Abstract
An Arabidopsis cDNA encoding the dihydrolipoamide S-acetyltransferase subunit of the plastid pyruvate dehydrogenase complex (E2) was isolated from a λPRL2 library. The cDNA is 1709 bp in length, with a continuous open reading frame of 1440 bp encoding a protein of 480 amino acids with a calculated molecular mass of 50,079 D. Southern analysis suggests that a single gene encodes plastid E2. The amino acid sequence has characteristic features of an acetyltransferase, namely, distinct lipoyl, subunit-binding, and catalytic domains, although it is unusual in having only a single lipoyl domain. The in vitro synthesized plastid E2 precursor protein has a relative molecular weight of 67,000 on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Upon incubation of the precursor with pea (Pisum sativum) chloroplasts, it was imported and processed to a mature-sized relative molecular weight of 60,000. The imported protein was located in the chloroplast stroma, associated with the endogenous pyruvate dehydrogenase. Catalytically active recombinant plastid E2 was purified as a glutathione S-transferase fusion protein. Analysis of plastid E2 mRNA by reverse transcriptase-polymerase chain reaction showed highest expression in flowers, followed by leaves, siliques, and roots. The results of immunoblot analysis indicate that protein expression was similar in roots and flowers, less similar in leaves, and even less similar in siliques. This is the first report, to our knowledge, describing a plastid E2.
The PDC contains multiple enzymes that, in concert, catalyze the oxidative decarboxylation of pyruvate, the transfer of an acetyl moiety to CoA, and the reduction of NAD+. These reactions occur sequentially on the E1 (EC 1.2.4.1), E2 (EC 2.3.1.12), and E3 (EC 1.8.1.4) component enzymes of the complex and support the overall reaction: pyruvate + CoASH + NAD+ → acetyl-CoA + NADH + CO2. Most mitochondrial PDCs also contain an E3BP as a component of the complex. These E3BPs are similar to E2 in that most contain lipoyl domains at the N terminus (Patel and Roche, 1990).
Plants are unique in that they possess two forms of PDC, one associated with mitochondria and one associated with plastids (for review, see Luethy et al., 1996). The mitochondrial form is important in controlling the entry of carbon into the citric acid cycle (Randall et al., 1996), whereas the plastid form provides acetyl-CoA and NADH for fatty acid biosynthesis (Camp and Randall, 1985). The component enzymes of the two complexes show amino acid homology (especially within defined domains), but it is clear that the complexes are different both enzymatically and antigenically (Camp and Randall, 1985; Miernyk et al., 1985; Luethy et al., 1995). Activity of the mitochondrial complex is controlled by the associated E1 kinase and phosphatase enzymes, whereas the plastid PDC is not (for review, see Luethy et al., 1996).
In eukaryotes and some bacteria, E2 forms the core of the multienzyme complex by associating 20 trimers into a pentagonal dodecahedron. The E1 and E3 enzymes bind to specific regions of E2 (Rahmatullah et al., 1989). The E2 subunit of mitochondrial PDC from mammalian sources can be divided into four distinct domains: two lipoyl domains, a subunit-binding domain, and an inner catalytic domain (Reed and Hackert, 1990; Perham, 1991). These domains occur sequentially from the N to the C termini of the protein. Until now, only one E2 subunit has been described from plants, an Arabidopsis mitochondrial isoform, which is similar to the human and bovine mitochondrial E2 subunits (Guan et al., 1995). The cloning of the plastid E2 subunit is described here. The genomic organization was examined by Southern analysis, and the expression of the gene was examined by RT-PCR and immunoblotting. The intracellular location of the protein translated from the cDNA was determined by in vitro chloroplast import assays. Integration of imported protein into a complex with the E1 enzyme was also examined using immunoprecipitation of imported E2 with antibodies raised to the E1α subunit of plastid PDC. Purified recombinant plastid E2 was catalytically active in acetyltransferase assays. To our knowledge, this is the first description of a plastid E2 sequence, which differs from the previously described mitochondrial isoform both in primary sequence and in the number of domains (i.e. it has a single lipoyl domain).
MATERIALS AND METHODS
cDNA Screening
Selection of Arabidopsis EST cDNA clones (Newman et al., 1994) was accomplished by searching the EST database using the BLASTP program of the National Center for Biotechnology Information. One EST cDNA clone (accession no. W43179) was found to have significant homology to a Synechocystis sp. sequence, which was designated as a dihydrolipoamide acetyltransferase subunit, making this a potential candidate for plastid E2. This partial cDNA clone was obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus) and used as a probe to screen an Arabidopsis λZIPLOX cDNA library (λPRL2, also from the Arabidopsis Biological Resource Center). Two rounds of screening were used to isolate independent candidate clones. A partial sequence was obtained for three candidate clones, and the longest of the three was sequenced completely.
Southern Analysis
Genomic DNA was prepared from young green leaves of Arabidopsis var Columbia using the sarkosyl method, digested with appropriate restriction enzymes, and Southern blotting was carried out as described by Ausubel et al. (1995).
Protein Expression, Purification, and Antibody Preparation
The catalytic domain of the plastid E2 was expressed in Escherichia coli using the pET28c expression vector (Novagen, Madison, WI). Antibodies were raised against the purified recombinant protein in New Zealand White rabbits, as described by Harlow and Lane (1988). Immunoblotting (Towbin et al., 1979) was used to evaluate the resulting antiserum.
