Abstract
The exponential increase in the use of mobile communication has triggered public concerns about the potential adverse effects of radiofrequency electromagnetic fields (RF-EMF) emitted by mobile phones on the central nervous system (CNS). In this study, we explored the relationship between calcium channels and apoptosis or autophagy in the hippocampus of C57BL/6 mice after RF-EMF exposure with a specific absorption rate (SAR) of 4.0 W/kg for 4 weeks. Firstly, the expression level of voltage-gated calcium channels (VGCCs), a key regulator of the entry of calcium ions into the cell, was confirmed by immunoblots. We investigated and confirmed that pan-calcium channel expression in hippocampal neurons were significantly decreased after exposure to RF-EMF. With the observed accumulation of autolysosomes in hippocampal neurons via TEM, the expressions of autophagy-related genes and proteins (e.g., LC3B-II) had significantly increased. However, down-regulation of the apoptotic pathway may contribute to the decrease in calcium channel expression, and thus lower levels of calcium in hippocampal neurons. These results suggested that exposure of RF-EMF could alter intracellular calcium homeostasis by decreasing calcium channel expression in the hippocampus; presumably by activating the autophagy pathway, while inhibiting apoptotic regulation as an adaptation process for 835 MHz RF-EMF exposure.
Keywords: Apoptosis, Autophagy, Calcium channels, Hippocampus, Radiofrequency electromagnetic field
INTRODUCTION
As the frequency and duration of mobile phone communication has increased, public concern about possible adverse biological health effects of exposure to radiofrequency electromagnetic fields (RF-EMF) have been growing. In 2011, the WHO's International Agency for Research on Cancer classified RF-EMF from cell phones as a ‘possible human carcinogen (Class 2B). Many scientists have recognized RF-EMF as a potential cause to serious adverse risks to human health [1]. The main concern is the central nervous system (CNS), due to the close proximity between mobile phones and the brain during communication. Research on RF radiation has demonstrated a negative impact on neurological factors; such as neurodevelopment, the blood-brain barrier, neurite outgrowth, neurotransmitter release, cognitive impairment, and ultimately, behavior [2,3,4,5,6]. Additional cellular effects of RF-EMF exposure included alterations of intracellular and molecular pathways; such as, the extracellular signal-regulated kinase pathway, apoptosis, and regulation of the cell cycle [5,7,8,9]. In addition, autophagy was induced in mammalian cells after exposures of 835 MHz and 1800 MHz RF-EMF [10,11]. Autophagy is a destructive mechanism in which unnecessary proteins or damaged organelles are encapsulated by an autophagosome, which then fuses with a lysosome [12]. Therefore, autophagy may play a protective role in various disease states and may aid in the maintenance of cellular homeostasis in the presence of various stressors [13].
The hippocampus belongs to the limbic system and plays a crucial role in the formation of new memories and spatial navigation, and is also related to learning and emotions [14,15,16]. Importantly, dysfunction of the hippocampus may lead to early Alzheimer's disease [17,18]. Shrinkage of the hippocampus is involved in severe mental disorders such as schizophrenia and major depressive disorders [19,20]. Exposure to RF-EMF leads to impairment of intracellular calcium homeostasis in the murine hippocampus [21]. A number of studies have highlighted the importance of calcium entry into the cell for neuronal survival. Alterations in intracellular calcium concentrations stimulate a variety of intracellular pathogenic events, including activation of apoptosis [22]. Therefore, calcium influx through voltage-gated calcium channels (VGCCs) is an important factor for the initial stages of apoptosis through the regulation of calcium ion entry across the plasma membrane [23]. A high level of calcium in mitochondria is essential to trigger apoptosis [22,24].
Previously, we reported that 12-week exposure to 835 MHz RF-EMF activates autophagy in the striatum and hypothalamus in mice brain; whereas, 4-week exposure to RF-EMF does not induce the autophagy pathways in these specific brain regions in mice [11]. However, we found that 4-week exposure to RF-EMF does lead to strong autophagic responses in the hippocampus of brain of mice [11]. In the present study, we aimed to investigate the molecular mechanisms (apoptosis and autophagy) that are affected by the calcium concentration in the mouse hippocampus after RF-EMF exposure. We detected the α1 subunits of all types of voltage-gated calcium channels (VGCCs) to quantify their expression. In addition, expressions of Bcl2 and Bax were also quantified. Transmission electron microscopy (TEM) was used to examine the fine structure of morphologic changes after autophagy induction in hippocampal neurons following 4-week exposure of RF-EMF.
METHODS
Animals
6-Week-old mice (C57BL/6, male weighing 25–30 g) were purchased from Daehan Bio Link (DBL, Chungbuk, South Korea). The mice were maintained under specifically controlled conditions (ambient temperature 23±2℃, 12-h light/dark cycle). Food pellets (DBL, Chungbuk, South Korea) and water were supplied ad libitum. After a week adaptation period, the mice were assigned to sham exposure or RF-EMF exposure for 4 weeks.
The study was generated by 2 independent batches of RF-EMF exposure experiment under the same condition. Thus, we collected the samples of hippocampus of mice twice from two different batches of RF-EMF exposed experiments. In total, 13 independent mice were used in each condition's group for qPCR, WB and TEM. On one pair of hippocampus, one was used for qRT-PCR (n=10) and the other was used for WB (n=10). The ultrastructure images by TEM were from the mice hippocampus of the first batch. We used 3 mice brain sample for TEM in both sham control and RF-EMF exposure group (n=3). Thus, 10 mice for qPCR/WB and 3 mice for TEM in each condition (total n=13). All procedures complied with National Institutes of Health guidelines of the NIH for animal research and were approved by Dankook University Institutional Animal Care and Use Committee (IACUC; DKU-15-001), which adheres to the guidelines issued by the Institution of Laboratory of Animal Resources (ILAR).
Exposure system of RF-EMF signal
After the adaption week, mice were exposed to 835 MHz RF-EMF using our RF generator. The frequencies of the 800MHz band are technically advantageous in the area obstructed by buildings due to the long distances the wavelength can reach. Therefore, this bandwidth frequency is used intensively and broadly for telecommunication networks in many countries. Among these various bands, 829-839 MHz (835 MHz, middle point) are considered the main frequencies and are actively used by telecommunication companies. The everyday use and predominant exposure of 835 MHz RF-EMF, has made the finding of any biological effects to be of paramount importance. The whole body exposure was at a specific absorption rate (SAR) value of 4.0 W/kg for 5 h daily for 4 weeks. Additionally, in the safety level, 4.0 W/kg SAR value is the maximum permitted exposure to human limbs but whole body is allowed up to 0.08 W/kg SAR for general public person in accordance with the International Commission on Non-Ionizing Radiation Protection (ICNIRP) guidelines. However, we used 4.0 W/kg SAR value for our experimental purposes in our mice model. The SAR generated by our RF generator was evaluated by measuring E-field at the phantom position in the air; while simultaneously by considering the ratio of E-field in the liquid to E-field in the air at the same position in the same environment. Following calculation of SAR, the value is 4.14 W/kg which is proximity of setting up output power at SAR 4.0 W/kg of horn antenna of the exposure apparatus. As a result, measurement of RF signal and SAR value generated from our RF-EMF generator produce 835 MHz RF-EMF with 4.0 W/kg SAR. More information about the exposure system has been described in details in our previous reports [25]. The mice in control group received sham exposure in parallel for 4 weeks. Sham exposure group was maintained under same environmental conditions, except for the exposure of RF-EMF. Both groups were treated in the same circular pattern from housing cage to exposure cage throughout the experiments. The mice could move freely and were not restricted to access food and water during the exposure. The cage for RF-EMF exposure was placed in the RF-EMF generator. 835 MHz RF-EMF was delivered to the mice from a horn antenna, installed above the mouse cage. The bottom and walls of the cage were covered by ceramic wave absorption material. All the animal experiments were completed under an automatic lighting system and air conditioning facility, which was maintained in constant temperature.
