Abstract
Lung cancer is the first leading cause of cancer deaths worldwide. Non‐small cell lung cancer (NSCLC) is the most common type of lung cancer. Increasing evidence shows that long noncoding RNA (lncRNA) are capable of modulating tumor initiation, proliferation and metastasis. In the present study, we aimed to evaluate whether circulating lncRNA could be used as biomarkers for diagnosis and prognosis of NSCLC. Expression profiles of 14 lncRNA selected from other studies were validated in 20 pairs of tissues by quantitative real‐time PCR, and the dysregulated lncRNA thus identified were further validated in serum samples from two independent cohorts along with three tumor makers (CEA, CYFRA21‐1, and SCCA). Receiver‐operating characteristic analysis was utilized to estimate the diagnostic efficiency of the candidate lncRNA and tumor markers. Importantly, we observed an association between lncRNA expression and overall survival (OS) rate of NSCLC. The expressions of SOX2 overlapping transcript (SOX2OT) and ANRIL were obviously upregulated in NSCLC tissues and serum samples compared with normal controls (P < 0.01). Based on the data from the training set, we next used a logistic regression model to construct an NSCLC diagnostic panel consisting of two lncRNA and three tumor markers. The area under the curve of this panel was 0.853 (95% confidence interval = 0.804–0.894, sensitivity = 77.1%, specificity = 79.2%), and this was distinctly superior to any biomarker alone (all at P < 0.05). Similar results were observed in the validation set. Intriguingly, Kaplan–Meier analysis demonstrated that low expressions of SOX2OT and ANRIL were both associated with higher OS rate (P = 0.008 and 0.017, respectively), and SOX2OT could be used as an independent prognostic factor (P = 0.036). Taken together, our study demonstrated that the newly developed diagnostic panel consisting of SOX2OT, ANRIL, CEA, CYFRA21‐1, and SCCA could be valuable in NSCLC diagnosis. LncRNA SOX2OT and ANRIL might be ideal biomarkers for NSCLC prognosis.
Keywords: diagnosis, long noncoding RNA, non‐small cell lung cancer, prognosis, serum, tumor biomarker
Abbreviations
- AUC
area under the curve
- CEA
carcinoembryonic antigen
- CI
confidence interval
- CYFRA21‐1
cytokeratin 19 fragment
- lncRNA
long noncoding RNA
- NSCLC
non‐small cell lung cancer
- OS
overall survival
- qRT‐PCR
quantitative real‐time PCR
- ROC
receiver‐operating characteristic
- SCCA
squamous cell carcinoma antigen
- SOX2OT
SOX2 overlapping transcript
1. Introduction
As one of the most common fatal tumors, lung cancer is the leading cause of cancer‐related dysthanasia around the world (Chen et al., 2016; Fu et al., 2016; Siegel et al., 2017). Non‐small cell lung cancer (NSCLC) is the most common subtype of lung cancer, accounting for nearly 80% of lung cancers (Travis et al., 2015). Although patients diagnosed at an early stage can receive favorable prognosis from surgical therapy in combination with chemotherapy, radiotherapy, or targeted drug therapy, the 5‐year survival rate for most NSCLC patients remains poor with the 5‐year survival rate of 21% (Miller et al., 2016). Therefore, effective means for early diagnosis are imperative to reduce death. In general, individual symptoms are not evident at the early stage, imaging technologies, such as chest X‐ray and computed tomography (CT), are not definitive, and overdiagnosis and systemic cumulative radiation injuries occur occasionally (Bach et al., 2012; Gilbert, 2017). Although bronchoscopy in combination with pathological examination is the gold standard to diagnose NSCLC, it is not an appropriate approach to screen all individuals with respiratory diseases owing to the invasiveness and poor compliance. Currently, some serum tumor markers, such as carcinoembryonic antigen (CEA), squamous cell carcinoma antigen (SCCA) and cytokeratin 19 fragment (CYFRA21‐1), are being employed extensively to diagnose NSCLC due to the rapid detection methods available (Chen et al., 2015; Zhao et al., 2015b). However, these serum markers exhibit very low sensitivity and specificity (I and Cho, 2015). Therefore, it is urgently necessary to develop innovative and noninvasive diagnostic biomarkers that have high sensitivity and specificity.
Long noncoding RNA (lncRNA) has been the topic of debate in the realm of neoplasms as accumulating evidence has shown that lncRNA drive tumor initiation, proliferation, and metastasis (Reis and Verjovski‐Almeida, 2012; Ricciuti et al., 2016; Schmitt and Chang, 2016; Sun et al., 2017; Yang et al., 2015). Numerous studies have noted the differential expression of lncRNA in NSCLC and non‐neoplastic tissues/cells (Han et al., 2015; Yang et al., 2014; Zhao et al., 2015b). In addition, many studies have highlighted that some lncRNA steadily subsist in human serum/plasma (Qi et al., 2016; Shi et al., 2016), facilitating subsequent investigation on the role of serum/plasma lncRNA in the diagnosis and prognosis of NSCLC. However, only few serum lncRNA, such as MALAT1, HOTAIR, and GAS5, exhibit significantly different serum expression levels between NSCLC patients and healthy controls (Li et al., 2017; Liang et al., 2016; Zhang et al., 2017). Moreover, technical issues, such as using single or undercounted lncRNA, an insufficient specimen size, and a lack of independent validation, are often noted in such studies.
In the present study, we validated the expressions of lncRNA in two independent serum cohorts of NSCLC patients in order to identify a panel of diagnostic lncRNA. We then assessed the diagnostic efficacy of this candidate lncRNA panel. Moreover, we evaluated the association between lncRNA and prognosis of NSCLC in order to validate the feasibility of detecting NSCLC using circulating lncRNA.