Chloroplast Import
Chloroplasts were isolated from green pea (Pisum sativum L. var Little Marvel) seedlings grown for 10 d. Import was conducted as described previously (Bruce et al., 1994) with the following modifications: (a) the plastid E2 precursor protein was transcribed and translated from the cDNA using the T7 TNT quick-coupled system (Promega) incorporating [35S]Met (DuPont-NEN); (b) postimport protease treatment was conducted by adding thermolysin to 0.2 mg mL−1 (in import buffer containing 1 mm CaCl2) directly to the import reaction, which was then incubated on ice for 30 min; and (c) the chloroplasts were then washed twice with 1 mL of import buffer (50 mm Hepes-KOH, pH 8.0, and 300 mm sorbitol) supplemented with 2.5 mm each EDTA and EGTA.
Postimport fractionation of chloroplasts was conducted as described previously by Bruce et al. (1994). Immunoprecipitation of imported E2 was conducted as described previously by Harlow and Lane (1988) using antibodies to plastid E1α (Johnston, 1998).
Expression of Recombinant Plastid E2
The proposed mature portion of plastid E2 (from Ile-58) was expressed in E. coli using the pGEX-2T expression vector (Pharmacia Biotech) modified to include additional restriction enzyme sites. A PreScission protease site (Pharmacia Biotech) was included 5′ to the plastid E2-coding sequence to allow removal of the GST fusion protein. Recombinant plastid E2 protein was purified by glutathione-Sepharose 4B affinity chromatography, according to the manufacturer's instructions (Pharmacia Biotech). Polyclonal antibodies against plastid E2 and an anti-GST monoclonal antibody (Pharmacia Biotech) were used to identify the purified proteins.
Catalytic Activity
Purified plastid E2 was assayed for dihydrolipoamide acetyltransferase activity by quantitating the formation of a [14C]acetyl adduct of reduced dihydrolipoamide from [1-14C]acetyl-CoA, according to the method of Reid et al. (1977). Specific activities were calculated as micromoles of 14C product formed per minute per milligram of recombinant protein.
Expression Analyses
Total RNA was isolated from roots, leaves, flowers, and siliques of Arabidopsis by the method of Chomczynski and Sacchi (1987). RT-PCR was conducted according to the manufacturer's instructions using the Access RT-PCR system (Promega). Clarified protein lysates of the four organs examined were separated by SDS-PAGE, followed by immunoblotting (Towbin et al., 1979), using antibodies raised against recombinant plastid E2.
DNA Sequencing and Analysis
The cDNA clone was sequenced separately on each strand by a primer-walking approach. Automated Dye-Deoxy terminator cycle-sequencing using ABI 373 and ABI 377 instruments (ABI, Columbia, MD) was performed at the DNA core facility at the University of Missouri (Columbia). DNA and deduced amino acid sequences were compiled and analyzed using the Lasergene computer program (DNASTAR, Madison, WI). Sequence alignments and phylogenetic analyses were conducted using the GeneWorks computer program (IntelliGenetics, Mountain View, CA).
RESULTS AND DISCUSSION
cDNA Screening and Southern Analysis
Kaneko et al. (1996) recently reported the complete sequence of the genome of the unicellular cyanobacterium Synechocystis sp. This genome contains an open reading frame that was assigned as a dihydrolipoamide S-acetyltransferase subunit based on 21.3% identity with the rat E2 sequence, and this homology was predominantly in the lipoyl domain. This Synechocystis sp. sequence also exhibits 25% amino acid identity with the human E3BP. Based on this sequence an Arabidopsis EST clone (accession no. W43179) was identified as potentially encoding either a plastid PDC E2 or E3BP. This clone was used as a probe to screen an Arabidopsis λZIPLOX cDNA library. Thirteen positive plaques were obtained during primary screening of the cDNA library, and the results of Southern analysis suggested that three of these were potentially full-length clones (data not presented). One of these cDNA clones was sequenced completely on both strands using a primer-walking approach. The cDNA is 1709 bp in length, with a continuous open reading frame of 1440 bp encoding a protein of 480 amino acids, and it has a calculated molecular mass of 50,079 D.
The genomic organization of the plastid E2 was examined by Southern blotting. Genomic DNA from Arabidopsis leaves was digested with five restriction enzymes, and a Southern blot was prepared by hybridizing with the plastid E2 EST clone as a probe (Fig. 1). A single hybridizing band was present in all lanes. The smear beneath the band in the BamHI lane probably indicates that the DNA had degraded slightly during the overnight digestion with this enzyme. The hybridizing band in the HindIII lane was considerably smaller than the bands in the other lanes. HindIII cut internally in the cDNA (at nucleotide 638). The EST cDNA clone starts at position 781 of the plastid E2 sequence and, as a result, is specific for the C-terminal portion of the full-length clone. The absence of a second hybridizing band in the HindIII lane, therefore, is probably due to the probe not binding to the other fragment. The presence of a single hybridizing band in each lane is consistent with plastid E2 being encoded by a single gene in Arabidopsis.
Figure 1.
Southern analysis of genomic DNA from Arabidopsis. DNA (20 μg) was digested separately with the restriction enzymes indicated. The blot was probed with the plastid E2 EST cDNA clone, washed at high stringency, and exposed to film for 1 week at −70°C. The positions of DNA standards (λDNA digested with HindIII) are indicated on the left.
