Abstract
Despite being some of the most important crops globally, there has been limited research on forages when compared with cereals, fruits, and vegetables. This review summarizes the literature highlighting the significance of forage crops, the current improvements and some of future directions for improving yield and nutritional quality. We make the point that the knowledge obtained from model plant and grain crops can be applied to forage crops. The timely development of genomics and bioinformatics together with genome editing techniques offer great scope to improve forage crops. Given the social, environmental and economic importance of forage across the globe and especially in poorer countries, this opportunity has enormous potential to improve food security and political stability.
Keywords: forage, nutritional enhancement, grass production, legumes, breeding, management
Introduction
Forage grasslands are used to feed livestock and globally it has been estimated that they represent 26% of the land area, and 70% of agricultural area (FAO, 2010). Such crops are significant economically, as the European example shows (see Figure 1). Forage crops are usually grasses (Poaceae) or herbaceous legumes (Fabaceae). Some tree legumes such as mulga (Acacia aneura) and leadtree (Leucaena leucocephala) are also grown in desert and tropical grasslands (Muir et al., 2011). In the tropics, popular grasses include Napier grass (Pennisetum purpureum), Brachiaria, and Panicum species. In the poorest parts of the world livestock production is critically important for smallholders’ livelihoods. Sub-Saharan Africa is an example and frequently women maintain the livestock production systems (Njuki and Sanginga, 2013). In temperate climates, the main grasses include bentgrass (Agrostis spp.), fescue (Festuca spp.), ryegrass (Lolium spp.) and orchard grass (Dactylis spp.) or hybrids of these. For example, Festuca and Lolium hybrids has been developed from 1970s (Ghesquière et al., 2010) giving rise to crops such as Festulolium pabulare which combines the superior forage quality of Lolium multiflorum with the persistence and stress tolerance of Festuca arundinacea. Some maize (Zea mays) cultivars have been specifically bred for forage. The commonly cultivated herbaceous legumes are trefoil (Lotus corniculatus), medics (Medicago spp.), clover (Trifolium spp.) and vetches (Vicia spp.). Brassica forage species include cultivars of oilseed rape (Brassica napus) and kale (Brassica oleracea). Fodder beet (Beta vulgaris) is another temperate forage. The combination of forage crops grown in any country varies depending on climate and livestock needs, however, the perennial legume lucerne or alfalfa (Medicago sativa) is the most widely cultivated as it can be grown with both temperate and tropical grasses, or as a standalone crop. This is a huge topic to review as there are so many species grown across the world, therefore we have chosen to focus on a few examples, the tropical grasses Pennisetum and Brachiaria, and more prominently the temperate crops Lolium and alfalfa.
FIGURE 1.
Total European Union output for all crops in 2016 from Eurostat data (Helminger et al., 2016). This includes a substantial portion for forage crops, comparable to the production of fruits and wine.
In an ideal world, we would all eat pulses rather than the animal products generated from them, as grain legumes are the food that offers the most sustainable future (Foyer et al., 2016). There is continued pressure from many groups to lower human consumption of animal products due to livestock efficiency issues and for human health (Cramer et al., 2017). There is a lack of reliable statistics for the proportion of adults adopting a plant-based diet, but it is estimated to be between 1 and 10% of the population in developed western countries such as within the European and United States (Mcevoy and Woodside, 2010) and studies support these diets as healthy and nutritionally adequate (American Dietetic Association and Dietitians of Canada, 2003). However, the consumption of livestock products can be regarded as important to a healthy diet due to their high nutrient density (CAST, 2013) regardless of the numerous efficiency and environment concerns (Di Paola et al., 2017), particularly true in developing countries where undernourishment incidences are estimated as ∼4–22% of the population (Alexandratos et al., 2006). Livestock production can convert non-edible crops such as the forages into human food, with sustainable intensification possible when inputs and outputs of the system are balanced (Derner et al., 2017).
Moreover, the cultural and social significance of livestock cannot be underestimated and the trend of increased global production is set to continue (Thornton, 2010). Livestock feature prominently across all cultures both in cuisine, but also music and literature. Additionally, in many developing countries the rearing of livestock such as cattle and goats are vital in times of hardship; many view animals as living ‘piggy-banks,’ that can for example pay the family school fees (Herrero et al., 2013). Therefore, in practice livestock production is set to continue throughout the world and forage crops will be grown for coming decades. Plant research has chiefly focussed on grain crops, but here we argue that there is enormous potential for improving forages. Improving the yield and nutritional quality of forage crops can help mitigate the unsustainable negative impacts of livestock production.
