Abstract
Background
The EML4‐ALK fusion gene has recently been identified as a driver mutation in a subset of non‐small cell lung cancers. In subsequent studies, EML4‐ALK has been detected in a low percentage of patients, and was associated with a lack of EGFR or KRAS mutations, younger age, and adenocarcinoma with acinar histology. Cases with the EML4‐ALK fusion gene were examined to clarify the clinicopathological characteristics of young adenocarcinoma patients.
Methods
Between December 1998 and May 2009, 85 patients aged ≤ 50 with lung adenocarcinoma were treated at our hospital. We examined 49 samples from adenocarcinoma patients who underwent surgical resection, chemotherapy, and/or radiotherapy for the EML4‐ALK gene. None of the patients received ALK inhibitors because these drugs had not been approved in Japan before 2012. EML4‐ALK fusion genes were screened using multiplex reverse‐transcription PCR assay, and were confirmed by direct sequencing.
Results
The EML4‐ALK fusion gene was detected in five tumors (10.2%). One patient had stage IB disease, one had stage IIIA, and three had stage IV. Histologically, there was one solid adenocarcinoma, two acinar adenocarcinomas, and two papillary adenocarcinomas. EML4‐ALK fusion genes were mutually exclusive to EGFR and KRAS mutations. The five‐year survival rate was 59.4% in patients without EML4‐ALK fusion and was not reached in patients with EML4‐ALK fusion.
Conclusion
The EML4‐ALK fusion gene may be a strong oncogene in younger patients with lung adenocarcinoma.
Keywords: Adenocarcinoma, EML4‐ALK, non‐small cell lung cancer, young patient
Introduction
Lung cancer is one of the most prevalent cancers worldwide, and the mortality rate is expected to remain very high for several decades. Although a combination of surgery, chemotherapy, and/or radiotherapy can be used to treat non‐small cell lung cancer (NSCLC), the prognosis for patients remains dismal. While the histologic subtype is an important factor for choosing between standard cytotoxic chemotherapies, tyrosine kinase‐based therapeutics also play a key role, particularly in genetically defined subsets of patients. Following the discovery of activating mutations in EGFR associated with sensitivity to EGFR‐tyrosine kinase inhibitors (TKIs),1 therapy with gefitinib, erlotinib, or afatinib has become a first‐line treatment for patients with EGFR mutations.2, 3
In 2007, Soda et al. identified another type of tyrosine kinase with accelerated activity in a fusion gene formed between EML4 and ALK located within chromosome 2p.4 Previous studies have reported that 1.6–13.5% of lung tumors harbor EML4‐ALK fusions.4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15 Large‐scale screening using reverse‐transcription (RT)‐PCR in 7344 NSCLC specimens showed EML4‐ALK fusion genes in 200 cases (2.7%), with 94% of such cases involving adenocarcinoma.8 ALK fusion genes, including fusion to EML4, KIF5B, TFG, and KLC1, have been reported to be associated with a history of light/never smoking, young age, lack of EGFR or KRAS mutations, and adenocarcinoma with an acinar histology.5, 6, 7, 11, 12
ALK kinase inhibitors have been developed and are reported to suppress the growth of EML4‐ALK fusion‐positive cells.4, 7 Thus, treatment with ALK inhibitors can be effective for NSCLC patients whose tumors contain an EML4‐ALK fusion.16 Clinical trials for EML4‐ALK positive lung cancer with ALK‐TKI crizotinib have demonstrated that TKI treatment is superior to standard chemotherapy in patients with previously untreated advanced NSCLC associated with ALK fusion genes.17
In this study, we determined the frequency of EML4‐ALK fusion genes to clarify the clinicopathological characteristics of patients aged ≤ 50 years with lung adenocarcinoma and EML4‐ALK fusion to identify useful information regarding patient selection for ALK‐TKI therapy.
Methods
Patients and sample collection
Between December 1998 and May 2009, 85 patients (male/female: 38/47) aged ≤ 50 were diagnosed with lung adenocarcinoma at the National Kyushu Cancer Center Hospital. We examined 17 frozen and 32 formalin‐fixed samples available for RNA analysis (male/female: 23/26) from patients who underwent resection, chemotherapy, or radiotherapy for the presence of the EML4‐ALK gene. Biopsy specimens were obtained before chemotherapy or radiotherapy. Histological diagnosis of the tumors was based on World Health Organization (WHO) criteria, and tumor node metastasis (TNM) stage was determined according to Union for International Cancer Control TNM criteria version 7. Our institutional review board approved the genetic analyses conducted in the present study. All specimens were subjected to hematoxylin‐eosin staining in the Department of Diagnostic Pathology of our hospital. Two board‐certified pathologists independently reviewed the slides and made the diagnoses according to the WHO classification of lung tumors.