2. Materials and methods
2.1. Ethical statement
All experiments were performed in accordance with relative regulations and manners. The study was approved by the Clinical Research Ethics Committee of Qilu Hospital of Shandong University. All clinical samples were collected from the Qilu Hospital of Shandong University, and written informed consent was obtained from each participant before sample collection.
2.2. Study design
Our study had three progressive stages: the screening stage, the training stage, and the validation stage. In the screening stage, we selected 14 lncRNA that are reportedly dysregulated in NSCLC tissue (Feng et al., 2015; Han et al., 2015; Hou et al., 2014; Li et al., 2016; Lu et al., 2013; Nie et al., 2015, 2016; Qiu et al., 2014; Thai et al., 2013; Wang et al., 2015; Weber et al., 2013; Wu et al., 2015; Zhang et al., 2014b; Zhao et al., 2015a) and measured their expressions in 20 paired NSCLC and matched nontumorous tissues. Next, lncRNA with aberrant expression in the tissue samples thus identified were measured in 92 paired samples (46 NSCLCs and 46 healthy controls). The lncRNA with aberrant serum expressions were further evaluated in a large cohort (140 NSCLC patients and 120 healthy controls) during the training stage. Data thus collected were combined with the currently available lung tumor markers (CEA, CYFRA21‐1, and SCCA) to construct a diagnostic panel based on a logistic regression model. The validity and diagnostic efficiency of the panel were assessed using receiver‐operating characteristic (ROC) analysis. In the validation stage, the diagnostic lncRNA panel built in the training stage was verified in another independent cohort of 100 NSCLC patients and 100 healthy controls. A Kaplan–Meier survival analysis was also conducted at the validation stage to evaluate the correlation between certain lncRNA and the overall survival (OS) rate of NSCLC patients in order to assess the prognostic power of the identified lncRNA.
2.3. Patients and specimens
All participants were enrolled from the Qilu Hospital of Shandong University between January 2010 and February 2012. All enrolled patients met the following inclusion criteria: Primary NSCLC was confirmed through histological examination of available samples, and no chemotherapy or radiotherapy was received before tissue/serum collection. Table S1 summarizes the clinicopathological characteristics of the patients enrolled in the training and validation stages, including sex, age, tumor size, lymph node metastasis, and TNM stage. Control samples were collected from 220 healthy subjects without any malignant tumor as indicated by normal levels of tumor markers. The demographic information of the controls is also available in Table S1. The tumors were staged according to the 8th Union of International Control of Cancer (UICC) classification (Kay et al., 2017).
Following surgical tumor resection, patients included in the validation set were followed up every 3 months for the first 2 years and were thereafter followed up every 6 months until March 31, 2017. Five patients were excluded during this period due to imperfect follow‐ups, and the median follow‐up time was 46 (range, 5–72) months.
Peripheral blood samples were collected from all participants before surgical resection and pharmacological intervention. Serum isolated from whole blood was immediately transferred to a 1.5‐mL Eppendorf tube and subjected to a two‐step centrifugation (1500 g for 10 min at 4 °C, and then 13 800 g for 15 min at 4 °C) to eliminate cell sediments. One aliquot of the serum sample was used within 2 h of collection to assess the expressions of tumor markers (CEA, CYFRA21‐1, and SCCA), while a second aliquot was stored at −80 °C in an RNase‐free tube until total RNA extraction.
2.4. Detection of common tumor markers
Serum CEA and CYFRA 21‐1 levels were detected using Elecsys and Cobas e analyzers (Roche Diagnostics, Mannheim, Germany) on Roche Cobas 6000 analyzer. Serum SCCA level was measured by ARCHITECT SCC Reagent Kit (Abbott Laboratories, Niigata, Japan) using an Abbott I2000 analyzer.
2.5. RNA isolation and quantitative real‐time PCR (qRT‐PCR)
Total RNA was extracted from frozen tissue by TRIzol (Invitrogen, Carlsbad, CA, USA), while RNA extraction from serum samples was accomplished using the nucleic acid purification reagent from the diagnostic kit for quantification of hepatitis C virus RNA (QIAGEN, Shenzhen, China) according to the manufacturer's instructions. RNA concentration was measured by NanoDrop spectrophotometer. The purified RNA was reversely transcribed into cDNA using PrimeScript™ RT Reagent Kit (Takara, Dalian, Liaoning, China) on a SimpliAmp™ Thermal Cycler (ABI, Singapore, Singapore). Briefly, cDNA synthesis was carried out in a 20‐μL reaction system consisting of 1 μg template RNA, 4 μL 5× PrimeScript buffer mix, 1 μL PrimeScript RT enzyme mix I, 1 μL oligo dT primer and RNase‐free dH2O. The reaction mixture was sequentially incubated at 37 °C for 30 min, at 85 °C for 5 s, and then at 4 °C for 60 min. qRT‐PCR was performed in a 25‐μL reaction system containing 2 μL diluted cDNA, 12.5 μL SYBR Premix Ex Taq (Takara), 0.5 μL DyeII, 0.75 μL each of the forward and reverse primers, and 8.5 μL nuclease‐free water. Amplifications were conducted on a CFX96™ Real‐Time System (Bio‐Rad Laboratories, Hercules, CA, USA). Briefly, after an initial denaturation step at 95 °C for 30 s, 42 amplification cycles were carried out with a melting temperature of 95 °C for 5 s and an annealing temperature of 58 °C for 30 s. This was followed by a melting curve analysis to ensure the specificity of the PCR products. Each experiment was performed in triplicate. GAPDH was selected as the housekeeping gene, and the relative expressions of lncRNA were calculated using the method. The amplicons of target genes were identified after gel extraction and sequencing using PubMed gene blast.