Quantitative real-time PCR and semi-quantitative RTPCR
In order to determine whether autophagy is also triggered in the hippocampus of mice brain following the 4 week exposure to RF-EMF, selected autophagy genes such as Atg4A/B, Atg5, Atg9A, Beclin1/2 and LC3A/B were examined. Expressional levels of these genes were found by using either quantitative real time-PCR or semi-quantitative RT-PCR, and changes were recorded. Mice were euthanized the day following the completion of the RF-EMF exposure for 4 weeks (28 days). After the mouse was euthanized by cervical dislocation and head was decapitated with scissors, the whole hippocampus was isolated from the brain. The dissected hippocampus was immediately frozen and stored at −80℃. Total RNA was extracted from the whole hippocampus by using TRIzol reagent (Thermo Fisher Scientific, USA). Purified RNA was reverse transcribed to cDNA using MMLV Reverse-Transcriptase (Bioneer, South Korea) and an oligo-d(T)18 primer. Quantitative RT-PCR reactions were carried out with Rotor-gene SYBR Green supermix Kit (QIAgen, Germany) and fluorescence was measured using Rotor Gene PCR Cycler (QIAgen, Germany). The expression levels of the genes were normalized to that of GAPDH, as a housekeeping gene. GAPDH primer was purchased from QIAgen (Germany). The primers used for qRT-PCR and sqRT-PCR (Table 1) were synthesized from Bioneer or Cosmogenetech (South Korea). The five biologically independently prepared samples by pooling were tested and each PCR reaction was done in triplicate. The relative levels of specific mRNA were calculated by normalizing to the expression of GAPDH by the 2−ΔΔCt method (n=10). Also, the expression values of the RF-EMF exposed groups were normalized to those of the sham-exposed group. Semi-quantitative RT-PCR reactions were performed using PCR PreMix (Bioneer, South Korea). Subsequently, sqPCR product of each gene was electrophoresed in 1.5% agarose gel and signal intensity of PCR product was visualized the Syto 60 (Li-Cor, USA)-stained DNA using the Odyssey infrared imaging system (Li-Cor, USA).
Table 1. Sequences of oligonucleotide used for RT-PCR.
Western blot analysis
The hippocampus was quickly dissected from brain of mice and one side of hippocampus from each mouse (n=10) was quickly lysed with RIPA lysis buffer (ATTO, Japan), which was supplemented with protease inhibitor and phosphate inhibitor cocktail (ATTO, Japan). The hippocampal lysates were then homogenized and sonicated briefly in ice. For Western blot, pooling samples were used depending on protein concentration. Protein concentration was determined by Lowry protein assay (Bio-Rad, USA). Lysated samples were separated by electrophoresis on 10–15% SDS-polyacrylamide gel and transferred to polyvinylidene difluoride (PVDF) membrane (ATTO, Japan). Membranes were blocked in Odyssey blocking buffer (Li-COR, Lincoln, NE, USA) for an hour at room temperature, then incubated in a specific primary antibody solution. Primary antibodies (Abs) were used: (i) anti-LC3B Ab (Cell Signaling Technology, USA), (ii) anti-eclin2 Ab (Cell Signaling Technology, USA), (iii) anti-Bcl2 Ab (Santa Cruz Biotechnology, USA), (iv) anti-Bax Ab (Santa Cruz Biotechnology, USA), and (v) anti-Pan calcium channel Ab (Sigma-Aldrich, USA); binding all types' α1 subunits of voltagegated calcium channels. The goat anti-rabbit 800CW or goat anti-mouse 680RD (Li-Cor, USA), were used as secondary antibodies. The membrane was washed thoroughly with tris-buffed saline containing 0.1% Tween-20 (200 mM Tris, 137 mM NaCl, 0.1% Tween20, pH 7.4). Protein bands were quantitated by using Odyssey infrared imaging system (Li-Cor, USA) and normalized by using α-tubulin band.
Transmission electron microscopy (TEM)
The dissected hippocampus' were fixed immediately in 2% glutaraldehyde-2% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) for 2 h at 4℃. Following three washes in the phosphate buffer, the brain tissues were post-fixed with 1% osmium tetroxide on ice for 2 h and washed three times, all in the phosphate buffer. The tissues were then embedded in Epon 812 mixture after dehydration in an ethanol and propylene oxide series. Polymerization was conducted with pure resin at 70℃ for 24 h. Ultrathin sections (~70 nm) were obtained with an ultramicrotome (MT-X, RMC, Tucson, AZ, USA) and then collected on 100 mesh copper grids. After staining with 2% uranyl acetate (15 min) and lead citrate (5 min), the sections were visualized using the transmission electron microscopy (TEM) (Technai G2 Spirit Twin, FEI, Hillsboro, OR, USA) at 120 kV. The whole pair of hippocampus was used for TEM study (n=3).
Statistical analysis
All data is presented as the mean±SEM. The significances of all data assessed by two-tailed, Student's t-test with probability values of p<0.05 considered statistically significant. We used GraphPad Prism 4 program (GraphPad Software, Inc., La Jolla, CA) for the statistical analysis.
RESULTS
Decreased expression of VGCCs in the hippocampus
To investigate the effects of RF-EMF exposure on calcium homeostasis in the hippocampus, we focused on VGCCs, which controls intracellular calcium concentrations [26]. Total protein extracted from the hippocampus was used to perform western blots using an anti-Pan calcium channel antibody (Sigma, St. Louis, USA), which recognizes all types of α1 subunits of VGCCs. The data showed that 4-week exposure to RF-EMF decreased the expression of VGCCs in the mouse hippocampus (Fig. 1). These data strongly suggested that exposure to RF-EMF could induce alterations to calcium homeostasis in the mouse hippocampus.
Fig. 1. The expression of calcium channels in the mouse hippocampus following a 4-week exposure to RF-EMF.
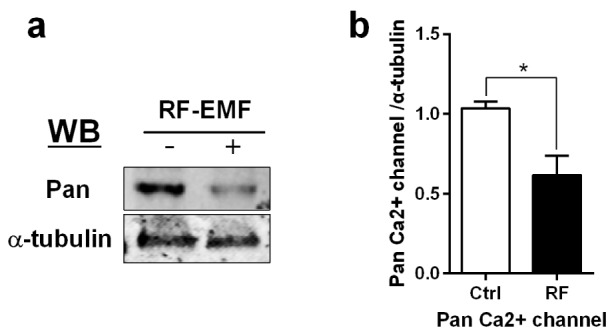
Total hippocampus was isolated from sham-exposed mice (Ctrl) and RF-exposed mice (RF). (a) Representative immunoblot of pan-calcium channel antibody. (b) The expression level of calcium channels in the hippocampus is significantly decreased after 835 MHz RF-EMF exposure for 4 weeks. The bar represents the mean±SEM. *p<0.05 vs. Ctrl (n=10).