Nucleic acid extraction
Total RNA was extracted from frozen and formalin‐fixed paraffin‐embedded (FFPE) tissues using an RNeasy Kit (Qiagen, Valencia, CA, USA). Genomic DNA from frozen tissues was extracted using the phenol‐chloroform method. Genomic DNA from FFPE tissues was extracted using TakaRa DEXPAT (TaKaRa Bio.Inc., Kusatsu, Shiga, Japan). Quantification of extracted nucleic acids and measurement of the A260/A280 ratio were performed using an ultraviolet spectrophotometer (DU800, Beckman‐Coulter, Tokyo, Japan).
ALK fusion analysis by multiplex reverse transcription‐PCR and sequencing
Complementary (c)DNA synthesis from total RNA was performed using random primers and SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). To detect EML4‐ALK fusion cDNA, multiplex PCR was performed using the Amplitaq Gold DNA 360 Master Mix (Applied Biosystems, Foster City, CA, USA). Primer sets for variants of EML4‐ALK fusion were used as reported previously.18, 19 Amplification of EML4‐ALK fusion cDNA was performed for 35 cycles (1 minute at 94°C, 1 minute at 64°C, and 1 minute at 72°C) using the TGRADIENT system (Biometra, East Lyme, CT, USA). GAPDH cDNA was amplified by PCR with the primers 5′‐TGTCAGTGGTGGACCTGACC‐3′ and 5′‐TGAGCTTGACAAAGTGGTCG‐3′ using TaKaRa Ex‐Taq (TaKaRa Bio. Inc.) Amplification of GAPDH cDNA was performed for 35 cycles (30 seconds at 94°C, 30 seconds at 60°C, and 1 minute at 72°C) using the TGRADIENT system (Biometra). Agarose gel electrophoresis was performed to detect PCR products, and the results were observed using Gel Doc 2000 (Bio‐Rad, Hercules, CA, USA). PCR products were purified and labeled for sequencing using the BigDye v1.1 kit (Applied Biosystems) according to the manufacturer's protocol. Sequencing was performed using a 310 Genetic Analyzer (Applied Biosystems).
Mutation analysis for EGFR and KRAS by sequencing
Genomic DNA from each sample was used for sequencing analysis of EGFR exons 19 and 21 and KRAS exon 1. The sequencing primers used for PCR were: EGFR exon 19: 5′‐TGGCACCATCTCACAATTGC‐3′(forward), 5′‐GAAAAGGTGGGCCTGAGGTTC‐3′(reverse); EGFR exon 21: 5′‐CATGAACTACTTGGAGGACC‐3′ (forward), 5′‐CAGGAAAATGCTGGCTGACC‐3′ (reverse); and KRAS exon 1: 5′‐GACTGAATATAAACTTGTGG‐3′ (forward) 5′‐CTATTGTTGGATCATATTCG‐3′ (reverse). Each PCR was run for 35 cycles, and the annealing temperatures were 64°C (EGFR exon 19), 60°C (EGFR exon 21), and 56°C (KRAS exon 1) using TaKaRa Ex‐Taq (TaKaRa Bio. Inc.).
Statistical analysis
The overall survival (OS) duration was calculated from the date of initial therapy of the patients. Survival curves were prepared using the Kaplan–Meier method, and comparisons among the survival curves were made using the log‐rank test. The Cox proportional hazards model was used to assess the following factors: age, gender, smoking history, pathology, stage, and EGFR and KRAS mutation status. Data were considered significant at P ≤ 0.05.
Results
Identification of the EML4‐ALK fusion gene
Eighty‐five patients aged ≤ 50 with lung adenocarcinomas were treated at our hospital during the study period. We examined 49 samples (17 frozen and 32 formalin‐fixed samples) available for RNA analysis for the presence of the EML4‐ALK fusion gene. Using multiplex RT‐PCR and direct sequencing, EML4‐ALK transcripts were detected in 5 of the 49 tumors (10.2%). Table 1 shows the clinical and pathological profiles of all patients with the EML4‐ALK fusion gene. There were four cases of variants with fusion points between EML4 exon 20 and ALK exon 20 (variant 2). One tumor involved EML4 exon 6, which included a splice form that differed by 32 nucleotides from intron 6 of EML4 (variant 3b).
Table 1.
