Abstract
Heart valves are organized connective tissues of high mechanical demand. They open and close over 100,000 times a day to preserve unidirectional blood flow by maintaining structure-function relationships throughout life. In affected individuals, structural failure compromises function and often leads to regurgitant blood flow and progressive heart failure. This is most common in degenerative valve disease due to age-related wear and tear, or congenital malformations. At present, the only effective treatment of valve disease is surgical repair or replacement and this is often impermanent and requires anti-coagulation therapy throughout life. Therefore, there is a critical need to discover new alternatives. A promising therapeutic area is tissue regeneration and in non-valvular tissues this requires a tightly regulated genetic “growth program” involving cell proliferation. To explore this in heart valves, we performed RNA-seq analysis to compare transcriptional profiles of aortic valve tissue isolated from mice during stages of growth (postnatal day (PND) 2) and adult maintenance (4 months). Data analysis reveals distinct mRNA profiles at each time point and pathway ontology identifies associated changes in biological functions. The PND2 aortic valve is characterized by extensive cell proliferation and expression of mRNAs related to the extracellular matrix (ECM). At 4 months, proliferation is not significant and a differential set of ECM-related genes are expressed. Interestingly there is enrichment of the defense response biological process at this later time point. Together, these data highlight the unique transcriptome of the postnatal valve during stages of growth and maturation, as well as biological functions associated with adult homeostatic valves. These studies create a platform for future work exploring the molecular programs altered in the onset of heart valve disease after birth and provide insights for the development of mechanistic-based therapies.
Keywords: aortic valve, RNA-sequencing, mRNA, cell proliferation, extracellular matrix, postnatal, adult
Introduction
The average heart beats over a billion times during one lifespan to continuously provide blood to every part of the body. Crucial to this task are the four heart valves (aortic, pulmonic, tricuspid and mitral) that function to maintain the unidirectional blood flow. Distinct from the cardiac muscle, the mature valve leaflets are highly organized structures comprised of three layers of extracellular matrix (ECM) components including collagens, proteoglycans, and elastin (1). Formation and maintenance of the valve ECM is mediated by a heterogeneous population of valve interstitial cells (VICs) that are fibroblast-like in phenotype (2). Surrounding the VICs and ECM is a single layer of valve endothelial cells (VECs) that physically protect the valve from external stimuli, and molecularly communicate with underlying VICs to regulate homeostasis of the ECM (3–5). The complex relationship between valve cell populations and the ECM is critical for establishing and maintaining structure-function relationships throughout life. This relationship begins during embryonic development, as mesenchymal precursor cells in the endocardial cushions transition towards an activated VIC phenotype and degrade primitive ECM within the cushions while secreting more diverse ECM components. Elongation and remodeling of the immature valve structures continues for a short time during the postnatal period until around day 10 in the mouse when the ECM components are more defined. Once valve formation is complete, VICs convert to a quiescent phenotype and in the absence of disease, maintain physiological turnover of the ECM to provide efficient function throughout life [reviewed (2, 6)]. While the regulation of valve development is well established, the mechanisms that regulate postnatal valve growth and remodeling, as well as adult homeostasis are poorly understood. Despite constant mechanical demand on the valve leaflets, turnover of valve cell populations in adult valves is relatively low (4). Therefore, it remains unclear how valve cell populations and structure-function relationships are maintained throughout life in healthy individuals, yet dysregulation of these relationships likely underlie the onset and progression of valve dysfunction and disease.
Heart valve disease is a growing public health problem that can affect both adult and pediatric patients. Significant defects during embryonic valve development lead to congenital malformations which compromise the typical structure of the valve, often resulting in reduced ability to function correctly [reviewed (7)]. Distinct from valve disease present at birth, pathology can also be acquired and is most prevalent in the aging population, with up to 13% of people aged over 75 affected by diseases including calcification or myxomatous degeneration (8). Currently, the only effective treatment for valve disease is surgical repair or replacement, resulting in over 90,000 valve replacement surgeries performed in the US each year (9). Surgical treatment comes with many complications including the need for repeat surgeries due to low valve durability and high thrombogenicity, in addition to the large personal and societal economic burdens (10). Therefore, there is a critical need for the development of alternate therapeutics.
A promising therapeutic area is emerging in the field of self-repair and regenerative medicine. Common to both congenital and age-related valve disease is the damage and consequent loss of healthy cell populations alongside the development of pathological cell populations that are therefore unable to preserve valve structure-function relationships (11). The field of cardiac regeneration has recently made significant advances in elucidating the molecular mechanisms of regeneration, and it has been reported that the neonatal myocardium has remarkable regenerative capacity during the first seven days of life (12–14). Furthermore, several pathways have been identified as key players, and the ability to recapitulate these neonatal programs in adults has proven successful in promoting myocardial regeneration after injury and in disease models (15–18).
Neonatal, adult and potential regenerative programs have not been examined in heart valves and therefore the goal of this current study is to initiate this discovery. To do this, we used RNA-seq analysis to explore differential molecular profiles between postnatal and adult valve cell populations. This analysis will help define potential regeneration indicators that in the future might be reintroduced in diseased or aging adult valves to increase their self-repair capacity and improve structure-function relationships. Our study has defined transcriptional differences between postnatal day 2 (PND2) and adult (4 months) aortic valves and identified significant changes in key biological functions related to cell proliferation, ECM, and defense response that may be important for determining the regenerative capacity of the valve to aid in the future development of alternative therapeutics.
Material and Methods
Mice
C57BL/6J mice were fed regular chow mix and housed in a controlled environment with 12 h light/dark cycles at 21°C and 23% humidity and water ad libitum. Animals were euthanized by CO2 exposure followed by secondary euthanasia by cervical dislocation (adult mice) or decapitation (pups). All animal procedures were approved by The Research Institute at Nationwide Children’s Hospital Institutional Animal Care and Use Committee (Protocol # AR13-00054).
Tissue Preparation
Hearts were collected from postnatal day 2 (PND2) and 4 month old C57BL/6J mice and fixed in 4% paraformaldehyde/1xPBS overnight at 4°C. For paraffin sections, tissue was embedded in paraffin wax and sectioned at 10 µm. Paraffin was removed in xylene, and tissue sections were re-hydrated through a graded ethanol series and rinsed in 1xPBS as previously described (19). Tissue sections containing aortic valves were then subjected to Movat’s Pentachrome staining, EdU staining, or immunohistochemistry/immunofluorescence (described below). For cryo sections, tissue was embedded in OCT and frozen, then sectioned at 7 µm. Prior to staining, tissue was permeabilized using 0.1% Triton-X 100 in 1xPBS and then subjected to immunofluorescence staining.
Immunohistochemistry/Immunofluorescence
Whole hearts from PND2 and 4 month old C57BL/6J mice were collected and prepared according to above methods. Movat’s Pentachrome staining was performed on paraffin tissue sections at each time point according to the manufacturer’s instructions (Russel Movat, American MasterTech, #KTRMP), then mounted using VectaMount Permanent Mounting Medium (Vector Laboratories, H-5000). For antibody detection, fixed paraffin tissue sections were subjected to antigen retrieval by boiling for 10 min in unmasking solution (Vector Laboratories), and both cryo and paraffin sections were subjected to blocking for 1 h at room temperature (1% BSA, 1% cold water fish skin gelatin, 0.1% Tween-20/PBS) as described (20). Tissue sections were then incubated overnight at 4°C or 1 h at room temperature with primary antibodies against Mmp3 (rabbit, 1:100 paraffin, Abcam ab53015), Nid2 (rabbit, 1:200 cryo, Abcam ab14513), Ptgs2 (Rabbit, 1:100 paraffin, Cell Signaling 12282), and Rarres2 (Mouse, 1:100 paraffin, Santa Cruz sc-373797). For immunofluorescent primary antibody detection of Mmp3, Nid2, and Ptgs2, sections were incubated for 1 h at room temperature with Donkey anti-rabbit or Goat anti-rabbit Alexa-Fluor IgG secondary antibodies (1:500) (LifeTechnologies), then mounted in Vectashield anti-fade medium with DAPI (Vector Laboratories) to detect cell nuclei. For diaminobenzidine (DAB) staining of Rarres2, sections were stained using Mouse and Rabbit Specific HRP/DAB (ABC) Detection IHC kit (Abcam, ab64264), counterstained with hematoxylin (Vector Laboratories, H-3404), and mounted using VectaMount Permanent mounting medium (Vector Laboratories, H-5000). Images were visualized using an Olympus BX51 microscope and captured using an Olympus DP71 camera and CellSens software. Image brightness and contrast were edited using Adobe Photoshop CC.
EdU Staining and Quantification
PND2 and 4 month old C57BL/6J mice were injected subcutaneously with 10 µg/g body weight EdU (Invitrogen) dissolved in 1xPBS. 24 h later, mice were sacrificed and hearts were collected and prepared according to above methods. Fixed tissue sections were blocked for 1 h at room temperature (1% BSA, 0.1% Cold water fish skin gelatin, 0.1% Tween 20 in PBS with 0.05% NaN3), followed by use of Click-it EdU Kit (Invitrogen) to detect presence of EdU according to the manufacturer’s instructions. Sections were then mounted in Vectashield anti-fade medium with DAPI (Vector Laboratories) to detect cell nuclei. The total number of cell nuclei in one leaflet were counted using ImageJ cell counter. The number of EdU + cells were then counted and calculated as a percentage of total cells. An average of 9 leaflets were counted and averaged for each mouse, with a total n = 3. Statistical analysis was performed in GraphPad Prism 7.0a.