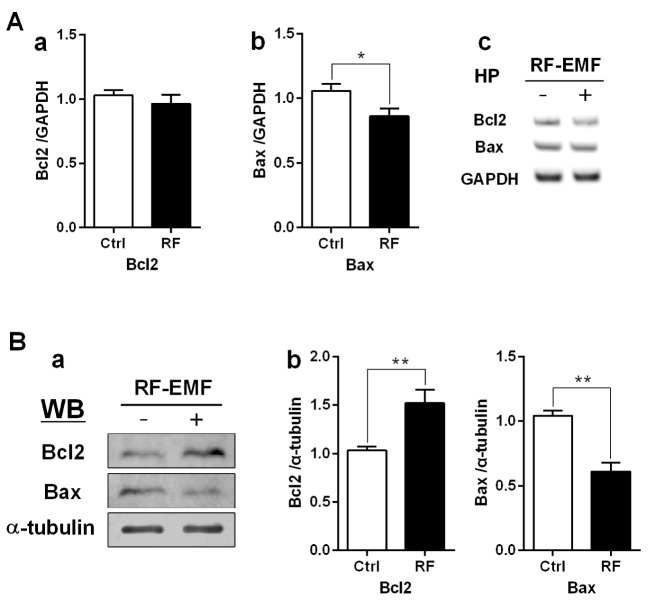
The expression level of Bax is down but that of Bcl2 is up in the hippocampus after RF-EMF exposure
To elucidate the effects of 4-week RF-EMF exposure on apoptosis, the anti-apoptotic factor Bcl2 and the pro-apoptotic factor Bax were analyzed in the hippocampus; with both RT-PCR and western blotting. The RT-PCR results revealed that the transcript level of the pro-apoptotic gene Bax slightly declined in the hippocampus of RF-EMF-exposed mice (Fig. 2A). Similarly, immunoblotting analysis indicated that the level of anti-apoptotic Bcl2 protein was significantly increased; while Bax protein expression was significantly decreased, by nearly half in the hippocampus following RF-EMF exposure (Fig. 2B). These results indicated that the apoptotic pathway may be hindered following a 4-week exposure to RF-EMF.
Fig. 2. Regulation of apoptosis in the mouse hippocampus following RF-EMF exposure.
Hippocampal mRNA (A) and proteins (B) were analysed for the expression level of apoptotic genes/proteins. (Aa, Ab) Quantification of Bcl2 and Bax mRNA transcripts by qRT-PCR. (Ac) Representative gel images of Bcl2 and Bax by sqRT-PCR. The expression levels of the hippocampus of RF-EMF-exposed mice were normalized to those of the sham-exposed mice. The relative mRNA levels of each gene were calculated by normalizing to GAPDH expression by the 2−ΔΔCt method (n=10). (Ba) Representative immunoblots of hippocampus from sham-control and RF-EMF group. (Bb) The expression level of Bcl2 was significantly increased, but the expression level of Bax was significantly decreased after 835 MHz RF-EMF exposure for 4 weeks. *p<0.05 and **p<0.01, vs. Ctrl. Average is the mean±SEM (n=10).
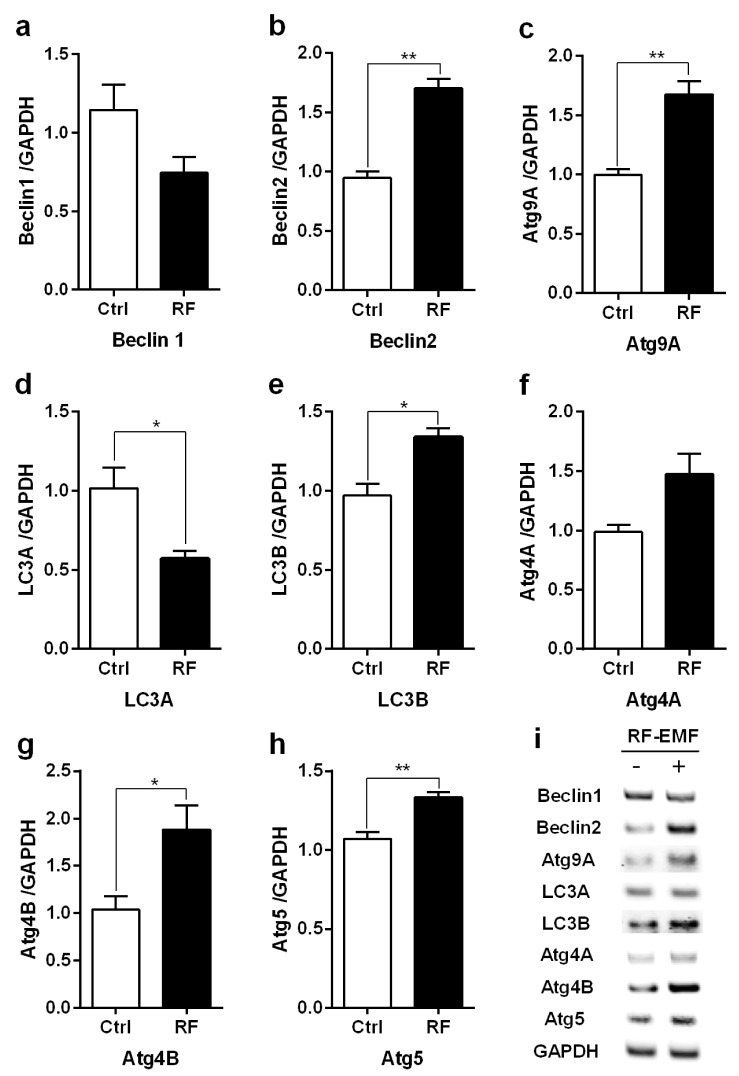
Autophagy-related genes are significantly upregulated in the hippocampus following RF-EMF exposure
To determine whether autophagy is triggered in the mouse hippocampus following 4-week exposure to RF-EMF, we examined the expression of selected autophagy genes (Atg4A/B, Atg5, Atg9A, Beclin1/2, and LC3A/B) using quantitative real time-PCR (qRT-PCR) and semi-quantitative RT-PCR (sqRT-PCR). As shown in Fig. 3, the qRT-PCR results indicated that the expression of most of the genes related to autophagy (Atg4B, Atg5, Atg9A, Beclin2, and LC3B) were significantly increased at about 1.5-2.0 fold, as compared to controls in the hippocampus (Fig. 3). However, the expression of Atg4A, Beclin1 did not differ from that in control animals following RF-EMF exposure (Fig. 3). Of interest, the expression of LC3A was decreased in RF group (Fig. 3d). In addition, sqRT-PCR was carried out to further validate the changes in gene expression found with qRT-PCR. Generally, the results from sqPCR were consistent with the patterns seen with the qRT-PCR data, indicating that Atg4B, Atg5, Atg9A, Beclin2, and LC3A/B showed higher intensity in RF-EMF group compared to controls (Fig. 3i). Analysis of qRT-PCR indicated that there is upregulation of genes related to autophagy in the hippocampus in response to 4-week exposure of RF-EMF.
