Abstract
Background
PIK3CA mutations frequently occur in breast cancer patients. This study was conducted to evaluate the relationship between PIK3CA mutations and neoadjuvant treatment response and to analyze the clinical implications.
Methods
PubMed, Embase, and the Cochrane database were searched for relevant studies in September 2017. The pooled risk ratio (RR) was estimated using fixed effects or random effects models according to heterogeneity among studies.
Results
This meta‐analysis included 20 studies with 4392 patients. The pooled RR showed that PIK3CA mutation is correlated to lower pathological complete response (pCR) in unselected HER2+ patients (RR = 0.73; 95% confidence interval [CI] 0.66–0.81), thus the predictive value of PIK3CA status may be stronger in HER2+/HR+ patients (RR = 0.50; 95% CI 0.27–0.93) and those administered dual‐targeting treatment (RR = 0.55; 95% CI 0.39–0.78). In contrast with wild type, either exon 9 (RR = 0.55; 95% CI 0.39–0.78) or exon 20 (RR = 0.71; 95% CI 0.58–0.89) mutations were significantly associated with lower pCR. The predictive value of exon 9 mutations was not significantly greater than exon 20 mutations (RR = 0.76; 95% CI 0.51–1.13).
Conclusion
In early breast cancer, PIK3CA mutations seem to identify HER2+ patients who are less likely to reach pCR. The clinical implications of PIK3CA mutations tend to vary between exon 9 and exon 20. This mechanism should be explored in further studies.
Keywords: Breast cancer, neoadjuvant treatment, PI3K pathway, PIK3CA
Introduction
Neoadjuvant treatment (NAT) is a conventional treatment for locally advanced breast cancer.1 It has been accepted as an important option for early stage breast cancer patients and achieves similar long‐term clinical outcomes as adjuvant treatment.2 The achievement of pathological complete response (pCR) is a valid predictor of good prognosis, especially for triple negative breast cancer (TNBC) and HER2+ patients.3, 4 Although many studies have explored the predictive biomarkers of NAT response, there is no current method to screen patients that may be sensitive to NAT. Promising biomarkers, such as tumor‐infiltrating lymphocytes (TILs), TP53, and the germline BRCA mutation, are under investigation.5, 6, 7
Activation of the PI3K pathway is common breast cancer,8 and results from PIK3CA mutation or PTEN loss.9 It has been reported that PIK3CA status impacts solid cancer prognosis.10, 11 More than 90% of PIK3CA mutations in breast tumors appear in exons 9 and 20.12
A number of studies of PIK3CA mutation in HER2+ breast cancer have been reported, but have mainly focused on the prognostic value to advanced stage breast cancer. Recently, a pooled analysis of 967 HER2+ breast cancer patients from five randomized trials was conducted.13 The authors found a significantly lower pCR rate in PIK3CA mutant (MT) compared to wild‐type (WT) tumors after neoadjuvant chemotherapy.
While the pCR rate is significantly lower in HER2+ patients, it remains uncertain in hormone receptor positive (HR+) and HR negative (HR‐)/HER2‐ subtypes. The biological functions of exon 9 and 20 mutations may be different,14 and whether such discrepancies could affect the response to NAT has not been fully elucidated. We conducted a systematic review and meta‐analysis of PIK3CA related studies of NAT to clarify the possible association between PIK3CA mutation and response to breast cancer NAT. Exon 9 or 20 mutations lead to PIK3CA mutation; therefore, we conducted subgroup analyses of relevant studies to determine pCR rates between exon 9 and 20 MT and WT tumors.
Methods
Search strategy
Online databases including PubMed, Embase, and the Cochrane database were searched to identify relevant literature published up to September 2017. The following key word combinations were used: “breast cancer,” “neoadjuvant,” and “PIK3CA.” Published studies were included based on the following criteria: (i) English publications; (ii) studies focusing on early stage breast cancer patients and NAT; and (iii) studies with clinical or pathological response outcomes. Studies were excluded if they were: (i) reviews or mechanism studies; or (ii) duplicate studies.
Data extraction
Two reviewers independently extracted the information from all eligible studies. Pathological or clinical response was the end point of interest. The following information was extracted: first author, region, population, sample size, PIK3CA mutation incidence, NAT regime, and PIK3CA sequence.
Statistical analysis
Fixed effects (Mantel–Haenszel) or random effects (DerSimonian–Laird) models were used to pool risk ratio and 95% confidence interval (CI), according to heterogeneity. The heterogeneity test was verified using Higgins–I2 statistics. If significant heterogeneity was observed (I2 > 50%), a random effects model was used; otherwise, the fixed effects model was used. Publication bias was estimated using an Egger’s test with a funnel plot. All P values were calculated using a two‐sided test and P < 0.05 was considered statistically significant. Statistical analyses in our study were carried out using Stata 12.0 (Stata Corp LP, College Station, TX, USA).