Forage Crops in Livestock Diets
Forage crops can be feed directly to livestock or can be processed by partial drying or pre-digestion. Because of this processing, animal feeds can be categorized as either bulky feeds or concentrates. Bulky feeds are also termed forage and are produced from grass, cereal and legume cropping as described above, such as alfalfa, Lolium or a mixture of the two. This forage can be provided to animals directly through grazing pasture land or in a processed form, such as hay (where water content is >15%) or dried (pelleted) biomass. Concentrates are generally cereal, oilseed and legumes seeds, or bi-products of their preparation for human food, biofuel and textile. They can also include high energy feedstuffs such as sugar-rich crop molasses and fats of animal origin, for example fish by-catch discards. In industrialized countries, production of both these categories of feed can surpass the amount produced for plant-based food for human consumption; in United States over double dry matter per-capita per year (DM cap/yr.) is produced for animal feed than for foodstuffs (Krausmann et al., 2008).
Livestock diet can therefore be exclusively forage or largely forage with concentrate supplementation. Concentrate supplementation is used to compensate nutritional deficiencies in the forage supply, increase animal performance such as milk production or at particularly challenging periods of development, for example calving. Due to most livestock diet being of forage this review focuses on the main crops used worldwide and will not discuss concentrates. The amount plant science has contributed to improvements in concentrates has been underappreciated and undervalued in literature, however, the role these crops have on livestock production has been reviewed previously (Erb et al., 2012).
Forage crops can be grown in mixed species cultivation to provide nutritional and environmental benefits. By offering livestock mixed grazing pastures or blending feeds, nutritional quality can be enhanced. For example, alfalfa is the highest-yielding perennial forage legume and produces more protein per unit area than other forage legumes and so can be grown alone or in combination with a range of different grass species. Well-managed alfalfa is normally grown successively for 3 or more years, but if harvested too late in the season the crop cannot survive the winter (Bélanger et al., 2006).
Forage Nutritional Content
Digestibility
The nutritional status of a forage crop depends upon the concentration (and ratios) of carbohydrates, proteins, and lipids. The composition of these organic nutrients determines the digestibility (D-value) of each crop which along with mineral and vitamins provides the amount of energy which can be derived by the animal (ME measured in MJ/kg DM) (Osbourn, 1980). Such calculations are becoming increasingly prevalent when growers are deciding which crop to grow based and particularly dependent on if the animal is non-ruminant or ruminant.
In forage crops 50–80% of DM is carbohydrate; if this percentage is too low then supplements of grains can be added. The primary types of carbohydrate are the insoluble structural saccharides cellulose and hemicellulose, or the storage forms such as starch and water-soluble polymers (e.g., fructans). These are degraded into simple sugars through cleavage of glycosidic bonds, either by the animal itself (non-ruminant and ruminants) or via microbial digestion and subsequent animal absorption (ruminants only). Different ratios of carbohydrates within the forage crop will have altered downstream digestibility for the animal, especially if the cell-wall structure constrains digestion by the microbial population or limits plant cell wall penetration (Weimer, 1996). Although lignin, a polyphenolic compound within forage, is not a carbohydrate, it has a dramatic impact on the digestibility of cellulose hemicellulose; lignin binds with structural carbohydrates and cell wall proteins and reduces nutrient availability. For forages increased lignin concentration in the growing crop will increase the percentage of indigestible DM. Of the major forage crops grown globally grasses, particularly Lolium perenne, have high digestibility due to high soluble sugar content alongside low lignin content (Ruckle et al., 2017).
Animal digestion of simple carbohydrates produces monosaccharides which can be readily metabolized. In ruminants, only microbial digestion of structural carbohydrates produces simple sugars which are subsequently metabolized to pyruvate. Pyruvate is absorbed by the animal and is metabolized further into volatile fatty acids (VFAs) which are a major energy source, (Bergman, 1990). Ruminants absorb VFAs in their rumen, and the rate of this is dependent on the concentration of individual VFAs, rumen pH and the absorptive area in the ruminal lining.