Fig. 3. Expression levels of autophagyrelated genes in mouse hippocampus.
Total RNA was extracted from the hippocampus of sham-exposed mice (Ctrl) and RF-EMF exposed mice (RF). (a–h) Quantification of Beclin1/2, Atg9A, LC3A/B, Atg4A/B, Atg5 mRNA was analysed by qRT-PCR. (i) Representative gel images of autophagy related genes, amplified by sqRT-PCR. The relative expression levels of each gene were calculated by normalizing GAPDH expression by the 2−ΔΔCt method. Each bar represents the mean±SEM. *p<0.05 and **p<0.01 vs. Ctrl (n=10).
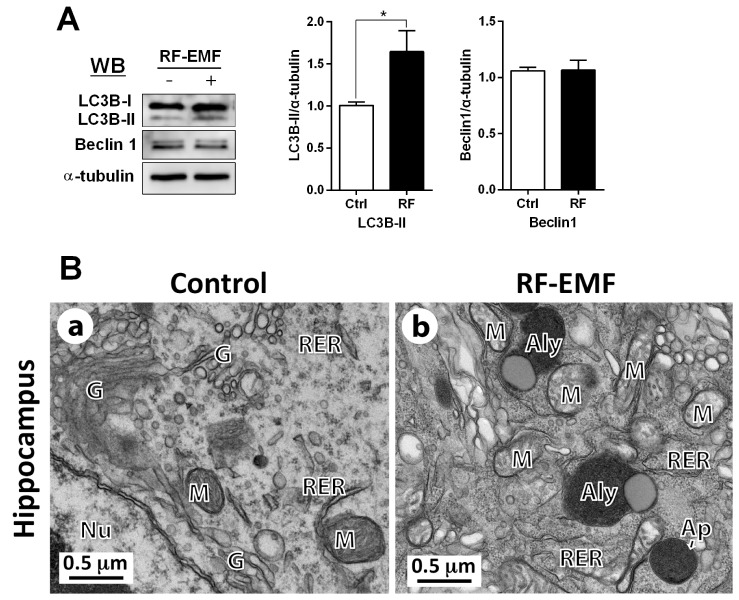
Autophagy structures are accumulated in the hippocampus
In parallel with the upregulation of mRNA level of autophagy in the hippocampus depending on RF-EMF exposure, we further explored the levels of autophagy-related proteins such as LC3B-II and Beclin1; which play a key role in the formation of the autophagosome in response to environmental cellular stress [27,28,29]. The LC3B-II protein level was significantly increased by 1.7-fold compared to control in the hippocampus, but the expression of Beclin1 was not changed (Fig. 4A). Ultrastructural comparisons between sham and RF-EMF-exposed hippocampal neurons was conducted using TEM. The results indicated that the augmented number of autophagy structures, such as autophagosomes and autolysosomes, were observed in hippocampal neurons following RF-EMF exposure (Fig. 4B). These results indicated that 4-week exposure to RF-EMF could induce higher LC3B-II expression and accumulate more autophagy structures, in comparison to the sham control group in the hippocampus. The data strongly suggested that activation of the autophagy pathway is triggered in the mouse hippocampus by RF-EMF exposure.
Fig. 4. LC3B-II and beclin1 expression and ultrastructure of autophagy in the hippocampal neurons.
(A) The expression level of LC3B was significantly increased by RF-EMF exposure, but beclin1 was not changed. Average is mean±SEM. *p<0.05 vs Ctrl. (n=10). (B) Representative TEM micrographs were acquired from the hippocampal neurons of sham exposed control (Ba) and RF-EMF exposed mice (n=3) (Bb). In the hippocampus of RF-EMF-exposed mice (Bb), numerous autophagosome (Ap) and autolysosome (Aly) were observed compared to sham control mice. Abbreviations are: Ap, autophagosome; Aly, autolysosome; G, Golgi apparatus; M, mitochondria; Nu, nucleus; RER, rough endoplasmic reticulum. Size bars: 0.5 µm.
DISCUSSION
This study evaluated the effects of RF-EMF exposure on calcium channel expression. Intracellular calcium concentrations are regulated by calcium channels and affect various neuronal processes [30]. Exposure to RF-EMF caused impairment of intracellular calcium homeostasis in the murine hippocampus [21]. The calcium ion is an essential component for the regulation of a variety of cellular processes, including apoptosis [31]. The impairment of calcium homeostasis in a cell may lead to activation of apoptosis [23,31]. In this study, we showed that exposure to RF-EMF may alter the calcium homeostasis in the hippocampus of mice by decreasing the level of pan-calcium channels, which regulate calcium influx (Fig. 1). Calcium homeostasis can be regulated by several types of calcium channels, including VGCCs [32]. VGCCs are responsible for fast calcium influx into the cell, which controls the entry of calcium ions across the plasma membrane [32].
Intracellular calcium overload may activate pro-apoptotic genes, which cause the degeneration of neurons and ultimately cell death [31,33]. It has also been suggested that capacitive calcium influx through calcium release-activated calcium channels causes apoptosis [22]. However, inhibition of calcium channels in neurons, which causes a reduction in the level of intracellular calcium, may serve a neuroprotective function to promote neuronal survival [23]. This is consistent with our finding that, in response to RF-EMF exposure, apoptosis might be inhibited (Fig. 2) and the expression of VGCCs was also significantly reduced (Fig. 1). Further studies are required to gain more evidence on the direct relationship between calcium signaling and apoptosis in neurons in response to RF-EMF exposure.
In the present study, we found significant augmentation of both autophagy-related genes and proteins in the hippocampus following a 4-week exposure to 835 MHz RF-EMF with 4.0 W/kg SAR (Figs. 3 and 4A). Several studies previously suggested that intracellular calcium is an important regulator for initiation of autophagy processes [34,35]. Additionally, the autophagic flux was attenuated by increased cytosolic calcium concentration, due to prevent the fusion between autophagosomes and lysosomes in hepatocytes [36]. Prevention of cytosolic calcium influx by calcium channel blockers effectively restored the autophagic flux, presenting by accumulation of autophagosomes and increase in LC3-II level in the obesity-induced autophagy arrest model [36]. As calcium channel blockers can prevent the cytosolic calcium influx, inhibition of calcium channels could be a molecular mechanism for initiation of the autophagic processes in autophagy defects. In this study, we found that the expression of calcium channels was decreased and the processes of autophagy were induced by exposure of RF-EMF. These results suggested that decreased level of calcium channel expression could interfere with the intracellular calcium influx, therefore, autophagy processes can be activated in hippocampus of mice.
