technologies) as previously described8,93. Genomic DNA was used as a reference channel and was labeled by random priming using Klenow fragments (New England Biolabs), aminoallyl-dUTP and random primers as described previously93. The cDNA and gDNA were subsequently coupled to the succinimidyl ester fluorescent dye (Life Technologies) AlexaFluor 647 or AlexaFluor 546, respectively, following the manufacturer’s protocols. The microarray design (GPL19458) and the protocol for hybridization, data acquisition and data analysis have been published previously8. Statistical analyses were performed using an unpaired one-tailed Student’s t-test. Genes were considered differentially expressed if they demonstrated a ratio-to-control value of ± 2-fold with a P < 0.05.
qPCR validation
One replicate of the WT, ∆letS and the complemented strain induced (ON) or not (OFF) with IPTG exposed to water as described above was used to validate the microarray results using qPCR. RNA was extracted as described above. 1 μg of total RNA was transcribed to cDNA (Supercript II, Life Technologies) using random hexamers. qPCR was performed as described previously using the primers described in Supplementary Table S2. Ct values were normalized to the 16 S rRNA.
Northern Blotting
Lp strains grown on CYE agar for 3 days at 37 °C were used to inoculate AYE broth. All strains were grown to exponential phase (OD600 0.4–0.7) at 37 °C shaking (200 rpm). For each strain, 10 ml of exponential phase bacterial culture was centrifuged for 10 minutes at 4500 rpm, the supernatant removed and the pellet re-suspended in 10 ml of Fraquil. Water-exposed bacteria were incubated at 42 °C for 2 hours, after which cells were pelleted and suspended in 1 ml of TRIzol reagent (Invitrogen). RNA was extracted according to the manufacturer’s protocol. RNA from the WT was also extracted from 10 ml of exponential phase culture and 5 ml of post-exponential phase culture (OD600 > 3). 1 ug of RNA was loaded and migrated on a 6% Tris-borate-EDTA-urea polyacrylamide gel (Ambion) at 180 mV. The RNA was transferred onto a positively charged nylon membrane (Thermo Scientific) using a semidry gel blotting system (BioRad) for 20 minutes at 200 mA. The membranes were pre-hybridized in ULTRAhyb-Oligo Hybridization Buffer (Ambion) for 1 hour at 37 °C before hybridization with 5′ biotinylated RsmY and RsmZ probes (Integrated DNA Technologies). Hybridization was performed overnight in a rotating chamber at 37 °C. Blots were washed twice with 2X SSC (0.15 M NaCl and 0.015 M sodium citrate) and 0.5% SDS for 30 minutes. The biotinylated probed were detected using the Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific) as per the manufacturer’s instructions. The exposure time used for image acquisition was 1 second.
Bioinformatic analyses
NCBI Conserved Domain Database (CDD https://www.ncbi.nlm.nih.gov/cdd/) was used to search for conserved protein domains that may be implicated in signal sensing within LetS. The Accession number YP_095929.1 representing Lpg1912 of Legionella pneumophila Philadelphia-1 strain was queried. The TMHMM server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and TOPCONS (http://topcons.net/) were used to predict the transmembrane helices on the N-terminal of LetS94,95.
Availability of materials and data
Microarray data generated during the current study are available from Gene Expression Omnibus (GSE98743).
Electronic supplementary material
Acknowledgements
This study was founded by Discovery Grant 418289–2012 from the National Sciences and Engineering Research Council of Canada (NSERC) and a John R. Evans Leaders Fund – Funding for research infrastructure from the Canadian Foundation for Innovation to SPF. NM was the recipient of a PhD scholarship from Fond de Recherche du Québec – Nature et Technologie. PM was the recipient of an Undergraduate Student Research Award from NSERC. JS is supported by a PhD scholarship from Fond de Recherche du Québec – Nature et Technologie.