Results
A total of 313 studies were retrieved. After preliminary screening, 263 were excluded by title, abstract, and duplication. Studies with no response data (n = 15), overlapping data (n = 7), no full text article (n = 2), and review articles (n = 4) were also excluded. A total of 22 articles referring to 20 studies were included in our meta‐analysis (Fig 1).
Figure 1.
Flow diagram of the systematic search and selection process.
Study characteristics
As shown in Table 1, 20 studies including 4392 patients were included in our meta‐analysis.13, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 Overall, PIK3CA mutation incidence in our meta‐analysis was 22.4% (range 7.7–39.0%). pCR was 28% for PIK3CA MT and 38% for PIK3CA WT. Seven studies were conducted in the United States,17, 18, 22, 24, 25, 26, 28 nine in Europe,13, 15, 16, 19, 20, 21, 29, 30, 31 and four in Asia.23, 32, 33, 34 Two studies included objective response rate,18, 19 while the others reported pCR as the endpoint in WT versus MT PIK3CA tumors. Most of the included studies (12/20) used formalin‐fixed paraffin embedded breast samples. Most studies sequenced PIK3CA exons 9 and 20, while the remainder also analyzed one or more of exons 1, 4, 7, 9, and 20. Other information, such as NAT regime, first author, study name, and population are illustrated in Table 1. In each subgroup, the pCR rate was higher in PIK3CA WT than in MT patients, as illustrated in Table 2.
Table 1.
A summary of study characteristics
| Author year | Country | Study name | Population | Number of patients | Number of PIK3CA mutated patients | Endpoints | Sample type | NAT regime | Sequenced PIK3CA |
|---|---|---|---|---|---|---|---|---|---|
| Barbareschi et al. 201215 | Italy | N/A | HER2+ | 26 | 4 (15.4%) | pCR | FFPE | AH → TH → CMFH | Exon 9/20 |
| Bianchini et al. 201716 | Italy | NeoSphere | HER2+ | 417 | 81 (19.4%) | pCR | FFPE | (i) TH; (ii) TPH; (iii) PH; (iv) TP | Exon 7/9/20 |
| Dave et al. 201117 | USA | HER2+ | 80 | 15 (18.8%) | pCR | FFPE | (i) H; (ii) L | NR | |
| Ellis et al. 201018 | USA | P024, RAD 2222, ROL | HR+ | 235 | 76 (32.3%) | OR | FFPE | Tamoxifen + Letrozole | Exon 7/9/20 |
| Guarneri et al. 201419 | Italy | CONSORT | HR+/HER2‐ | 92 | 34 (37.0%) | OR | FFPE | (i) Letrozole; (ii) Letrozole + L | Exon 9/20 |
| Hanusch et al. 201520 | Germany | GBG‐70 | HER2+ | 61 | 13 (21.3%) | pCR | NR | Afatinib → TH Afatinib → ACH | Exon 9/20 |
| Harbeck et al. 201621 | Germany | WSG‐ADAPT | HR+/HER2+ | 114 | 18 (15.8%) | pCR | NR | (i) T‐DM1; (ii) T‐DM1 + Tamoxifen or AI; (iii) H + Tamoxifen or AI | NR |
| Haas et al. 201722 | USA | KRISTINE | HER2+ | 425 | 114 (26.8%) | pCR | NR | (i) T‐DM1 + P; (ii) TCbPH | NR |
| Huang 201523 | China | N/A | HER2+ | 77 | 30 (39.0%) | pCR | FFPE | (i) TCH; (ii) TAH | Exon 4/9/20 |
| Hoadley et al. 201524 | USA | CALGB 40601 | HER2+ | 181 | 14 (7.7%) | pCR | NR | (i) TL; (ii) TH; (iii) THL | Exon 9/20 |
| Loibl et al. 201625, 26 | USA | GeparSepto | HER2+ | 291 | 63 (21.6%) | pCR | FFPE | THP | Exon 9/20 |
| Loibl et al. 201627 | USA | GeparTrio | HER2+ | 82 | 31 (37.8%) | pCR | FFPE | (i) TAC; (ii) TAC → NX | NR |
| Loibl et al. 201613, 27 | Germany | GeparQuattro GeparQuinto GeparSixto NeoALTTO CHERLOB |