Protein
Nitrogen (N) availability to animals is predominantly from forage proteins and are estimated using crude total protein Kjeldahl measurements. Protein is usually abundant in the major form of Ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), although relative amounts vary between species (Wallace et al., 1997). This is especially true when comparing content in grasses with herbaceous legumes, with red clover (Trifolium pratense), white clover (Trifolium repens) and lucerne (Medicago sativa) grown widely due to their high protein value (Ruckle et al., 2017). Again, lignin will severely affect the digestibility of protein. Some micronutrients like proanthocyanidins or condensed tannins also change the digestibility of protein, but they inhibit protein degradation through binding. This can be advantageous as rapid protein degradation is causative of bloat, however, too high a tannin content will mean protein passing through the digestive track is unabsorbed and therefore a loss in nutrition value (Lees, 1992; Piluzza et al., 2014). This means there is a balance between reduced bloat and animal productivity (Mueller-Harvey, 2006). All grasses contain little or no proanthocyanidins, whereas many legumes especially big trefoil (Lotus pedunculatus) and Sericea lespedeza (Lespedeza cuneata) can have levels as high as 18% DM (Barry and Manley, 1984; Mueller-Harvey, 2006). Other N-containing compounds can be found in forage such as nucleic acids, nitrate and ammonia (Wallace et al., 1997).
Lipids
Lipids in forage crops are mostly found as polyunsaturated fatty acids (PUFAs) in the range of 10 – 30 g kg-1 (Hatfield et al., 2007) of which the most abundant is α-linolenic acid [62% total lipids (Clapham et al., 2005)], with linolenic and palmitic acid also being present (Harfoot and Hazlewood, 1988). These dietary lipids are important in final animal product quality; forage diets with lower PUFA levels than cereal diets can produce leaner meat (Wood et al., 2004; Van Elswyk and McNeill, 2014). Moreover, fresh forage has been shown through numerous studies to produce milk with lowered PUFA content and increased trans-fatty acids (Elgersma et al., 2006; Chilliard et al., 2007). Studies have been used to profile PUFAs across forage species, with grasses tending to have more α-linolenic acid when compared to legumes and legumes in turn having higher linolenic acid content (Boufaïed et al., 2003). Striking differences in PUFA content can be seen within species through profiling cultivars, and moreover the harvest period and its environment (Elgersma et al., 2003; Clapham et al., 2005). For example Lolium perenne, Festuca pratensis (meadow fescue), and Festulolium hybrids of the two have been shown to vary not only between species at the beginning of their growth season, but more prominently between individual cutting regimes (Dewhurst et al., 2001).
Trace Elements
Minerals and trace elements from forages are important for maintaining livestock health. As there is a move toward using fewer antibiotics in animal production the nutritional balance of feed takes on additional importance. Zinc is particularly important for the immune system and supplements can be added to animal feed, but addition of too much results in wasteful excretion, reviewed in Brugger and Windisch (2015). Contrastingly, avoiding accumulation of toxic minerals can also be important for forage crops. Getting the balance right is crucial as low levels of selenium can be beneficial for livestock, but high concentrations are toxic (Zhu et al., 2009). Some elements accumulated in plants can make them unpalatable for livestock, but the ability of forage crops to grow fast and quickly recover from cutting makes them ideal crops for phytoremediation [e.g., Napier grass, (Ishii et al., 2015)].
Biomass Production
Probably the most important trait of any forage crop is rapid biomass production, as crops are either cut or grazed directly, and nutritional quality depends on the rate of biomass production. Intensive production with faster growth often decreases this nutritional, but this depends on the species grown and some cultivars have better recovery from defoliation. Plant height correlates well with biomass for most crops (e.g., maize) and this factor together with ground area cover are the criteria underpinning methods to assess yields (Freeman et al., 2007).
Many plant species can be grown for forage production, but the ability of the shoot meristem to respond with increased growth after cutting is essential. In some forage species, aboveground grazing or cutting has been correlated with increased root exudation (Paterson and Sim, 1999). This flush of carbon release by roots can stimulate rhizosphere microbes that in turn help to mobilize soil nutrients to sustain aboveground regrowth. Maintaining an optimal nutrient and water supply is very important for forage biomass production. For example, the importance of N supply for re-growth after cutting grass has been demonstrated (Dawson et al., 2004). Furthermore, the previous N status of alfalfa influences its regrowth ability (Meuriot et al., 2004, 2005).