Aortic Valve Isolation and RNA-Sequencing
Aortic valves from wild type PND2 and 4 month old C57BL/6J mice were isolated with minimal myocardial contamination and immediately flash frozen in liquid nitrogen. Frozen samples were sent to Ocean Ridge Biosciences LLC (Deerfield Beach, FL), where RNA isolation and sequencing was performed as follows. Total RNA was extracted using the TRI Reagent® (Molecular Research Center; Part #: TR118) method, and isolated RNA was quantified using chip-based capillary electrophoresis (Agilent 2100 Bioanalyzer Pico Chip). RNA was digested with RNase free DNase I (Epicentre; Part # D9905K) and purified through minElute columns (Qiagen; Part #: 74204). Final RNA samples were quantified by O.D. measurement and re-quantified using chip-based capillary electrophoresis. Amplified cDNA libraries were prepared from 200 ng on DNA-free total RNA using TruSeq Stranded Total mRNA Library Prep Kit LT (Illumina Inc.; Part #s: RS-122-2101 and RS-122-2102). Chip-based capillary electrophoresis was used to assess quality and size distribution of the libraries. KAPA Library Quantification Kit (Kapa Biosystems, Boston, MA) was used to quantify the libraries. Libraries were pooled at equimolar concentrations and were clustered on an illumina cBot cluster station. Clustering was performed with the HiSeq PE cluster kit v4 and sequenced on an Illumina HiSeq Flow Cell v4 with 50 nt paired-end reads plus dual index reads using the Illumina HiSeq SBS Kit v4. An average of approximately 48.3 million passed-filter 50 nucleotide paired-end reads were obtained per sample (24.1M per direction).
Raw FASTQs were split into files containing 4,000,000 reads and checked for quality using the FASTX-Toolkit. The reads were filtered (removing sequences that did not pass Illumina’s quality filter) and trimmed based on the quality results (3 nucleotides at the left end of the R1 reads and 1 nt at the left end of the R2 reads). Sequence alignment was performed using TopHat v2.1.0 to the mm10 genome. BAM files were merged on a per sample. Exon and gene level counting were performed using the easyRNASeq version 2.4.7 package. A binary annotation file, built using the annotation file generation function of EasyRNASeq, was used for this analysis; the Ensembl release 83 GTF file was used as input. Annotation was performed using a Gene Transfer Format (GTF) annotation file for Mus musculus, which was downloaded on February 11, 2016 and contains the current Ensembl Mouse release 83. Filtering of the RPKM values was performed to retain a list of genes with a minimum of approximately 50 mapped reads in 25% or more samples. The threshold of 50 mapped reads is considered the Reliable Quantification Threshold, as the RPKM values for a gene represented by 50 reads should be reproducible in technical replicates. To avoid reporting large fold changes due to random variation of counts from low abundance mRNA, RPKM values equivalent to a count of ≤10 reads per gene were replaced with the average RPKM value equivalent to 10 reads/gene across all the samples in the experiment.
An unpaired two-sample heteroscedastic t-test was performed on the log2 RPKM values to compare the overall effects of age (PND2 or 4 month) on gene expression. Fold changes were also calculated for 4 month / PND2 using the mean of each group being compared. If the mean of both groups considered in the fold change comparison was below RQT, “NA” is reported. All statistical analysis was performed using R version 3.2.2 statistical computing software. A total of 7,496 genes were determined to have a low FDR-value (FDR <0.1) for the unpaired t-test. Full dataset is available through NCBI GeoDatasets, accession code GSE108083, “RNA-seq analysis of aortic heart valves in mice”.
RNA-Sequencing Data Analysis
A heatmap was generated from 23,303 differentially expressed genes. Log2 transformed RPKM values were utilized and hierarchical clustering analysis was performed with Cluster 3.0 software (21). Genes and samples were clustered using centered correlation as the similarity measure and average linkage as the clustering method. A volcano plot was generated utilizing ggplot2 and is plotted as the -Log10(p-value) vs. Log2 Fold Change. The volcano plot highlights the differential gene expression between postnatal day 2 and 4 month aortic valves. A Venn diagram was generated based only on genes with a low t-test p value (p < 0.05), a fold change >2, and RPKM values above the Reliable Quantification Threshold for all biological replicate samples from either group. If at least one of the gene reads from a triplicate set was proven undetectable while all gene reads in the comparative sample set was proven detectable, the gene was considered to be uniquely expressed. If the gene read from both triplicate sample sets had detectable RPKM values about the Detection Threshold, the gene was considered common amongst sample groups. Genes with at least one triplicate below the Detection Threshold in both samples sets are not represented in the Venn diagram.
Functional annotation was performed through the utilization of Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6.8 (22). 2,082 differentially expressed genes with an FDR <0.05 and fold change >2, were assessed utilizing Gene Ontology (GO) FAT terms, which were employed to filter out broad GO categories based on a measured specificity of each term. Visualization of GO term analysis was performed using the GOPlot R package version 1.0.2 (23). To reduce the redundancy of GO terms, the reduce overlap function was used, with the threshold set to 0.75, which removes GO terms that have a gene overlap greater than or equal to the set threshold. Bubble plots were generated for the reduced GO term list using the GoBubble function, and the top 15 GO terms from biological processes, cellular component, and molecular function were visualized. Each bubble represents a term, where the size of the bubble correlates to the number of genes within the term, and it is plotted as –log (FDR) vs. z-score. The z-score is a crude measurement, predicting if a term will be upregulated or downregulated, and is calculated by taking the number of upregulated genes and subtracting the number of downregulated genes and then dividing this number by the square root of the number of genes in each pathway. The circle plot was generated using the GoCircle function, and highlights the gene expression changes within each of the selected terms. The circle plot highlights the overall gene expression change by showing increased expression in red and decreased expression in blue. The circle plot also highlights the p-value of the GO term by the height of the inner rectangle, which is also colored by z-score. A chord plot was generated using the GoChord function, and it represents 59 differentially expressed genes and their correlation to the following associated terms: extracellular matrix, cell proliferation, cell cycle, mitotic cell cycle process, defense response, and regulation of immune system processes. The chord plot also highlights the log fold change of each differentially expressed gene that is shown.
qRT-PCR
RNA was extracted from isolated aortic valves from PND2 and 4 month old C57BL/6J mice to validate RNA-seq findings. Briefly, Trizol reagent (Invitrogen) was used to extract RNA according to manufacturer’s instructions, and cDNA and PCR reactions were performed as previously described by our lab (24). Primers for genes selected for validation were designed in NCBI Primer-BLAST based on FASTA sequence and shown below:
| Gene name | Forward primer sequence (5′ to 3′) | Reverse primer sequence (5′ to 3′) |
|---|---|---|
| Nid2 | AGGAGTGAGCATGTTTCGG | AGGGGTATTGCCAGCTTCAC |
| Mmp3 | TGCATGACAGTGCAAGGGAT | ACACCACACCTGGGCTTATG |
| Marckls1 | CCCGTGAACGGAACAGATGA | CCCACCCTCCTTCCGATTTC |
| Gsn | GGGACGGCCGGTTACTTAAA | CTTCAGGAATTCGGGGTGCT |
| Filip1l | AGGCTCCACTGCTGGATTTC | GACTTCTCTGACACGGGACG |
| Myoc | ACGACACTAAAACGGGGACC | TTCTGGCCTTTGCTGGTAGG |
| Retnla | GGAACTTCTTGCCAATCCAGC | CAGTGGTCCAGTCAACGAGT |
| Npdc | GCACTCCCGACACTTTTCTC | GGTACCCACTCCGGGAACT |
| Sfrp4 | CCTGGCAACATACCTGAGCA | AGCATCATCCTTGAACGCCA |
| Mki67 | AGAGCTAACTTGCGCTGACT | ACTCCTTCCAAACAGGCAGG |
| Nrg1 | CCATCTCTCGATGGGCTTCC | ATGCAGAGGCAGAGGCTTAC |
| Nrep | GCATGATGCCCTTTTTCATCCA | TCCTTAGGCACGGGAAGTCT |
| Acta2 | CCTTCGTGACTACTGCCGAG | GAAGGTAGACAGCGAAGCCA |
| Dlk1 | AGAGTACCCCTCTCCTCACC | CGCCGCTGTTATACTGCAAC |
| Cfd | TACATGGCTTCCGTGCAAGT | GGGTGAGGCACTACACTCTG |
Quantitative real-time PCR using a Step One Plus Real Time PCR system (Applied Biosystems) was used to detect changes in gene expression with Sybr Green reagents. Cycle counts for each target gene were normalized to β-actin expression and differences in gene expression were reported as a fold change from the 4 month time point. Statistical analysis was performed in GraphPad Prism 7.0a.