Chloroplast Import
Chloroplast transit peptides are characterized by an Ala following the initiating Met, an abundance of Ser and Thr, and few acidic residues (von Heijne et al., 1989). In addition, von Heijne et al. (1989) also identified a region in chloroplast transit peptides consisting of the last 10 residues before the cleavage site, which is rich in Arg residues, lacks Leu residues, and has a potential amphipathic β-strand structure. Using these criteria, we propose that the transit peptide of the plastid E2 consists of the first 57 amino acids and that cleavage of the transit peptide occurs between residues Glu-57 and Ile-58. The assignation of this cleavage site is based purely on amino acid sequence characteristics. However, because there is no consensus sequence for chloroplast transit-peptide cleavage, only microsequencing of the native protein will determine the true cleavage site. Based on this proposed cleavage site, the deduced amino acid sequence encoding the mature protein would have a molecular mass of 43.9 kD. When a plastid E2 chloroplast import reaction was subjected to size-exclusion chromatography using a Superose 12 column, a number of peaks were obtained. One of these peaks contained [35S]Met-labeled E2 with a Mr of 44,200, which would correspond to the E2 monomer. This is approximately the same as the molecular mass of 43.9 kD calculated by the primary sequence if cleavage of the transit peptide occurs between residues Glu-57 and Ile-58.
Because the N terminus of the deduced amino acid sequence of the isolated clone has the characteristics of a chloroplast transit peptide, plastid targeting was confirmed by in vitro import into isolated pea chloroplasts. The E2 clone was transcribed and translated from the original cloning vector (pZL1), which contains a T7 promoter sequence, in a rabbit reticulocyte lysate system. The lysate containing the radiolabeled plastid E2 protein was then incubated with freshly isolated pea seedling chloroplasts (Fig. 2A).
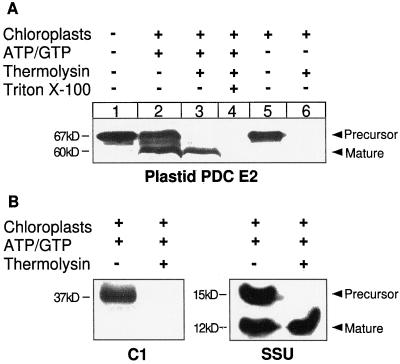
Figure 2.
Chloroplast import. A, Plastid E2 import. Pea chloroplasts were incubated at 25°C for 30 min with in vitro translated plastid E2 in the presence (+) or absence (−) of MgATP and NaGTP (3 mm each). After import, the chloroplasts were treated with thermolysin to digest any nonimported protein. The positions of the precursor and mature forms of plastid E2 are indicated. B, Control imports. The mitochondrial protein Cyt c1 (C1) and the plastid protein small subunit of Rubisco (SSU) were incubated with isolated chloroplasts in the presence of MgATP and NaGTP for 30 min, followed by treatment with thermolysin. The positions of precursor and mature forms are indicated.
The precursor form of plastid E2 (480 amino acids) was translated by the reticulocyte lysate as a single protein with an Mr of 67,000 on SDS-PAGE (Fig. 2A, lane 1). Although the calculated Mr of the deduced amino acid sequence for the precursor protein is 50,079, the apparent molecular mass on SDS-PAGE is always considerably higher than predicted; this size anomaly is seen with all E2s. The interdomain linker regions of E2 proteins are rich in turn-inducing and charged amino acid residues (Guest et al., 1989; Reed and Hackert, 1990; Perham, 1991), which contributes to these proteins migrating at a higher-than-predicted Mr on SDS-PAGE (Guest et al., 1985).
When the precursor was incubated with isolated chloroplasts, it was imported and processed to the mature form (Fig. 2A, lanes 2 and 3). Thermolysin treatment digests all proteins on the outside of chloroplasts but does not penetrate the envelope membranes (Bruce et al., 1994). Therefore, proteins resistant to thermolysin digestion were imported to a protected location within the chloroplasts. After thermolysin treatment mature E2 was the only form remaining (Fig. 2A, lane 3). The precursor form was completely digested. When the chloroplasts were lysed with Triton X-100, mature E2 was thermolysin sensitive (Fig. 2A, lane 4). In control experiments in which import was conducted in the absence of ATP (chloroplasts incubated on ice, in the dark, and without exogenous ATP), the precursor was not processed to the mature form (Fig. 2A, lane 5). Treatment of these control imports with thermolysin resulted in complete digestion of the precursor (Fig. 2A, lane 6); therefore, no import had occurred.
The in vitro binding of precursor proteins to chloroplasts requires low ATP levels, whereas translocation requires 1 mm ATP (for review, see Archer and Keegstra, 1990). The ATP-dependent import of the protein encoded by the cDNA verified that it encodes the plastid form of dihydrolipoamide S-acetyltransferase. Control incubations using the mitochondrial precursor protein Cyt c1 (Fig. 2B, C1) and the plastid small subunit of Rubisco (Fig. 2B, SSU) showed that only chloroplast proteins were imported, i.e. the chloroplast preparation was free of any mitochondrial contamination and the chloroplasts were competent for import.
Suborganellar Destination of Imported E2
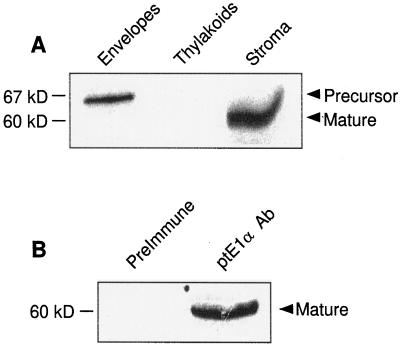
Plastid PDC activity is present in a 100,000g (stromal) fraction from lysed chloroplasts (Camp and Randall, 1985). Imported and processed E2 was localized in the stromal fraction of pea chloroplasts that were subfractionated after import. The precursor form of E2 was present in the envelope fraction (Fig. 3A, Envelopes). This represents E2 protein, which is probably associated with the import complex of the outer and/or inner envelopes. This protein was not exposed to the stromal protease, suggesting that the protein had not crossed the inner envelope. The thylakoid fraction contained no labeled protein (Fig. 3A, Thylakoids), and the stromal fraction contained mature E2 (Fig. 3A, Stroma).