Author Contributions
N.M. contributed to experimental design, conducted the survival experiments at 4 °C and 25 °C, constructed SPF291, performed and analyzed the microarray data, analyzed qPCR results, conducted other phenotypic characterizations (microscopic observation, heat shock and pigment production), Northern Blots and wrote the manuscript. P.M. conducted the survival experiments, assisted in constructing SPF291, performed the microarray, performed qPCR, and analyzed the transcriptomic and microscopic data. T.M. performed qPCR and analyzed results. J.S. performed Northern blots. S.P.F. constructed SPF41, contributed to the experimental design, writing and editing the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-24263-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fliermans CB, et al. Ecological distribution of Legionella pneumophila. Appl Environ Microbiol. 1981;41:9–16. doi: 10.1128/aem.41.1.9-16.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buse HY, Ashbolt NJ. Differential growth of Legionella pneumophila strains within a range of amoebae at various temperatures associated with in-premise plumbing. Lett Appl Microbiol. 2011;53:217–224. doi: 10.1111/j.1472-765X.2011.03094.x. [DOI] [PubMed] [Google Scholar]
- 3.Tyson JY, et al. Multiple Legionella pneumophila Type II Secretion Substrates, Including a Novel Protein, Contribute to Differential Infection of the Amoebae Acanthamoeba castellanii, Hartmannella vermiformis, and Naegleria lovaniensis. Infect Immun. 2013;81:1399–1410. doi: 10.1128/IAI.00045-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prashar A, Terebiznik MR. Legionella pneumophila: homeward bound away from the phagosome. Curr Opin Microbiol. 2015;23:86–93. doi: 10.1016/j.mib.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Fields BS, Benson RF, Besser RE. Legionella and Legionnaires’ Disease: 25 Years of Investigation. Clin Microbiol Rev. 2002;15:506–526. doi: 10.1128/CMR.15.3.506-526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zimmerman L. Aerosol Infectivity of Legionella pneumophila. N Engl J Med. 1979;301:1004–1004. doi: 10.1056/NEJM197912133012402. [DOI] [PubMed] [Google Scholar]
- 7.Borella P, Guerrieri E, Marchesi I, Bondi M, Messi P. Water Ecology of Legionella and Protozoan: Environmental and Public Health Perspectives. Biotechnol Annu Rev. 2005;11:355–380. doi: 10.1016/S1387-2656(05)11011-4. [DOI] [PubMed] [Google Scholar]
- 8.Li L, Mendis N, Trigui H, Faucher SP. Transcriptomic changes of Legionella pneumophila in water. BMC Genomics. 2015;16:637. doi: 10.1186/s12864-015-1869-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bandyopadhyay P, Xiao H, Coleman HA, Price-Whelan A, Steinman HM. Icm/Dot-Independent Entry of Legionella pneumophila into Amoeba and Macrophage Hosts. Infect Immun. 2004;72:4541–4551. doi: 10.1128/IAI.72.8.4541-4551.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Söderberg MA, Dao J, Starkenburg SR, Cianciotto NP. Importance of Type II Secretion for Survival of Legionella pneumophila in Tap Water and in Amoebae at Low Temperatures. Appl Environ Microbiol. 2008;74:5583–5588. doi: 10.1128/AEM.00067-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, Faucher SP. The Membrane Protein LasM Promotes the Culturability of Legionella pneumophila in Water. Front Cell Infect Microbiol. 2016;6:113. doi: 10.3389/fcimb.2016.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abbott ZD, Yakhnin H, Babitzke P, Swanson MS. csrR, a Paralog and Direct Target of CsrA, Promotes Legionella pneumophila Resilience in Water. mBio. 2015;6:e00595–00515. doi: 10.1128/mBio.00595-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li L, Faucher SP. Role of the LuxR family transcriptional regulator Lpg2524 in the survival of Legionella pneumophila in water. Can J Microbiol. 2017 doi: 10.1139/cjm-2016-0780. [DOI] [PubMed] [Google Scholar]
- 14.Trigui H, Dudyk P, Oh J, Hong J-I, Faucher SP. A regulatory feedback loop between RpoS and SpoT supports the survival of Legionella pneumophila in water. Appl Environ Microbiol. 2014;81:918–928. doi: 10.1128/AEM.03132-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krell T, et al. Bacterial Sensor Kinases: Diversity in the Recognition of Environmental Signals. Annu Rev Microbiol. 2010;64:539–559. doi: 10.1146/annurev.micro.112408.134054. [DOI] [PubMed] [Google Scholar]