Results
Postnatal Valve Maturation and Adult Maintenance Are Associated with Distinct Transcriptional Profiles
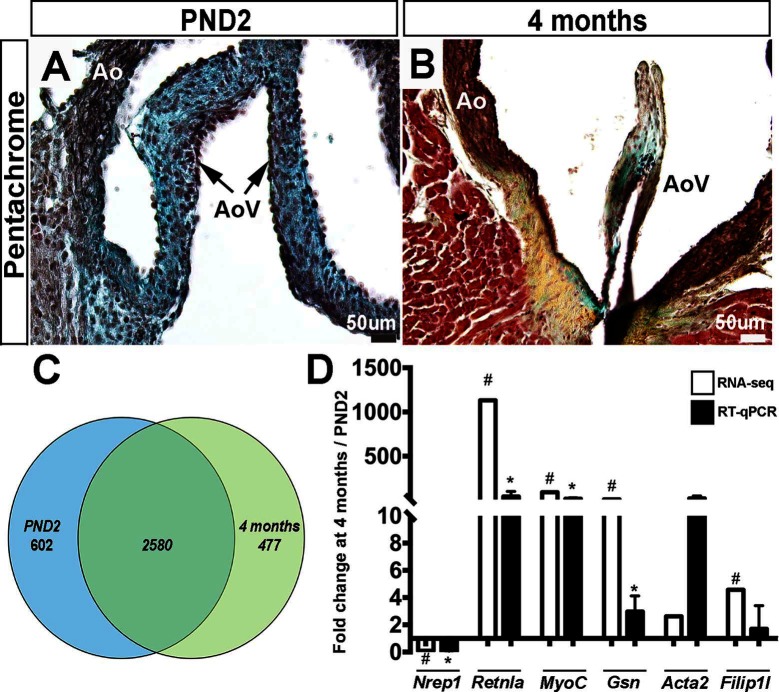
As previously described, the valve structures continue to grow and remodel after birth (1). As shown by Movat’s Pentachrome stain, murine aortic valve structures at postnatal day 2 (PND2) are thick and composed of predominantly proteoglycan (blue), with less extensive collagen and elastin (Figure 1A). By 4 months of age, the leaflets have elongated and display distinct layers of collagen (fibrosa, yellow), proteoglycan (blue), and elastin (black) (Figure 1B).
Figure 1.
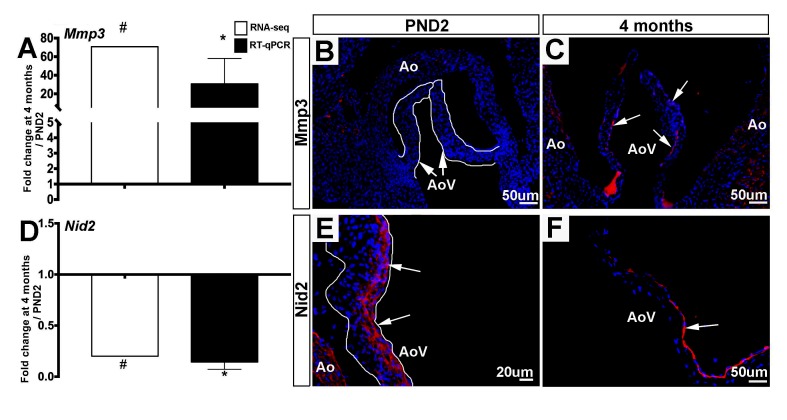
Postnatal and adult aortic vales display differences in ECM composition and gene expression. Movat’s Pentachrome staining to show extracellular matrix composition at PND2 (A) and 4 months (B) (black: elastic fibers and cell nuclei, blue: proteoglycans, yellow: collagen, red: muscle and fibrinoids). (C) Venn diagram to show distribution of the detected mRNAs that were uniquely or commonly expressed at PND2 or 4 months. (D) RT-qPCR validation (black bars) of RNA-seq findings (white bars) (n = 3, *:p < 0.05; two-tailed unpaired t-test, #:FDR <0.05). Ao, aorta; AoV, aortic valve.
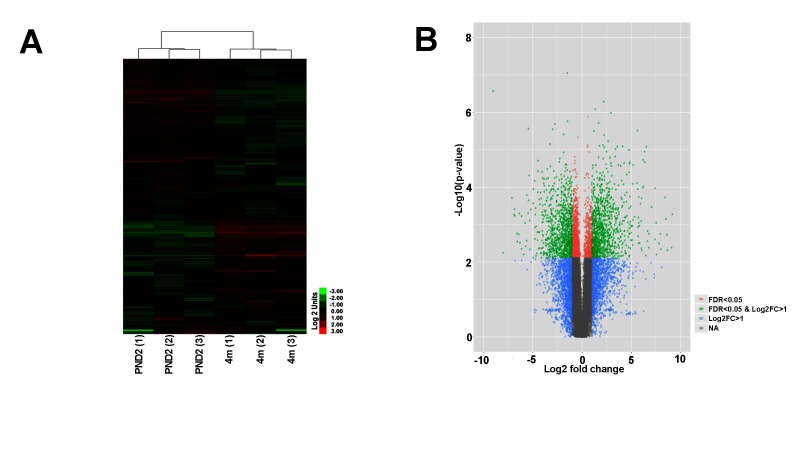
In order to further define molecular profiles associated with the structural changes in postnatal and 4 month old aortic valves, we performed RNA-sequencing on isolated samples. Overall, RNA expression for samples consistently clustered by time point, as shown several ways including a Pearson’s correlation matrix (data not shown), principal component analysis (PCA) (data not shown) and hierarchical heatmap (Figure 2A). Of 23,303 detectable genes, 3,659 genes were found to have a p-value < 0.05 and a fold change >2, and include 1,858 upregulated and 1,801 downregulated transcripts. Of these 3,659 differentially expressed genes, 602 were unique to the PND2 time point and include Dlk1, Hif3a, Agtr2 and S100A9, while 477 were only expressed at 4 months (Cfd, Rtn1a, Clec3a, Adipoq, etc.), leaving 2,580 common to both groups (Figure 1C). Table 1 includes the top 20 mRNAs uniquely expressed at each time point based on RPKM value, which is indicative of mRNA abundance. Additional RT-qPCR analysis of independent cDNA samples validated RNA-seq findings in 10 out of 12 genes (85%) at a significance threshold of p < 0.05 (Figures 1D, 4A and 5A,D and data not shown).
Figure 2.
RNA-seq analysis reveals transcriptional differences between PND2 and 4 month old murine aortic valves. (A) Hierarchical heatmap cluster shows clustering analysis of biological replicates (n = 3). (B) Volcano plot of differentially expressed genes sorted according to fold change and significance (FDR). Green dots represent genes with log2 fold change >1 and FDR <0.05, pink dots represent genes with FDR < 0.05, and blue dots represent genes with log2 fold change >1.
Table 1.
Top 20 unique genes at PND2 and 4 month time points.
| PND2 | 4 Month |
| Dlk1 | Cfd |
| Hif3a | 2210407C18Rik |
| Agtr2 | Retnla |
| S100a9 | Clec3a |
| Slc38a5 | Adipoq |
| Col24a1 | Ces1d |
| Bmp7 | Thrsp |
| Vash2 | Pck1 |
| Igf2bp3 | Mgl2 |
| Stfa1 | C7 |
| S100a8 | Inmt |
| Cited1 | Cidec |
| Gm5483 | Fmo3 |
| Frem2 | Angpt4 |
| Gipr | Hamp |
| Ube2c | Art1 |
| Dctd | Tmem45b |
| 1110032F04Rik | Olfr224 |
| C1qtnf3 | Plin1 |
| Cdkn3 | Rpl3l |
Genes are listed in order of RPKM with the most highly expressed at the top of the list. Only genes with a low t-test p value (t-test P: Age < 0.05), a Fold Change: 4mo/P2 >2 up/down, and above the Reliable Quantification Threshold in all samples from either group (i.e., the gene RPKM values were >RQT in all three P2 samples or in all three 4mo samples) were retained for further filtering.
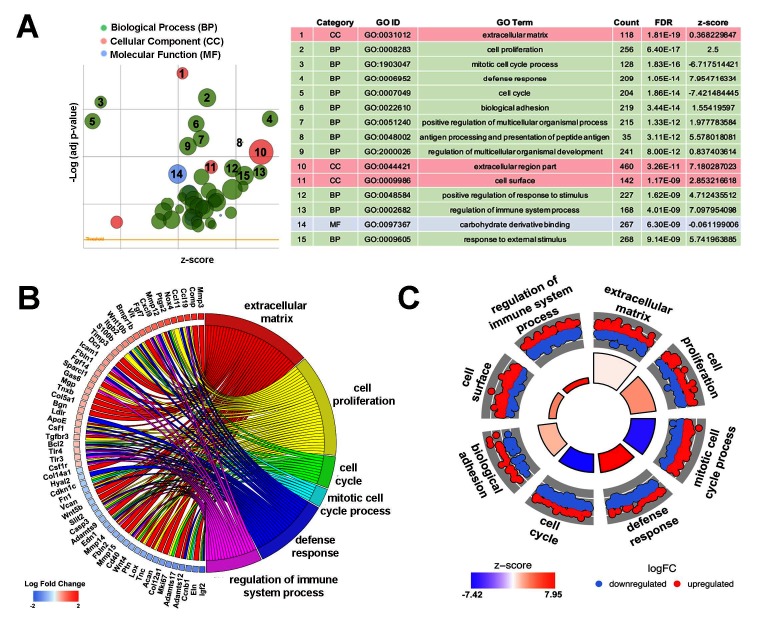
Figure 3.
Pathway analysis identifies diverse key biological processes, cellular components and molecular functions between PND2 and 4 month old murine aortic valves. (A) Gene ontology (GO) analysis reveals top differentially regulated GO terms, graphically displayed according to significance (p-value) and z-score, a measure of overall up or down regulation for the category. Data represented as a bubble plot (left). The size of each circle represents the number of differentially expressed genes in that GO term, while color represents the category (green: biological process pink: cellular component, blue: molecular function). Table (right) lists the top GO terms along with count, FDR, and z-score. (B) Chord plot showing 59 differentially expressed genes previously associated with healthy and diseased valves, and their overlap between significant GO terms as determined in (A). Color below each gene corresponds to log fold change, with red representing increased expression at the 4 month time point and blue representing decreased expression at 4 months. (C) Circular plot highlighting gene expression differences within each selected GO term, with each red dot depicting a gene upregulated at 4 months in that category, and each blue dot showing a gene downregulated at 4 months. The height of the inner rectangle represents the p-value of the GO term and is colored according to z-score, with red being increased at the 4 month time point and blue being decreased at 4 months.