HER2+ | 967 | 210 (21.7%) | pCR | FFPE | (i) ACH → TH; (ii) ACL → TL; (iii) THL; (iv) TH; (v) TL; (vi) THB; (vii) TCbHB; (viii) TH → CAFH; (ix) TL → CAFL; (x) THL → CAFH | Exon 9/20 |
| Liedtke et al. 200828 | USA | N/A | ALL | 140 | 23 (16.4%) | pCR | NR | (i) FAC; (ii) T → FAC | Exon 1/9/20 |
| Lips et al. 201529 | Netherlands | N/A | TNBC | 140 | 23 (16.4%) | pCR | FTS | (i) AC; (ii) AC → TX; (iii) AC → XCb + Thiotepa | Exon 9/20 |
| Toomey et al. 201730 | Ireland | TCHL (ICORG10–05) | HER2+ | 74 | 18 (24.3%) | pCR | FFPE | (i) TCbL; (ii) TCbH; (iii) TCbHL | Exon 1/4/7/9/20 |
| Schneeweiss et al. 201431 | Germany | TRYPHAENA | HER2+ | 126 | 39 (31.0%) | pCR | NR | (i) FECHP → THP; (ii) FEC → THP; (iii) TCbHP | Exon 7/9/20 |
| Sueta et al. 201432 | Japan | N/A | HER2+ | 42 | 7 (16.7%) | pCR | FFPE | (i) FAC → T; (ii) TC | Exon 9/20 |
| Yuan et al. 201533 | China | N/A | ALL | 729 | 142 (19.5%) | pCR | FTS | (i) CAF; (ii) AC; (iii) A → T; (iv) A → TC; (v) A → TCb | Exon 9/20 |
| Zhang et al. 201434 | China | N/A | ALL | 93 | 30 (32.3%) | pCR | FFPE | TA | Exon 9/20 |
Pathological complete response (pCR) was based on Miller and Payne histopathology scoring system. Objective response (OR) was evaluated according Response Evaluation Criteria in Solid Tumors and was defined as complete + partial response. A, anthracycline; AI, aromatase inhibitors; ALL, all subtypes of breast cancer patients; B, bevacizumab; C, cyclophosphamide; Cb, carboplatin; FFPE, formalin‐fixed, paraffin‐embedded; F, fluorouracil; FTS, frozen tissue sample; G, gemcitabine; H, trastuzumab; HR, hormone receptor; L, lapatinib; M, methotrexate; N, vinorelbine; N/A, not applicable; NR, not reported; P, pertuzumab; T, taxanes; TNBC, triple negative breast cancer; X, capecitabine.
Table 2.
A summary of pCR incidence among different subgroups
| PIK3CA status | pCR | Non‐pCR | pCR rate (%) | |
|---|---|---|---|---|
| Overall | MT | 323 | 841 | 28 |
| WT | 1252 | 2052 | 38 | |
| HR+ | MT | 39 | 268 | 13 |
| WT | 276 | 731 | 27 | |
| HR‐ | MT | 50 | 107 | 32 |
| WT | 247 | 351 | 41 | |
| HER2+ | MT | 287 | 636 | 31 |
| WT | 1068 | 1482 | 42 | |
| HER2‐ | MT | 17 | 132 | 11 |
| WT | 94 | 317 | 23 | |
| Exon 9 | MT | 28 | 175 | 14 |
| Exon 20 | MT | 76 | 320 | 19 |
HR, hormone receptor; MT, mutant; pCR, pathological complete response; WT, wild type.
Meta‐analysis
PIK3CA mutations and pathological complete response (pCR) in HER2+ patients
A total of 13 studies of unselected HER2+ patients were used for analysis (Table 3).13, 15, 17, 20, 22, 23, 24, 25, 26, 30, 31, 32, 33 In this study, unselected HER2+ patients are defined as the entire HER2+ population with no restriction to HR status or NAT regime. The fixed effects model was used because of low heterogeneity, except in the HER2+/HR+ subgroup. WT unselected HER2+ patients achieved a higher rate of pCR (RR = 0.73; 95% CI 0.66–0.81) (Fig 2a). There were significant statistical differences in pCR between PIK3CA MT and WT after single‐targeting trastuzumab treatment (RR = 0.71; 95% CI 0.54–0.94) (Fig 2b), but not after single‐targeting lapatinib treatment (RR = 0.76; 95% CI 0.42–1.37). The trend remained significant in the HER2+/HR+ (RR = 0.50; 95% CI 0.27–0.93) and trastuzumab dual‐targeting (RR = 0.71; 95% CI 0.62–0.80) subgroups (Fig 2c).
Table 3.