2.6. Statistical analysis
All lncRNA expression data were assessed by spss statistics 22.0 (IBM, Chicago, IL, USA) to ascertain normal distribution. Nonparametric Mann–Whitney U‐test was used to compare the differences in lncRNA expression between the tumor and control groups. Scatter diagrams were generated with graphpad prism 5 (San Diego, CA, USA). Logistic regression analysis was performed utilizing matlab software (MATLAB, R2014a, Natick, MA, USA) to establish the lncRNA panel. ROC curves were generated using medcalc 15.2.2 (MedCalc, Mariakerke, Belgium), and the area under the curve (AUC) was utilized to evaluate the feasibility of using the lncRNA to detect NSCLC. Survival curves were generated using the Kaplan–Meier method, and survival differences were estimated by a log‐rank test. Multivariate Cox regression analysis was performed to identify significantly independent prognostic factors. A P‐value < 0.05 was considered statistically significant.
3. Results
3.1. Discovery and validation of dysregulated lncRNA in tissue and serum
Based on previous studies, we selected 14 aberrant NSCLC‐related lncRNA as potential diagnostic candidates in the present study. A total of 20 paired NSCLC and matched adjacent normal tissues were used to validate the relative expressions of these 14 NSCLC‐related lncRNA by qRT‐PCR. Our data showed that 12 lncRNA were significantly dysregulated in NSCLC tissues compared with adjacent normal tissues (Table S2). Next, we assessed the expressions of the 12 lncRNA in 92 serum samples (46 NSCLCs and 46 healthy controls) and identified two lncRNA [SOX2 overlapping transcript (SOX2OT) and ANRIL] that were overexpressed in tumor serum samples compared with healthy controls (Table S3, all at P < 0.01).
3.2. Verification of expression of the selected lncRNA in serum
We then increased the sample size to include 260 serum specimens (140 NSCLC patients and 120 healthy controls) in the training set to verify the expressions of SOX2OT and ANRIL by qRT‐PCR. We confirmed the overexpression of SOX2OT and ANRIL in the serum of NSCLC patients compared with controls (Fig. 1A,B). Concurrently, we evaluated the expression levels of three tumor markers (CEA, CYFRA21‐1, and SCCA) in the training set (Fig. 1C–E). ROC was used to evaluate the diagnostic efficacy of the two lncRNA and the three tumor markers currently employed for NSCLC detection. The AUCs of SOX2OT, ANRIL, CEA, CYFRA21‐1, and SCCA were 0.745 [95% confidence interval (CI) = 0.688–0.797], 0.723 (95% CI = 0.665–0.777), 0.631 (95% CI = 0.570–0.690), 0.620 (95% CI = 0.558–0.680), and 0.612 (95% CI = 0.550–0.672), respectively (Fig. 2).
Figure 1.
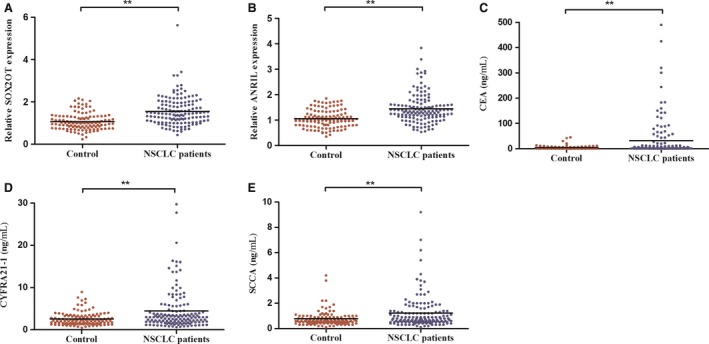
Expression levels of SOX2OT, ANRIL, CEA, CYFRA21‐1, and SCCA in serum samples of NSCLC patients and healthy controls during the training stage. SOX2OT and ANRIL were upregulated in the serum of NSCLC patients compared to healthy controls, as tested by qRT‐PCR (A, B). CEA, CYFRA21‐1, and SCCA were overexpressed in NSCLC patients compared with healthy controls (C–E). **P < 0.01.
Figure 2.
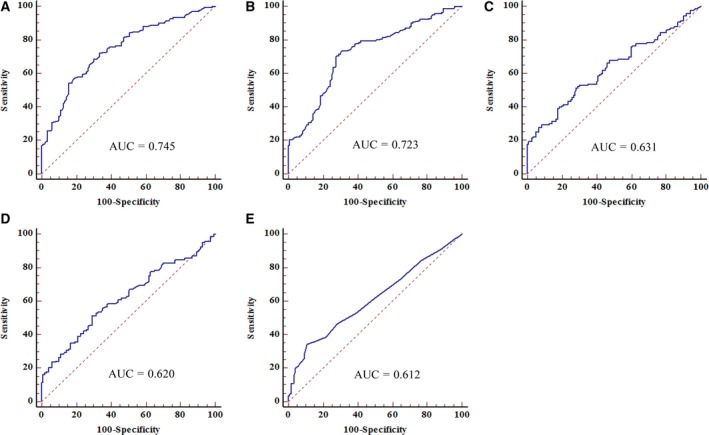
ROC analyses of SOX2OT, ANRIL, CEA, CYFRA21‐1, and SCCA during the training stage for diagnosis of NSCLC. The AUCs of SOX2OT (A) and ANRIL (B) were greater than those of CEA (C), CYFRA21‐1 (D), and SCCA (E), P < 0.05.