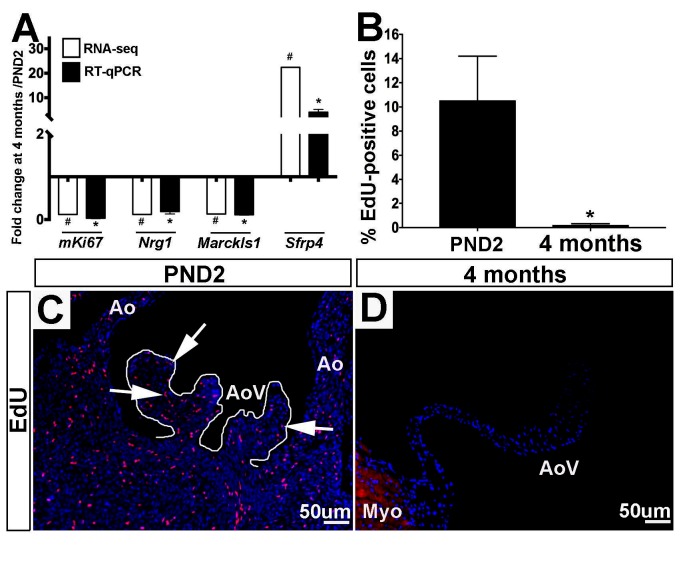
Figure 4.
Positive regulators of cell proliferation are enriched at PND2 in murine aortic heart valves. (A) RT-qPCR performed on independent samples (black bars) validates the RNA-seq fold change (white bars) trends for proliferation-related genes. Expression is normalized to 1, indicated by the x-axis. (n=3, *:p<0.05; two-tailed unpaired t-test, #:FDR<0.05). (B) Quantification of 5-ethynyl-2′-deoxyuridine (EdU) positive cells as a percentage of total cell nuclei (indicated by DAPI, blue) at PND2 and 4 month time points. (n=3, *:p<0.05, two-tailed unpaired t-test). (C) Representative images of staining for incorporation of EdU into replicating DNA (white arrows point to EdU-positive cells) reveals high levels of proliferation in PND2 AoV as compared to (D) EdU staining in 4 month old aortic valve. Ao, aorta; AoV, aortic valve; Myo, myocardium.
Figure 5.
ECM molecular profiles between PND2 and 4 month old murine aortic valves are distinct. (A) RT-qPCR performed on independent samples (black bars) validates the trends seen in RNA-seq data (white bars). Expression is normalized to 1, indicated by the x-axis (n=3, *:p<0.05; two-tailed unpaired t-test, #:FDR<0.05). (B) Immunofluorescence staining for Mmp3 reveals low expression at PND2 compared to (C) 4 months (white arrows indicate Mmp3-positive cells). (D) RT-qPCR performed on independent samples (black bars) validates the trends seen in RNA-seq data (white bars) for ECM gene Nid2. Expression is normalized to 1, indicated by the x-axis (n=3, *:p<0.05; two-tailed unpaired t-test, #:FDR<0.05). (E) Immunofluorescence staining for Nid2 reveals high expression atPND2 (white arrows indicate Nid2-positive cells) compared to (F) Nid2 staining at the 4 month timepoint. Ao, aorta; AoV, aortic valve.
Transcriptional Analysis Identifies Age-Dependent Transcriptional Profiles and Biological Functions
Heatmap hierarchical clustering analysis, where 23,300 differentially expressed genes and samples were clustered using center correlation as the similarity measurement and average linkage as the clustering method, revealed molecular similarities between biological replicates at each time point and distinct differences between PND2 and 4 months (Figure 2A). Additional volcano plot analysis graphically displays the differential expression of 23,300 individual transcripts based on significance and fold change (Figure 2B). To determine functions associated with differential gene expression changes at each time point, Gene Ontology (GO) pathway analysis was performed. The bubble plot in Figure 3A visualizes the biological processes, cellular components, and molecular functions enriched by the differential data set and the table highlights the top 15 GO terms represented. These include biological processes such as cell proliferation, mitotic cell cycle, and defense response, along with cellular components such as extracellular matrix (ECM), indicating that valve maturation involves considerable changes in cell proliferation, ECM composition, and immune system programs. This is further highlighted in Figure 3B circle plot displaying genes which are known to be expressed in the heart valves based on previous publications, and their association with each GO term. More specific trends in these GO terms are shown in Figure 3C, with individually upregulated and downregulated genes in each category shown as red and blue dots, respectively. The inner rectangles are sized to positively correlate with the significance of each GO term, and colored to represent the overall direction of change in expression of each individual term. For example, the term “mitotic cell cycle process” has an overall down regulation at 4 months of age, while the “defense response” has an overall upregulation. In contrast, the “extracellular matrix” GO term is overall neither up-, nor downregulated, but the change in many individual transcripts is significant. Together these genomic analyses have defined transcriptional profiles of PND2 and 4 month aortic valve structures, and identified changes in functions associated with these mRNA patterns.
Proliferation Programs Are Downregulated in 4 month Old Aortic Valves
Based on enrichment of cell proliferation-related genes from GO analysis (Table 2), we first validated the fold change trends observed by RNA-seq using RT-qPCR (Figure 4A) on independent biological samples. These validated genes include three positive regulators of proliferation mKi67, Nrg1 and Marcksl1, which all decreased in 4 month old aortic valves, and an anti-proliferative gene, Sfrp4, found to have increased expression at this time point (Figure 4A) (25–28). To further validate mRNA findings, we utilized 5-ethynyl-2′-deoxyuridine (EdU) to visualize and compare the number of cells actively undergoing mitosis in the aortic valve at PND2 and 4 months of age (Figure 4B-D). At the earlier time point, ~10.5% of cells were found to be EdU-positive, while only 0.16% of cells were proliferating at 4 months (Figure 4B). These data are consistent with transcriptional changes and previous reports from our lab showing that ~6.2% of VECs (of the total VEC population) and ~3.3% of VICs (of the total VIC population) are proliferative at post natal stages, while in the young adult, proliferation rates of both VECs (~2%) and VICs (~1.1%) were significantly lower (4). Together, these observations suggest that a decline in cell division at 4 months is due to a combination of decreasing expression of postnatal proliferation programs while simultaneously increasing adult programs which inhibit proliferation.
Table 2.
Genes included in the “Cell Proliferation” GO term.
| Downregulated at 4 months | Upregulated at 4 months |
| AGTR2 | ADIPOQ |
| H19 | HSPA1A |
| BMP7 | CD74 |
| VASH2 | PLA2G2D |
| IGF2 | RBP4 |
| IGF2BP1 | H2-AA |
| FAM83D | H2-AB1 |
| CTHRC1 | VSIG4 |
| CRH | CCL19 |
| CDK1 | CRLF1 |
| NRK | CCL11 |
| BEX1 | NOX4 |
| BUB1 | SFRP4 |
| IL31RA | CD209A |
| MELK | PTGFR |
| SCUBE2 | BCL6 |
| CCNB1 | ITGAX |
| FIGNL1 | KCNA1 |
| AURKB | LGI4 |
| UHRF1 | ATF3 |
| AGER | PTGS2 |
| CDCA7L | AR |
| SHCBP1 | ESR1 |
| WISP1 | CFB |
| NRG1 | CEBPA |
| MKI67 | APOD |
| HELLS | MMP12 |
| ASPM | IL2RA |
| MCM10 | RUNX3 |
| IQGAP3 | NR4A1 |
| KIF20B | HSF4 |
| MARCKSL1 | CD274 |
| BIRC5 | ALDH3A1 |
| RACGAP1 | IFIT3 |
| ROR2 | FGF7 |
| CHEK1 | IL7R |
| TNC | CLEC11A |
| CDH3 | WFDC1 |
| SFRP2 | AGAP2 |
| E2F8 | FLT3L |
| E2F7 | FOLR2 |
| SOX4 | LIMS2 |
| CDC20 | GAPT |
| CENPF | SERPINF1 |
| PTN | NR1D1 |
| WNT4 | FGF16 |
| IRF6 | BMPR1B |
| F2RL1 | SERPINE2 |
| FOXM1 | IGHD |
| HMGA2 | F3 |
| MYCN | WNT10B |
| CD40 | TRPV2 |
| CDC6 | CD28 |
| MMP14 | MLXIPL |
| TACC3 | SIX1 |
| EDN1 | CPEB1 |
| HMGB2 | CYR61 |
| SMARCA1 | PTGIR |
| LAMC2 | ECM1 |
| GPC3 | THPO |
| PROX1 | CX3CL1 |
| VASH1 | ITGB2 |
| CD276 | ABCB1A |
| RNASEH2B | THRB |
| NASP | H2-M3 |
| BLM | TOB2 |
| ORC1 | CHD5 |
| NKX2-5 | MUSTN1 |
| CDK2 | SAMD9L |
| TFRC | FTH1 |
| LOXL2 | PODN |
| CXADR | PTPRC |
| CASP3 | FCGR2B |
| SLIT2 | S100B |
| CAV3 | FOSL2 |
| DBN1 | ATF5 |
| EFNB2 | COL18A1 |
| FN1 | BTG2 |
| CDH5 | PTPRU |
| SMYD2 | COL4A3 |
| TNFRSF13C | IL15 |
| CDKN1C | NOV |
| DOT1L | FBLN1 |
| RPS6KA2 | JUN |
| SNAI2 | PTAFR |
| MEG3 | ADRB2 |
| RIAN | SPN |
| RRM1 | HAVCR2 |
| HAS2 | IL33 |
| GM13275 | RORA |
| DDR1 | LRP1 |
| ATPIF1 | NTN1 |
| GATA6 | ITGAM |
| ERBB2 | LEFTY1 |
| MCM7 | PID1 |
| DISC1 | GAS6 |
| SCARB1 | CST3 |
| CTPS | EGR3 |
| PTPRK | NAMPT |
| PTPRF | PDGFD |
| EDNRA | CD37 |
| CD24A | CD9 |
| SLC25A5 | SLC11A1 |
| TEK | CNN1 |
| TRIM35 | MVP |
| CNOT6 | ANGPT1 |
| KDM5B | H2-T23 |
| PICALM | DOCK8 |
| TSPO | |
| CD86 | |
| NACC2 | |
| SPHK2 | |
| HCLS1 | |
| IRS2 | |
| ZFP36 | |
| CDKN1A | |
| TRF | |
| SLFN1 | |
| IGFBP4 | |
| RHBDD1 | |
| DPT | |
| TNS2 | |
| DOCK2 | |
| NCF1 | |
| PIK3CB | |
| APOE | |
| PAWR | |
| CD46 | |
| CSF1 | |
| IRF1 | |
| CNTFR | |
| TACC2 | |
| TGFBR3 | |
| NUPR1 | |
| BCL2 | |
| VIPR2 | |
| PTTG1 | |
| TLR4 | |
| RUNX2 | |
| HYAL1 | |
| IGFBP5 | |
| CSF1R | |
| NCOA3 | |
| PAK1 | |
| PDE5A | |
| BRIP1 | |
| CORO1A | |
| IFITM3 |
Genes are sorted according to fold change with the highest fold change at the top of the list.