Figure 3.
Localization and assembly of imported plastid E2. A, Localization of imported and processed plastid E2. After import, chloroplasts were reisolated and fractionated to obtain envelope, thylakoid membrane, and stromal fractions. The precursor and mature forms of E2 are indicated. B, Immunoprecipitation of imported plastid E2. After import, a stromal fraction was prepared and immunoprecipitated with preimmune serum or anti-plastid E1α antibodies.
When antibodies raised against recombinant plastid E1α (Johnston, 1998) were used to immunoprecipitate an E2 import reaction (chloroplast stromal fraction), only the band corresponding to the mature form of E2 was immunoprecipitated (Fig. 3B, ptE1α Ab). Preimmune serum did not precipitate any protein (Fig. 3B, Preimmune). That imported E2 was immunoprecipitated by antibodies to E1α provides good evidence that the imported E2 assembled into a complex with the endogenous E1 enzyme. Because E2 binding to the E1 enzyme is mediated by the E1β-subunit (Wynn et al., 1992), immunoprecipitation by antibodies to E1α suggests that imported E2 was integrated into the pea plastid PDC. Further work will be undertaken to determine whether imported E2 is capable of forming a high-molecular-weight complex composed of a number of E2 subunits, similar to the 60-subunit core seen in mammals, and whether a 60-mer E2 core forms before the binding of the E1 heterotetramer.
Purification and Catalytic Activity of Plastid E2
To examine the activity of recombinant plastid E2, the full-length protein was expressed in E. coli using a modified pGEX-2T vector. This allowed expression of plastid E2 as a GST fusion protein. Inclusion of the PreScission protease site introduced two additional amino acid residues (Gly and Pro) N terminal to Ile-58.
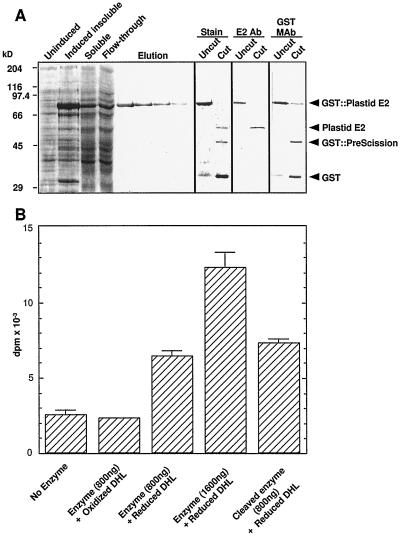
The recombinant protein was purified from a soluble extract of BL21 E. coli cells (Fig. 4A). Isopropylthio-β-galactoside induced expression of a protein with an Mr of 90,000 (Fig. 4A, GST::plastid E2). This protein was purified to homogeneity by glutathione-Sepharose chromatography (Fig. 4A, Elution). The PreScission protease cleaved >95% of the recombinant protein within 4 h (Fig. 4A, Stain, Cut). The apparent difference in abundance of the cleaved plastid E2 relative to the uncut E2 in the stained panel likely reflects a difference in affinity of the Coomassie Blue dye for these two proteins. The three bands in the cut sample correspond to the plastid E2 (60 kD), the PreScission protease (46 kD), and GST (29 kD). Antibodies against recombinant plastid E2 specifically recognized the upper band (Fig. 4A, E2 Ab), whereas the two other bands were immunodecorated by an anti-GST monoclonal antibody (Fig. 4A, GST MAb). The PreScission protease is a GST fusion protein, which accounts for the signal obtained with the GST monoclonal antibody.
Figure 4.
Expression and catalytic activity of plastid E2. A, Expression of plastid E2. Mature plastid E2 was expressed in E. coli. Isopropylthio-β-galactoside induced the expression of plastid E2 (GST::Plastid E2). Plastid E2 was eluted from a glutathione-Sepharose affinity column in four fractions (Elution). The purified protein was then digested with the PreScission protease. Uncut and cut samples were separated by SDS-PAGE in triplicate, followed by staining with Coomassie Brilliant Blue (Stain), or transfer to nitrocellulose and immunolabeling with plastid E2 antibody (E2 Ab) or GST monoclonal antibody (GST MAb). The products of digestion are indicated. B, Activity of purified recombinant plastid E2. Controls were no enzyme and enzyme plus oxidized dihydrolipoamide (DHL). Catalytic activity of recombinant GST::plastid E2 and cleaved enzyme was measured at the protein concentrations indicated in the presence of reduced dihydrolipoamide. Error bars indicate the sd of triplicate assays; absence of error bars indicates negligible sd.