A summary of pooled RRs of patients with PIK3CA WT and MT
| Categories by PIK3CA and NAT | No. of studies | PIK3CA MT | PIK3CA WT | Pooled RR | P | Heterogeneity (I2) (%) | ||
|---|---|---|---|---|---|---|---|---|
| pCR | Non‐pCR | pCR | Non‐pCR | |||||
| Unselected breast cancer | 3 | 35 | 223 | 138 | 563 | 0.70 (0.49–0.98) | 0.036 | 0.0 |
| Unselected HER2+ | 13 | 286 | 636 | 1068 | 1482 | 0.73 (0.66–0.81) | 0.00 | 0.9 |
| HER2+/HR+ | 3 | 30 | 147 | 237 | 447 | 0.50 (0.27–0.93) | 0.028 | 64.4 |
| HER2+/HR‐ | 2 | 38 | 78 | 172 | 223 | 0.72 (0.55–0.95) | 0.02 | 0.0 |
| HER2+ with single trastuzumab | 7 | 46 | 117 | 164 | 280 | 0.71 (0.54–0.94) | 0.016 | 0.0 |
| HER2+ with single laptinib | 2 | 10 | 57 | 47 | 168 | 0.76 (0.42–1.37) | 0.363 | 0.0 |
| HER2+ with dual‐targeting treatment | 7 | 196 | 370 | 740 | 794 | 0.71 (0.62–0.80) | 0.00 | 42.1 |
| Unselected HR+ | 2 | 9 | 121 | 39 | 284 | 0.74 (0.22–2.44) | 0.615 | 52.1 |
| Unselected HR‐ | 1 | 2 | 5 | 15 | 28 | 1.01 (0.29–3.51) | 0.99 | NA |
| HR‐/HER2‐ | 2 | 10 | 24 | 60 | 90 | 0.77 (0.44–1.34) | 0.353 | 0.0 |
| HR+ with neoadjuvant endocrine therapy | 2 | 69 | 40 | 138 | 67 | 1.03 (0.63–1.70) | 0.901 | 86.0 |
P was used to estimate the difference when P < 0.05. Unselected were defined irrespective of HER2 status or therapy regime. Pathological complete response (pCR) was based on Miller and Payne histopathology scoring system. Objective response (OR) was evaluated according Response Evaluation Criteria in Solid Tumors and was defined as complete + partial response. HR, hormone receptor; MT, mutant; NA, not applicable; RR, risk ratio; WT, wild type.
Figure 2.
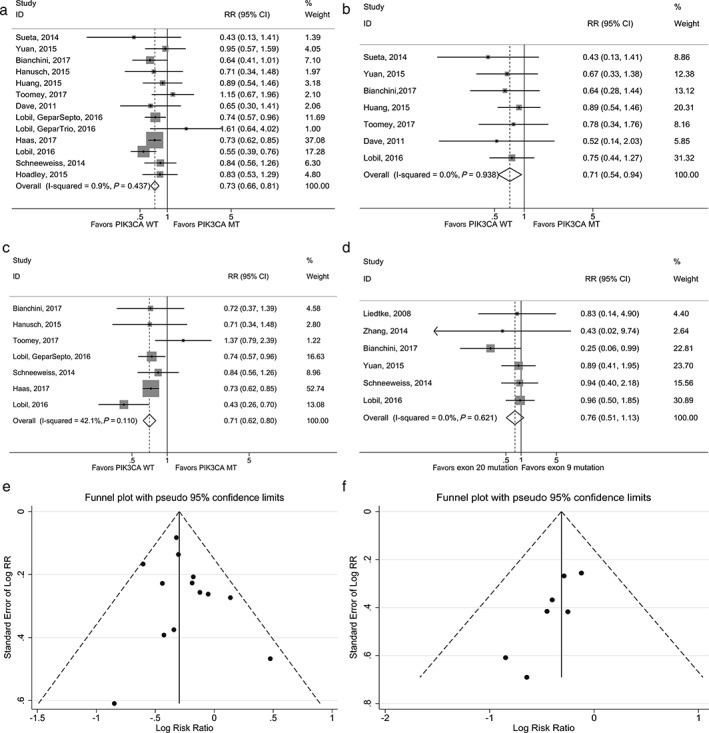
Forest plot of pathological complete response (pCR) of risk ratio (RR) with PIK3CA mutation (MT) versus wild type (WT) in (a) HER2+ patients, (b) in HER2+ patients with restriction to single‐targeting trastuzumab treatment, and (c) in HER2+ patients with restriction to dual‐targeting treatment. (d) Forest plot of pCR of RR with exon 9 versus exon 20. Funnel plot for meta‐analysis of pCR with PIK3CA MT versus WT (e) in unselected HER2+ patients (13 studies) and (f) in HER2+ patients with restriction to single‐targeting trastuzumab treatment (7 studies). P value was used to estimate the difference when P < 0.05. CI, confidence interval.
PIK3CA mutations and pCR in unselected hormone receptor positive (HR+) patients
We identified two studies investigating pCR in unselected HR+ patients regarding PIK3CA status (Table 3).28, 33 Pooled RR was 0.74 (95% CI 0.22–2.44). The random effects model was used because heterogeneity between the studies (I2 = 52.1%) was found. PIK3CA status was not associated with pCR in HR+ patients.