Postnatal and Adult Aortic Valves Have Distinct Extracellular Matrix mRNA Programs
As indicated by the GO term z-score in Figure 3A, the overall expression of ECM transcripts does not significantly increase or decrease with age, yet there are considerable differences in the specific ECM-related mRNAs that are expressed between the two time points (Table 3). Matrix metalloproteinases, or Mmps, are known to be expressed in both healthy and diseased valves and are associated with physiological and pathological remodeling of the ECM respectively (29–31). Also known as stromelysin-1, Mmp3 targets degradation of proteoglycans, collagens, and elastins (32) and this Mmp family member has been described in cancer (33, 34), but little is known about Mmp3 in mouse valves. In this study, Mmp3 increased from 1.22 reads per million kilobases (RPKM) at PND2 to 86.14 RPKM at 4 months. This significant increase was confirmed by RT-qPCR (Figure 5A,B) and immunofluorescence of aortic valve tissue sections, where Mmp3 is localized primarily to the sub-endothelial region of the leaflet (Figure 5B, Figure S1). In our previous VEC RNA-seq study, Mmp3 transcript was not detected in the VEC population at any time point (4), and this is consistent with predominant expression of the protein in VICs; largely those close to the sub-endothelial location at 4 months (arrows, Figure 5C, Figure S1). In this study we show that the basement membrane ECM protein Nidogen 2 (Nid2) was more highly detected in PND2 samples at 76 RPKM, while only 15 RPKM were detected at 4 months. This expression pattern was confirmed by RT-qPCR (Figure 5D) and immunofluorescence data shows broad Nid2 localization in both VECs and VICs at PND2, but more localized within the endothelial cell layer at 4 months (Figure 5E,F, Figure S1), and this pattern of decreased expression with maturation is consistent at the RNA level in VECs as previously described (4). These data show that PND2 and 4 month old aortic valves have distinct ECM-related transcriptional profiles associated with growth and maintenance respectively.
Table 3.
Genes included in the “Extracellular Matrix” GO term.
| Downregulated at 4 months | Upregulated at 4 months |
| S100A9 | MYOC |
| COL24A1 | MMP3 |
| BMP7 | GLDN |
| 2010005H15RIK | CHAD |
| CTHRC1 | MMP10 |
| FREM2 | COMP |
| COL26A1 | ENTPD2 |
| ELN | LAMC3 |
| COL9A1 | CILP2 |
| FREM1 | SOD3 |
| ADAMTS12 | FBLN7 |
| FRAS1 | NPNT |
| MFAP2 | PRELP |
| ADAMTS17 | MMP12 |
| WISP1 | COL4A6 |
| HMCN1 | VIT |
| LAMA1 | CCBE1 |
| COL12A1 | CPXM2 |
| ACAN | OPTC |
| TNC | SERPINF1 |
| FREM3 | SERPINE2 |
| LAMB1 | F3 |
| LOX | CRISPLD2 |
| EMILIN3 | WNT10B |
| TFPI2 | EPYC |
| NID2 | AEBP1 |
| PTN | CYR61 |
| WNT4 | ECM1 |
| MMP15 | ECM2 |
| FBLN2 | PODN |
| MMP14 | SMOC2 |
| TPSB2 | COL5A3 |
| PXDN | TIMP3 |
| LAMC2 | SMOC1 |
| GPC3 | COL18A1 |
| ADAMTS9 | DCN |
| GPC2 | HIST1H4C |
| NID1 | COL4A3 |
| LOXL2 | NOV |
| ANGPTL4 | FBLN1 |
| SLIT2 | SPARCL1 |
| WNT5B | SPN |
| VCAN | NTN1 |
| CMA1 | RARRES2 |
| FN1 | CST3 |
| COL5A1 | LGALS3BP |
| COL5A2 | IGFBP7 |
| LAMA3 | VWA1 |
| P3H1 | MGP |
| TUBB5 | TNXB |
| AGRN | COL15A1 |
| SFPQ | BGN |
| ITGA6 | THSD4 |
| LAMC1 | HIST1H4A |
| SLC25A5 | TRF |
| CD93 | LAMB2 |
| COL4A1 | DPT |
| LMCD1 | |
| APOE | |
| ADAMTSL5 | |
| TGFBR3 |
Genes are sorted according to fold change with the highest fold change at the top of the list.
Gene Ontology Defense Response Markers Are Highly Enriched in 4 month Old Aortic Valves
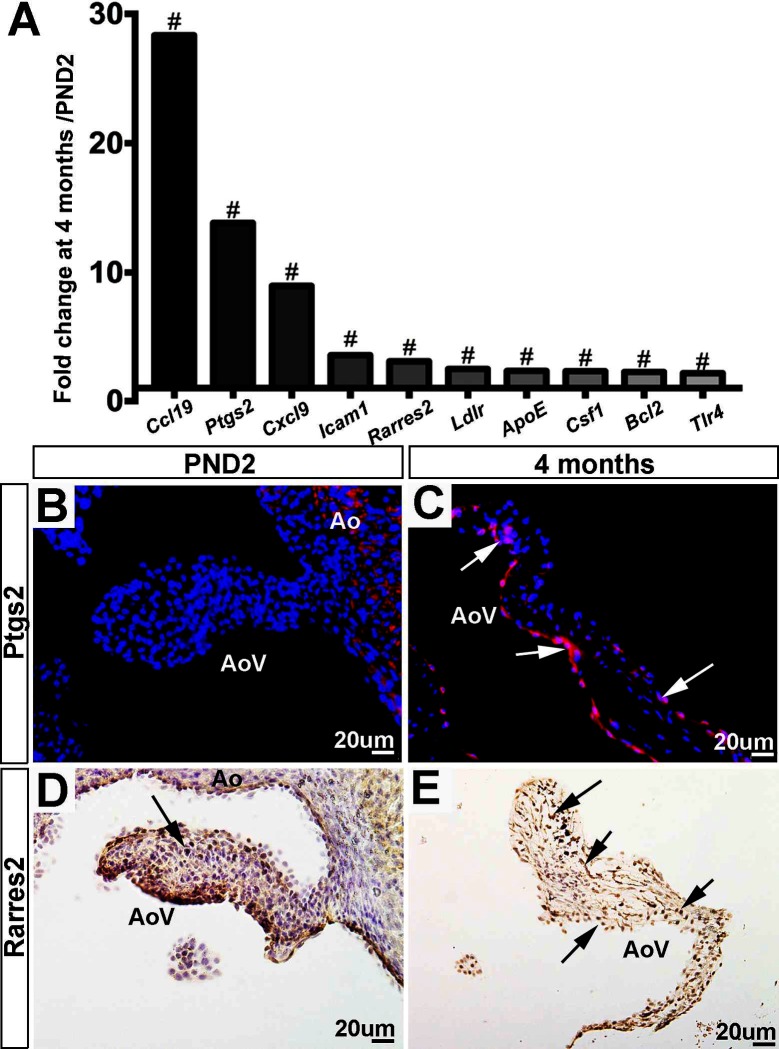
As shown in Figure 6A and Table 4, a large majority (77%) of defense response genes are most highly expressed in aortic valve structures at 4 months of age and include Ccl19, Ptgs2 and Cxcl9. The increased expression of Ptgs2 (also known as Cox2) (Figure 6B,C) and Rarres2 (also known as Tig2) (Figure 6D,E) are confirmed here by immunofluorescence. Ptgs2 is an enzyme involved in the synthesis of prostaglandins which are known to mediate pain and inflammation responses (35), and has been described in the valve as a pro-osteogenic marker (36). Consistent with this previous valve study, we observed expression towards the endothelium at 4 months of age (Figure 6C, Figure S1), and this is consistent with RNA expression in the VEC population (4). Like Ptgs2, Rarres2 is also known to regulate inflammation and has been linked with hypertension (37), a known risk factor of aortic valve stenosis (38). By immunofluorescence, Rarres2 is widely expressed throughout the valve leaflet including VECs and VICs at 4 months of age, which is a significant increase compared to PND2 (4). Overall, our RNA-seq data shows that expression of defense response-related genes increases with age in the murine aortic valve.
