Abstract
Background
Signal transducer and activator of transcription 3 (STAT3) is constitutively activated in several malignancies. Here, we define the correlation between STAT3 expression and lymph node micrometastasis of early‐stage non‐small cell lung cancer. Then we highlight some possibilities associated with developing a way to detect tumor micrometastasis and an anticancer drug that might therapeutically inhibit the STAT3 signaling pathway.
Methods
The samples were collected from 50 patients with early‐stage non‐small cell lung cancer and 50 patients with benign lung tumors. Mucin 1 mRNA expression was evaluated to determine lymph node micrometastasis status. STAT3 mRNA, STAT3 protein, and phosphorylated STAT3 protein expression were evaluated through reverse transcription polymerase chain reaction, western blot, and immunohistochemistry, respectively. Measurement data was represented as mean ± standard deviation, and the t‐rest or F‐test were used. The χ2‐test was used in enumeration data. Logistic regression analysis was carried out to determine the independent risk factors influencing lymph node micrometastasis.
Results
STAT3 mRNA and proteins expression were correlated with lymph node micrometastasis (P < 0.05). Logistic regression analysis revealed STAT3 protein overexpression and the differentiation degree of tumors were independent risk factors for lymph node micrometastasis.
Conclusion
Overexpression of STAT3 might promote lymphatic micrometastasis of early‐stage non‐small cell lung cancer and might be a clinical predictor of lymph node micrometastasis.
Keywords: Lymph node micrometastasis, mucin 1, non‐small cell lung cancer, phosphorylated signal transducer and activator of transcription 3, signal transducer and activator of transcription 3
Introduction
Lung cancer is the leading cause of tumor‐specific death in many countries nowadays, and its incidence rate is rising every year.1 The main pathological type of lung cancer, which covers >80%, is non‐small cell lung cancer (NSCLC). It appears that lymph node metastasis of lung cancer may occur early, and latter lung cancer can metastasize to other distant organs. Metastatic spread constitutes the primary source of morbidity and mortality, and influences the effect of the patients’ treatment and prognosis.2 Although the regional development of NSCLC is not as aggressive as small‐cell lung cancer, micrometastasis may occur in NSCLC and it will seriously influence the patients’ prognosis. Accumulating evidence suggests that constitutively activated signal transducer and activator of transcription 3 (STAT3) has been found to be related to tumor development and progression in numerous forms of primary cancers. Persistent phosphorylation of STAT3 has been shown in 22~65% of NSCLC. The constantly activated STAT3 contributes to oncogenesis by upregulation of genes encoding bcl‐xl, bcl‐2, cyclinD1, c‐myc, survivin, mcl‐1, vascular endothelial growth factor, interleukin‐10, and so on, which can protect apoptosis, enhance cell proliferation, promote angiogenesis, and evade immune surveillance.3, 4
There are several methods to test tumor micrometastasis, such as polymerase chain reaction (PCR), immunohistochemistry, flow cytometry, western blot, and so on.5 PCR is considered to be an ideal method, and mucin 1 (MUC1) mRNA is a selective target gene for tumor micrometastasis detection using PCR.
Therefore, we wanted to explore whether STAT3 can be related to lymph node micrometastasis of NSCLC. To address this question, we evaluated the expression of MUC1 mRNA in the lymph node samples of NSCLC to determine micrometastasis. Then, we evaluated what role STAT3 overexpression plays in lymph node micrometastasis of NSCLC.
Methods
Patients
The samples of cancer tissues, lymph nodes from 50 patients with early‐stage NSCLC (20 stage IA samples; 20 stage IB samples; 10 stage II samples without lymph node metastasis) and 50 normal lung tissue samples, and lymph node samples from 50 patients with benign lung tumors who underwent complete resection in Provincial Hospital Affiliated to Shandong University, Jinan, China, from June 2008 to January 2010, were collected. Our study was approved by the ethics committee in our hospital, and we obtained consent from the patients. All lung cancer patients underwent complete resection of tumors and systemic lymph node dissection (according to Naruke lymph node profile).
The study group consisted of 26 men and 24 women, ranging in age from 36 to 67 years (mean 51.3 years). There were 21 squamous cell carcinomas and 29 adenocarcinomas; 31 well‐moderately differentiated cases, 19 poor differentiated cases (Table 1).