Clinicopathological profile of patients with the EML4‐ALK fusion gene
| Case No. | EML4‐ALK | Gender | Age | Smoking history | T factor | N factor | M factor | Stage | Treatment | Pathology | EGFR | KRAS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Variant 2 | F | 47 | S | 4 | 1 | 0 | III A | C + R, Surg | Acinar | WT | WT |
| 2 | Variant 2 | F | 49 | NS | 1 | 3 | 1 | IV | C | Acinar | WT | WT |
| 3 | Variant 2 | M | 37 | S | 4 | 3 | 1 | IV | C | Solid | WT | WT |
| 4 | Variant 3 | M | 50 | NS | 2 | 0 | 0 | I B | Surg | Papillary | WT | WT |
| 5 | Variant 2 | F | 39 | NS | 2 | 3 | 1 | IV | C | Papillary | WT | WT |
C, chemotherapy; NS, nonsmoker; R, radiotherapy; S, smoker; Surg, surgery; WT, wild type.
Clinicopathological characteristics of patients with EML4‐ALK fusion genes
Table 2 summarizes the clinicopathological characteristics of these patients in relation to EML4‐ALK status. The five patients with EML4‐ALK fusion genes included three women and two men, ranging in age from 37 to 50 years. One patient had stage IB disease, one had stage IIIA, and three had stage IV disease with N3 lymph node metastases. The EML4‐ALK fusion gene in these younger (≤ 50 years) patients with lung adenocarcinoma was associated with higher stage tumors. The EML4‐ALK fusion gene was mutually exclusive to EGFR and KRAS mutations. Histologically, there was one solid adenocarcinoma, two acinar adenocarcinomas, and two papillary adenocarcinomas (Fig 1). None of the tumors with EML4‐ALK fusion genes had a lepidic predominant adenocarcinoma component.
Table 2.
Relationship between EML4‐ALK gene fusion and clinicopathological profiles in younger (≤ 50 years) patients with lung adenocarcinoma
| Total | EML4‐ALK | ||||||
|---|---|---|---|---|---|---|---|
| (n = 49) | Positive (n = 5) | Negative (n = 44) | P | ||||
| Variable | No. | (%) | No. | (%) | No. | (%) | |
| Age, years | |||||||
| Median | 48 | 47 | 48 | 0.560 | |||
| Range | 31–50 | 37–50 | 31–50 | ||||
| Gender | |||||||
| Male | 23 | (47) | 2 | (40) | 21 | (48) | 0.743 |
| Female | 26 | (53) | 3 | (60) | 23 | (52) | |
| Smoking history | |||||||
| Non‐smoker | 27 | (55) | 3 | (60) | 24 | (55) | 0.816 |
| Ever smoker | 22 | (45) | 2 | (40) | 20 | (45) | |
| Pathology | |||||||
| With lepidic growth | 16 | (33) | 0 | (0) | 16 | (36) | 0.100 |
| Without lepidic growth | 33 | (67) | 5 | (100) | 27 | (64) | |
| Stage | |||||||
| I | 23 | (47) | 1 | (20) | 22 | (50) | 0.002 |
| II | 4 | (8) | 0 | (0) | 4 | (9) | |
| III | 15 | (31) | 1 | (20) | 14 | (32) | |
| IV | 7 | (14) | 3 | (60) | 4 | (9) | |
| EGFR | |||||||
| Wild type | 36 | (73) | 5 | (100) | 31 | (70) | 0.156 |
| Mutation | 13 | [27] | 0 | [0] | 13 | [30] | |
| KRAS | |||||||
| Wild type | 48 | [98] | 5 | [100] | 43 | [98] | 0.733 |
| Mutation | 1 | [2] | 0 | [0] | 1 | [2] | |
Stages I–III versus IV.
Figure 1.