PIK3CA mutations and pCR in unselected HR‐ patients
Little data of pCR in unselected HR‐ and PIK3CA mutated patients was available. Liedtke et al. reported that PIK3CA MT did not influence pCR rate in unselected HR‐ patients (RR = 1.01; 95% CI 0.29–3.51) (Table 3).28
PIK3CA mutations and pCR in HR‐/HER2‐ patients
Two studies investigated pCR in HR‐/HER2‐ patients to PIK3CA mutation status (Table 3).29, 33 Pooled RR was 0.77 (95% CI 0.44–1.34). The fixed effects model was used because of low heterogeneity (I2 = 0.0%). PIK3CA status was not associated with pCR in HR‐/HER2‐ patients.
PIK3CA mutations and response in HR+ patients with neoadjuvant endocrine therapy
Two studies investigated neoadjuvant endocrine therapy, with objective response rate (partial and complete response by Response Evaluation Criteria in Solid Tumors) as their outcome (Table 1).18, 19 PIK3CA status was not related to objective response (RR = 1.03; 95% CI 0.63–1.70), with significant heterogeneity (I2 = 86.0%) (Table 3), thus the random effects model was used (Table 4).
Table 4.
A summary of pooled RRs of patients with exon 9/20 and WT
| Categories by mutation region | No. of studies | PIK3CA MT (exon 9 or 20) | PIK3CA WT (exon 9 or 20) | Pooled RR | P | Heterogeneity (I2) (%) | ||
|---|---|---|---|---|---|---|---|---|
| pCR | Non‐pCR | pCR | Non‐pCR | |||||
| Exon 9 | 6 | 28 | 175 | 494 | 1354 | 0.55 (0.39–0.78) | 0.001 | 0.0 |
| Exon 20 | 6 | 76 | 320 | 494 | 1354 | 0.71 (0.58–0.89) | 0.002 | 6.4 |
P was used to estimate the difference when P < 0.05. MT, mutant; pCR, pathological complete response; RR, risk ratio; WT, wild type.
Exon 9 and 20 mutations in PIK3CA and pCR
Six studies separately reported pCR between PIK3CA exon 9 and PIK3CA exon 20 mutations.16, 27, 28, 31, 33, 34 There was no heterogeneity among studies. Both PIK3CA exon 9 and 20 mutations were significantly associated with lower pCR compared to WT. A comparison between exon 9 and exon 20 mutations was conducted. PIK3CA exon 20 mutations may yield a lower pCR (RR = 0.76; 95% CI 0.51–1.13) (Fig 2d, Table 5).
Table 5.
A summary of pooled RRs of patients between exons 9 and 20
| Categories by mutation region | No. of studies | Exon 9 | Exon 20 | Pooled RR | P | Heterogeneity (I2) (%) | ||
|---|---|---|---|---|---|---|---|---|
| pCR | Non‐pCR | pCR | Non‐pCR | |||||
| Exon 9 and Exon 20 | 6 | 28 | 175 | 76 | 320 | 0.76 (0.51–1.13) | 0.169 | 0.0 |
P was used to estimate the difference when P < 0.05. MT, mutant; pCR, pathological complete response; RR, risk ratio.
Sensitivity analysis and publication bias
After excluding two studies by Loibl et al., the pooled RR (RR = 0.82; 95% CI 0.63–1.07) was insignificant.13, 27 The other results were significant, suggesting that no single study had any influence on the pooled RR. The funnel plot and Egger’s test (P = 0.014) showed publication bias in the HER2+ subgroup of single‐targeting trastuzumab therapy (Fig 2f), but not in unselected HER2+ patients (Fig 2e).
Discussion
To our knowledge, this is the first systematic review and meta‐analysis to determine a relationship between PIK3CA mutation and NAT response in early stage breast cancer. Previous preclinical and clinical studies suggest that exon 9 and 20 mutations may differ. However, the predictive value of pCR between exon 9 and 20 mutations is not definitive.
Preclinical studies suggest that PIK3CA mutation might result in abnormal PI3K pathway activation, which leads to resistance to trastuzumab.35 Our analysis confirms these results. In all HER2+ patients, PIK3CA MT appears to play a relevant role in defining the likelihood of lower pCR in NAT.
There was obvious publication bias among seven subgroup studies of single‐targeting trastuzumab therapy; however, neither heterogeneity nor sensitivity analysis was obvious in this subgroup. Four of the studies were funded by national/academic funding,17, 23, 30, 33 one was industry‐funded,13 one was funded by both national/academic and industry funding,16 and one received no funding.32 Improved access to unpublished data is needed to overcome the problem of potential bias in results.
Hormone receptor and HER2 subtypes represent different diseases that differ in clinical behavior as well as in sensitivity to chemotherapy.36 The predictive value of PIK3CA status in unselected HR+ and HR‐ patients is unclear. Our pooled analysis of seven studies proved that pCR in the HR+/HER2+ subgroup might be significantly related to PIK3CA status. This result indicates a potential interaction between HR and HER2 pathways.