Assays for dihydrolipoamide S-acetyltransferase activity established that the recombinant plastid E2 was catalytically active (Fig. 4B). When radiolabeled acetyl-CoA was incubated with recombinant plastid E2, there was rapid production of the 14C product. A no-enzyme control was included to account for any [14C]acetyl-CoA that partitioned into the benzene phase (Fig. 4B, column 1). Oxidized dihydrolipoamide (thioctic acid) was used as an additional control to ensure that bona-fide acetyltransferase activity was occurring (Fig. 4B, column 2). This confirmed that oxidized dihydrolipoamide was not a substrate for plastid E2. When enzyme and reduced dihydrolipoamide were used in reactions, there was a significant production of 14C product relative to controls (Fig. 4B, column 3). Doubling the enzyme concentration resulted in greater than double the activity above control levels (Fig. 4B, column 4). The enzyme activity data are expressed as disintegrations per minute × 10−3 purely to illustrate the control reactions. The specific activity of the recombinant GST::plastid E2 fusion protein was 3.89 μmol min−1 mg−1 E2. Reid et al. (1977) reported pea mitochondrial dihydrolipoamide S-acetyltransferase-specific activities of 0.084 in an acetone-powder fraction and 1.0 in a 136,000g PDC enzyme pellet. Compared with these values, the recombinant plastid E2 enzyme had a much higher specific activity. After the GST moiety was removed by PreScission protease cleavage, the free plastid E2 (Fig. 4B, column 5) had a higher specific activity of 4.09 μmol min−1 mg−1. This specific activity was similar to that of recombinant human E2 (4.79 μmol min−1 mg−1) reported by Yang et al. (1997). The acetyltransferase activity of the recombinant protein supports our conclusion that the isolated cDNA encodes a dihydrolipoamide S-acetyltransferase and not an E3BP. In addition, that recombinant E2 has a high specific activity suggests that the proposed mature start site of Ile-58 is close enough to the native start site not to interfere with the correct folding and hence the catalytic activity of the protein.
Expression Analyses of Plastid E2
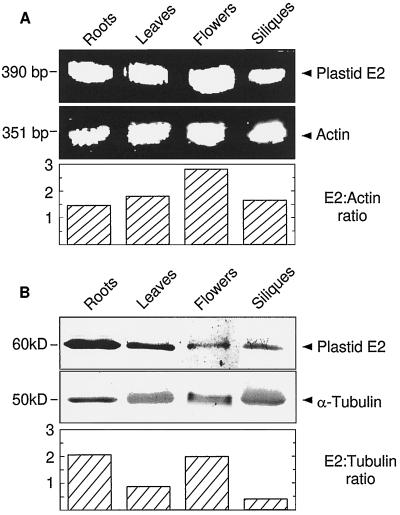
RT-PCR was used to examine the expression pattern of mRNA for plastid E2. The relative levels of plastid E2 mRNA were examined in roots, green leaves, flowers, and siliques of Arabidopsis. Two sequence-specific primers were used to yield an amplification product of 390 bp.
The plastid E2 mRNA was present in the four organs examined in RT-PCR (Fig. 5A). The expression levels differed slightly among the four organs examined. Expression was lowest in roots, 10% higher in siliques, 16% higher in leaves, and 86% higher in flowers (Fig. 5A, compare ratios of E2 to actin). Actin (ACT1 gene) was used as a control for basal expression level. Actin was expressed similarly in all four organs. When RNA was used as the template in PCR (without RT), no products were observed (data not presented). Therefore, the RNA was free of contaminating genomic DNA and the products amplified by RT-PCR represent mRNA transcript levels in the four organs. However, although an equal amount of total RNA from each organ was used, it is possible that individual RT-PCR reactions were not completely linear during the number of cycles used.
Figure 5.
RT-PCR and immunoblot analyses of the expression pattern of plastid E2. A, RT-PCR yielded a 390-bp product from the plastid E2 mRNA. A 351-bp actin fragment was the control. The total optical density of each band was determined using the QS30 program (PDI, Huntington Station, NY) and the ratio of E2 to actin was calculated. B, Immunoblotting, using antibodies raised against the catalytic domain of plastid E2, was used to examine plastid E2 protein expression from various organs. A monoclonal antibody against α-tubulin was used as a basal expression control. The graph of the ratio of E2 to tubulin was produced by scanning blots and determination of the optical density of individual bands as described above. Each lane contained 10 μg of protein.
The relative expression of the plastid E2 protein was also examined using antibodies raised to recombinant plastid E2 (Fig. 5B). As with the mRNA, the plastid E2 protein was present in all organs studied; however, when normalized to α-tubulin, a differential expression pattern was observed. The highest E2 protein expression was seen in roots and flowers, followed by leaves at about 40% of that of roots and flowers, and siliques at about 20% of that of roots and flowers. The asynchronous expression patterns between RT-PCR and immunoblot data could reflect organ-specific posttranscriptional control. In summary, the plastid E2 gene was expressed in all of the organs studied, with minor variations in the abundance of mRNA and protein between organs.