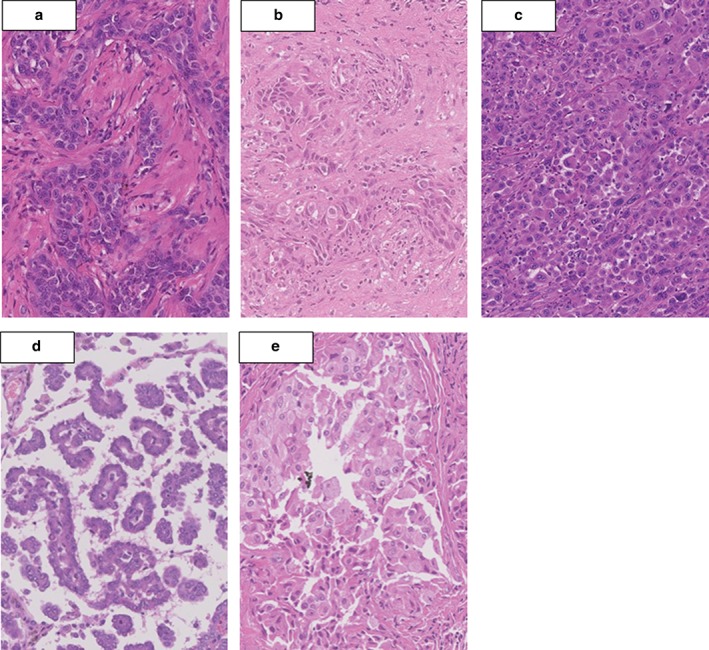
Histopathological results of EML4‐ALK fusion‐harboring tumors: (a,b) two acinar adenocarcinomas (cases 1 and 2), (c) one solid adenocarcinoma (case 3), and (d,e) two papillary adenocarcinomas (cases 4 and 5).
Clinical outcome of patients with and without EML4‐ALK
Of the five patients with EML4‐ALK fusion genes, three patients with stage IV disease received platinum‐based chemotherapy, such as carboplatin + paclitaxel, cisplatin + gemcitabine + vinorelbine, or cisplatin + S1. None of the patients received ALK inhibitors because these drugs were not approved in Japan before 2012. The overall response to chemotherapy was progressive disease in two cases (cases 3 and 5), and stable disease in one case (case 2). One patient (case 1) received preoperative chemoradiotherapy with cisplatin + S1, and achieved stable disease. The patient (case 4) with the variant 3‐ALK fusion received left lower lobectomy. Four months after surgery, multiple pulmonary metastases appeared, however, he was not treated for the recurrence because of his poor performance status.
Overall, the five‐year survival rate of the 49 patients was 54.9%. The five‐year survival rate was 59.4% in the patients without EML4‐ALK fusion. In the patients with EML4‐ALK fusion, the five‐year survival rate was not reached, while the one‐year survival rate was 60% and the two‐year survival rate was 40% (Fig 2). After univariate analysis of eight factors, subgroups consisting of pathological features without lepidic growth, higher stage, and positive status of EML4‐ALK fusion showed significantly shorter survival, with P values of 0.0315, 0.0003, and 0.0037, respectively. Although gender and EGFR status were likely to affect survival, no significance was observed in this analysis. Multivariate analysis identified that stage was the only significant prognostic factor, with a hazard ratio of 4.975, and EML4‐ALK fusion was not identified as significant (Table 3).
Figure 2.
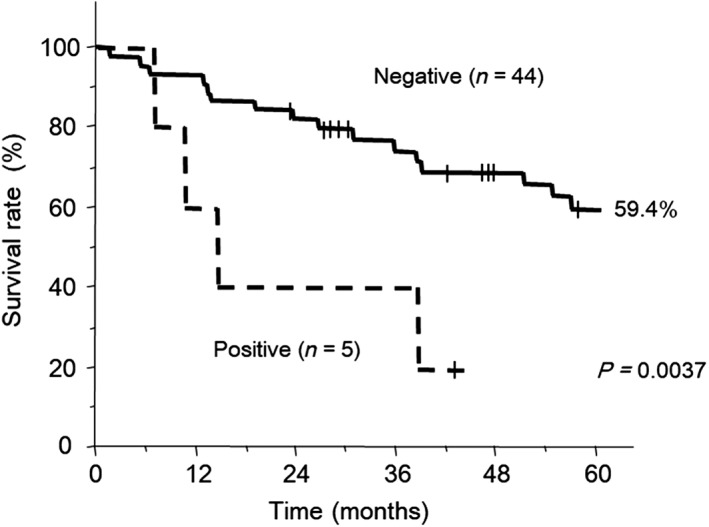
Kaplan–Meier plots of the overall survival of EML4‐ALK‐positive compared to EML4‐ALK‐negative patients. Overall survival was calculated from the date of initial therapy of the patients.
Table 3.