3.3. Construction of a diagnostic lncRNA panel for NSCLC
Based on the training set, a logistic regression model was established to diagnose NSCLC. The probability of predicting NSCLC from the 5‐molecule panel was enumerated using following formula: Logit (P) = 1.6487 − (0.002 × CEA) − (0.0249 × CYFRA21‐1) − (0.095 × SCCA) − (0.5474 × SOX2OT) − (0.6174 × ANRIL). We constructed the diagnostic 5‐molecule panel from the training set using this formula and obtained an AUC of 0.853 (95% CI = 0.804–0.894, sensitivity = 77.1%, specificity = 79.2%, Fig. 3A). The predictive capability of the panel was significantly higher compared with that of any molecule alone (all at P < 0.05).
Figure 3.

ROC analyses of the diagnostic panel consisting of SOX2OT, ANRIL, CEA, CYFRA21‐1, and SCCA for the diagnosis of NSCLC. The AUCs of the diagnostic panel for the diagnosis of NSCLC in the training set (A) and validation set (B) were calculated by ROC analysis. The AUCs of the diagnostic panel for patients with TNM stage I (C), II (D), and III (E) in validation set were performed by ROC analysis.
3.4. Validation of the constructed lncRNA panel
We further validated the expression pattern of SOX2OT and ANRIL lncRNA and the efficiency of the 5‐molecule panel estimated during the training stage by employing another independent validation set (100 NSCLC patients and 100 controls). No significant differences were observed in the distribution of age, sex, and tumor characteristics for the NSCLC and control samples between the training and validation sets (Table S1). Similarly, SOX2OT and ANRIL were clearly upregulated in the validation set (Fig. S1A,B). The expressions of CEA, CYFRA21‐1, and SCCA in the validation set are shown in Fig. S1C–E.
The AUCs of SOX2OT, ANRIL, CEA, CYFRA21‐1, and SCCA in the validation set were 0.740 (95% CI = 0.673–0.799), 0.723 (95% CI = 0.656–0.784), 0.622 (95% CI = 0.551–0.689), 0.622 (95% CI = 0.551–0.690), and 0.607 (95% CI = 0.536–0.675), respectively (Fig. S2). The AUC of the 5‐molecule panel in the validation set was 0.883 (95% CI = 0.830–0.924, sensitivity = 91.0%, specificity = 70.0%), which was statistically higher than that of any single molecule (all at P < 0.05) (Fig. 3B). In addition, we calculated the diagnostic performance of this panel in distinguishing NSCLC patients with different TNM stages from healthy individuals in the validation set. The AUCs of the panel for patients with TNM stages I, II, and III were 0.862 (95% CI = 0.790–0.916), 0.875 (95% CI = 0.808–0.925), and 0.917 (95% CI = 0.855–0.958), respectively (Fig. 3C–E).
3.5. Stability of SOX2OT and ANRIL in serum
The serum from NSCLC patients was subjected to harsh conditions to evaluate the stability of SOX2OT and ANRIL. To this end, five serum samples were incubated at room temperature for various durations (0, 3, 6, 12, and 24 h) or were subjected to repetitive freeze–thaw cycles (1, 4, 6, 8 and 10). RNA was then isolated, and qRT‐PCR was performed to test the expressions of SOX2OT and ANRIL. The data showed imperceptible changes in the relative expressions of SOX2OT and ANRIL under the two conditions tested (Fig. S3).
3.6. Correlation between the two serum lncRNA and clinicopathological characteristics
Table 1 lists the correlation between SOX2OT and ANRIL expression levels in serum and the clinicopathological characteristics of the NSCLC patients in the validation set. Higher levels of serum SOX2OT were remarkably correlated with lymph node metastasis (P < 0.05). However, we did not observe any association between the expressions of the lncRNA and patient age, sex, tumor size, or TNM stage.
Table 1.
Correlation between serum lncRNA concentrations and clinicopathological characteristics of patients with NSCLC in validation set [median (interquartile range)]
| Parameters | Total cases | SOX2OT | P value | ANRIL | P value |
|---|---|---|---|---|---|
| Age (years) | |||||
| ≤ 61 | 57 | 1.27 (1.01–1.50) | 0.05 | 1.38 (1.04–1.87) | 0.71 |
| > 61 | 43 | 1.34 (1.19–1.74) | 1.32 (1.04–1.72) | ||
| Sex | |||||
| Male | 65 | 1.33 (1.17–1.63) | 0.15 | 1.32 (1.05–1.75) | 0.90 |
| Female | 35 | 1.18 (0.95–1.62) | 1.36 (1.01–1.83) | ||
| Tumor size | |||||
| ≤ 3 cm | 44 | 1.27 (1.01–1.62) | 0.54 | 1.24 (0.97–1.72) | 0.17 |
| > 3 cm | 56 | 1.31 (1.16–1.62) | 1.43 (1.10–1.85) | ||
| Lymph node metastasis | |||||
| Negative | 52 | 1.22 (0.97–1.52) | 0.01 | 1.21 (0.97–1.72) | 0.08 |
| Positive | 48 | 1.40 (1.22–1.70) | 1.41 (1.12–1.87) | ||
| TNM stage | |||||
| I | 31 | 1.22 (0.96–1.58) | 0.33 | 1.18 (0.96–1.72) | 0.30 |
| II | 39 | 1.28 (1.05–1.53) | 1.25 (1.04–1.69) | ||
| III | 30 | 1.38 (1.22–1.64) | 1.58 (1.14–1.91) | ||
3.7. Association between serum lncRNA and OS rate
A total of 100 NSCLC patients (including five patients who were excluded during the follow‐up period) were followed up for a mean duration of 46 (range 5–72) months. Of the 95 NSCLC patients, 60 (63.2%) died within 5 years of surgery, and the 5‑year OS rate was 36.8%. Kaplan–Meier survival analysis indicated that patients with low expressions of SOX2OT (P = 0.008, Fig. 4A) and ANRIL (P = 0.017, Fig. 4B) achieved a higher 5‐year OS rate. Univariate Cox proportional hazards regression model analysis revealed a statistically significant correlation between the OS rate of NSCLC patients and SOX2OT level (P = 0.009), ANRIL level (P = 0.019), tumor size (P < 0.001), lymph node metastasis (P < 0.001), and TNM stage (P < 0.001) (Table 2). Above‐mentioned parameters were further analyzed using a multivariate analysis, and SOX2OT level (P = 0.036) and TNM stage (P = 0.023) were identified as statistically significant prognostic factors (Table 2).