- 16.Mascher T, Helmann JD, Unden G. Stimulus Perception in Bacterial Signal-Transducing Histidine Kinases. Microbiol Mol Biol Rev. 2006;70:910–938. doi: 10.1128/MMBR.00020-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capra EJ, Laub MT. The Evolution of Two-Component Signal Transduction Systems. Annu Rev Microbiol. 2012;66:325–347. doi: 10.1146/annurev-micro-092611-150039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stock AM, Robinson VL, Goudreay PN. Two-Component Signal Transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 19.Hammer BK, Tateda ES, Swanson MS. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol Microbiol. 2002;44:107–118. doi: 10.1046/j.1365-2958.2002.02884.x. [DOI] [PubMed] [Google Scholar]
- 20.Pernestig AK. O., M. & Georgellis, D. Identification of UvrY as the cognate response regulator for the BarA sensor kinase in. Escherichia coli J Biol Chem. 2001;276:225–231. doi: 10.1074/jbc.M001550200. [DOI] [PubMed] [Google Scholar]
- 21.Marutani M, et al. Gac two-component system in Pseudomonas syringae pv. tabaci is required for virulence but not for hypersensitive reaction. Mol Genet Genomics. 2007;279:313. doi: 10.1007/s00438-007-0309-y. [DOI] [PubMed] [Google Scholar]
- 22.Dupré E, et al. Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS. PLo Pathog. 2015;11:e1004700. doi: 10.1371/journal.ppat.1004700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards RL, Jules M, Sahr T, Buchrieser C, Swanson MS. The Legionella pneumophila LetA/LetS Two-Component System Exhibits Rheostat-Like Behavior. Infect Immun. 2010;78:2571–2583. doi: 10.1128/IAI.01107-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotter PA, Miller JF. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol Microbiol. 1997;24:671–685. doi: 10.1046/j.1365-2958.1997.3821741.x. [DOI] [PubMed] [Google Scholar]
- 25.Deora R, Bootsma HJ, Miller JF, Cotter PA. Diversity in the Bordetella virulence regulon: transcriptional control of a Bvg-intermediate phase gene. Mol Microbiol. 2001;40:669–683. doi: 10.1046/j.1365-2958.2001.02415.x. [DOI] [PubMed] [Google Scholar]
- 26.Zere TR, et al. Genomic Targets and Features of BarA-UvrY (-SirA) Signal Transduction Systems. PLoS One. 2015;10:e0145035. doi: 10.1371/journal.pone.0145035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teplitski M, Goodier RI, Ahmer BMM. Pathways Leading from BarA/SirA to Motility and Virulence Gene Expression in. Salmonella. J Bacteriol. 2003;185:7257–7265. doi: 10.1128/JB.185.24.7257-7265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kay E, Dubuis C, Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci USA. 2005;102:17136–17141. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller CL, et al. RsmW, Pseudomonas aeruginosa small non-coding RsmA-binding RNA upregulated in biofilm versus planktonic growth conditions. BMC Microbiol. 2016;16:155. doi: 10.1186/s12866-016-0771-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol. 2005;58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki K, et al. Regulatory Circuitry of the CsrA/CsrB and BarA/UvrY Systems of Escherichia coli. J Bacteriol. 2002;184:5130–5140. doi: 10.1128/JB.184.18.5130-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altier C, Suyemoto M, Lawhon SD. Regulation of Salmonella enterica Serovar Typhimurium Invasion Genes by csrA. Infect Immun. 2000;68:6790–6797. doi: 10.1128/IAI.68.12.6790-6797.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pessi G, et al. The Global Posttranscriptional Regulator RsmA Modulates Production of Virulence Determinants and N-Acylhomoserine Lactones in Pseudomonas aeruginosa. J Bacteriol. 2001;183:6676–6683. doi: 10.1128/JB.183.22.6676-6683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martínez LC, et al. Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol. 2011;80:1637–1656. doi: 10.1111/j.1365-2958.2011.07674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmer BMM, Van Reeuwijk J, Watson PR, Wallis TS, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- 36.Marcus SL, Brumell JH, Pfeifer CG, Finlay BB. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2000;2:145–156. doi: 10.1016/S1286-4579(00)00273-2. [DOI] [PubMed] [Google Scholar]