Results of univariate and multivariate analyses of the prognostic factors for overall survival in younger (≤ 50 years) patients with lung adenocarcinoma (n = 49)
| Variable | No. | (%) | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|---|---|
| Five‐year survival (%) | P | Hazard ratio | 95% confidence interval | P | ||||
| Age, years | ||||||||
| ≤ 40 | 7 | (14) | 42.9 | |||||
| > 40 | 42 | (86) | 56.9 | 0.4053 | ||||
| Gender | ||||||||
| Male | 23 | (47) | 39.0 | |||||
| Female | 26 | (53) | 67.5 | 0.0777 | 0.410 | 0.146–1.157 | 0.0922 | |
| Smoking history | ||||||||
| Non‐smoker | 27 | (55) | 62.9 | |||||
| Ever smoker | 22 | (45) | 45.1 | 0.1379 | ||||
| Pathology | ||||||||
| With lepidic growth | 16 | (33) | 76.9 | |||||
| Without lepidic growth | 33 | (67) | 44.4 | 0.0315 | 1.965 | 0.495–7.813 | 0.3369 | |
| Stage | ||||||||
| I–III | 42 | (86) | 62.1 | |||||
| IV | 7 | (14) | 14.3 | 0.0003 | 4.975 | 1.534–16.129 | 0.0075 | |
| EML4‐ALK fusion | ||||||||
| Negative | 44 | (90) | 59.4 | |||||
| Positive | 5 | (10) | NR | 0.0037 | 2.215 | 0.514–9.537 | 0.2856 | |
| EGFR | ||||||||
| Wild type | 36 | (73) | 46.6 | |||||
| Mutation | 13 | (27) | 82.1 | 0.0625 | 2.058 | 0.405–0.417 | 0.3843 | |
| KRAS | ||||||||
| Wild type | 48 | (98) | 54.6 | |||||
| Mutation | 1 | (2) | NR | NE | ||||
NE, not evaluable; NR, not reached.
Discussion
It remains controversial whether younger patients with NSCLC have a better or worse prognosis than older patients.20, 21, 22, 23 In the present study conducted to detect EML4‐ALK fusion genes in 49 samples from patients aged ≤ 50 with lung adenocarcinomas, five adenocarcinomas (10.2%) proved positive for fusion messenger RNA. Previous studies have reported that between 1.6% and 13.5% of lung tumors harbor EML4‐ALK fusions (Table 4).4, 6, 7, 9, 10, 11, 12, 13, 14, 15 The frequency of EML4‐ALK‐positive patients in our study was very high compared to the results of previous studies. EML4‐ALK fusions may be more common in younger patients with lung adenocarcinomas. Inamura et al. reported that 4 out of 16 patients (25%) aged < 50 had EML4‐ALK fusions, while seven of 237 patients (3%) aged ≥ 50 had EML4‐ALK fusions.5 Shaw et al. demonstrated that the median age of NSCLC patients with EML4‐ALK fusion, EGFR mutation, and wild type genes was 52, 66, and 64 years, respectively.11
Table 4.
Studies evaluating the frequency of EML4‐ALK gene rearrangements in lung cancer
| First author | Histological characteristics | Detection method | Population | Total number of patients | Number of EML4‐ALK positive patients | Percentage |
|---|---|---|---|---|---|---|
| Rodig et al.9 | Adenocarcinoma | IHC, FISH | American (US) | 358 | 20 | 5.6 |
| Koivunen et al.7 | NSCLC | RT‐PCR | American (US) (138), Korean (167) | 305 | 8 | 2.6 |
| Sequist et al.10 | NSCLC | Multiplex RT‐PCR | White (503), Black (7), Asian (22) | 546 | 27 | 4.9 |
| Shaw et al.11 | Enriched NSCLC | FISH | Non‐Asian (132), Asian (9) | 141 | 19 | 13.5 |
| Soda et al.4 | NSCLC | RT‐PCR | Japanese | 75 | 5 | 6.7 |
| Inamura et al.6 | Adenocarcinoma | RT‐PCR | Japanese | 149 | 5 | 3.4 |
| Takeuchi et al.14 | Adenocarcinoma | Multiplex RT‐PCR | Japanese | 253 | 11 | 4.3 |
| Shinmura et al.12 | NSCLC | RT‐PCR | Japanese | 77 | 2 | 2.6 |
| Takahashi et al.13 | NSCLC | RT‐PCR | Japanese | 313 | 5 | 1.6 |
| Takeuchi et al.15 | Adenocarcinoma | IHC, Multiplex RT‐PCR | Japanese | 130 | 7 | 5.4 |
| Present study | Adenocarcinoma in patients aged ≤50 years | Multiplex RT‐PCR | Japanese | 49 | 5 | 10.2 |
FISH, fluorescence in situ hybridization; IHC, immunohistochemistry; NSCLC, non‐small cell lung cancer; RT, reverse‐transcription.