Comparison of the Arabidopsis Plastid E2 Amino Acid Sequence with Those of Other PDC E2 Proteins
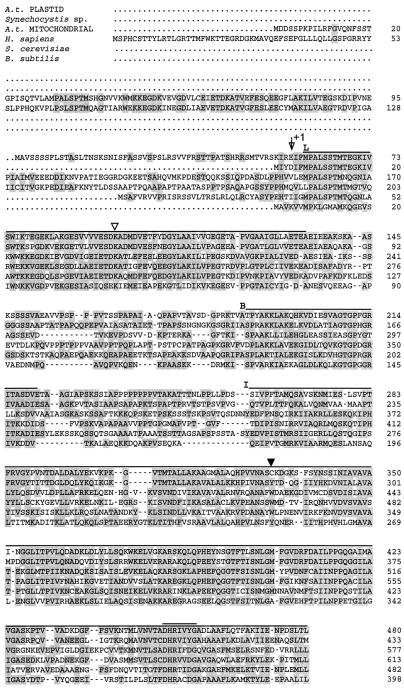
Alignment of the deduced amino acid sequence of the isolated clone with acetyltransferase subunits from Synechocystis sp., Arabidopsis (mitochondrial), Homo sapiens, Saccharomyces cerevisiae, and Bacillus subtilis PDCs shows areas of conserved residues (Fig. 6; shading indicates amino acid identity of at least two sequences). There are three distinct regions of homology: the lipoyl (Fig. 6, L, overlined), subunit-binding (Fig. 6, B, overlined), and inner catalytic (Fig. 6, I, overlined) domains. The plastid sequence has one lipoyl domain, as do the Synechocystis sp., S. cerevisiae, and B. subtilis sequences. The Arabidopsis mitochondrial and H. sapiens PDC E2 sequences both contain two lipoyl domains. The lack of a second lipoyl domain in the plastid sequence does not mean that the cDNA is not full length. There are no amino acid sequence motifs 5′ to the coding region, which suggest the presence of an additional lipoyl domain. The presence of a single lipoyl domain is sufficient to support catalytic function of the α-ketoglutarate dehydrogenase complex and the branched-chain α-keto acid dehydrogenase complexes (for review, see Reed and Hackert, 1990). The E. coli acetyltransferase subunit has three lipoyl domains, but deletion of one or two of these domains did not significantly affect the activity (Guest et al., 1985). Therefore, there may be some redundancy with the extra lipoyl domains (Reed and Hackert, 1990).
Figure 6.
Sequence comparison of six E2 subunits. The first sequence shown is the Arabidopsis plastid PDC E2 sequence (A. t. PLASTID) reported here. Shading of amino acid residues denotes identity between at least two sequences. Hyphens denote alignment gaps in the sequences. Three conserved domains are denoted by their corresponding letters and are overlined after their designation. L, Lipoyl domain; B, subunit-binding domain; I, inner catalytic domain. The “arrow +1” indicates the proposed mature start of the Arabidopsis plastid protein. ▿, Position of the conserved Lys involved in lipoic acid binding. ▾, The only Cys residue in the plastid E2 sequence. The conserved motif of the active site of acetyltransferases is double overlined. The accession numbers of the sequences are as follows: Synechocystis sp., D90915; Arabidopsis (mitochondrial), Z46230; H. sapiens, J03866; S. cerevisiae, J04096; and B. subtilis, M57435.
There are a number of conserved motifs in the lipoyl domain in which amino acid residues are identical in at least five of the six sequences, most strikingly the MPALSxTM-67, ExDKA-97, and GYLAxI-112 motifs (residue numbers are based on the Arabidopsis plastid E2 amino acid sequence described here). The amino acid residue denoted as x corresponds to nonidentity between the conserved residues in the plastid and Synechocystis sp. sequences and the conserved residue in at least three of the four other sequences, e.g. GYLA(A/K)I-112. The lipoyl domain also contains a Lys conserved in all six sequences, part of the ExDKA motif, (Fig. 6, ▿), which is likely the binding site of the lipoic acid moiety (Reed, 1974; Russell and Guest, 1991; Dardell et al., 1993).
The only Cys residue in the plastid E2 sequence is at position 331 (Fig. 6, ▾), close to the center of the catalytic domain. This position is occupied by a conserved Trp residue in the mitochondrial sequences. There is evidence that Cys-alkylating agents, such as N-ethylmaleimide, cause a pronounced inhibition of the mitochondrial PDC but have no effect on the plastid PDC activity (Johnston, 1998). The paucity of Cys residues in the plastid E2 sequence, compared with, for example, the four Cys residues in Arabidopsis mitochondrial E2 (Guan et al., 1995), might contribute to the observed differences. It suggests further that Cys-331 is not exposed or is not involved in catalysis.
The catalytic domain of Arabidopsis plastid E2 contains an amino acid sequence motif similar to the conserved DHRxxDG-457 motif (Fig. 6, double overlined) characteristic of the active site of acetyltransferases (Guest, 1987). However, in the plastid E2 and Synechocystis sp. E2 sequences, the second Asp is replaced by a Tyr. This is a striking difference because all other reported E2 sequences have the second conserved Asp, and site-directed mutagenesis of this Asp to Ala in S. cerevisiae inactivated the enzyme (Reed and Hackert, 1990). The difference in the plastid E2 sequence might simply reflect further divergence between plastid and mitochondrial sequences. The Arabidopsis plastid E1α active-site sequence also diverges greatly from other eukaryotic PDC E1α sequences in one respect. A highly conserved Cys residue proposed to be an essential component of the active site is replaced by a Val residue (Johnston et al., 1997). This demonstrates further the divergence of plastid and mitochondrial PDC subunits. Whether the amino acid substitution in the active site of the plastid E2 contributes to a difference in catalytic properties of the plastid and mitochondrial E2 subunits remains to be determined.