Figure 4.

Survival analyses of NSCLC patients stratified by median lncRNA expression level. Kaplan–Meier curves show that patients with low SOX2OT (A) or low ANRIL (B) expression had higher survival rate in the validation set.
Table 2.
Univariate and multivariate analyses for OS prediction in validation cohort. HR, hazard ratio
| Parameters | Categories | Univariate analysis | Multivariate analysis | ||
|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | ||
| Age (years) | ≤ 61, > 61 | 1.335 (0.803–2.220) | 0.265 | ||
| Sex | Male, female | 0.586 (0.330–1.040) | 0.068 | ||
| Tumor size | ≤ 3 cm, > 3 cm | 3.071 (1.720–5.481) | < 0.001 | 1.375 (0.725–2.607) | 0.329 |
| Lymph node metastasis | Negative, positive | 4.530 (2.570–7.983) | < 0.001 | 1.638 (0.745–3.601) | 0.220 |
| TNM stage | I, II, III | 2.788 (1.961–3.966) | < 0.001 | 1.859 (1.089–3.175) | 0.023 |
| SOX2OT expression | Low, high | 1.988 (1.187–3.330) | 0.009 | 1.793 (1.040–3.093) | 0.036 |
| ANRIL expression | Low, high | 1.856 (1.108–3.109) | 0.019 | 1.507 (0.874–2.598) | 0.140 |
4. Discussion
In this study, we initially selected 14 lncRNA from previously published NSCLC‐related studies and then validated their dysregulated expressions in NSCLC tissue samples. Among 14 lncRNA tested, we confirmed the dysregulation of 12 lncRNA in NSCLC tissues, corroborating that our results were consistent with previous studies. We next assessed the expression levels of the 12 aberrant lncRNA in serum and observed that two lncRNA (SOX2OT and ANRIL) were significantly upregulated in NSCLC patients compared with healthy controls. Intriguingly, the AUCs of SOX2OT and ANRIL were both greater than those of the currently used tumor markers (CEA, CYFRA21‐1, and SCCA), indicating remarkable efficacy of SOX2OT and ANRIL in distinguishing NSCLC patients from controls. Based on this result, we constructed a diagnostic panel consisting of the two lncRNA and the three tumor markers, which could distinguish NSCLC patients from healthy controls with significantly higher AUC. In addition, circulating SOX2OT was identified as an independent factor for NSCLC prognosis.
To the best of our knowledge, we are the first to report the expression profile of serum lncRNA in a large sample set (460 specimens) with two independent sets. A previous study by Hu et al. (2016) has reported the expression profile of lncRNA in NSCLC plasma; however, their sample size was relatively small (140 specimens). Moreover, most of the previous studies of NSCLC‐related lncRNA focus on single molecules when exploring the potential of lncRNA as novel tumor biomarkers. However, multiple factors may jointly portray their roles in tumor initiation, development, and metastasis progress. Importantly, one lncRNA is too inadequate to act as a novel malignant tumor biomarker; nevertheless, the commonly used tumor markers have lower sensitivity in diagnosing NSCLC. In our study, lncRNA (SOX2OT and ANRIL) and tumor markers (CEA, CYFRA21‐1, and SCCA) were combined into a single diagnostic panel after a logistic regression analysis in the training and validation sets. Our subsequent analysis showed that in the same cohort, this diagnostic panel possessed a superior diagnostic capacity compared with the use of the five molecules separately. Therefore, this panel‐based, integrated analysis provided a multidimensional interpretation to recognize new cases of NSCLC. Besides, our results clearly depicted the excellent sensitivity and specificity of this panel in diagnosing NSCLC, especially at an early stage based on the AUC of the panel in NSCLC patients at different TNM stages.
In this study, we demonstrated a significant association between SOX2OT expression and lymph node metastasis which is a vital prognostic feature for NSCLC patients. Furthermore, NSCLC patients with low expressions of SOX2OT and ANRIL had worse OS rate than those with high expression levels. To exclude the mutual impact of the various factors associated with prognosis, multivariate survival analysis revealed that only TNM stage and SOX2OT could act as independent prognostic factors for NSCLC. Interestingly, SOX2OT and ANRIL expressions have previously been linked with cancers by others. Hou et al. (2014) have reported SOX2OT to be a prognostic indicator of poor survival in lung cancer. ANRIL has been recently identified as an independent prognostic factor for disease‐free survival and OS in patients with gastric cancer (Zhang et al., 2014a). SOX2OT and ANRIL are considered as potential prognosis biomarkers in these studies.