Table 1.
Clinical characteristics of 50 patients with early‐stage non‐small cell lung cancer
| Characteristics | No. of patients | Ratio (%) |
|---|---|---|
| Gender | ||
| Male | 26 | 52.0 |
| Female | 24 | 48.0 |
| Age (years) | ||
| ≤60 | 31 | 62.0 |
| >60 | 19 | 38.0 |
| Histological type | ||
| SCC | 21 | 42.0 |
| ADC | 29 | 58.0 |
| Differentiation | ||
| Well + moderate | 31 | 62.0 |
| Poor | 19 | 38.0 |
| Clinical stage | ||
| IA | 20 | 40.0 |
| IB | 20 | 40.0 |
| II | 10 | 20.0 |
| Total | 50 | 100 |
ADC, adenocarcinoma; SCC, squamous cell carcinoma.
Isolation of total RNA and reverse transcription PCR
RNA was extracted from lymph node samples, lung cancer samples, and normal lung tissue samples by using Trizol Reagent (Invitrogen, Carlsbad, CA, USA). Cellular RNA (1 μg) was used for cDNA synthesis. For PCR, we used the specific kit from Applied Biosystems (Foster City, CA, USA). PCR primers for detecting mRNA for MUC1 in lymph node samples, STAT3 in cancer and normal lung tissue samples, and β‐actin were synthesized by Shandong Academy of Medical Sciences, Shandong, China. Primer sequences were as follows: β‐actin, sense, 5′‐GTGGGGCGCCCCAGGCACCA‐3′, antisense, 5′‐CTCCTTAATGTCACGCACGATTTC‐3′; MUC1, sense, 5′CGTCGTGGACATTGATGGTACC3′, antisense, 5′GGTACCTCCTCTCACCTCCTCCAA3′; STAT3, sense, 5′‐TTGCCAGTTGTGGTGATC‐3′, antisense, 5′‐ CCAGACCCAGAAGGAGAAGC ‐3′.
Western blot
The tissues were lysed in lysing buffer and total protein was estimated by using the bichloroacetic acid method. The whole cell extracts (20 μg/lane) were separated by sodium dodecyl sulfate‐polyacrylamide gel electrophoresis and transferred to nitrocellulose membrane. After blocked in Tris‐buffered saline with 5% (w/v) non‐fat dry milk, membranes were incubated with primary antibodies (1:100 dilution) according to the manufacturer’s instructions, and then incubated with horseradish peroxidase conjugated secondary antibody. The protein bands were recorded on X‐ray film and examined.
Immunohistochemistry
Immunohistochemical staining for STAT3 and phosphorylated STAT3 (pSTAT3) protein was performed using the streptavidin‐peroxidase method. Slides were deparaffinized and rehydrated with xylene and graded alcohol. Optimal antigen retrieval was carried out in citrate buffer (pH = 6.0) for 10 min with a steam oven to enhance the immunoreactivity. The protein expression was scored semiquantitatively by the proportion of the stained tumor nuclei or cytoplasm as follows: staining proportion was classified as 0 to 3 (0 = 0~5% positive cells, 1 = 5~25% positive cells, 2 = 25~50%, 3 ≥50%). Two independent observers did the semiquantitative scoring of the expression levels of STAT3 and pSTAT3 proteins, and both observers re‐examined the immunostained slides to determine a consensus score.
Statistical analysis
All statistical analyses were performed with SPSS 11.5 statistical software (SPSS Inc., Chicago, IL, USA). Measurement data were represented as mean ± standard deviation, and the t‐rest or F‐test were used. The χ2‐test was used in enumeration data. Logistic regression analysis was performed to determine the independent risk factors influencing lymph node micrometastasis of early‐stage NSCLC. Differences were considered significant when the P‐value was <0.05.