PIK3CA mutations were associated with a lower pCR rate in the HR‐/HER2‐ subgroup, although the difference was insignificant. This might be a result of the relatively small sample size of the HR‐/HER2‐ subgroup, with a relatively low occurrence of PIK3CA mutations.37
Activation of the PI3K pathway might lead to anti‐estrogen resistance.38 We found no difference between PIK3CA mutation status and neoadjuvant endocrine therapy response. Heterogeneity was found between two studies.18, 19 Results of a study by Guarneri et al. indicated that PIK3CA MT might lead to a favorable objective response to endocrine therapy,19 while Ellis et al. reached a different conclusion.18 The disparity may result from the different regimes used. In the study by Guarneri et al., HR+/HER2‐ patients were likely to benefit from additional lapatinib, particularly those with PIK3CA mutations; however, neoadjuvant endocrine therapy is still at an early stage.
Prognostic association between PIK3CA status and survival among studies remains controversial. Yang et al. reported that the prognostic role of PIK3CA may differ between various subgroups.39 PIK3CA mutations are associated with favorable outcomes in HR+ patients after endocrine therapy.40, 41, 42 In HER2+ patients, some studies have reported that PI3KCA mutations are not related to prognosis;13, 43 however others suggest that PI3KCA mutations are associated with poorer outcomes.44
In vitro studies found that PIK3CA exon 9 and 20 mutations may differ14, therefore, the clinical implications of exon 9 and 20 mutations on pCR require explanation. pCR was the same between exons 9 and 20 MT. The possible reasons for this result are as follows: (i) exon 9 and 20 mutations were often combined for analysis and some studies did not report the number of PIK3CA exon 9 and 20 mutations, which may generate selection bias; (ii) insignificant results between exon 9 and 20 mutations may have resulted from the small sample size of only 203 exon 9 and 396 exon 20 mutations, which is relatively low; and (iii) heterogeneity among patients. The frequency of PIK3CA mutation and pCR may vary among different subtypes.
There are some limitations to this analysis. First, because we chose English‐based articles we may have overlooked important information published in other languages. Second, clinical heterogeneity may exist among studies, such as age, race, NAT regime, and test method. Different NAT might have a significant impact on pCR, but this could not be concluded as a result of the small study sample. Third, clinical and methodological heterogeneity existed among the studies. Finally, the mutation detection methods were different across the studies, including direct, Sanger, pyrosequencing, and DNA sequencing platforms.
In early stage breast cancer, PIK3CA mutations seem to identify HER2+ patients who are likely to achieve a low pCR. The clinical implications of PIK3CA mutations might vary between exon 9 and exon 20 mutations after NAT. This mechanism should be explored by further study.
Disclosure
No authors report any conflict of interest.
Acknowledgments
The study was sponsored and funded by the National Natural Science Foundation of China (81573504/81673509), a Precision Medical Project Grant by the National Key Research and Development Program (2016YFSF090494), and Beijing Municipal Natural Science Foundation (7171012).
References
- 1. Fisher B, Bryant J, Wolmark N et al. Effect of preoperative chemotherapy on the outcome of women with operable breast cancer. J Clin Oncol 1998;16:2672–85. [DOI] [PubMed] [Google Scholar]
- 2. Mauri D, Pavlidis N, Ioannidis JPA. Neoadjuvant versus adjuvant systemic treatment in breast cancer: A meta‐analysis. J Natl Cancer Inst 2005; 97: 188–94. [DOI] [PubMed] [Google Scholar]
- 3. Kong X, Moran MS, Zhang N, Haffty B, Yang Q. Meta‐analysis confirms achieving pathological complete response after neoadjuvant chemotherapy predicts favourable prognosis for breast cancer patients. Eur J Cancer 2011; 47: 2084–90. [DOI] [PubMed] [Google Scholar]
- 4. von Minckwitz G, Untch M, Blohmer JU et al Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol 2012; 30: 1796–804. [DOI] [PubMed] [Google Scholar]
- 5. Loi S, Michiels S, Salgado R et al Tumor infiltrating lymphocytes (TILs) indicate trastuzumab benefit in early‐stage HER2‐positive breast cancer (HER2+ BC). Cancer Res 2013; 73 (24 Suppl): Abstract S1‐05. [Google Scholar]
- 6. Bonnefoi H, Piccart M, Bogaerts J et al TP53 status for prediction of sensitivity to taxane versus non‐taxane neoadjuvant chemotherapy in breast cancer (EORTC 10994/BIG 1‐00): A randomized phase 3 trial. (Published erratum appears in Lancet Oncol 2013;14:e47. Lancet Oncol 2011; 12: 527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. von Minckwitz G, Hahnen E, Fasching PA et al Pathological complete response (pCR) rates after carboplatin‐containing neoadjuvant chemotherapy in patients with germline BRCA (gBRCA) mutation and triple‐negative breast cancer (TNBC): Results from GeparSixto. J Clin Oncol 2014; 32 (15 Suppl): Abstract 1005. [Google Scholar]