Further evidence for autophagy induction after RF-EMF exposure is that although hippocampal structures (CA1-CA4 and DG) were not distinguished in our situation. Due to the difficulty of hippocampal staining, autophagic structures; such as autophagosome and autolysosomes were accumulated in hippocampal neurons of brain of mice exposed to RF-EMF as a whole. Additionally, we obtained continuously consistent results in randomly cut all area of hippocampus by TEM experiment. Previously, we have reported that 12-week exposure to RF-EMF activates autophagy in the striatum and hypothalamus in mice brain, whereas 4-week exposure to RF-EMF does not induce the autophagy pathways in these brain regions in mice [11]. However, we found that 4-week exposure to RF-EMF leads to strong autophagic responses in the hippocampus of mice brain. Our results showed that autophagy can be triggered in the hippocampus following RF-EMF exposure, as a physiologic means of adaptation to cellular stress to 835 MHz radiofrequency at 4.0 W/kg SAR for 4 weeks. However, the processes of autophagy induced by RF-EMF exposure were not observed at hypothalamus in previous study [11]. It is not clear yet why different region of brain response differentially to RF-EMF. We speculated that the structure or function of the hippocampus are more sensitive than hypothalamus to the stress from radiofrequency electromagnetic field. The hippocampus belongs to the limbic system and plays important roles in the consolidation of information from memory; but the hypothalamus is also responsible for the regulation of certain metabolic processes and other activities to link the nervous system to the endocrine system. In addition, the location of brain regions to RF-EMF exposure or the different tissue configurations could contribute the differential responses. Further researches will be required to investigate the differential responses among brain regions.
In this study, we also found that the apoptosis pathway was inhibited, while autophagy was increased in the hippocampus after 4 weeks exposure of RF-EMF (Fig. 3). RF-EMF exposure was reported to upregulate the caspase-3-dependent apoptotic pathway in primary cultured rodent brain neurons [8,37]. Similar cellular stressors can cause either autophagy or apoptosis in a cell; the method triggered depends on the threshold of sensitivity to the specific stressor [38]. Both mechanisms are tightly regulated by the inhibition of mediator proteins Bcl2 and Beclin1 [39]. Activation of autophagy blocks the apoptosis pathway through inhibition of the Bax protein [38]. Anti-apoptotic Bcl-2 protein was upregulated, thereby inhibiting the pro-apoptotic Bax protein in the hippocampus after RF-EMF exposure (Fig. 2).
Autophagy is an essential cellular mechanism that processes selective degradation of organelles or unusual protein aggregates [40]. Importantly, dysregulation of autophagy may cause various neurodegenerative diseases [41,42,43]. The analysis of the expression of autophagy-related genes in this study suggests that they may play a crucial role for autophagosomal formation [44]. Activity of endoplasmic reticulum (ER)-associated class III PI3K (also known as Vps34) drives vesicle nucleation by the formation of a multi-complex with Beclin1/2, which initiates autophagosomal formation [40]. The trans-membrane protein Atg9A/B may recruit lipids to the phagophore [38,45]. During the autophagy process, LC3 is cleaved by cysteine protease Atg4A/B and undergoes post-translational modifications [46]. A lipidated form of LC3, microtubule-associated protein 1 light chain 3 (MAP1LC3)-II (LC3-II) is associated with autophagic vesicles [27,47]. Importantly, a complex of an E3 ubiquitin ligase-like enzyme Atg5 with Atg12-Atg16L1 regulates autophagosomal elongation and this complex is essential for LC3-I combination with phosphatidylethanolamine (PE) to form LC3-II (LC3-PE) [27,38,46,48]. Eventually, mature autophagosomes that engulfed intracellular organelles fused with lysosome generate autolysosome [27,38]. Therefore, the level of LC3B-II might reflect the relative amount of autophagosomes in hippocampal neurons [29]. RF-EMF exposure significantly increased the protein level of LC3B-II, a protein that is used for autophagy markers as well as represents overall autophagic flux [29], in the hippocampus (Fig. 3A). As shown in Fig. 3B, exposure of RF-EMF resulted in the accumulation of autophagic vesicles; such as autophagosomes and autolysosomes, which might also represent active autophagy in the hippocampus. Thus, RF-EMF exposure likely induces autophagy in mouse hippocampal neurons.
The hippocampus is crucial for its role in the formation of new memories and spatial navigation, and is also involved in learning and emotions [14,15,16]. Importantly, dysfunction of the hippocampus may lead to early Alzheimer's disease (AD) [17,18]. AD is mainly caused by extracellular amyloid beta (Aβ) deposits, but autophagy could provide neuroprotection through the disruption of Aβ aggregates [49,50]. This study may provide evidence that autophagy offers neuroprotection in the hippocampus in response to RF-EMF exposure. Moreover, autophagy may prevent neuronal cell death through the down-regulation of apoptosis, which can lead to neurodegenerative diseases.
Calcium signaling plays a key role in the regulation of neurotransmitter release, as well as cell death [51,52]. Changes in calcium homeostasis may have significant effects on neurotransmitter release from synaptic terminals. Therefore, RM-EMF exposure may cause alterations to neurotransmission in the hippocampus, leading to abnormal behavior and/or disease states, such as learning impairments, anxiety, and depression [53,54]. Our previous study demonstrated that RF-EMF exposure to mice may lead to hyperactivity-like behavior by generation of demyelination in the cerebral cortical neurons [25]. However, more detailed studies are necessary to elucidate the scope of impact of RF-EMF on the brain.
In summary, we found that the level of calcium channels, which play a key role in calcium homeostasis, were decreased in the mouse hippocampus following a 4-week exposure to 835 MHz RF-EMF at 4.0 W/kg SAR, 5 h per day. RF-EMF exposure likely alters intracellular calcium by decreasing the expression of calcium channels, activating the autophagy pathway and suppressing the apoptotic pathway. The expression of genes and proteins related to autophagy were significantly increased, in parallel with the accumulation of autolysosomes in hippocampal neurons. Our results suggest that RF-EMF is acting as a stressor to the central nervous system and causing increases in autophagy and down-regulation of apoptosis.
ACKNOWLEDGEMENTS
Dr. Huh, Y.H. (Center for Electron Microscopy Research, Korea Basic Science Institute, Ochang, Chungbuk) for transmission of electron microscopy. This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2017R1D1A1B03029527).
Footnotes
Author contributions: J.H.K., H.R.K.; conception and design of study, acquisition of data, J.H.K., U.D.S., H.G.K., H.R.K.; analysis and/or interpretation of data, J.H.K., H.R.K.; drafting the manuscript, U.D.S., H.G.K., H.R.K.; revising the manuscript critically for important intellectual content.
CONFLICTS OF INTEREST: The authors declare no conflicts of interest.