- 37.Herren CD, et al. The BarA-UvrY Two-Component System Regulates Virulence in Avian Pathogenic Escherichia coli O78:K80:H9. Infect Immun. 2006;74:4900–4909. doi: 10.1128/IAI.00412-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palaniyandi S, et al. BarA-UvrY Two-Component System Regulates Virulence of Uropathogenic E. coli CFT073. PLoS One. 2012;7:e31348. doi: 10.1371/journal.pone.0031348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitra A, Palaniyandi S, Herren CD, Zhu X, Mukhopadhyay S. Pleiotropic Roles of uvrY on Biofilm Formation, Motility and Virulence in Uropathogenic Escherichia coli CFT073. PLoS One. 2013;8:e55492. doi: 10.1371/journal.pone.0055492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heurlier K, et al. Positive Control of Swarming, Rhamnolipid Synthesis, and Lipase Production by the Posttranscriptional RsmA/RsmZ System in Pseudomonas aeruginosa PAO1. J Bacteriol. 2004;186:2936–2945. doi: 10.1128/JB.186.10.2936-2945.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coggan KA, Wolfgang MC. Global regulatory pathways and cross-talk control Pseudomonas aeruginosa environmental lifestyle and virulence phenotype. Curr Issues Mol Biol. 2012;14:47. [PMC free article] [PubMed] [Google Scholar]
- 42.Reimmann C, et al. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 43.Mey AR, Butz HA, Payne SM. Vibrio cholerae CsrA Regulates ToxR Levels in Response to Amino Acids and Is Essential for Virulence. mBio. 2015;6:e01064–01015. doi: 10.1128/mBio.01064-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pernestig A-K, et al. The Escherichia coli BarA-UvrY Two-Component System Is Needed for Efficient Switching between Glycolytic and Gluconeogenic Carbon Sources. J Bacteriol. 2003;185:843–853. doi: 10.1128/JB.185.3.843-853.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frangipani E, et al. The Gac/Rsm and cyclic-di-GMP signalling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ Microbiol. 2014;16:676–688. doi: 10.1111/1462-2920.12164. [DOI] [PubMed] [Google Scholar]
- 46.Sahr T, et al. Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila. Mol Microbiol. 2009;72:741–762. doi: 10.1111/j.1365-2958.2009.06677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sahr T, et al. Deep sequencing defines the transcriptional map of L. pneumophila and identifies growth phase-dependent regulated ncRNAs implicated in virulence. RNA Biol. 2012;9:503–519. doi: 10.4161/rna.20270. [DOI] [PubMed] [Google Scholar]
- 48.Molofsky AB, Swanson MS. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol Microbiol. 2003;50:445–461. doi: 10.1046/j.1365-2958.2003.03706.x. [DOI] [PubMed] [Google Scholar]
- 49.Forsbach-Birk V, McNealy T, Shi C, Lynch D, Marre R. Reduced expression of the global regulator protein CsrA in Legionella pneumophila affects virulence-associated regulators and growth in Acanthamoeba castellanii. Int J Med Microbiol. 2004;294:15–25. doi: 10.1016/j.ijmm.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 50.Lynch D, Fieser N, Glöggler K, Forsbach-Birk V, Marre R. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol Lett. 2003;219:241–248. doi: 10.1016/S0378-1097(03)00050-8. [DOI] [PubMed] [Google Scholar]
- 51.Mendis N, McBride P, Faucher SP. Short-term and long-term survival and virulence of Legionella pneumophila in the defined freshwater medium Fraquil. PLoS One. 2015;10:e0139277. doi: 10.1371/journal.pone.0139277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fettes PS, Forsbach-Birk V, Lynch D, Marre R. Overexpresssion of a Legionella pneumophila homologue of the E. coli regulator csrA affects cell size, flagellation, and pigmentation. Int J Med Microbiol. 2001;291:353–360. doi: 10.1078/1438-4221-00141. [DOI] [PubMed] [Google Scholar]
- 53.Bouchard F, Veilleux J, De Blois C, Murray G, Joly JR. Nosocomial Legionnaires’ disease in the Quebec City area. Can Med Assoc J. 1985;132:159–160. [PMC free article] [PubMed] [Google Scholar]
- 54.Bédard E, et al. Combination of Heat Shock and Enhanced Thermal Regime to Control the Growth of a Persistent Legionella pneumophila Strain. Pathogens. 2016;5:35. doi: 10.3390/pathogens5020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farhat M, et al. Effects of Disinfection on Legionella spp., Eukarya, and Biofilms in a Hot Water System. Appl Environ Microbiol. 2012;78:6850–6858. doi: 10.1128/AEM.00831-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plouffe JF, Webster LR, Hackman B. Relationship between colonization of hospital building with Legionella pneumophila and hot water temperatures. Appl Environ Microbiol. 1983;46:769–770. doi: 10.1128/aem.46.3.769-770.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammer BK, Swanson MS. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol Microbiol. 1999;33:721–731. doi: 10.1046/j.1365-2958.1999.01519.x. [DOI] [PubMed] [Google Scholar]