- 8. Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt proto‐oncogene product by phosphatidylinositol‐3,4‐bisphosphate. Science 1997; 275: 665–8. [DOI] [PubMed] [Google Scholar]
- 10. Cicenas J, Urban P, Vuaroqueaux V et al Increased level of phosphorylated akt measured by chemiluminescence‐linked immunosorbent assay is a predictor of poor prognosis in primary breast cancer overexpressing ErbB‐2. Breast Cancer Res 2005; 7 (4): R394–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mei ZB, Duan CY, Li CB, Cui L, Ogino S. Prognostic role of tumor PIK3CA mutation in colorectal cancer: A systematic review and meta‐analysis. Ann Oncol 2016; 27: 1836–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Samuels Y, Wang Z, Bardelli A et al High frequency of mutations of the PIK3CA gene in human cancers. Science 2004; 304: 554. [DOI] [PubMed] [Google Scholar]
- 13. Loibl S, Majewski I, Guarneri V et al PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2‐positive breast cancer: Pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab. Ann Oncol 2016; 27: 1519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao L, Vogt PK. Helical domain and kinase domain mutations in p110alpha of phosphatidylinositol 3‐kinase induce gain of function by different mechanisms. Proc Natl Acad Sci U S A 2008; 105: 2652–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Barbareschi M, Cuorvo LV, Girlando S et al PI3KCA mutations and/or PTEN loss in Her2‐positive breast carcinomas treated with trastuzumab are not related to resistance to anti‐Her2 therapy. Virchows Arch 2012; 461: 129–39. [DOI] [PubMed] [Google Scholar]
- 16. Bianchini G, Kiermaier A, Bianchi GV et al Biomarker analysis of the NeoSphere study: Pertuzumab, trastuzumab, and docetaxel versus trastuzumab plus docetaxel, pertuzumab plus trastuzumab, or pertuzumab plus docetaxel for the neoadjuvant treatment of HER2‐positive breast cancer. Breast Cancer Res 2017; 19 (1): 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dave B, Migliaccio I, Gutierrez MC et al Loss of phosphatase and tensin homolog or phosphoinositol‐3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2‐overexpressing locally advanced breast cancers. J Clin Oncol 2011; 29: 166–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ellis MJ, Lin L, Crowder R et al Phosphatidyl‐inositol‐3‐kinase alpha catalytic subunit mutation and response to neoadjuvant endocrine therapy for estrogen receptor positive breast cancer. Breast Cancer Res Treat 2010; 119: 379–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guarneri V, Generali DG, Frassoldati A et al Double‐blind, placebo‐controlled, multicenter, randomized, phase IIb neoadjuvant study of letrozole‐lapatinib in postmenopausal hormone receptor‐positive, human epidermal growth factor receptor 2‐negative, operable breast cancer. J Clin Oncol 2014; 32: 1050–7. [DOI] [PubMed] [Google Scholar]
- 20. Hanusch C, Schneeweiss A, Loibl S et al Dual blockade with AFatinib and Trastuzumab as NEoadjuvant treatment for patients with locally advanced or operable breast cancer receiving taxane‐anthracycline containing chemotherapy DAFNE (GBG‐70). (Published erratum appears in Clin Cancer Res 2016;22:4536. Clin Cancer Res 2015; 21: 2924–31. [DOI] [PubMed] [Google Scholar]
- 21. Harbeck N, Gluz O, Christgen M et al Abstract S5‐03: Final analysis of WSG‐ADAPT HER2+/HR+ phase II trial: Efficacy, safety, and predictive markers for 12‐weeks of neoadjuvant TDM1 with or without endocrine therapy versus trastuzumab+endocrine therapy in HER2‐positive hormone‐receptor‐positive early breast cancer. Cancer Res 2016; 76 (4 Suppl): Abstract S5‐03. [Google Scholar]
- 22. de Haas SL , Hurvitz SA, Martin M et al Abstract P6‐07‐09: Biomarker analysis from the neoadjuvant KRISTINE study in HER2‐positive early breast cancer (EBC), Cancer Res 2017;77 (4 Suppl): Abstract P6‐07‐09. [Google Scholar]
- 23. Huang L, Chen S, Yang W et al Efficacy and safety analysis of trastuzumab and paclitaxel based regimen plus carboplatin or epirubicin as neoadjuvant therapy for clinical stage II‐III, HER2‐positive breast cancer patients: A phase 2, open‐label, multicenter, randomized trial. Oncotarget 2015; 6: 18683–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoadley KA, Barry WT, Pitcher BN et al Abstract S3‐06: Mutational analysis of CALGB 40601 (Alliance), a neoadjuvant phase III trial of weekly paclitaxel (T) and trastuzumab (H) with or without lapatinib (L) for HER2‐positive breast cancer. Cancer Res 2015; 75 (9 Suppl): Abstract S3‐06. [Google Scholar]