This study utilized serum which is easily obtainable. Some studies have hypothesized that lncRNA can be released into various body fluids, including serum, saliva, and urine, as a result of cancer cell excretion. We next performed assays to assess the stability of the serum lncRNA. Our results indicated that the expressions of lncRNA did not change significantly even when serum samples were exposed to harsh conditions. Researchers have proposed at least three mechanisms that can explain the stability of lncRNA in serum: (a) lncRNA may be selectively enclosed into small, membrane‐covered vesicles, such as exosomes, microparticles, and apoptotic bodies; (b) lncRNA may fold into complex secondary and tertiary structures; or (c) lncRNA may bind with protein to avoid degradation (Hewson and Morris, 2016; Shi et al., 2016). Our results and these possible stabilizing mechanisms further underscored that serum SOX2OT and ANRIL may serve as potential novel biomarkers for diagnosis and prognosis of NSCLC.
Previous functional studies on lncRNA provide strong evidence that SOX2OT and ANRIL are potential biomarkers. SOX2OT is involved in various stages of progression of a malignant tumor. Splice variants (SOX2OT‐S1 and SOX2OT‐S2) have been identified to co‐upregulate with SOX2 and OCT4 (two key regulators of pluripotency) in esophageal squamous cell carcinoma (Shahryari et al., 2014). SOX2OT has also been found to be concomitantly overexpressed along with SOX2 and OCT4A in lung tumor samples (Saghaeian Jazi et al., 2016). SOX2OT may also be involved in cell differentiation and may thus influence tumorigenesis. Furthermore, SOX2OT is also involved in other biological characteristics of multiple pernicious tumors, including development and metastasis (Askarian‐Amiri et al., 2014; Shi, 2015; Zhang et al., 2016). Similarly, ANRIL also plays a crucial role in tumor evolution. Nie et al. (2015) have suggested that ANRIL controls the proliferation, apoptosis, and migration of NSCLC cells by silencing KLF2 and P21 in the EZH2 pathway. Additionally, Sun et al. (2016) have reported that ANRIL knockdown significantly suppresses cell migration and invasion, tumor growth, and the capacity for lymphatic metastasis, suggesting that ANRIL is a potential curative target in colorectal cancer. Qiu et al. (2016) have shown that ANRIL can decrease P15 expression and increase Bcl‐2 expression, thereby accelerating cell growth and suppressing apoptosis and senescence, eventually leading to the proliferation of epithelial ovarian cancer cells. These reports further strengthen our finding that circulating lncRNA SOX2OT and ANRIL have immense potential to serve as NSCLC biomarkers.
In our study, only two lncRNA (SOX2OT and ANRIL) were significantly overexpressed in NSCLC serum, which was inconsistent with other tissue/cell studies. We attributed this discrepancy to the sample type (tissue versus serum), sample size, and the demographic characteristics of the study population (ethnicity, region, etc.), as well as the testing methods employed. Most participants in our study were enrolled from the same hospital, which makes a limitation of our study. Further multicenter cohort should be carried out in future to further strengthen these results. As SOX2OT has a variety of transcript isoforms that play different roles in tumor progression, sequencing should be performed to further identify the functional isoforms. We validated the diagnostic potential of a 5‐molecule panel to distinguish NSCLC patients from healthy controls. However, we did not assess the two lncRNA in serum samples of patients with other malignant diseases or benign respiratory diseases. This will be important to explore the specificity of SOX2OT and ANRIL and warrant further investigation.
5. Conclusions
We established a diagnostic panel consisting of lncRNA (SOX2OT and ANRIL) and tumor markers (CEA, CYFRA21‐1, and SCCA), which could diagnose NSCLC with superior sensitivity and specificity. In addition, we identified SOX2OT as an independent predictor of NSCLC survival rate. Further studies on samples from diverse regions and ethnic groups using standardized detection methods should be conducted to confirm the usefulness of lncRNA as promising potential biomarkers for diagnosis and prognosis of NSCLC.
Author contributions
CW designed this study. YX, YZ, LD, XJ, SY, and YZ performed all the experiments. YX, LW, JL, WD, LW, and SZ evaluated the data. CW, SX, SL, YX, and YZ wrote the manuscript. All authors read and approved the final manuscript.
Supporting information
Fig. S1. Expression levels of SOX2OT, ANRIL, CEA, CYFRA21‐1, and SCCA in serum samples of NSCLC patients and healthy controls during the validation stage.
Fig. S2. ROC analyses of SOX2OT, ANRIL, CEA, CYFRA21‐1, and SCCA during the validation stage for NSCLC diagnosis.
Fig. S3. Stability of SOX2OT and ANRIL in serum.
Table S1. Clinicopathological characteristics of patients and demographic information of controls in training set and validation set.
Table S2. The selected lncRNA concentration in 20 paired NSCLC and adjacent normal tissues [median (interquartile range)].
Table S3. The selected lncRNA concentration in NSCLC serum of 46‐samples compared to the controls [median (interquartile range)].
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81472025, 81400029 and 81502164); Scientific Research Award Foundation for Outstanding Young and Middle‐Aged Scientists of Shandong Province (BS2014YY011, BS2015YY004); Shandong Medical and Health Science and Technology Development Programs (2014WS0143); Shandong Technological Development Project (2016CYJS01A02, 2015GSF118052, 2016GSF201055, and 2016GSF201165); and Taishan Scholar Program of Shandong Province.