- 58.Byrne B, Swanson MS. Expression of Legionella pneumophila virulence traits in response to growth conditions. Infect Immun. 1998;66:3029–3034. doi: 10.1128/iai.66.7.3029-3034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dalebroux ZD, Edwards RL, Swanson MS. SpoT governs Legionella pneumophila differentiation in host macrophages. Mol Microbiol. 2009;71:640–658. doi: 10.1111/j.1365-2958.2008.06555.x. [DOI] [PubMed] [Google Scholar]
- 60.Dalebroux ZD, Swanson M. S. ppGpp: magic beyond RNA polymerase. Nat Rev Micro. 2012;10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 61.Molofsky AB, Swanson MS. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol Microbiol. 2004;53:29–40. doi: 10.1111/j.1365-2958.2004.04129.x. [DOI] [PubMed] [Google Scholar]
- 62.Reimmann C, Valverde C, Kay E, Haas D. Posttranscriptional Repression of GacS/GacA-Controlled Genes by the RNA-Binding Protein RsmE Acting Together with RsmA in the Biocontrol Strain Pseudomonas fluorescens CHA0. J Bacteriol. 2005;187:276–285. doi: 10.1128/JB.187.1.276-285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valverde C, Heeb S, Keel C, Haas D. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol Microbiol. 2003;50:1361–1379. doi: 10.1046/j.1365-2958.2003.03774.x. [DOI] [PubMed] [Google Scholar]
- 64.Kay E, et al. Two GacA-Dependent Small RNAs Modulate the Quorum-Sensing Response in Pseudomonas aeruginosa. J Bacteriol. 2006;188:6026–6033. doi: 10.1128/JB.00409-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hovel-Miner G, et al. σS Controls Multiple Pathways Associated with Intracellular Multiplication of Legionella pneumophila. J Bacteriol. 2009;191:2461–2473. doi: 10.1128/JB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rasis M, Segal G. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol Microbiol. 2009;72:995–1010. doi: 10.1111/j.1365-2958.2009.06705.x. [DOI] [PubMed] [Google Scholar]
- 67.Bruggemann H, et al. Virulence strategies for infecting phagocytes deduced from in vivo transcriptional program of Legionella pneumophila. Cell Microbiol. 2006;8:1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- 68.Hayashi T, et al. Proteomic Analysis of Growth Phase-Dependent Expression of Legionella pneumophila Proteins Which Involves Regulation of Bacterial Virulence Traits. PLoS One. 2010;5:e11718. doi: 10.1371/journal.pone.0011718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bachman MA, Swanson MS. The LetE Protein Enhances Expression of Multiple LetA/LetS-Dependent Transmission Traits by Legionella pneumophila. Infect Immun. 2004;72:3284–3293. doi: 10.1128/IAI.72.6.3284-3293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gal-Mor O, Segal G. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb Pathog. 2003;34:187–194. doi: 10.1016/S0882-4010(03)00027-5. [DOI] [PubMed] [Google Scholar]
- 71.Chatfield CH, Cianciotto NP. The Secreted Pyomelanin Pigment of Legionella pneumophila Confers Ferric Reductase Activity. Infect Immun. 2007;75:4062–4070. doi: 10.1128/IAI.00489-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Flydal MI, et al. Phenylalanine Hydroxylase from Legionella pneumophila Is a Thermostable Enzyme with a Major Functional Role in Pyomelanin Synthesis. PLoS One. 2012;7:e46209. doi: 10.1371/journal.pone.0046209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cianciotto NP. An update on iron acquisition by Legionella pneumophila: new pathways for siderophore uptake and ferric iron reduction. Future Microbiol. 2015;10:841–851. doi: 10.2217/fmb.15.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beier, D. & Gross, R. In Bacterial Signal Transduction: Networks and Drug Targets (ed. Ryutaro Utsumi) 149–160 (Springer New York, 2008). [PubMed]
- 75.Edwards RL, et al. Nicotinic Acid Modulates Legionella pneumophila Gene Expression and Induces Virulence Traits. Infect Imm. 2013;81:945–955. doi: 10.1128/IAI.00999-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Romeo T, Vakulskas CA, Babitzke P. Posttranscriptional regulation on a global scale: Form and function of Csr/Rsm systems. Environ Microbiol. 2013;15:313–324. doi: 10.1111/j.1462-2920.2012.02794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sahr T, et al. The Legionella pneumophila genome evolved to accommodate multiple regulatory mechanisms controlled by the CsrA-system. PLOS Gen. 2017;13:e1006629. doi: 10.1371/journal.pgen.1006629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sharma UK, Chatterji D. Transcriptional switching in Escherichia coli during stress and starvation by modulation of σ70 activity. FEMS Microbiol Rev. 2010;34:646–657. doi: 10.1111/j.1574-6976.2010.00223.x. [DOI] [PubMed] [Google Scholar]