- 25. Loibl S, Budczies J, Weichert W et al Abstract P3‐07‐03: PIK3CA mutations predict resistance to trastuzumab/pertuzumab and nab‐paclitaxel in primary HER2‐positive breast cancer – Massive parallel sequencing analysis of 293 pretherapeutic core biopsies of the GeparSepto study. Cancer Res 2016; 76 (4 Suppl): Abstract P3‐07‐03. [Google Scholar]
- 26. Loibl S, Budczies J, Weichert W et al Abstract 5010: Prediction of therapy resistance by targeted massive‐parallel sequencing in primary HER2‐positive breast cancer. Cancer Res 2016; 76 (14 Suppl): Abstract 5010. [Google Scholar]
- 27. Loibl S, Majewski I, Guarneri V e a. Correlation of PIK3CA mutation with pathological complete response in primary HER2‐positive breast cancer: Combined analysis of 967 patients from three prospective clinical trials. 2015 ASCO Annual Meeting Proceedings. J Clin Oncol 2015; 33 (15 Suppl): Abstract 511. [Google Scholar]
- 28. Liedtke C, Cardone L, Tordai A et al PIK3CA‐activating mutations and chemotherapy sensitivity in stage II‐III breast cancer. Breast Cancer Res 2008; 10 (2): R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lips EH, Michaut M, Hoogstraat M et al Next generation sequencing of triple negative breast cancer to find predictors for chemotherapy response. Breast Cancer Res 2015; 17 (1): 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Toomey S, Eustace AJ, Fay J et al Impact of somatic PI3K pathway and ERBB family mutations on pathological complete response (pCR) in HER2‐positive breast cancer patients who received neoadjuvant HER2‐targeted therapies. Breast Cancer Res 2017; 19 (1): 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schneeweiss A, Chia S, Hegg R et al Evaluating the predictive value of biomarkers for efficacy outcomes in response to pertuzumab‐ and trastuzumab‐based therapy: An exploratory analysis of the TRYPHAENA study. Breast Cancer Res 2014; 16 (4): R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sueta A, Yamamoto Y, Yamamoto‐Ibusuki M et al An integrative analysis of PIK3CA mutation, PTEN, and INPP4B expression in terms of trastuzumab efficacy in HER2‐positive breast cancer. PLoS One 2014; 9: e116054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yuan H, Chen J, Liu Y et al Association of PIK3CA mutation status before and after neoadjuvant chemotherapy with response to chemotherapy in women with breast cancer. Clin Cancer Res 2015; 21: 4365–72. [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y, Liu M, Yang H et al PIK3CA mutations are a predictor of docetaxel plus epirubicin neoadjuvant chemotherapy clinical efficacy in breast cancer. Neoplasma 2014; 61: 461–7. [DOI] [PubMed] [Google Scholar]
- 35. Hanker AB, Pfefferle AD, Balko JM et al Mutant PIK3CA accelerates HER2‐driven transgenic mammary tumors and induces resistance to combinations of anti‐HER2 therapies. Proc Natl Acad Sci U S A 2013; 110: 14372–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sørlie T, Perou CM, Tibshirani R et al Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001; 98: 10869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer 2006; 94: 455–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Miller TW, Balko JM, Arteaga CL. Phosphatidylinositol 3‐kinase and antiestrogen resistance in breast cancer. J Clin Oncol 2011; 29: 4452–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang SX, Polley E, Lipkowitz S. New insights on PI3K/AKT pathway alterations and clinical outcomes in breast cancer. Cancer Treat Rev 2016; 45: 87–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Loi S, Haibe‐Kains B, Majjaj S et al PIK3CA mutations associated with gene signature of low mTORC1 signaling and better outcomes in estrogen receptor‐positive breast cancer. Proc Natl Acad Sci U S A 2010; 107: 10208–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ellis MJ, Lin L, Crowder R et al Phosphatidyl‐inositol‐3‐kinase alpha catalytic subunit mutation and response to neoadjuvant endocrine therapy for estrogen receptor positive breast cancer. Breast Cancer Res Treat 2010; 119: 379–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kalinsky K, Jacks LM, Heguy A et al PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res 2009; 15: 5049–59. [DOI] [PubMed] [Google Scholar]
- 43. Loi S, Michiels S, Lambrechts D et al Somatic mutation profiling and associations with prognosis and trastuzumab benefit in early breast cancer. J Natl Cancer Inst 2013; 105: 960–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Baselga J, Cortés J, Im SA et al Abstract S5‐1: Biomarker analyses in CLEOPATRA: A phase III, placebo‐controlled study of pertuzumab in HER2‐positive, first‐line metastatic breast cancer (MBC). Cancer Res 2012; 72 (24 Suppl): Abstract S5‐1. [DOI] [PubMed] [Google Scholar]