Yujiao Xie and Yi Zhang contributed equally to this article
References
- Askarian‐Amiri ME, Seyfoddin V, Smart CE, Wang J, Kim JE, Hansji H, Baguley BC, Finlay GJ and Leung EY (2014) Emerging role of long non‐coding RNA SOX2OT in SOX2 regulation in breast cancer. PLoS One 9, e102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach PB, Mirkin JN, Oliver TK, Azzoli CG, Berry DA, Brawley OW, Byers T, Colditz GA, Gould MK, Jett JR et al (2012) Benefits and harms of CT screening for lung cancer: a systematic review. JAMA 307, 2418–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Wang XY, Han XH, Wang H and Qi J (2015) Diagnostic value of Cyfra21‐1, SCC and CEA for differentiation of early‐stage NSCLC from benign lung disease. Int J Clin Exp Med 8, 11295–11300. [PMC free article] [PubMed] [Google Scholar]
- Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ and He J (2016) Cancer statistics in China, 2015. CA Cancer J Clin 66, 115–132. [DOI] [PubMed] [Google Scholar]
- Feng J, Sun Y, Zhang E, Lu X, Jin S and Guo R (2015) A novel long noncoding RNA IRAIN regulates cell proliferation in non small cell lung cancer. Int J Clin Exp Pathol 8, 12268–12275. [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Lu Z, Li Y, Zhang J, Zhang G, Chen X, Chu J, Ren J, Liu H and Guo X (2016) Cancer incidence and mortality in Shandong province, 2012. Chin J Cancer Res 28, 263–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert WH (2017) Cancer screening, overdiagnosis, and regulatory capture. JAMA Intern Med 177, 915–916. [DOI] [PubMed] [Google Scholar]
- Han L, Zhang EB, Yin DD, Kong R, Xu TP, Chen WM, Xia R, Shu YQ and De W (2015) Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non‐small cell lung cancer and affects cell apoptosis by regulating Bcl‐2. Cell Death Dis 6, e1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson C and Morris KV (2016) Form and function of exosome‐associated long non‐coding RNAs in cancer. Curr Top Microbiol Immunol 394, 41–56. [DOI] [PubMed] [Google Scholar]
- Hou Z, Zhao W, Zhou J, Shen L, Zhan P, Xu C, Chang C, Bi H, Zou J, Yao X et al (2014) A long noncoding RNA Sox2ot regulates lung cancer cell proliferation and is a prognostic indicator of poor survival. Int J Biochem Cell Biol 53, 380–388. [DOI] [PubMed] [Google Scholar]
- Hu X, Bao J, Wang Z, Zhang Z, Gu P, Tao F, Cui D and Jiang W (2016) The plasma lncRNA acting as fingerprint in non‐small‐cell lung cancer. Tumour Biol 37, 3497–3504. [DOI] [PubMed] [Google Scholar]
- I H and Cho JY (2015) Lung cancer biomarkers. Adv Clin Chem 72, 107–170. [DOI] [PubMed] [Google Scholar]
- Kay FU, Kandathil A, Batra K, Saboo SS, Abbara S and Rajiah P (2017) Revisions to the tumor, node, metastasis staging of lung cancer (8th edition): rationale, radiologic findings and clinical implications. World J Radiol 9, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Wang Y, Liu X, Luo P, Jing W, Zhu M and Tu J (2017) Identification of circulating long noncoding RNA HOTAIR as a novel biomarker for diagnosis and monitoring of non‐small cell lung cancer. Technol Cancer Res Treat 2017, 1533034617723754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Zhang G, Li J, Yang R, Chen S, Wu S, Zhang F, Bai Y, Zhao H and Wang Y (2016) Long noncoding RNA RGMB‐AS1 indicates a poor prognosis and modulates cell proliferation, migration and invasion in lung adenocarcinoma. PLoS One 11, e0150790. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liang W, Lv T, Shi X, Liu H, Zhu Q, Zeng J, Yang W, Yin J and Song Y (2016) Circulating long noncoding RNA GAS5 is a novel biomarker for the diagnosis of nonsmall cell lung cancer. Medicine 95, e4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu WQ, Xie WP and Hou YY (2013) Long non‐coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression. BMC Cancer 13, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R and Jemal A (2016) Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin 66, 271–289. [DOI] [PubMed] [Google Scholar]
- Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, Chen WS and Li B (2016) LncRNA‐UCA1 exerts oncogenic functions in non‐small cell lung cancer by targeting miR‐193a‐3p. Cancer Lett 371, 99–106. [DOI] [PubMed] [Google Scholar]
- Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH et al (2015) Long noncoding RNA ANRIL promotes non‐small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther 14, 268–277. [DOI] [PubMed] [Google Scholar]
- Qi P, Zhou XY and Du X (2016) Circulating long non‐coding RNAs in cancer: current status and future perspectives. Mol Cancer 15, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu JJ, Liu YL, Zhang Y, Ding JX and Hua KQ (2016) The long non‐coding RNA ANRIL promotes proliferation and cell cycle progression and inhibits apoptosis and senescence in epithelial ovarian cancer. Oncotarget 7, 32478–32492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L and Yin R (2014) CCAT2 is a lung adenocarcinoma‐specific long non‐coding RNA and promotes invasion of non‐small cell lung cancer. Tumour Biol 35, 5375–5380. [DOI] [PubMed] [Google Scholar]
- Reis EM and Verjovski‐Almeida S (2012) Perspectives of long non‐coding RNAs in cancer diagnostics. Front Genet 3, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciuti B, Mencaroni C, Paglialunga L, Paciullo F, Crino L, Chiari R and Metro G (2016) Long noncoding RNAs: new insights into non‐small cell lung cancer biology, diagnosis and therapy. Med Oncol 33, 18. [DOI] [PubMed] [Google Scholar]