References
- 1.Baan R, Grosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Islami F, Galichet L, Straif K WHO International Agency for Research on Cancer Monograph Working Group. Carcinogenicity of radiofrequency electromagnetic fields. Lancet Oncol. 2011;12:624–626. doi: 10.1016/s1470-2045(11)70147-4. [DOI] [PubMed] [Google Scholar]
- 2.Aldad TS, Gan G, Gao XB, Taylor HS. Fetal radiofrequency radiation exposure from 800-1900 mhz-rated cellular telephones affects neurodevelopment and behavior in mice. Sci Rep. 2012;2:312. doi: 10.1038/srep00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hao D, Yang L, Chen S, Tong J, Tian Y, Su B, Wu S, Zeng Y. Effects of long-term electromagnetic field exposure on spatial learning and memory in rats. Neurol Sci. 2013;34:157–164. doi: 10.1007/s10072-012-0970-8. [DOI] [PubMed] [Google Scholar]
- 4.Chen C, Ma Q, Liu C, Deng P, Zhu G, Zhang L, He M, Lu Y, Duan W, Pei L, Li M, Yu Z, Zhou Z. Exposure to 1800 MHz radiofrequency radiation impairs neurite outgrowth of embryonic neural stem cells. Sci Rep. 2014;4:5103. doi: 10.1038/srep05103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang J, Zhang Y, Yang L, Chen Q, Tan L, Zuo S, Feng H, Chen Z, Zhu G. Exposure to 900 MHz electromagnetic fields activates the mkp-1/ERK pathway and causes blood-brain barrier damage and cognitive impairment in rats. Brain Res. 2015;1601:92–101. doi: 10.1016/j.brainres.2015.01.019. [DOI] [PubMed] [Google Scholar]
- 6.Kim JH, Kim HJ, Yu DH, Kweon HS, Huh YH, Kim HR. Changes in numbers and size of synaptic vesicles of cortical neurons induced by exposure to 835 MHz radiofrequency-electromagnetic field. PLoS One. 2017;12:e0186416. doi: 10.1371/journal.pone.0186416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buttiglione M, Roca L, Montemurno E, Vitiello F, Capozzi V, Cibelli G. Radiofrequency radiation (900 MHz) induces Egr-1 gene expression and affects cell-cycle control in human neuroblastoma cells. J Cell Physiol. 2007;213:759–767. doi: 10.1002/jcp.21146. [DOI] [PubMed] [Google Scholar]
- 8.Liu YX, Tai JL, Li GQ, Zhang ZW, Xue JH, Liu HS, Zhu H, Cheng JD, Liu YL, Li AM, Zhang Y. Exposure to 1950-MHz TD-SCDMA electromagnetic fields affects the apoptosis of astrocytes via caspase-3-dependent pathway. PLoS One. 2012;7:e42332. doi: 10.1371/journal.pone.0042332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gherardini L, Ciuti G, Tognarelli S, Cinti C. Searching for the perfect wave: the effect of radiofrequency electromagnetic fields on cells. Int J Mol Sci. 2014;15:5366–5387. doi: 10.3390/ijms15045366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu K, Zhang G, Wang Z, Liu Y, Dong J, Dong X, Liu J, Cao J, Ao L, Zhang S. The protective effect of autophagy on mouse spermatocyte derived cells exposure to 1800 MHz radiofrequency electromagnetic radiation. Toxicol Lett. 2014;228:216–224. doi: 10.1016/j.toxlet.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Kim JH, Huh YH, Kim HR. Induction of autophagy in the striatum and hypothalamus of mice after 835 MHz radiofrequency exposure. PLoS One. 2016;11:e0153308. doi: 10.1371/journal.pone.0153308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 13.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konkel A, Cohen NJ. Relational memory and the hippocampus: representations and methods. Front Neurosci. 2009;3:166–174. doi: 10.3389/neuro.01.023.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deng W, Aimone JB, Gage FH. New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? Nat Rev Neurosci. 2010;11:339–350. doi: 10.1038/nrn2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eichenbaum H, Cohen NJ. Can we reconcile the declarative memory and spatial navigation views on hippocampal function? Neuron. 2014;83:764–770. doi: 10.1016/j.neuron.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Zang Y, He Y, Liang M, Zhang X, Tian L, Wu T, Jiang T, Li K. Changes in hippocampal connectivity in the early stages of Alzheimer's disease: evidence from resting state fMRI. Neuroimage. 2006;31:496–504. doi: 10.1016/j.neuroimage.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 18.Del Campo M, Hoozemans JJ, Dekkers LL, Rozemuller AJ, Korth C, Müller-Schiffmann A, Scheltens P, Blankenstein MA, Jimenez CR, Veerhuis R, Teunissen CE. BRI2-BRICHOS is increased in human amyloid plaques in early stages of Alzheimer's disease. Neurobiol Aging. 2014;35:1596–1604. doi: 10.1016/j.neurobiolaging.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Campbell S, Marriott M, Nahmias C, MacQueen GM. Lower hippocampal volume in patients suffering from depression: a metaanalysis. Am J Psychiatry. 2004;161:598–607. doi: 10.1176/appi.ajp.161.4.598. [DOI] [PubMed] [Google Scholar]
- 20.Videbech P, Ravnkilde B. Hippocampal volume and depression: a meta-analysis of MRI studies. Am J Psychiatry. 2004;161:1957–1966. doi: 10.1176/appi.ajp.161.11.1957. [DOI] [PubMed] [Google Scholar]
- 21.Maskey D, Kim M, Aryal B, Pradhan J, Choi IY, Park KS, Son T, Hong SY, Kim SB, Kim HG, Kim MJ. Effect of 835 MHz radiofrequency radiation exposure on calcium binding proteins in the hippocampus of the mouse brain. Brain Res. 2010;1313:232–241. doi: 10.1016/j.brainres.2009.11.079. [DOI] [PubMed] [Google Scholar]
- 22.Rizzuto R, Pinton P, Ferrari D, Chami M, Szabadkai G, Magalhães PJ, Di Virgilio F, Pozzan T. Calcium and apoptosis: facts and hypotheses. Oncogene. 2003;22:8619–8627. doi: 10.1038/sj.onc.1207105. [DOI] [PubMed] [Google Scholar]
- 23.Sendrowski K, Rusak M, Sobaniec P, Iłendo E, Dąbrowska M, Boćkowski L, Koput A, Sobaniec W. Study of the protective effect of calcium channel blockers against neuronal damage induced by glutamate in cultured hippocampal neurons. Pharmacol Rep. 2013;65:730–736. doi: 10.1016/s1734-1140(13)71052-1. [DOI] [PubMed] [Google Scholar]
- 24.Mattson MP, Chan SL. Calcium orchestrates apoptosis. Nat Cell Biol. 2003;5:1041–1043. doi: 10.1038/ncb1203-1041. [DOI] [PubMed] [Google Scholar]
- 25.Kim JH, Yu DH, Huh YH, Lee EH, Kim HG, Kim HR. Long-term exposure to 835 MHz RF-EMF induces hyperactivity, autophagy and demyelination in the cortical neurons of mice. Sci Rep. 2017;7:41129. doi: 10.1038/srep41129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreno Davila H. Molecular and functional diversity of voltagegated calcium channels. Ann N Y Acad Sci. 1999;868:102–117. doi: 10.1111/j.1749-6632.1999.tb11281.x. [DOI] [PubMed] [Google Scholar]
- 27.Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang C, Kaushal V, Shah SV, Kaushal GP. Autophagy is associated with apoptosis in cisplatin injury to renal tubular epithelial cells. Am J Physiol Renal Physiol. 2008;294:F777–F787. doi: 10.1152/ajprenal.00590.2007. [DOI] [PubMed] [Google Scholar]
- 29.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algül H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Bánhegyi G, Bao H, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolomé A, Bassham DC, Bassi MT, Bast RC, Jr, Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR, Jr, Becker C, Beckham JD, Bédard PA, Bednarski PJ, Begley TJ, Behl C, Behrends C, Behrens GM, Behrns KE, Bejarano E, Belaid A, Belleudi F, Bénard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S, Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B, Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Böckler S, Boes M, Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouché M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N, Braga VM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultynck G, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Bütikofer P, Caberlotto L, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao L, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Ceña V, Cenci S, Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EY, Chan MT, Chandra D, Chandra P, Chang CP, Chang RC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che Y, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen G, Chen H, Chen JW, Chen JK, Chen M, Chen M, Chen P, Chen Q, Chen Q, Chen SD, Chen S, Chen SS, Chen W, Chen WJ, Chen WQ, Chen W, Chen X, Chen YH, Chen YG, Chen Y, Chen Y, Chen Y, Chen YJ, Chen YQ, Chen Y, Chen Z, Chen Z, Cheng A, Cheng CH, Cheng H, Cheong H, Cherry S, Chesney J, Cheung CH, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GN, Cho DH, Cho SG, Cho WC, Cho YY, Cho YS, Choi AM, Choi EJ, Choi EK, Choi J, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung H, Chung T, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Clària J, Clarke PG, Clarke R, Clementi E, Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EE, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT, Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Cossarizza A, Costelli P, Costes S, Cotman SL, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui T, Cutillas PR, Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai C, Dai W, Dai Y, Dalby KN, Dalla Valle L, Dalmasso G, D'Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Dávila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P, de Figueiredo RC, de la Fuente J, De Martino L, De Matteis A, De Meyer GR, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LM, Deldicque L, Delorme-Axford E, Deng Y, Dengjel J, Denizot M, Dent P, Der CJ, Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Díaz-Araya G, Díaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding Z, Dini L, Distler JH, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RC, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong X, Dong Z, Donohue TM, Jr, Doran KS, D'Orazi G, Dorn GW, 2nd, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du L, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA, Jr, Dupont N, Dupuis L, Durán RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L, Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJ, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R, Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martínez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Færgeman NJ, Faggioni A, Fairlie WD, Fan C, Fan D, Fan J, Fang S, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng Y, Feng Y, Ferguson TA, Fernández ÁF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernández-López A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, François A, Frankel LB, Fraser ID, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu D, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannagé M, Gao FB, Gao F, Gao JX, García Nannig L, García Véscovi E, Garcia-Macía M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge P, Ge S, Gean PW, Gelmetti V, Genazzani AA, Geng J, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F, Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E, Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM, Jr, Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gómez-Sánchez R, Gonçalves DA, Goncu E, Gong Q, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, González-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo C, Guo L, Guo M, Guo W, Guo XG, Gust AA, Gustafsson ÅB, Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hammond EM, Han F, Han W, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U, Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He C, He CY, He F, He G, He RR, He XH, He YW, He YY, Heath JK, Hébert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her C, Herman PK, Hernández A, Hernandez C, Hernández-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Höglinger GU, Höhfeld J, Holz MK, Hong Y, Hood DA, Hoozemans JJ, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu G, Hu HM, Hu H, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang C, Huang HL, Huang KH, Huang KY, Huang S, Huang S, Huang WP, Huang YR, Huang Y, Huang Y, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SN, Hussain S, Hwang JJ, Hwang S, Hwang TI, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe K, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AK, Izquierdo JM, Izumi Y, Izzo V, Jäättelä M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia K, Jia L, Jiang H, Jiang H, Jiang L, Jiang T, Jiang X, Jiang X, Jiang X, Jiang Y, Jiang Y, Jiménez A, Jin C, Jin H, Jin L, Jin M, Jin S, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GV, Johnson JD, Jonasch E, Jones C, Joosten LA, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju D, Ju J, Juan HF, Juenemann K, Juhász G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kågedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut Ido C, Khambu B, Khan MM, Khandelwal VK, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim JH, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knævelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Köhler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong D, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou Y, Koukourakis MI, Koumenis C, Kovács AL, Kovács T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar A, Kumar D, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJ, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BY, Law HK, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SJ, Lee SY, Lee SH, Lee SS, Lee SJ, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei J, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng S, Lenz G, Lenzi P, Lerman LO, Lettieri Barbato D, Leu JI, Leung HY, Levine B, Lewis PA, Lezoualc'h F, Li C, Li F, Li FJ, Li J, Li K, Li L, Li M, Li M, Li Q, Li R, Li S, Li W, Li W, Li X, Li Y, Lian J, Liang C, Liang Q, Liao Y, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin F, Lin FC, Lin K, Lin KH, Lin PH, Lin T, Lin WW, Lin YS, Lin Y, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu J, Liu JJ, Liu JL, Liu K, Liu L, Liu L, Liu Q, Liu RY, Liu S, Liu S, Liu W, Liu XD, Liu X, Liu XH, Liu X, Liu X, Liu X, Liu Y, Liu Y, Liu Z, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, López-Otín C, López-Vicario C, Lorente M, Lorenzi PL, Lõrincz P, Los M, Lotze MT, Lovat PE, Lu B, Lu B, Lu J, Lu Q, Lu SM, Lu S, Lu Y, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo H, Luo J, Luo S, Luparello C, Lyons T, Ma J, Ma Y, Ma Y, Ma Z, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magariños M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manié SN, Manzoni C, Mao K, Mao Z, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Mariño G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martín-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martínez-Velázquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, McNeish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei Y, Meier UC, Meijer AJ, Meléndez A, Melino G, Melino S, de Melo EJ, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng L, Meng LH, Meng S, Menghini R, Menko AS, Menna-Barreto RF, Menon MB, Meraz-Ríos MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Molinari M, Møller AB, Mollereau B, Mollinedo F, Mongillo M, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CE, Mpakou VE, Mukhtar H, Mulcahy Levy JM, Muller S, Muñoz-Moreno R, Muñoz-Pinedo C, Münz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VC, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nürnberger T, O'Donnell VB, O'Donovan T, O'Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O'Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JH, Outeiro TF, Ouyang DY, Ouyang H, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palková Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan H, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KB, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstädt H, Pavone F, Pedrozo Z, Peña FJ, Peñalva MA, Pende M, Peng J, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJ, Pereira PC, Pérez-de la, Pérez-Pérez ME, Pérez-Rodríguez D, Pérez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carle V, Pietrocola F, Pimentel-Muiños FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Pöggeler S, Poirot M, Polčic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu Z, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren X, Renna M, Reusch JE, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodrigues CM, Rodríguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MI, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth KA, Roué G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB, 3rd, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AM, Sánchez-Alcázar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, Dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schönenberger MJ, Schönthal AH, Schorderet DF, Schröder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Seguí-Simarro JM, Segura-Aguilar J, Seki E, Sell C, Seiliez I, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CK, Shen CC, Shen HM, Shen S, Shen W, Sheng