Results
Results of reverse transcription PCR
We examined MUC1 mRNA expression by reverse transcription PCR in all samples of lymph nodes, and STAT3 mRNA expression in all NSCLC and normal lung tissue samples (Figs 1, 2). MUC1 mRNA expression was detected in 33 lymph node samples that belong to 19 NSCLC patients and 0 in the lymph node samples from patients with benign lung tumors (Table 2). STAT3 mRNA expression values in NSCLC and normal lung tissue samples were 0.8019 ± 0.1462 and 0.3303 ± 0.0236, respectively (t′ = 22.105, P = 0.000). In NSCLC with lymph node micrometastasis, the STAT3 mRNA expression value was 0.9502 ± 0.1004 and that of NSCLC without lymph node micrometastasis was 0.7110 ± 0.0800. STAT3 mRNA overexpression in NSCLC samples correlated with lymph node micrometastasis status (t = 9.308, P = 0.000), tumor differentiation degree (t = −6.936, P = 0.000) and the pathological TNM stage (F = 36.001, P = 0.000), but not correlated with age, gender, and histological type (Table 3).
Figure 1.
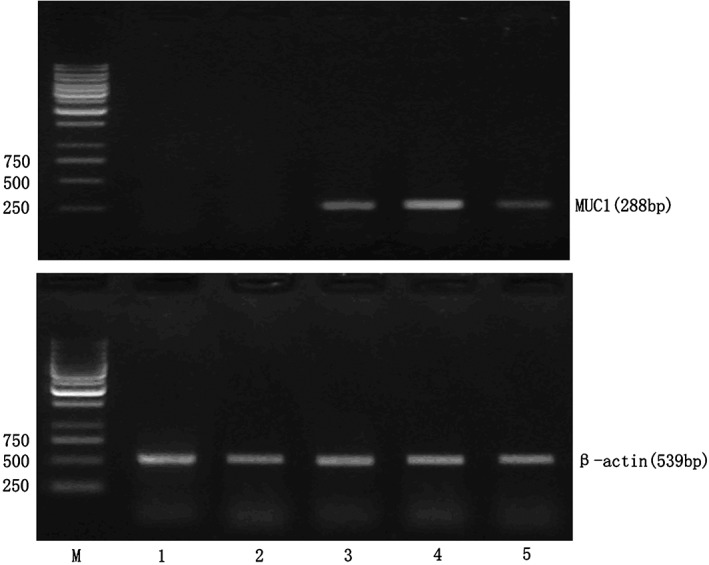
Mucin 1 (MUC1) mRNA expression in lymph node samples. M, marker; 1: negative control lymph node sample; 2: lymph node sample without micrometastasis; 3~5: lymph node samples with micrometastasis.
Figure 2.
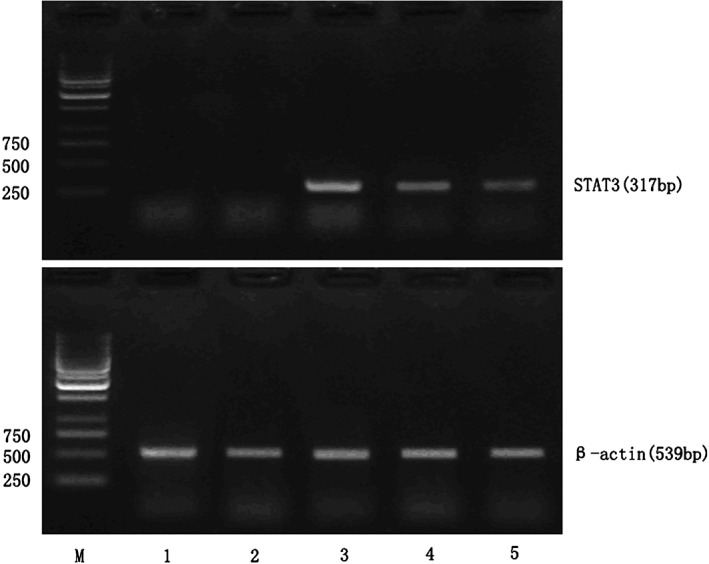
Signal transducer and activator of transcription 3 (STAT3) mRNA expression in lung cancer samples and normal lung tissue samples. M, marker; 1~2: normal lung tissue samples; 3~4: non‐small cell lung cancer samples with lymph node micrometastasis; 5: non‐small cell lung cancer samples without lymph node micrometastasis.
Table 2.