Figure 6.
Expression of defense-related genes increases with valve maturation. (A) RNA-sequencing fold changes of defense-related genes. Expression is normalized to 1, indicated by the x-axis (#:FDR < 0.05). (B) Immunofluorescence staining for Ptgs2 validates RNA-seq trends, showing low expression at PND2 compared to (C) Ptgs2 staining at the 4 month time point (white arrows indicate Ptgs2-positive cells). (D) Immunofluorescence staining for Rarres2 validates RNA-seq trends, showing low expression at PND2 compared to (E) Rarres2 staining at the 4 month time point. Ao, aorta; AoV, aortic valve.
Table 4.
Genes included in the “Defense Response” GO term.
| Downregulated at 4 months | Upregulated at 4 months |
| AGTR2 | CFD |
| S100A9 | ADIPOQ |
| CITED1 | HAMP |
| IGF2 | HP |
| COLEC10 | CD74 |
| NGP | CCL8 |
| C1QTNF3 | H2-AA |
| CRH | H2-EB1 |
| S100A8 | H2-AB1 |
| IL31RA | CLEC10A |
| ADAMTS12 | GM2564 |
| CHAF1B | CCL19 |
| AGER | CCL11 |
| PBK | C4B |
| ULBP1 | C4A |
| RNASEL | PTGFR |
| RAET1E | BCL6 |
| ELF3 | ITGAX |
| BRINP1 | PTGS2 |
| RAET1D | ESR1 |
| F2RL1 | CFB |
| CD40 | APOD |
| LCK | IL2RA |
| MASP1 | H2-Q7 |
| TPSB2 | FFAR4 |
| MDK | CXCL9 |
| EDN1 | ITIH4 |
| HMGB2 | SLAMF7 |
| SLFN9 | C1S1 |
| CPA3 | IFIT3 |
| CD276 | C1S2 |
| TSPAN6 | GBP2 |
| TYRO3 | FGF7 |
| CASP6 | C1RA |
| CCR1 | C1RB |
| SLIT2 | WFDC1 |
| FN1 | OAS2 |
| GM13275 | GPR17 |
| GGT5 | PELI3 |
| HYAL2 | MYLK |
| EDNRA | CFH |
| CD24A | ISG20 |
| FANCA | H2-K1 |
| TRIM35 | SERPINF1 |
| SUSD4 | NR1D1 |
| CD93 | CLEC7A |
| SLC35B3 | BMPR1B |
| C1RL | |
| F3 | |
| LYZ1 | |
| TGTP1 | |
| SLAMF8 | |
| TGTP2 | |
| ITK | |
| HRH1 | |
| ALOX5 | |
| CD28 | |
| NR1H3 | |
| PSTPIP1 | |
| IRAK3 | |
| LYZ2 | |
| H2-D1 | |
| TAP2 | |
| APOBEC1 | |
| GNG7 | |
| PTGIR | |
| ECM1 | |
| CX3CL1 | |
| ITGB2 | |
| CD14 | |
| CCRL2 | |
| HIST2H3C2 | |
| GBP6 | |
| GBP10 | |
| H2-M3 | |
| C3 | |
| MILL2 | |
| SERPING1 | |
| CYBB | |
| PTPRC | |
| FCGR2B | |
| RAB7B | |
| HIST1H2BE | |
| TRIM30A | |
| NFKBIZ | |
| ICAM1 | |
| SLFN8 | |
| IFIH1 | |
| HIST1H2BJ | |
| FAS | |
| IL15 | |
| NOV | |
| PTAFR | |
| MAP1A | |
| CASP1 | |
| MYO1F | |
| ADRB2 | |
| FGF14 | |
| HIST1H2BC | |
| SPN | |
| HAVCR2 | |
| IL33 | |
| B2M | |
| COLEC12 | |
| RORA | |
| ITGAM | |
| TAP1 | |
| RARRES2 | |
| CST3 | |
| ZC3H12A | |
| IRGM2 | |
| NAIP5 | |
| HIST1H2BL | |
| IGTP | |
| PNMA1 | |
| HIST1H2BK | |
| CD37 | |
| TLR8 | |
| SLC11A1 | |
| MGLL | |
| IL17RE | |
| HIST1H2BN | |
| CADM1 | |
| H2-T23 | |
| HERC6 | |
| CD86 | |
| SETD6 | |
| ZFP36 | |
| THEMIS2 | |
| LDLR | |
| HFE | |
| C1QA | |
| IGFBP4 | |
| LMCD1 | |
| ALOX5AP | |
| NCF1 | |
| GBP8 | |
| APOE | |
| TNFRSF25 | |
| SERPINB9 | |
| CD46 | |
| CSF1 | |
| IRF1 | |
| NUPR1 | |
| BCL2 | |
| STAR | |
| DRD1 | |
| PIK3AP1 | |
| IRF8 | |
| TLR4 | |
| TLR3 | |
| BIRC3 | |
| HYAL1 | |
| CSF1R | |
| C1QB | |
| UNC93B1 | |
| TRIM21 | |
| PDE5A | |
| PTGIS | |
| CORO1A | |
| AOAH | |
| IFITM3 |
Genes are sorted according to fold change with the highest fold change at the top of the list.
Discussion
This current study explores transcriptional differences in PND2 and 4 month old murine aortic valve expression profiles with the goal of identifying genetic programs representative of valve growth and valve maintenance, respectively. Long term, this may be important for the development of alternative therapies; specifically, those exploring the growth or regenerative capacity of adult diseased valves. Our results indicate that at PND2, dynamic leaflet growth is associated with a unique transcriptional profile compared to homeostatic adult valves at 4 months. Of the 23,303 detectable genes in our RNA-seq data, the number of differentially expressed transcripts found to be up and down regulated at each time point were approximately equal, suggesting that gene transcription patterns were not overtly altered, but rather transitioned from a postnatal to adult expression profile. Such profiles are related to enriched biological functions including cell proliferation at PND2 and defense response at 4 months of age. Interestingly, ECM components were enriched at both time points, but the associated mRNA profiles were unique. Together these analyses contribute to the current knowledge and further advance our understanding of the molecular signatures and biological functions characteristic of the whole aortic valve structure at PND2 and 4 months. provide critical information related to genetic programs in the growing and homeostatic heart valve.
PND2 murine aortic valves are defined by a specific transcriptional profile including the unique expression of 602 genes (Figure 1C, Table 1) that were not detected at 4 months of age. In addition, 1,801 transcripts were upregulated at PND2, while 1,858 were decreased compared to the adult homeostatic valve. According to Gene Ontology analysis, many of the transcripts enriched at PND2 suggest an overall upregulation of active cell proliferation by a specific set of genes. These include increased expression of those associated with active cell division (Ki67, Aurkb), pro-proliferation markers (Bub1, Cdk1, Foxm1, Nrg1) (27, 39, 40) and decreased expression of proliferation inhibitors (Nox4, Sfrp) (41–44). This specific expression profile of cell proliferation markers supports EdU observations and represents active elongation of the immature valve structure. It will become important to understand how this molecular signature of cell proliferation is downregulated after maturation is complete and explore the potential of re-introducing key regulators to stimulate cell division and replenish dysfunctional cell populations in the adult following injury or disease.
In addition to high levels of cell proliferation, the PND2 valve is characterized by a specific ECM mRNA profile (Table 3) which likely corresponds to the mechanical demands during the postnatal period. When comparing PND2 to 4 months, RNA-seq analysis reveals significant enrichment of highly expressed fibrillar collagens including Col24a1, Col9a1, and Col5a1, indicating the need for additional stability as the growing postnatal valve adapts to hemodynamic changes in response to closing of the foramen ovale (45, 46). RNA-seq analysis also uncovered higher PND2 levels of ECM proteins such as Frem1/Frem2 and Nidogen2 (Nid2), which have been shown to stabilize basement membranes underlying endothelial cells and may provide further structural integrity to the developing valve (47, 48). Besides identifying the enrichment of differentially expressed fibrillary collagens and specific proteoglycans, RNA-seq analysis shows that the PND2 valve also expresses a distinct profile of ECM enzymes, such as Mmp15 and Adamts17, indicating the need for physiological remodeling during growth and maturation
The 4 month adult valve is physically and molecularly distinct from the postnatal valve, with elongated leaflets containing distinct layers of collagen, proteoglycan, and elastin (Figure 1B). At this time point, 477 transcripts were found to be uniquely expressed, with the most abundant unique mRNAs including Cfd, Retnla, and Clec3a. In contrast to the PND2 aortic valve, the adult valve displays significantly decreased levels of cell proliferation, likely due to increased expression of proliferation inhibitors and lack of enrichment of positive regulators. At 4 months, the valve ECM is diverse compared to PND2 (Table 3) and likely reflects differences in biomechanical demand in response to the adult circulatory system (49, 50). Similar to the PND2 valve, collagens and proteoglycans are predominant. However, the most highly differentially expressed collagens are those associated with basement membranes, including Col4a6 and Col18a1, which act as a cell scaffold to maintain current cell populations and cell integrity as opposed to providing support for high cell turnover. In addition, the proteoglycan profile is moved towards enrichment of decorin (DCN) and biglycan (BGN) consistent with previous studies in aging pigs (51). Furthermore, the contribution of ECM remodeling enzymes is shifted to Mmp3 and its inhibitor Timp3 at 4 months, possibly indicating a differential need of the ECM to sustain homeostasis. Previous studies have suggested correlations between VIC phenotype and ECM composition (52, 53) and therefore we anticipate that our findings at 4 months are related to the quiescent VICs, while the diversity at PND2 is dictated by proliferative and active VICs.