Phylogenetic Analysis of PDC E2 Sequences
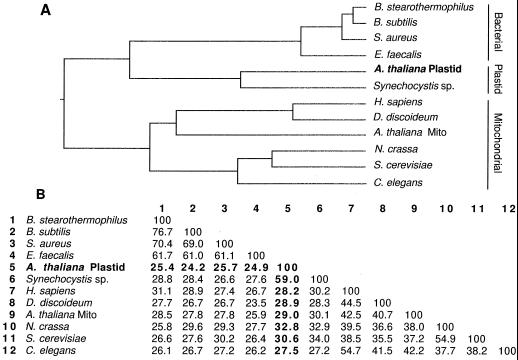
Twelve PDC E2 sequences, obtained using the BLASTP program, were subjected to phylogenetic analysis (Fig. 7A). The dendrogram shows that the plastid PDC E2 sequence (Fig. 7A, boldface type) is most closely related to the proposed Synechocystis sp. E2 and is more distantly related to other eubacterial PDC E2 sequences. It is least related to the mitochondrial sequences, including the only other published plant sequence, Arabidopsis. The relatedness of the Arabidopsis plastid and Synechocystis sp. sequences adds to the evidence that cyanobacteria may be similar to the progenitors of chloroplasts (Giovannoni et al., 1988; Gray, 1989; Sugita et al., 1997).
Figure 7.
Comparison of amino acid sequences of 12 E2s. A, The length of each branch of the dendrogram is proportional to sequence identity. The cDNA clone described here is in boldface (A. thaliana Plastid). The sequences segregate into three groups denoted on the right as Bacterial, Plastid, and Mitochondrial. Mito, Mitochondria. B, Amino acid identity matrix for the sequences in A. The identities of the plastid sequence with the other E2 sequences are in boldface. GenBank accession numbers for the sequences not included in Figure 6 are as follows: Bacillus stearothermophilus, X53560; Staphylococcus aureus, X58434; Dictyostelium discoideum, U06634; Neurospora crassa, J04432; and Caenorhabditus elegans, Z77659. The Enterococcus faecalis sequence can be accessed in the PIR database, accession no. S16989.
Figure 7B shows an amino acid identity matrix for the sequences compared in the dendrogram. The overall amino acid identity between the Arabidopsis plastid sequence and the Synechocystis sp. sequence is 59%, whereas the Arabidopsis plastid and mitochondrial sequences have an overall amino acid identity of 29%. The Synechocystis sp. sequence had previously been designated as a dihydrolipoamide S-acetyltransferase based on, at best, 30% identity with various mitochondrial E2 sequences. Its 59% identity with the plastid E2 protein confirms that it is a dihydrolipoamide S-acetyltransferase.
The cDNA clone isolated and described here is very different from all reported mitochondrial sequences except within defined domains. The characteristic plastid transit peptide, the import competence of the in vitro synthesized protein, and the acetyltransferase activity obtained with the recombinant protein support our conclusion that the cDNA encodes a plastid-targeted PDC E2 protein.
ACKNOWLEDGMENTS
We thank Professor Kenneth Keegstra for the gift of the cDNA encoding the small subunit of Rubisco and Professor Matthew A. Harmey for the cDNA encoding Cyt c1. We also thank Nancy David and Sheri Neff for technical assistance and Dr. Patricia Coello for assistance with Southern blotting.
Abbreviations:
- E1
pyruvate dehydrogenase (consisting of α- and β-subunits) of the PDC
- E2
dihydrolipoamide S-acetyltransferase of the PDC
- E3
dihydrolipoamide dehydrogenase
- E3BP
E3-binding protein
- EST
expressed sequence tag
- GST
glutathione S-transferase
- PDC
pyruvate dehydrogenase complex
- RT
reverse transcriptase
The accession number for the sequence reported in this article is AF066079.
Footnotes
This research was supported by National Science Foundation grant no. IBN 94-19489. This is journal report no. 12,808 from the Missouri Agricultural Experiment Station.
LITERATURE CITED
- Archer EK, Keegstra K. Current views on chloroplast protein import and hypotheses on the origin of the transport mechanism. J Bioenerg Biomembr. 1990;22:789–810. doi: 10.1007/BF00786931. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology, Ed 3. Boston, MA: John Wiley & Sons; 1995. [Google Scholar]
- Bruce BD, Perry S, Froehlich J, Keegstra K. In vitro import into chloroplasts. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual, Ed 2, J1. Boston, MA: Kluwer Academic Publishers; 1994. pp. 1–15. [Google Scholar]
- Camp PJ, Randall DD. Purification and characterization of the pea chloroplast pyruvate dehydrogenase complex. A source of acetyl-CoA and NADH for fatty acid biosynthesis. Plant Physiol. 1985;77:571–577. doi: 10.1104/pp.77.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Dardel F, Davis AL, Laue ED, Perham RN. Three-dimensional structure of the lipoyl domain from Bacillus stearothermophilus pyruvate dehydrogenase complex. J Mol Biol. 1993;229:1037–1048. doi: 10.1006/jmbi.1993.1103. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Turner S, Olsen GJ, Barns S, Lane DJ, Pace NR. Evolutionary relationships among cyanobacteria and green chloroplasts. J Bacteriol. 1988;170:3584–3592. doi: 10.1128/jb.170.8.3584-3592.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. The evolutionary origins of organelles. Trends Genet. 1989;5:294–299. doi: 10.1016/0168-9525(89)90111-x. [DOI] [PubMed] [Google Scholar]
- Guan Y, Rawsthorne S, Scofield G, Shaw P, Doonan J. Cloning and characterization of a dihydrolipoamide acetyltransferase (E2) subunit of the pyruvate dehydrogenase complex from Arabidopsis thaliana. J Biol Chem. 1995;270:5412–5417. doi: 10.1074/jbc.270.10.5412. [DOI] [PubMed] [Google Scholar]