Reverse transcription polymerase chain reaction analysis of mucin 1 mRNA expression in lymph node samples belonging to non‐small cell lung cancer patients
| Characteristics | MUC1 | χ2 | P‐value | |
|---|---|---|---|---|
| (+) | (−) | |||
| Gender | ||||
| Male | 9 | 17 | 0.263 | 0.608 |
| Female | 10 | 14 | ||
| Age (years) | ||||
| ≤60 | 10 | 21 | 1.142 | 0.285 |
| >60 | 9 | 10 | ||
| Histological type | ||||
| SCC | 11 | 10 | 3.178 | 0.075 |
| ADC | 8 | 21 | ||
| Differentiation | ||||
| Well + moderate | 5 | 26 | 16.563 | 0.000 |
| Poor | 14 | 5 | ||
| pTNM | ||||
| IA | 4 | 16 | 7.131 | 0.028 |
| IB | 8 | 12 | ||
| II | 7 | 3 | ||
| Total | 19 | 31 | — | — |
ADC, adenocarcinoma; MUC1, mucin 1; pTNM, pathological TNM; SCC, squamous cell carcinoma.
Table 3.
Reverse transcription polymerase chain reaction analysis of signal transducer and activator of transcription 3 mRNA expression in non‐small cell lung cancer and normal lung tissue samples
| Characteristics | STAT3 mRNA value | t, t′ or F | P‐value |
|---|---|---|---|
| Gender | |||
| Male | 0.7817 ± 0.1349 | −1.017 | 0.314 |
| Female | 0.8237 ± 0.1575 | ||
| Age (years) | |||
| ≤60 | 0.7888 ± 0.1521 | −0.802 | 0.426 |
| >60 | 0.8231 ± 0.1373 | ||
| Histological type | |||
| SCC | 0.8393 ± 0.1564 | 1.563 | 0.125 |
| ADC | 0.7748 ± 0.1346 | ||
| Differentiation | |||
| Well + moderate | 0.7217 ± 0.0945 | −6.936 | 0.000 |
| Poor | 0.9327 ± 0.1191 | ||
| pTNM | |||
| IA | 0.6848 ± 0.0764 | 36.001† | 0.000 |
| IB | 0.8257 ± 0.1023 | ||
| II | 0.9885 ± 0.1073 | ||
| MUC1 | |||
| Positive | 0.9502 ± 0.1004 | 9.308 | 0.000 |
| Negative | 0.7110 ± 0.0800 | ||
| Type of tissue | |||
| NSCLC tissues | 0.8019 ± 0.1462 | 22.105‡ | 0.000 |
| Normal lung tissues | 0.3303 ± 0.0236 | ||
F‐test.
t′‐test.
ADC, adenocarcinoma; MUC1, mucin 1; NSCLC, non‐small cell lung cancer; pTNM, pathological TNM; SCC, squamous cell carcinoma; STAT3, signal transducer and activator of transcription 3.
Results of western blot
The expression value of STAT3 protein and pSTAT3 protein in NSCLC samples were 0.9459 ± 0.1375 and 0.6503 ± 0.1510, which were much higher than that expressed in normal lung tissue samples (0.3887 ± 0.0269 and 0.2192 ± 0.0215). The expression value of STAT3 protein and pSTAT3 protein in NSCLC samples with lymph node micrometastasis were 1.0860 ± 0.0783 and 0.7987 ± 0.0936, which were also higher than that expressed in lung cancer samples without lymph node micrometastasis (0.8600 ± 0.0846 and 0.5593 ± 0.0972). The result is shown in Figure 3. Through statistical analysis, STAT3 and pSTAT3 overexpression in NSCLC samples correlated with lymph node micrometastasis status (t = 9.427, P = 0.000; t = 8.572, P = 0.000), tumor differentiation degree (t = −6.876, P = 0.000; t = −8.003, P = 0.000), and the TNM stage (F = 32.589, P = 0.000; F = 29.031, P = 0.000), but had no significant correlation with age, gender, and histological type (Table 4).
Figure 3.