One of the most prominent differences in expression profiles between the PND2 and 4 month aortic valve is the considerable upregulation of defense-related transcripts at the older time point, indicating increased immune system activation with valve maturation (Figure 6, Table 4). Previous studies from other groups have shown that the appearance of immune markers such as Ptgs2 and Rarres2 precedes the onset of disease both in the heart valve and other cardiovascular systems (36, 37). In addition, there is increasing evidence to suggest that inflammation in the valve is an initial homeostatic repair mechanism activated in response to minor valve injuries sustained throughout life, but that this repair mechanism may become pathogenic if overactive or long-lasting [reviewed (54)]. Our study suggests that some level of activation of the immune system in the valve is present at 4 months of age under homeostatic conditions, however, it is not clear whether these defense markers are an early indication of valve degeneration, or a root cause of disease themselves. Further investigation into target genes such as Cfd and Adipoq will give insight into the possible role of defense response genes in valve disease therapeutics.
Our data shows that postnatal heart valves contain highly proliferative VICs producing a distinctive set of postnatal ECM proteins, while adult VICs are mainly quiescent and are associated with a very different ECM composition as well as increased defense response markers. A limitation of our study is that RNA-seq analysis was performed on whole valve tissue and therefore RNA profiles cannot be distinguished between VEC and VIC populations, however IHC studies shown here, in combination with a previous study from our group (4) support enrichment towards one cell type. Nonetheless, we have unveiled multiple conceivable processes that contribute to postnatal valve maturation and maintainence that pave the way for elucidiating mechanisms underlying valve defects present at birth and those acquired later in life. Furthermore, the basic priciples of cell proliferation and ECM remodeling may also be applied to valvulogenesis and congenital valve malformation. From here further research is needed to determine how findings in this study can be used to develop alternative therapeutic strategies to promote self-repair, replenishment and regeneration of valve cell populations in pathogenesis.
Ethics Statement
All animal procedures were approved by The Research Institute at Nationwide Children’s Hospital Institutional Animal Care and Use Committee (Protocol # AR13-00054).
Author Contributions
The experimental data was collected by EN, and RNA-seq performed and analyzed by CN. Additional analysis of RNA-seq data was undertaken by SL. EN and JL generated the manuscript with input from SL. The entire study was overseen by JL.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kaitlyn Thatcher for technical support and the Animal Resource Center staff at The Research Institute at Nationwide Children’s Hospital.
Footnotes
Funding. This work was supported by National Institute of Health (HL127033, JL) and The Research Institute Training Association at Nationwide Children’s Hospital (EN).
Supplementary material
The Supplementary Material for this article can be found online at:https://www.frontiersin.org/articles/10.3389/fcvm.2018.00030/full#supplementary-material
Cell localization of Mmp3, Nid2 and Ptgs2 in the aortic valve at 4 months of age. Immunohistochemistry to show Mmp3 (A), Nid2 (B) and Ptgs2 (C) expression (red) relative to CD31 (endothelial cells, green) in 4 month old aortic valve leaflets. Arrows indicate protein expression in VICs, arrowheads denote staining in CD31-positive VECs.
References
- 1.Hinton RB, Lincoln J, Deutsch GH, Osinska H, Manning PB, Benson DW, et al. Extracellular matrix remodeling and organization in developing and diseased aortic valves. Circ Res (2006) 98(11):1431–8. 10.1161/01.RES.0000224114.65109.4e [DOI] [PubMed] [Google Scholar]
- 2.Horne TE, Vandekopple M, Sauls K, Koenig SN, Anstine LJ, Garg V, et al. Dynamic heterogeneity of the heart valve interstitial cell population in mitral valve health and disease. J Cardiovasc Dev Dis (2015) 2(3):214–32. 10.3390/jcdd2030214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosse K, Hans CP, Zhao N, Koenig SN, Huang N, Guggilam A, et al. Endothelial nitric oxide signaling regulates Notch1 in aortic valve disease. J Mol Cell Cardiol (2013) 60:27–35. 10.1016/j.yjmcc.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anstine LJ, Bobba C, Ghadiali S, Lincoln J. Growth and maturation of heart valves leads to changes in endothelial cell distribution, impaired function, decreased metabolism and reduced cell proliferation. J Mol Cell Cardiol (2016) 100:72–82. 10.1016/j.yjmcc.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huk DJ, Austin BF, Horne TE, Hinton RB, Ray WC, Heistad DD, et al. Valve endothelial cell-derived Tgfβ1 signaling promotes nuclear localization of Sox9 in interstitial cells associated with attenuated calcification. Arterioscler Thromb Vasc Biol (2016) 36(2):328–38. 10.1161/ATVBAHA.115.306091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tao G, Kotick JD, Lincoln J. Heart valve development, maintenance, and disease: the role of endothelial cells. Curr Top Dev Biol (2012) 100:203–32. 10.1016/B978-0-12-387786-4.00006-3 [DOI] [PubMed] [Google Scholar]
- 7.Lincoln J, Yutzey KE. Molecular and developmental mechanisms of congenital heart valve disease. Birth Defects Res Part A Clin Mol Teratol (2011) 91(6):526–34. 10.1002/bdra.20799 [DOI] [PubMed] [Google Scholar]
- 8.Benjamin EJ, Blaha MJ, Chiuve SE, Cushman M, Das SR, Deo R, et al. Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation (2017) 135(10):e146–603. 10.1161/CIR.0000000000000485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bach DS, Radeva JI, Birnbaum HG, Fournier AA, Tuttle EG. Prevalence, referral patterns, testing, and surgery in aortic valve disease: leaving women and elderly patients behind? J Heart Valve Dis (2007) 16(4):362–9. [PubMed] [Google Scholar]
- 10.Etnel JR, Elmont LC, Ertekin E, Mokhles MM, Heuvelman HJ, Roos-Hesselink JW, et al. Outcome after aortic valve replacement in children: A systematic review and meta-analysis. J Thorac Cardiovasc Surg (2016) 151(1):143–52. 10.1016/j.jtcvs.2015.09.083 [DOI] [PubMed] [Google Scholar]
- 11.Bischoff J, Aikawa E. Progenitor cells confer plasticity to cardiac valve endothelium. J Cardiovasc Transl Res (2011) 4(6):710–9. 10.1007/s12265-011-9312-0 [DOI] [PubMed] [Google Scholar]
- 12.Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol (1996) 28(8):1737–46. 10.1006/jmcc.1996.0163 [DOI] [PubMed] [Google Scholar]
- 13.Soonpaa MH, Field LJ. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am J Physiol (1997) 272(1 Pt 2):H220–6. 10.1152/ajpheart.1997.272.1.H220 [DOI] [PubMed] [Google Scholar]
- 14.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, et al. Transient regenerative potential of the neonatal mouse heart. Science (2011) 331(6020):1078–80. 10.1126/science.1200708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heallen T, Zhang M, Wang J, Bonilla-Claudio M, Klysik E, Johnson RL, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science (2011) 332(6028):458–61. 10.1126/science.1199010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, Mcanally J, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci USA (2013) 110(34):13839–44. 10.1073/pnas.1313192110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foglia MJ, Poss KD. Building and re-building the heart by cardiomyocyte proliferation. Development (2016) 143(5):729–40. 10.1242/dev.132910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bassat E, Mutlak YE, Genzelinakh A, Shadrin IY, Baruch Umansky K, Yifa O, et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature (2017) 547(7662):179–84. 10.1038/nature22978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lincoln J, Alfieri CM, Yutzey KE. Development of heart valve leaflets and supporting apparatus in chicken and mouse embryos. Dev Dyn (2004) 230(2):239–50. 10.1002/dvdy.20051 [DOI] [PubMed] [Google Scholar]
- 20.Peacock JD, Lu Y, Koch M, Kadler KE, Lincoln J. Temporal and spatial expression of collagens during murine atrioventricular heart valve development and maintenance. Dev Dyn (2008) 237(10):3051–8. 10.1002/dvdy.21719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics (2004) 20(9):1453–4. 10.1093/bioinformatics/bth078 [DOI] [PubMed] [Google Scholar]