The acinar pattern is reported to be associated with ALK‐rearranged lung adenocarcinoma in Asian populations,5, 15 whereas the signet‐ring cell histology is reported most frequently in Western patients.11 We previously reported a case of signet ring carcinoma (SRC) of the lung with an EML4‐ALK fusion gene mimicking mucinous (colloid) adenocarcinoma.18 Ou et al. demonstrated that patients with SRC of the lung were significantly younger than patients with adenocarcinoma, with the proportion of patients with SRC < 40 years at 5.0% compared to 1.3% of patients with adenocarcinoma.24 However, there were no patients with primary SRC of the lung in the present study.
Limited data exist to date on the efficacy of the currently available therapies in patients with EML4‐ALK NSCLC. In a study by Shaw et al., 12 patients with ALK genomic alterations were treated with platinum‐based chemotherapy. The response rate, time to progression, and OS were similar to those of NSCLC patients harboring EGFR mutations or those that were wild type for both EML4‐ALK and EGFR.11 Camidge et al. demonstrated that ALK‐positive patients have significantly longer progression‐free survival (PFS) on pemetrexed compared to triple‐negative (EGFR, KRAS, ALK wild‐type) patients, whereas EGFR or KRAS mutant patients do not.25 ALK‐rearranged tumors demonstrate relatively high response rates to single‐agent treatment with pemetrexed, with an objective response rate of 29% observed in a phase 3 study of ALK‐positive patients, compared to ~10% in unselected NSCLC patients.26 Li et al. found that the median thymidylate synthase RNA level, a biomarker of pemetrexed sensitivity, was significantly lower in ALK‐positive than in ALK‐negative lung adenocarcinomas.8 In our study, although three ALK‐positive patients received platinum‐based chemotherapy, all were resistant to the treatment.
The prognosis and natural history of ALK‐rearrangements in NSCLC have been explored retrospectively. For example, Rodig et al. demonstrated that patients with ALK‐rearranged tumors often present at a higher stage, most commonly stage IV, compared to those with ALK germ‐line tumors.9 ALK‐positive patients have also been reported to have a higher propensity for pericardial and pleural disease than triple‐negative patients.27 Notably, Shaw et al. demonstrated one and two‐year OS rates of 74% and 54%, respectively, among 82 ALK‐positive patients treated with crizotinib. In that study, survival of the ALK‐positive controls did not differ significantly from that of the entire group of 252 wild‐type controls, with a median OS duration of 20 versus 15 months.16
In our present study, patients with EML4‐ALK fusion showed significantly shorter survival than those with negative status. The five‐year survival rate was 59.4% in patients without the EML4‐ALK fusion, although there were no five‐year survivors with the EML4‐ALK fusion. However, multivariate analysis identified that EML4‐ALK fusion was not a prognostic factor in young (≤ 50 years) patients with lung adenocarcinoma in our study, because of the small number of patients with EML4‐ALK fusions.
Several methods, including PCR, immunohistochemistry, and fluorescence in situ hybridization are currently being evaluated for the detection of EML4‐ALK NSCLC. In this study, we used the multiplex RT‐PCR method for screening because this method can rapidly identify ALK rearrangement. Of the five EML4‐ALK fusion samples, there was one frozen sample from a resected tumor (case 4) and four FFPE samples (cases 1, 2, 3, and 5) from resected tumor and biopsy specimens. The RNA extracted from FFPE is highly degraded, and in general, more difficult to use for PCR relative to fresh‐frozen tissue. In case 5, the commercially available chromosomal fluorescence in situ hybridization analysis showed split signals for ALK, which confirmed EML4‐ALK fusion.28 Immunohistochemical analysis of FFPE tissue specimens remains the mainstay of routine surgical pathology practice. Mino‐Kenudson et al. reported the use of an immunohistochemical test based on novel antibodies with increased sensitivity and specificity for detecting ALK protein expression in FFPE samples.29 Takeuchi et al. developed an intercalated antibody‐enhanced polymer method that incorporates an intercalating antibody between the primary antibody to ALK and dextran polymer‐based detection reagents.15 These methods should be used to detect EML4‐ALK fusion in lung cancer specimens.
Crizotinib is a selective adenosine triphosphate‐competitive small molecule oral inhibitor of ALK, c‐MET/hepatocyte growth factor receptor, and ROS1 receptor tyrosine kinases. Solomon et al. conducted an open‐label, phase 3 trial comparing crizotinib with chemotherapy in 343 patients with advanced ALK‐positive NSCLC who had received no previous systemic treatment. Consequently, crizotinib significantly prolonged PFS compared to the standard chemotherapy regimen, with a median PFS of 10.9 months in the crizotinib versus 7.0 months in the chemotherapy group and a response rate of 74% for crizotinib versus 45% for chemotherapy.17 To date, second‐generation ALK inhibitors, such as ceritinib and alectinib, have been developed, demonstrating significant clinical activity in ALK‐positive patients with NSCLC.30, 31, 32
In summary, the results of our study indicate that the EML4‐ALK fusion gene may be an oncogene in younger patients with lung adenocarcinoma.