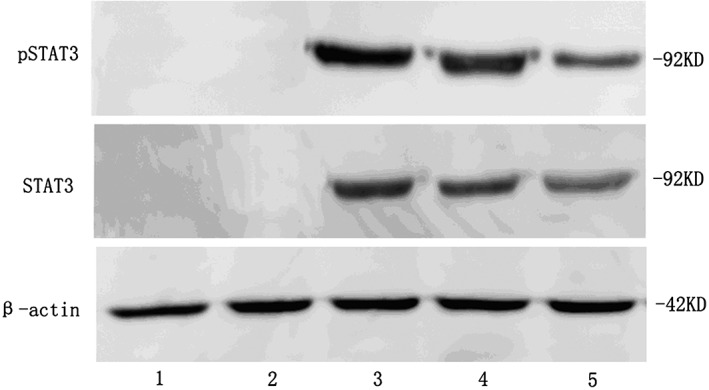
Signal transducer and activator of transcription 3 (STAT3), phosphorylated STAT3 (pSTAT3) proteins expression in lung cancer samples and normal lung tissue samples. 1~2: normal lung tissue samples; 3~4: non‐small cell lung cancer samples with lymph node micrometastasis; 5: non‐small cell lung cancer samples without lymph node micrometastasis.
Table 4.
Western blot analysis of signal transducer and activator of transcription 3 and signal transducer and activator of transcription 3 proteins expression in non‐small cell lung cancer and normal lung tissue samples
| Characteristics | STAT3 protein value | t, t′ or F | P‐value | pSTAT3 protein value | t, t′ or F | P‐value |
|---|---|---|---|---|---|---|
| Gender | ||||||
| Male | 0.9319 ± 0.1317 | −0.745 | 0.460 | 0.6304 ± 0.1395 | −0.966 | 0.339 |
| Female | 0.9610 ± 0.1418 | 0.6717 ± 0.1627 | ||||
| Age (years) | ||||||
| ≤60 | 0.9316 ± 0.1455 | −0.940 | 0.352 | 0.6374 ± 0.1562 | −0.763 | 0.449 |
| >60 | 0.9693 ± 0.1236 | 0.6711 ± 0.1436 | ||||
| Histological type | ||||||
| SCC | 0.9819 ± 0.1473 | 1.600 | 0.116 | 0.6769 ± 0.1642 | 1.064 | 0.293 |
| ADC | 0.9198 ± 0.1262 | 0.6309 ± 0.1404 | ||||
| Differentiation | ||||||
| Well + moderate | 0.8708 ± 0.0990 | −6.876 | 0.000 | 0.5618 ± 0.0972 | −8.003 | 0.000 |
| Poor | 1.0684 ± 0.0980 | 0.7946 ± 0.1041 | ||||
| pTNM | ||||||
| IA | 0.8314 ± 0.0813 | 32.589† | 0.000 | 0.5386 ± 0.0962 | 29.031† | 0.000 |
| IB | 0.9812 ± 0.1054 | 0.6667 ± 0.1105 | ||||
| II | 1.1043 ± 0.0757 | 0.8406 ± 0.1010 | ||||
| MUC1 | ||||||
| Positive | 1.0860 ± 0.0783 | 9.427 | 0.000 | 0.7987 ± 0.0936 | 8.572 | 0.000 |
| Negative | 0.8600 ± 0.0846 | 0.5593 ± 0.0972 | ||||
| Type of tissues | ||||||
| NSCLC tissues | 0.9459 ± 0.1375 | 27.370‡ | 0.000 | 0.6503 ± 0.1510 | 19.696‡ | 0.000 |
| Normal lung tissues | 0.3887 ± 0.0269 | 0.2192 ± 0.0215 | ||||
F‐test.
t′‐test.
ADC, adenocarcinoma; NSCLC, non‐small cell lung cancer; SCC, squamous cell carcinoma; STAT3, signal transducer and activator of transcription 3.
Results of immunohistochemistry
The expression rate of STAT3 protein and pSTAT3 protein in lung cancer samples with lymph node micrometastasis were 94.74% and 73.68% using the immunohistochemistry method, which were also higher than that in lung cancer samples without lymph node micrometastasis (both were 38.71%). No STAT3 protein and pSTAT3 protein were detected in normal lung tissue samples (Fig 4). STAT3 and pSTAT3 overexpression in NSCLC samples correlated with lymph node micrometastasis (χ2 = 15.407, P = 0.000; χ2 = 5.773, P = 0.016), tumor differentiation degree (χ2 = 11.092, P = 0.001; χ2 = 12.738, P = 0.000), and the TNM stage (χ2 = 8.750, P = 0.013; χ2 = 12.139, P = 0.002), but had no significant correlation with age, gender, and histological type (Table 5).