- 22.Huang Daw, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc (2009) 4(1):44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 23.Walter W, Sánchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics (2015) 31(17):2912–4. 10.1093/bioinformatics/btv300 [DOI] [PubMed] [Google Scholar]
- 24.Huk DJ, Hammond HL, Kegechika H, Lincoln J. Increased dietary intake of vitamin A promotes aortic valve calcification in vivo. Arterioscler Thromb Vasc Biol (2013) 33(2):285–93. 10.1161/ATVBAHA.112.300388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chien AJ, Conrad WH, Moon RT. A Wnt survival guide: from flies to human disease. J Invest Dermatol (2009) 129(7):1614–27. 10.1038/jid.2008.445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalwa H, Michel T. The MARCKS protein plays a critical role in phosphatidylinositol 4,5-bisphosphate metabolism and directed cell movement in vascular endothelial cells. J Biol Chem (2011) 286(3):2320–30. 10.1074/jbc.M110.196022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wadugu B, Kühn B. The role of neuregulin/ErbB2/ErbB4 signaling in the heart with special focus on effects on cardiomyocyte proliferation. Am J Physiol Heart Circ Physiol (2012) 302(11):H2139–H2147. 10.1152/ajpheart.00063.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juríková M, Danihel Ľ, Polák Š, Varga I. Ki67, PCNA, and MCM proteins: Markers of proliferation in the diagnosis of breast cancer. Acta Histochem (2016) 118(5):544–52. 10.1016/j.acthis.2016.05.002 [DOI] [PubMed] [Google Scholar]
- 29.Moesgaard SG, Aupperle H, Rajamäki MM, Falk T, Rasmussen CE, Zois NE, et al. Matrix metalloproteinases (MMPs), tissue inhibitors of metalloproteinases (TIMPs) and transforming growth factor-β (TGF-β) in advanced canine myxomatous mitral valve disease. Res Vet Sci (2014) 97(3):560–7. 10.1016/j.rvsc.2014.10.003 [DOI] [PubMed] [Google Scholar]
- 30.Jung JJ, Razavian M, Challa AA, Nie L, Golestani R, Zhang J, et al. Multimodality and molecular imaging of matrix metalloproteinase activation in calcific aortic valve disease. J Nucl Med (2015) 56(6):933–8. 10.2967/jnumed.114.152355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perrotta I, Sciangula A, Aquila S, Mazzulla S. Matrix Metalloproteinase-9 Expression in Calcified Human Aortic Valves: A Histopathologic, Immunohistochemical, and Ultrastructural Study. Appl Immunohistochem Mol Morphol (2016) 24(2):128–37. 10.1097/PAI.0000000000000144 [DOI] [PubMed] [Google Scholar]
- 32.Ye S, Eriksson P, Hamsten A, Kurkinen M, Humphries SE, Henney AM. Progression of coronary atherosclerosis is associated with a common genetic variant of the human stromelysin-1 promoter which results in reduced gene expression. J Biol Chem (1996) 271(22):13055–60. 10.1074/jbc.271.22.13055 [DOI] [PubMed] [Google Scholar]
- 33.Banik D, Netherby CS, Bogner PN, Abrams SI. MMP3-mediated tumor progression is controlled transcriptionally by a novel IRF8-MMP3 interaction. Oncotarget (2015) 6(17):15164–79. 10.18632/oncotarget.3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mehner C, Miller E, Nassar A, Bamlet WR, Radisky ES, Radisky DC. Tumor cell expression of MMP3 as a prognostic factor for poor survival in pancreatic, pulmonary, and mammary carcinoma. Genes Cancer (2015) 6(11-12):480–9. 10.18632/genesandcancer.90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narumiya S, Sugimoto Y, Ushikubi F. Prostanoid receptors: structures, properties, and functions. Physiol Rev (1999) 79(4):1193–226. 10.1152/physrev.1999.79.4.1193 [DOI] [PubMed] [Google Scholar]
- 36.Wirrig EE, Gomez MV, Hinton RB, Yutzey KE. COX2 inhibition reduces aortic valve calcification in vivo. Arterioscler Thromb Vasc Biol (2015) 35(4):938–47. 10.1161/ATVBAHA.114.305159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kunimoto H, Kazama K, Takai M, Oda M, Okada M, Yamawaki H. Chemerin promotes the proliferation and migration of vascular smooth muscle and increases mouse blood pressure. Am J Physiol Heart Circ Physiol (2015) 309(5):ajpheart.00820.2014–1028. 10.1152/ajpheart.00820.2014 [DOI] [PubMed] [Google Scholar]
- 38.Chu Y, Lund DD, Doshi H, Keen HL, Knudtson KL, Funk ND, et al. Fibrotic Aortic Valve Stenosis in Hypercholesterolemic/Hypertensive Mice. Arterioscler Thromb Vasc Biol (2016) 36(3):466–74. 10.1161/ATVBAHA.115.306912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ouyang B, Lan Z, Meadows J, Pan H, Fukasawa K, Li W, et al. Human Bub1: a putative spindle checkpoint kinase closely linked to cell proliferation. Cell Growth Differ (1998) 9(10):877–85. [PubMed] [Google Scholar]
- 40.Kalin TV, Ustiyan V, Kalinichenko VV. Multiple faces of FoxM1 transcription factor: lessons from transgenic mouse models. Cell Cycle (2011) 10(3):396–405. 10.4161/cc.10.3.14709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meloche S, Pouysségur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene (2007) 26(22):3227–39. 10.1038/sj.onc.1210414 [DOI] [PubMed] [Google Scholar]
- 42.Maganga R, Giles N, Adcroft K, Unni A, Keeney D, Wood F, et al. Secreted Frizzled related protein-4 (sFRP4) promotes epidermal differentiation and apoptosis. Biochem Biophys Res Commun (2008) 377(2):606–11. 10.1016/j.bbrc.2008.10.050 [DOI] [PubMed] [Google Scholar]
- 43.Schröder K, Wandzioch K, Helmcke I, Brandes RP. Nox4 acts as a switch between differentiation and proliferation in preadipocytes. Arterioscler Thromb Vasc Biol (2009) 29(2):239–45. 10.1161/ATVBAHA.108.174219 [DOI] [PubMed] [Google Scholar]
- 44.Perumal V, Pohl S, Keane KN, Arfuso F, Newsholme P, Fox S, et al. Therapeutic approach to target mesothelioma cancer cells using the Wnt antagonist, secreted frizzled-related protein 4: Metabolic state of cancer cells. Exp Cell Res (2016) 341(2):218–24. 10.1016/j.yexcr.2016.02.008 [DOI] [PubMed] [Google Scholar]
- 45.Cole-Jeffrey CT, Terada R, Neth MR, Wessels A, Kasahara H. Progressive anatomical closure of foramen ovale in normal neonatal mouse hearts. Anat Rec (2012) 295(5):764–8. 10.1002/ar.22432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steed E, Boselli F, Vermot J. Hemodynamics driven cardiac valve morphogenesis. Biochim Biophys Acta (2016) 1863(7 Pt B):1760–6. 10.1016/j.bbamcr.2015.11.014 [DOI] [PubMed] [Google Scholar]
- 47.Kohfeldt E, Sasaki T, Göhring W, Timpl R. Nidogen-2: a new basement membrane protein with diverse binding properties. J Mol Biol (1998) 282(1):99–109. 10.1006/jmbi.1998.2004 [DOI] [PubMed] [Google Scholar]
- 48.Pavlakis E, Chiotaki R, Chalepakis G. The role of Fras1/Frem proteins in the structure and function of basement membrane. Int J Biochem Cell Biol (2011) 43(4):487–95. 10.1016/j.biocel.2010.12.016 [DOI] [PubMed] [Google Scholar]
- 49.Erhart-Hledik JC, Favre J, Asay JL, Smith RL, Giori NJ, Mündermann A, et al. A relationship between mechanically-induced changes in serum cartilage oligomeric matrix protein (COMP) and changes in cartilage thickness after 5 years. Osteoarthr Cartil (2012) 20(11):1309–15. 10.1016/j.joca.2012.07.018 [DOI] [PubMed] [Google Scholar]
- 50.Batista MA, Nia HT, Önnerfjord P, Cox KA, Ortiz C, Grodzinsky AJ, et al. Nanomechanical phenotype of chondroadherin-null murine articular cartilage. Matrix Biol (2014) 38:84–90. 10.1016/j.matbio.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stephens EH, Chu CK, Grande-Allen KJ. Valve proteoglycan content and glycosaminoglycan fine structure are unique to microstructure, mechanical load and age: Relevance to an age-specific tissue-engineered heart valve. Acta Biomater (2008) 4(5):1148–60. 10.1016/j.actbio.2008.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aikawa E, Whittaker P, Farber M, Mendelson K, Padera RF, Aikawa M, et al. Human semilunar cardiac valve remodeling by activated cells from fetus to adult: implications for postnatal adaptation, pathology, and tissue engineering. Circulation (2006) 113(10):1344–52. 10.1161/CIRCULATIONAHA.105.591768 [DOI] [PubMed] [Google Scholar]
- 53.Balaoing LR, Post AD, Liu H, Minn KT, Grande-Allen KJ. Age-related changes in aortic valve hemostatic protein regulation. Arterioscler Thromb Vasc Biol (2014) 34(1):72–80. 10.1161/ATVBAHA.113.301936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li C, Xu S, Gotlieb AI. The response to valve injury. A paradigm to understand the pathogenesis of heart valve disease. Cardiovasc Pathol (2011) 20(3):183–90. 10.1016/j.carpath.2010.09.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cell localization of Mmp3, Nid2 and Ptgs2 in the aortic valve at 4 months of age. Immunohistochemistry to show Mmp3 (A), Nid2 (B) and Ptgs2 (C) expression (red) relative to CD31 (endothelial cells, green) in 4 month old aortic valve leaflets. Arrows indicate protein expression in VICs, arrowheads denote staining in CD31-positive VECs.