Disclosure
No authors report any conflict of interest.
Acknowledgment
The authors would like to thank Brian Quinn for his critical comments on the manuscript.
References
- 1. Lynch TJ, Bell DW, Sordella R et al Activating mutations in the epidermal growth factor receptor underlying responsiveness of non‐small‐cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129–39. [DOI] [PubMed] [Google Scholar]
- 2. Maemondo M, Inoue A, Kobayashi K et al Gefitinib or chemotherapy for non‐small‐cell lung cancer with mutated EGFR. N Engl J Med 2010; 362: 2380–8. [DOI] [PubMed] [Google Scholar]
- 3. Yang JC, Hirsh V, Schuler M et al Symptom control and quality of life in LUX‐Lung 3: A phase III study of afatinib or cisplatin/pemetrexed in patients with advanced lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013; 31: 3342–50. [DOI] [PubMed] [Google Scholar]
- 4. Soda M, Choi YL, Enomoto M et al Identification of the transforming EML4‐ALK fusion gene in non‐small‐cell lung cancer. Nature 2007; 448: 561–6. [DOI] [PubMed] [Google Scholar]
- 5. Inamura K, Takeuchi K, Togashi Y et al EML4‐ALK lung cancers are characterized by rare other mutations, a TTF‐1 cell lineage, an acinar histology, and young onset. Mod Pathol 2009; 22: 508–15. [DOI] [PubMed] [Google Scholar]
- 6. Inamura K, Takeuchi K, Togashi Y et al EML4‐ALK fusion is linked to histological characteristics in a subset of lung cancers. J Thorac Oncol 2008; 3: 13–7. [DOI] [PubMed] [Google Scholar]
- 7. Koivunen JP, Mermel C, Zejnullahu K et al EML4‐ALK fusion gene and efficacy of an ALK kinase inhibitor in lung cancer. Clin Cancer Res 2008; 14: 4275–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li T, Maus MK, Desai SJ et al Large‐scale screening and molecular characterization of EML4‐ALK fusion variants in archival non‐small‐cell lung cancer tumor specimens using quantitative reverse transcription polymerase chain reaction assays. J Thorac Oncol 2014; 9: 18–25. [DOI] [PubMed] [Google Scholar]
- 9. Rodig SJ, Mino‐Kenudson M, Dacic S et al Unique clinicopathologic features characterize ALK‐rearranged lung adenocarcinoma in the western population. (Published erratum appears in Clin Cancer Res 2009; 15: 7710). Clin Cancer Res 2009; 15: 5216–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sequist LV, Heist RS, Shaw AT et al Implementing multiplexed genotyping of non‐small‐cell lung cancers into routine clinical practice. Ann Oncol 2011; 22: 2616–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shaw AT, Yeap BY, Mino‐Kenudson M et al Clinical features and outcome of patients with non‐small‐cell lung cancer who harbor EML4‐ALK. J Clin Oncol 2009; 27: 4247–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shinmura K, Kageyama S, Tao H et al EML4‐ALK fusion transcripts, but no NPM‐, TPM3‐, CLTC‐, ATIC‐, or TFG‐ALK fusion transcripts, in non‐small cell lung carcinomas. Lung Cancer 2008; 61: 163–9. [DOI] [PubMed] [Google Scholar]
- 13. Takahashi T, Sonobe M, Kobayashi M et al Clinicopathologic features of non‐small‐cell lung cancer with EML4‐ALK fusion gene. Ann Surg Oncol 2010; 17: 889–97. [DOI] [PubMed] [Google Scholar]
- 14. Takeuchi K, Choi YL, Soda M et al Multiplex reverse transcription‐PCR screening for EML4‐ALK fusion transcripts. Clin Cancer Res 2008; 14: 6618–24. [DOI] [PubMed] [Google Scholar]
- 15. Takeuchi K, Choi YL, Togashi Y, Soda M et al KIF5B‐ALK, a novel fusion oncokinase identified by an immunohistochemistry‐based diagnostic system for ALK‐positive lung cancer. Clin Cancer Res 2009; 15: 3143–9. [DOI] [PubMed] [Google Scholar]