Figure 4.
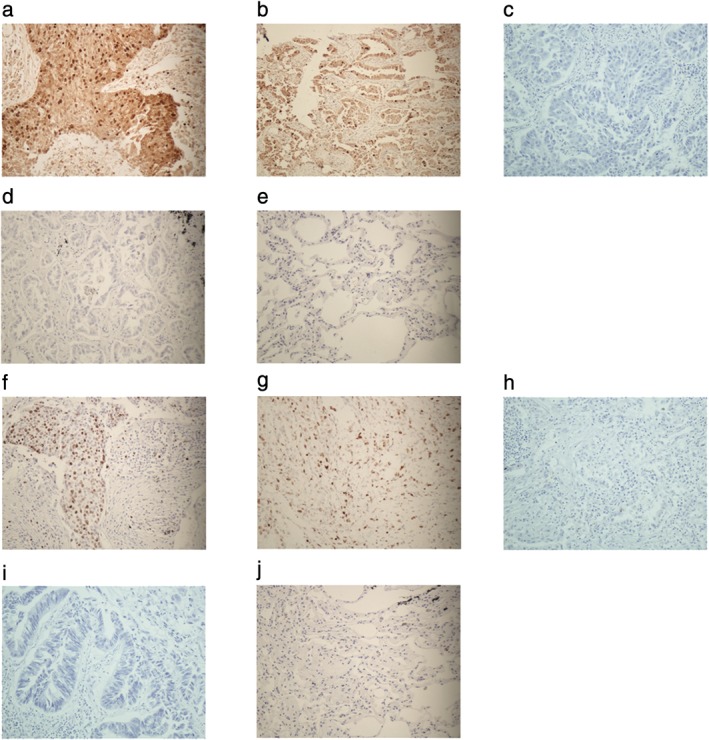
Signal transducer and activator of transcription 3 (STAT3) and phosphorylated STAT3 (pSTAT3) proteins expression in lung cancer samples and normal lung tissue samples (streptavidin‐peroxidase method, ×200). (a) STAT3 positive expression in squamous cell lung cancer, (b) STAT3 positive expression in lung adenocarcinoma, (c) STAT3 negative expression in squamous cell lung cancer, (d) STAT3 negative expression in lung adenocarcinoma, (e) STAT3 negative expression in normal lung tissue, (f) pSTAT3 positive expression in squamous cell lung cancer, (g) pSTAT3 positive expression in lung adenocarcinoma, (h) pSTAT3 negative expression in squamous cell lung cancer, (i) pSTAT3 negative expression in lung adenocarcinoma, and (j) pSTAT3 negative expression in normal lung tissue.
Table 5.
Immunohistochemistry analysis of signal transducer and activator of transcription 3 and phosphorylated signal transducer and activator of transcription 3 proteins expression in non‐small cell lung cancer samples
| Characteristics | STAT3 | χ2 | P‐value | pSTAT3 | χ2 | P‐value | ||
|---|---|---|---|---|---|---|---|---|
| (+) | (−) | (+) | (−) | |||||
| Gender | ||||||||
| Male | 15 | 11 | 0.120 | 0.729 | 12 | 14 | 0.742 | 0.389 |
| Female | 15 | 9 | 14 | 10 | ||||
| Age (years) | ||||||||
| ≤60 | 16 | 15 | 2.391 | 0.122 | 15 | 16 | 0.427 | 0.514 |
| >60 | 14 | 5 | 11 | 8 | ||||
| Histological type | ||||||||
| SCC | 15 | 6 | 1.970 | 0.160 | 12 | 9 | 0.384 | 0.536 |
| ADC | 15 | 14 | 14 | 15 | ||||
| Differentiation | ||||||||
| Well + moderate | 13 | 18 | 11.092 | 0.001 | 10 | 21 | 12.738 | 0.000 |
| Poor | 17 | 2 | 16 | 3 | ||||
| pTNM | ||||||||
| IA | 7 | 13 | 8.750 | 0.013 | 5 | 15 | 12.139 | 0.002 |
| IB | 15 | 5 | 12 | 8 | ||||
| II | 8 | 2 | 9 | 1 | ||||
| pSTAT3 | ||||||||
| Positive | 23 | 3 | 18.283 | 0.000 | — | — | — | — |
| Negative | 7 | 17 | ||||||
| MUC1 | ||||||||
| Positive | 18 | 1 | 15.407 | 0.000 | 14 | 5 | 5.773 | 0.016 |
| Negative | 12 | 19 | 12 | 19 | ||||
| Total | 30 | 20 | ‐ | ‐ | 26 | 24 | ‐ | ‐ |
ADC, adenocarcinoma; MUC1, mucin 1; NSCLC, non‐small cell lung cancer; pSTAT3, phosphorylated signal transducer and activator of transcription 3; pTNM, pathological TNM; SCC, squamous cell carcinoma; STAT3, signal transducer and activator of transcription 3.