- Saghaeian Jazi M, Samaei NM, Ghanei M, Shadmehr MB and Mowla SJ (2016) Overexpression of the non‐coding SOX2OT variants 4 and 7 in lung tumors suggests an oncogenic role in lung cancer. Tumour Biol 37, 10329–10338. [DOI] [PubMed] [Google Scholar]
- Schmitt AM and Chang HY (2016) Long noncoding RNAs in cancer pathways. Cancer Cell 29, 452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahryari A, Rafiee MR, Fouani Y, Oliae NA, Samaei NM, Shafiee M, Semnani S, Vasei M and Mowla SJ (2014) Two novel splice variants of SOX2OT, SOX2OT‐S1, and SOX2OT‐S2 are coupregulated with SOX2 and OCT4 in esophageal squamous cell carcinoma. Stem Cells 32, 126–134. [DOI] [PubMed] [Google Scholar]
- Shi X (2015) Up‐regulation of long non‐coding RNA Sox2ot promotes hepatocellular carcinoma cell metastasis and correlates with poor prognosis. Int J Clin Exp Pathol 8, 4008–4014. [PMC free article] [PubMed] [Google Scholar]
- Shi T, Gao G and Cao Y (2016) Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis Markers 2016, 9085195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel RL, Miller KD and Jemal A (2017) Cancer statistics, 2017. CA Cancer J Clin 67, 7–30. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jin SD, Zhu Q, Han L, Feng J, Lu XY, Wang W, Wang F and Guo RH (2017) Long non‐coding RNA LUCAT1 is associated with poor prognosis in human non‐small lung cancer and regulates cell proliferation via epigenetically repressing p21 and p57 expression. Oncotarget 8, 28297–28311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun ZQ, Ren WG, Xie X, Li XY and Li GY (2016) Downregulation of long non‐coding RNA ANRIL suppresses lymphangiogenesis and lymphatic metastasis in colorectal. Oncotarget 7, 47536–47555. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Thai P, Statt S, Chen CH, Liang E, Campbell C and Wu R (2013) Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol 49, 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis WD, Brambilla E, Nicholson AG, Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E, Flieder DB et al (2015) The 2015 World Health Organization classification of lung tumors: impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 10, 1243–1260. [DOI] [PubMed] [Google Scholar]
- Wang GC, Chen HB and Liu J (2015) The long noncoding RNA LINC01207 promotes proliferation of lung adenocarcinoma. Am J Cancer Res 5, 3162–3173. [PMC free article] [PubMed] [Google Scholar]
- Weber DG, Johnen G, Casjens S, Bryk O, Pesch B, Jöckel KH, Kollmeier J and Brüning T (2013) Evaluation of long noncoding RNA MALAT1 as a candidate blood‐based biomarker for the diagnosis of non‐small cell lung cancer. BMC Res Notes 6, 518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Liu H, Shi X, Yao Y, Yang W and Song Y (2015) The long non‐coding RNA HNF1A‐AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget 6, 9160–9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Xu E, Dai J, Liu B, Han Z, Wu J, Zhang S, Peng B, Zhang Y and Jiang Y (2015) A novel long noncoding RNA AK001796 acts as an oncogene and is involved in cell growth inhibition by resveratrol in lung cancer. Toxicol Appl Pharmacol 285, 79–88. [DOI] [PubMed] [Google Scholar]
- Yang YRZS, Zhong CL, Li YX, Zhao SS and Feng XJ (2014) Increased expression of the lncRNA PVT1 promotes tumorigenesis in non‐small cell lung cancer. Int J Clin Exp Pathol 7, 6929–6935. [PMC free article] [PubMed] [Google Scholar]
- Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W et al (2014a) Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR‐99a_miR‐449a. Oncotarget 5, 2276–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YY, Lian JC and Xu HY (2016) LncRNA Sox2ot overexpression serves as a poor prognostic biomarker in gastric cancer. Am J Transl Res 8, 5035–5043. [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Xia Y, Wang Z, Zheng J, Chen Y, Li X, Wang Y and Ming H (2017) Serum long non coding RNA MALAT‐1 protected by exosomes is upregulated and promotes cell proliferation and migration in non‐small cell lung cancer. Biochem Biophys Res Comm 490, 406–414. [DOI] [PubMed] [Google Scholar]
- Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS et al (2014b) P53‐regulated long non‐coding RNA TUG1 affects cell proliferation in human non‐small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis 5, e1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Hou X and Zhan H (2015a) Long non‐coding RNA PCAT‐1 over‐expression promotes proliferation and metastasis in non‐small cell lung cancer cells. Int J Clin Exp Med 8, 18482–18487. [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Yu HX, Han ZF, Gao N, Xue JR and Wang Y (2015b) Clinical significance of joint detection of serum CEA, SCCA, and bFGF in the diagnosis of lung cancer. Int J Clin Exp Pathol 8, 9506–9511. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Expression levels of SOX2OT, ANRIL, CEA, CYFRA21‐1, and SCCA in serum samples of NSCLC patients and healthy controls during the validation stage.
Fig. S2. ROC analyses of SOX2OT, ANRIL, CEA, CYFRA21‐1, and SCCA during the validation stage for NSCLC diagnosis.
Fig. S3. Stability of SOX2OT and ANRIL in serum.
Table S1. Clinicopathological characteristics of patients and demographic information of controls in training set and validation set.
Table S2. The selected lncRNA concentration in 20 paired NSCLC and adjacent normal tissues [median (interquartile range)].
Table S3. The selected lncRNA concentration in NSCLC serum of 46‐samples compared to the controls [median (interquartile range)].