- 16. Shaw AT, Yeap BY, Solomon BJ et al Effect of crizotinib on overall survival in patients with advanced non‐small‐cell lung cancer harbouring ALK gene rearrangement: A retrospective analysis. Lancet Oncol 2011; 12: 1004–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Solomon BJ, Mok T, Kim DW et al First‐line crizotinib versus chemotherapy in ALK‐positive lung cancer. (Pulished erratum appears in N Engl J Med 2015; 373: 1582). N Engl J Med 2014; 371: 2167–77. [DOI] [PubMed] [Google Scholar]
- 18. Ohba T, Sugio K, Kometani T et al Signet ring cell adenocarcinoma of the lung with an EML4‐ALK fusion gene mimicking mucinous (colloid) adenocarcinoma: A case report. Lung Cancer 2011; 73: 375–8. [DOI] [PubMed] [Google Scholar]
- 19. Ohba T, Toyokawa G, Osoegawa A et al Mutations of the EGFR, K‐ras, EML4‐ALK, and BRAF genes in resected pathological stage I lung adenocarcinoma. Surg Today 2016; 46: 1091–8. [DOI] [PubMed] [Google Scholar]
- 20. Mauri D, Pentheroudakis G, Bafaloukos D et al Non‐small cell lung cancer in the young: A retrospective analysis of diagnosis, management and outcome data. Anticancer Res 2006; 26: 3175–81. [PubMed] [Google Scholar]
- 21. Radzikowska E, Roszkowski K, Glaz P. Lung cancer in patients under 50 years old. Lung Cancer 2001; 33: 203–11. [DOI] [PubMed] [Google Scholar]
- 22. Sacher AG, Dahlberg SE, Heng J, Mach S, Jänne PA, Oxnard GR. Association between younger age and targetable genomic alterations and prognosis in non‐small‐cell lung cancer. JAMA Oncol 2016; 2: 313–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Subramanian J, Morgensztern D, Goodgame B et al Distinctive characteristics of non‐small cell lung cancer (NSCLC) in the young: A surveillance, epidemiology, and end results (SEER) analysis. J Thorac Oncol 2010; 5: 23–8. [DOI] [PubMed] [Google Scholar]
- 24. Ou SH, Ziogas A, Zell JA. Primary signet‐ring carcinoma (SRC) of the lung: A population‐based epidemiologic study of 262 cases with comparison to adenocarcinoma of the lung. J Thorac Oncol 2010; 5: 420–7. [DOI] [PubMed] [Google Scholar]
- 25. Camidge DR, Kono SA, Lu X et al Anaplastic lymphoma kinase gene rearrangements in non‐small cell lung cancer are associated with prolonged progression‐free survival on pemetrexed. J Thorac Oncol 2011; 6: 774–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shaw AT, Kim DW, Nakagawa K et al Crizotinib versus chemotherapy in advanced ALK‐positive lung cancer. N Engl J Med 2013; 368: 2385–94. [DOI] [PubMed] [Google Scholar]
- 27. Doebele RC, Lu X, Sumey C et al Oncogene status predicts patterns of metastatic spread in treatment‐naive nonsmall cell lung cancer. Cancer 2012; 118: 4502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Osoegawa A, Nosaki K, Miyamoto H et al Incidentally proven pulmonary "ALKoma". Intern Med 2010; 49: 603–6. [DOI] [PubMed] [Google Scholar]
- 29. Mino‐Kenudson M, Chirieac LR, Law K et al A novel, highly sensitive antibody allows for the routine detection of ALK‐rearranged lung adenocarcinomas by standard immunohistochemistry. Clin Cancer Res 2010; 16: 1561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hida T, Nokihara H, Kondo M et al Alectinib versus crizotinib in patients with ALK‐positive non‐small‐cell lung cancer (J‐ALEX): An open‐label, randomised phase 3 trial. Lancet 2017; 390: 29–39. [DOI] [PubMed] [Google Scholar]
- 31. Peters S, Camidge DR, Shaw AT et al Alectinib versus crizotinib in untreated ALK‐positive non‐small‐cell lung cancer. N Engl J Med 2017; 377: 829–38. [DOI] [PubMed] [Google Scholar]
- 32. Soria JC, Tan DSW, Chiari R et al First‐line ceritinib versus platinum‐based chemotherapy in advanced ALK‐rearranged non‐small‐cell lung cancer (ASCEND‐4): A randomised, open‐label, phase 3 study. Lancet 2017; 389: 917–29. [DOI] [PubMed] [Google Scholar]