Logistic regression analysis
Logistic regression analysis was carried out to determine the independent risk factors influencing lymph node micrometastasis of early‐stage NSCLC. The result revealed that STAT3 protein overexpression in tumors (OR 13.379, P = 0.033) and the differentiation degree of tumors (OR 0.131, P = 0.016) were independent risk factors for lymph node micrometastasis of early‐stage NSCLC (Table 6).
Table 6.
Logistic regression analysis
| B | SE | Wald | P | OR | 95% CI | |
|---|---|---|---|---|---|---|
| Gender | −0.063 | 0.862 | 0.005 | 0.942 | 0.939 | 0.173~5.084 |
| Age | −0.413 | 0.847 | 0.238 | 0.625 | 0.661 | 0.126~3.479 |
| Histological type | 0.872 | 0.856 | 1.037 | 0.308 | 2.392 | 0.447~12.814 |
| Differentiation | −2.035 | 0.841 | 5.856 | 0.016 | 0.131 | 0.025~0.679 |
| pTNM | −1.636 | 1.065 | 2.361 | 0.124 | 0.195 | 0.024~1.569 |
| STAT3 protein | 2.594 | 1.217 | 4.540 | 0.033 | 13.379 | 1.23~145.410 |
pTNM, pathological TNM; STAT3, signal transducer and activator of transcription 3.
Discussion
The involvement of lymph node metastasis is a very important prognostic factor in patients with potentially resectable NSCLC, and postoperative early recurrence often occurred in early‐stage NSCLC patients with lymph node micrometastasis. Accurate staging of individual patients remains a critical need. It guides the choice of therapy and stratifies patients appropriately for clinical trial of novel interventions. It also facilitates the comparison of treatment outcomes. Although patients with early‐stage NSCLC show an excellent prognosis, a few of them die of metastatic disease. In this subgroup of individuals, the search of occult metastasis might reveal that early dissemination of tumor cells could be the cause of cancer progression.
Disseminated tumor cells are regarded as a surrogate for early metastatic spread of disease. These cells can be detected in peripheral blood, bone marrow aspirates, and lymph nodes, where we refer to them as circulating tumor cells. Detection of disseminated tumor cells is a great technical challenge, and many different technologies have been developed to enhance the sensitivity and specificity of the testing for it. Different characteristics of tumor cells have been used to establish enrichment methods, including the differential expression of tumor‐specific markers on the surface of the cells, the size‐based selection of the cells, and other physical properties. The detection of circulating tumor cells has emerged in recent years as a biomarker with outstanding predictive and prognostic capacity in a number of malignancies including breast, prostate, lung, and colorectal cancer. However, the mechanism of micrometastasis is still unclear.
STAT3 has been reported to be correlated with lymph node metastasis of many human malignancies.3 Researchers reported that STAT3 expression was an independent risk factor for the prognosis of colorectal and gastric carcinoma6, 7 Abdulghani et al. reported that STAT3 promotes metastatic progression of prostate cancer8 Sato et al. reported that STAT3 promotes lymph node metastasis and correlates with breast cancer prognosis.9 So, what about the relationship between STAT3 expression and lymph node micrometastasis of early‐stage NSCLC? In order to test this hypothesis, we designed the present study, and we want to highlight some of the possibilities associated with developing a way to detect tumor micrometastasis and an anticancer drug that might therapeutically inhibit the STAT3 signaling pathway.