- 79.Nevo O, Zusman T, Rasis M, Lifshitz Z, Segal G. Identification of Legionella pneumophila Effectors Regulated by the LetAS-RsmYZ-CsrA Regulatory Cascade, Many of Which Modulate Vesicular Trafficking. J Bacteriol. 2014;196:681–692. doi: 10.1128/JB.01175-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Albert-Weissenberger C, et al. Control of Flagellar Gene Regulation in Legionella pneumophila and Its Relation to Growth Phase. J Bacteriol. 2009;192:446–455. doi: 10.1128/JB.00610-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jacobi S, Schade R, Heuner K. Characterization of the Alternative Sigma Factor σ54 and the Transcriptional Regulator FleQ of Legionella pneumophila, Which Are Both Involved in the Regulation Cascade of Flagellar Gene Expression. J Bacteriol. 2004;186:2540–2547. doi: 10.1128/JB.186.9.2540-2547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yura T, Nagai H, Mori H. Regulation of the Heat-Shock Response in Bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]
- 83.Schumann W. Regulation of bacterial heat shock stimulons. Cell Stress Chaperones. 2016;21:959–968. doi: 10.1007/s12192-016-0727-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Steinchen W, Bange G. The magic dance of the alarmones (p)ppGpp. Mol Microbiol. 2016;101:531–544. doi: 10.1111/mmi.13412. [DOI] [PubMed] [Google Scholar]
- 85.Potrykus K, Cashel M. (p)ppGpp: still magical? Annu Rev Microbiol. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 86.Battesti A, Majdalani N, Gottesman S. The RpoS-Mediated General Stress Response in Escherichia coli. Annu Rev Microbiol. 2011;65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Edwards RL, Dalebroux ZD, Swanson MS. Legionella pneumophila couples fatty acid flux to microbial differentiation and virulence. Mol Microbiol. 2009;71:1190–1204. doi: 10.1111/j.1365-2958.2008.06593.x. [DOI] [PubMed] [Google Scholar]
- 88.Schell U, et al. The α-hydroxyketone LAI-1 regulates motility, Lqs-dependent phosphorylation signalling and gene expression of Legionella pneumophila. Mol Microbiol. 2016;99:778–793. doi: 10.1111/mmi.13265. [DOI] [PubMed] [Google Scholar]
- 89.Sadosky AB, Wiater LA, Shuman HA. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun. 1993;61:5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Feeley JC, et al. Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol. 1979;10:437–441. doi: 10.1128/jcm.10.4.437-441.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sexton JA, Vogel JP. Regulation of Hypercompetence in Legionella pneumophila. J Bacteriol. 2004;186:3814–3825. doi: 10.1128/JB.186.12.3814-3825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schindelin J, Rueden CT, Hiner MC, Eliceiri KW. The ImageJ ecosystem: An open platform for biomedical image analysis. Mo Reprod Dev. 2015;82:518–529. doi: 10.1002/mrd.22489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Faucher, S. & Shuman, H. A. In Legionella: Methods and Protocols (eds Carmen Buchrieser & Hubert Hilbi) 567–582 (Humana Press, 2013).
- 94.Krogh A, Larsson G, von Heijne G, Sonnhammer LL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 95.Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 2015;43:W401–W407. doi: 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hales LM, Shuman HA. The Legionella pneumophila rpoS Gene Is Required for Growth within Acanthamoeba castellanii. J Bacteriol. 1999;181:4879–4889. doi: 10.1128/jb.181.16.4879-4889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kovach ME, et al. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 98.Faucher S, Mueller CA, Shuman HA. Legionella pneumophila Transcriptome during Intracellular Multiplication in Human Macrophages. Front Microbiol. 2011;2:60. doi: 10.3389/fmicb.2011.00060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen J, et al. Legionella Effectors That Promote Nonlytic Release from Protozoa. Science. 2004;303:1358–1361. doi: 10.1126/science.1094226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.