R, Sheng X, Sheng ZH, Shepherd TG, Shi J, Shi Q, Shi Q, Shi Y, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJ, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninová I, Slavov N, Smaili SS, Smalley KS, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song C, Song F, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, Clair DS, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Ström AL, Stromhaug P, Stulik J, Su YX, Su Z, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui X, Sukseree S, Sulzer D, Sun FL, Sun J, Sun J, Sun SY, Sun Y, Sun Y, Sun Y, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Swärd K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SW, Takacs-Vellai K, Takahashi Y, Takáts S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang D, Tang D, Tang G, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thomé MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TL, Tian L, Till A, Ting JP, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu S, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RF, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcátegui NL, Vaccari T, Vaccaro MI, Váchová L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJ, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, van Lookeren Campagne M, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PS, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira HL, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan X, Wang B, Wang C, Wang CY, Wang C, Wang C, Wang C, Wang D, Wang F, Wang F, Wang G, Wang HJ, Wang H, Wang HG, Wang H, Wang HD, Wang J, Wang J, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang X, Wang X, Wang Y, Wang Y, Wang Y, Wang YJ, Wang Y, Wang Y, Wang YT, Wang Y, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei J, Weide T, Weihl CC, Weindl G, Weis SN, Wen L, Wen X, Wen Y, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VK, Woodcock EA, Wright KL, Wu C, Wu D, Wu GS, Wu J, Wu J, Wu M, Wu M, Wu S, Wu WK, Wu Y, Wu Z, Xavier CP, Xavier RJ, Xia GX, Xia T, Xia W, Xia Y, Xiao H, Xiao J, Xiao S, Xiao W, Xie CM, Xie Z, Xie Z, Xilouri M, Xiong Y, Xu C, Xu C, Xu F, Xu H, Xu H, Xu J, Xu J, Xu J, Xu L, Xu X, Xu Y, Xu Y, Xu ZX, Xu Z, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang M, Yang PM, Yang P, Yang Q, Yang W, Yang WY, Yang X, Yang Y, Yang Y, Yang Z, Yang Z, Yao MC, Yao PJ, Yao X, Yao Z, Yao Z, Yasui LS, Ye M, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida K, Yoshimori T, Young KH, Yu H, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu Z, Yuan J, Yuan ZM, Yue BY, Yue J, Yue Z, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng J, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang H, Zhang H, Zhang H, Zhang J, Zhang J, Zhang J, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang L, Zhang L, Zhang MY, Zhang X, Zhang XD, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhao M, Zhao WL, Zhao X, Zhao YG, Zhao Y, Zhao Y, Zhao YX, Zhao Z, Zhao ZJ, Zheng D, Zheng XL, Zheng X, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou G, Zhou H, Zhou SF, Zhou XJ, Zhu H, Zhu H, Zhu WG, Zhu W, Zhu XF, Zhu Y, Zhuang SM, Zhuang X, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A, Zughaier SM. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berridge MJ. Neuronal calcium signaling. Neuron. 1998;21:13–26. doi: 10.1016/s0896-6273(00)80510-3. [DOI] [PubMed] [Google Scholar]
- 31.Berliocchi L, Bano D, Nicotera P. Ca2+ signals and death programmes in neurons. Philos Trans R Soc Lond B Biol Sci. 2005;360:2255–2258. doi: 10.1098/rstb.2005.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bootman MD, Collins TJ, Peppiatt CM, Prothero LS, MacKenzie L, De Smet P, Travers M, Tovey SC, Seo JT, Berridge MJ, Ciccolini F, Lipp P. Calcium signalling--an overview. Semin Cell Dev Biol. 2001;12:3–10. doi: 10.1006/scdb.2000.0211. [DOI] [PubMed] [Google Scholar]
- 33.Graham SH, Chen J. Programmed cell death in cerebral ischemia. J Cereb Blood Flow Metab. 2001;21:99–109. doi: 10.1097/00004647-200102000-00001. [DOI] [PubMed] [Google Scholar]
- 34.Høyer-Hansen M, Bastholm L, Szyniarowski P, Campanella M, Szabadkai G, Farkas T, Bianchi K, Fehrenbacher N, Elling F, Rizzuto R, Mathiasen IS, Jäättelä M. Control of macroautophagy by calcium, calmodulin-dependent kinase kinase-beta, and Bcl-2. Mol Cell. 2007;25:193–205. doi: 10.1016/j.molcel.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 35.Harr MW, Distelhorst CW. Apoptosis and autophagy: decoding calcium signals that mediate life or death. Cold Spring Harb Perspect Biol. 2010;2:a005579. doi: 10.1101/cshperspect.a005579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park HW, Park H, Semple IA, Jang I, Ro SH, Kim M, Cazares VA, Stuenkel EL, Kim JJ, Kim JS, Lee JH. Pharmacological correction of obesity-induced autophagy arrest using calcium channel blockers. Nat Commun. 2014;5:4834. doi: 10.1038/ncomms5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Köktürk S, Yardimoglu M, Celikozlu SD, Dolanbay EG, Cimbiz A. Effect of Lycopersicon esculentum extract on apoptosis in the rat cerebellum, following prenatal and postnatal exposure to an electromagnetic field. Exp Ther Med. 2013;6:52–56. doi: 10.3892/etm.2013.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salminen A, Kaarniranta K, Kauppinen A. Beclin 1 interactome controls the crosstalk between apoptosis, autophagy and inflammasome activation: impact on the aging process. Ageing Res Rev. 2013;12:520–534. doi: 10.1016/j.arr.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Pyo JO, Nah J, Jung YK. Molecules and their functions in autophagy. Exp Mol Med. 2012;44:73–80. doi: 10.3858/emm.2012.44.2.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19:983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 42.Nassif M, Valenzuela V, Rojas-Rivera D, Vidal R, Matus S, Castillo K, Fuentealba Y, Kroemer G, Levine B, Hetz C. Pathogenic role of BECN1/Beclin 1 in the development of amyotrophic lateral sclerosis. Autophagy. 2014;10:1256–1271. doi: 10.4161/auto.28784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shintani T, Klionsky DJ. Autophagy in health and disease: a doubleedged sword. Science. 2004;306:990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12:831–835. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 45.He C, Wei Y, Sun K, Li B, Dong X, Zou Z, Liu Y, Kinch LN, Khan S, Sinha S, Xavier RJ, Grishin NV, Xiao G, Eskelinen EL, Scherer PE, Whistler JL, Levine B. Beclin 2 functions in autophagy, degradation of G protein-coupled receptors, and metabolism. Cell. 2013;154:1085–1099. doi: 10.1016/j.cell.2013.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–170. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maskey D, Yousefi S, Schmid I, Zlobec I, Perren A, Friis R, Simon HU. ATG5 is induced by DNA-damaging agents and promotes mitotic catastrophe independent of autophagy. Nat Commun. 2013;4:2130. doi: 10.1038/ncomms3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy MP, LeVine H., 3rd Alzheimer's disease and the amyloidbeta peptide. J Alzheimers Dis. 2010;19:311–323. doi: 10.3233/JAD-2010-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho MH, Cho K, Kang HJ, Jeon EY, Kim HS, Kwon HJ, Kim HM, Kim DH, Yoon SY. Autophagy in microglia degrades extracellular β-amyloid fibrils and regulates the NLRP3 inflammasome. Autophagy. 2014;10:1761–1775. doi: 10.4161/auto.29647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neher E, Sakaba T. Multiple roles of calcium ions in the regulation of neurotransmitter release. Neuron. 2008;59:861–872. doi: 10.1016/j.neuron.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 52.Augustine GJ. How does calcium trigger neurotransmitter release? Curr Opin Neurobiol. 2001;11:320–326. doi: 10.1016/s0959-4388(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 53.Rubin RD, Watson PD, Duff MC, Cohen NJ. The role of the hippocampus in flexible cognition and social behavior. Front Hum Neurosci. 2014;8:742. doi: 10.3389/fnhum.2014.00742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mineur YS, Obayemi A, Wigestrand MB, Fote GM, Calarco CA, Li AM, Picciotto MR. Cholinergic signaling in the hippocampus regulates social stress resilience and anxiety- and depression-like behavior. Proc Natl Acad Sci U S A. 2013;110:3573–3578. doi: 10.1073/pnas.1219731110. [DOI] [PMC free article] [PubMed] [Google Scholar]