In this study, we found the STAT3 mRNA expression value, STAT3 protein and pSTAT3 protein expression values of lung cancer with lymph node micrometastasis were all much higher than that expressed in lung cancer samples without lymph node micrometastasis (P < 0.05). The expression rate of STAT3 protein and pSTAT3 protein in lung cancer samples with lymph node micrometastasis were also higher than that in lung cancer samples without lymph node micrometastasis (P < 0.05). Logistic regression analysis revealed that STAT3 protein overexpression in tumors and the differentiation degree of tumors were independent risk factors for lymph node micrometastasis of early‐stage NSCLC.
Metastasis is a complex process that consists of a few detailed steps including dissemination of tumor cells from primary sites, transportation of tumor cells within the lymphatics, settlement of tumor cells within lymph nodes, and re‐growth of tumors into a detectable size. The contribution of STAT3 to cancer has been widely confirmed by numerous studies demonstrating dysregulated activation in a variety of human tumors. Although the real mechanism of STAT3 on tumor metastasis is still unclear, it has been reported to promote tumor proliferation, survival, prevention of apoptosis and angiogenesis by upregulating the expression of its downstream genes such as survivin, bcl‐xl, cyclinD1, c‐myc, vascular endothelial growth factor, and so on. STAT3 can be activated through several ways, such as the JAK–STAT pathway and Ras–mitogen‐activated protein kinase pathway, and it can also be activated by many extracellular signals, such as interleukin‐6, interleukin‐11, leukemia inhibitory factor, epidermal growth factor, fibroblast growth factor, thyroid‐stimulating hormone, and so on.10
In this study, logistic regression analysis revealed that STAT3 protein overexpression in tumors and the differentiation degree of tumors were independent risk factors for lymph node micrometastasis of early‐stage NSCLC. It is anticipated that new anticancer strategies will be developed to inhibit lymph node metastasis and improve the patients’ prognosis.
Disclosure
No authors report any conflict of interest.
Contributor Information
Qian Zhao, Email: amber0629@126.com.
Zhi‐Ping Zhang, Email: zzpzx1971@163.com.
References
- 1. Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin 2013; 63: 11–30. [DOI] [PubMed] [Google Scholar]
- 2. Araki K, Kidokoro Y, Hosoya K et al Excellent prognosis of lepidic‐predominant lung adenocarcinoma: Low incidence of lymphatic vessel invasion as a key factor. Anticancer Res 2014; 34: 3153–6. [PubMed] [Google Scholar]
- 3. Kamran MZ, Patil P, Gude RP. Role of STAT3 in cancer metastasis and translational advances. Biomed Res Int 2013; 2013: 421821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen SH, Murphy DA, Lassoued W, Thurston G, Feldman MD, Lee WM. Activated STAT3 is a mediator and biomarker of VEGF endothelial activation. Cancer Biol Ther 2008; 7: 1994–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balic M, Williams A, Dandachi N, Cote RJ. Micrometastasis: Detection methods and clinical importance. Cancer Biomark 2011; 9: 397–419. [DOI] [PubMed] [Google Scholar]
- 6. Ji K, Zhang M, Chu Q et al The role of p‐STAT3 as a prognostic and clinicopathological marker in colorectal cancer: A systematic review and meta‐analysis. PLoS One 2016; 11: e0160125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wu L‐J, Li H‐X, Luo X‐T et al STAT3 activation in tumor cell‐free lymph nodes predicts a poor prognosis for gastric cancer. Int J Clin Exp Pathol 2014; 7: 1140–6. [PMC free article] [PubMed] [Google Scholar]
- 8. Abdulghani J, Gu L, Dagvadorj A et al Stat3 promotes metastatic progression of prostate cancer. Am J Pathol 2008; 172: 1717–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sato T, Neilson LM, Peck AR et al Signal transducer and activator of transcription‐3 and breast cancer prognosis. Am J Cancer Res 2011; 1: 347–55. [PMC free article] [PubMed] [Google Scholar]
- 10. Carpenter RL, Lo HW. STAT3 target genes relevant to human cancers. Cancers 2014; 6: 897–925. [DOI] [PMC free article] [PubMed] [Google Scholar]