- Guest JR. Functional implications of structural homologies between chloramphenicol acetyltransferase and dihydrolipoamide acetyltransferase. FEMS Microbiol Lett. 1987;44:417–422. [Google Scholar]
- Guest JR, Angier SJ, Russell GC. Structure, expression and protein engineering in the pyruvate dehydrogenase complex of Escherichia coli. Ann NY Acad Sci. 1989;573:76–99. doi: 10.1111/j.1749-6632.1989.tb14988.x. [DOI] [PubMed] [Google Scholar]
- Guest JR, Lewis HM, Graham LD, Packman LC, Perham RN. Genetic reconstruction and functional analysis of the repeating lipoyl domains in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. J Mol Biol. 1985;185:743–754. doi: 10.1016/0022-2836(85)90059-2. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Johnston ML (1998) Molecular and biochemical characterization of the plastid pyruvate dehydrogenase subunits from Arabidopsis thaliana. MS thesis, University of Missouri, Columbia
- Johnston ML, Luethy MH, Miernyk JA, Randall DD. Cloning and molecular analyses of the Arabidopsis thaliana plastid pyruvate dehydrogenase subunits. Biochim Biophys Acta. 1997;1321:200–206. doi: 10.1016/s0005-2728(97)00059-5. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S and others. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- Luethy MH, David NR, Elthon TE, Miernyk JA, Randall DD. Characterization of a monoclonal antibody recognizing the E1α subunit of the plant mitochondrial pyruvate dehydrogenase. J Plant Physiol. 1995;145:443–449. [Google Scholar]
- Luethy MH, Miernyk JA, David NR, Randall DD. Plant pyruvate dehydrogenase complexes. In: Patel MS, Roche TE, Harris RA, editors. Alpha-Keto Acid Dehydrogenase Complexes. Basel, Switzerland: Birkhauser Verlag; 1996. pp. 71–99. [Google Scholar]
- Miernyk JA, Camp PJ, Randall DD. Regulation of plant pyruvate dehydrogenase complexes. Curr Top Plant Biochem Physiol. 1985;4:175–190. [Google Scholar]
- Newman T, de Bruijn RJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikel N, Somerville S, Thomashow M and others. Genes galore. A summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MS, Roche TE. Molecular biology and biochemistry of pyruvate dehydrogenase complexes. FASEB J. 1990;4:3224–3233. doi: 10.1096/fasebj.4.14.2227213. [DOI] [PubMed] [Google Scholar]
- Perham RN. Domains, motifs and linkers in the 2-oxo acid dehydrogenase multienzyme complexes: a paradigm in the design of a multifunctional protein. Biochemistry. 1991;30:8501–8512. doi: 10.1021/bi00099a001. [DOI] [PubMed] [Google Scholar]
- Rahmattullah M, Gopalakrishnan S, Radke GA, Roche TE. Domain structures of the dihydrolipoamide transacetylase and the protein X components of mammalian pyruvate dehydrogenase complex. J Biol Chem. 1989;264:1245–1251. [PubMed] [Google Scholar]
- Randall DD, Miernyk JA, David NR, Gemel J, Luethy MH. Regulation of leaf mitochondrial pyruvate dehydrogenase complex activity by reversible phosphorylation. In: Shewry PR, Halford NG, editors. Protein Phosphorylation in Plants. Oxford, UK: Clarendon Press; 1996. pp. 87–103. [Google Scholar]
- Reed LJ. Multienzyme complexes. Acc Chem Res. 1974;7:40–46. [Google Scholar]
- Reed LJ, Hackert ML. Structure function relationships in dihydrolipoamide acetyltransferase. J Biol Chem. 1990;265:8971–8974. [PubMed] [Google Scholar]
- Reid EE, Thompson P, Lyttle CR, Dennis DT. Pyruvate dehydrogenase complex from higher plant mitochondria and proplastids. Plant Physiol. 1977;59:842–848. doi: 10.1104/pp.59.5.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell GC, Guest JR. Sequence similarity within the family of dihydrolipoamide acyltransferases and discovery of a previously unidentified fungal enzyme. Biochim Biophys Acta. 1991;1076:225–232. doi: 10.1016/0167-4838(91)90271-z. [DOI] [PubMed] [Google Scholar]
- Sugita M, Sugishita H, Fujishiro T, Tsuboi M, Sugita C, Endo T, Sugiura M. Organization of a large gene cluster encoding ribosomal proteins in the cyanobacterium Synechococcus sp. strain PCC 6301: comparison of gene clusters among cyanobacteria, eubacteria and chloroplast genomes. Gene. 1997;195:73–79. doi: 10.1016/s0378-1119(97)00169-8. [DOI] [PubMed] [Google Scholar]
- Towbin H, Straehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G, Steppuhn J, Herrmann RG. Domain structure of mitochondrial and chloroplast targeting peptides. Eur J Biochem. 1989;180:535–545. doi: 10.1111/j.1432-1033.1989.tb14679.x. [DOI] [PubMed] [Google Scholar]
- Wynn RM, Chuang JL, Davie JR, Fisher CW, Hale MA, Cox RP, Chuang DT. Cloning and expression in Escherichia coli of mature E1β subunit of bovine mitochondrial branched-chain α-keto acid dehydrogenase complex. J Biol Chem. 1992;267:1881–1887. [PubMed] [Google Scholar]
- Yang D, Song J, Wagenknecht T, Roche TE. Assembly and full functionality of recombinantly expressed dihydrolipoyl acetyltransferase component of the human pyruvate dehydrogenase complex. J Biol Chem. 1997;272:6361–6369. doi: 10.1074/jbc.272.10.6361. [DOI] [PubMed] [Google Scholar]