Improving Forage Crops
Cultivar Breeding
Due to the relatively recent cultivation of forage crops compared to other agricultural plant species, there were few improvements before 1900. Recently, agricultural trends and the global economic importance of forages, mean new cultivars have been bred. These improvements are helped by many closely related wild populations which can be used in development of new lines (Boller and Green, 2010). The most desirable improvements are increasing dry matter yield (DMY), crop durability and resistance to diseases particularly by pathogenic fungus and pests particularly nematodes, digestibility of DM, and nutritional content of this tissue. Arguably the greatest improvements have been made in breeding of Medicago spp., Trifolium spp., Lolium, and Festuca. Large scale breeding programs include testing of these crops, such as NE1010, a multistate cooperative effort of 15 institutes across 12 North-eastern states of United States and Canada (NIMSS, 2017). Similar tropical grass breeding programs include the Brachiaria partnership between the International Centre for Tropical Agriculture based in Colombia (C.I.A.T.), the Ugandan National Livestock Resources Research Institute (NaLIRI), the Tanzania Livestock Research Institute (TALIRI), the Institute of Agricultural Research of Mozambique (IIAM) and the Brazilian Agricultural Research Corporation (E.M.B.R.A.P.A.) (CIAT and CGIAR, 2015) which is being conducted across Eastern and Southern Africa.
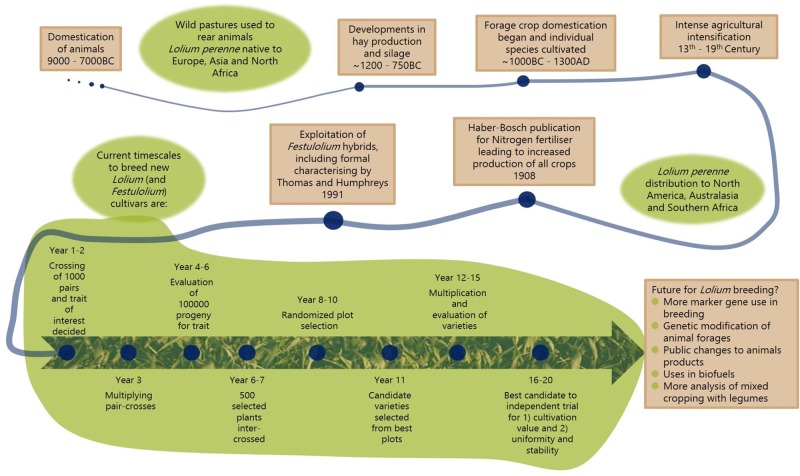
Breeding programs for forage crops are fraught with difficulties. Individual plants have high genotypic and phenotypic heterogeneity with many species being polyploid, a problem which is exacerbated by in-breeding across many grasses, and few agronomic traits being linked to distinct genes (Poehlman, 1987; Vogel and Pedersen, 1993). Studies have focussed on this problem in specific legumes (Jahufer et al., 2002; Riday and Brummer, 2007; Collins et al., 2012; Luo et al., 2016) and grasses (de Araüjo et al., 2002; Piano et al., 2007; Blackmore et al., 2016). Regardless of these problems there have been some major developments in breeding lines for forages, especially in Medicago and Lolium. Figure 2 shows a brief historical timeline of Lolium cultivation, and includes the current breeding regimes for grasses; future breeding possibilities are also included and discussed in later sections. One of the most interesting breeding developments is the exploitation of closely related species of Lolium and Festuca (Thomas and Humphreys, 1991; Humphreys et al., 2003) to create hybrid Festulolium cultivars. These cultivars have the high quality characteristics of Lolium combined with the stress tolerance and persistence found in Festuca (Ghesquière et al., 2010). Backcrossing of Festulolium have generated novel hybrids with more stable protein content when compared to parental lines (Humphreys et al., 2014). Advances in phenotyping are making it easier to include the quantification of characteristics in the field; such as high level imaging of growing crops to accurately determine later traits like biomass (Walter et al., 2012).
FIGURE 2.
Cultivation timeline of Lolium perenne (perennial ryegrass), including current typical breeding regime and future possibilities for breeding programs. The historical timeline is described in brown boxes (Haber, 2002; Hirata, 2011), and general trends in green areas, including modern grass breeding regime as used across Europe (Bruins, 2016).
New cultivars are being helped by advances in sequencing methods that can provide more transcriptomic data (Barrett et al., 2009; Pfeifer et al., 2013; Yates et al., 2014), including the identification of SNPs which may be investigated to improve Lolium (Blackmore et al., 2016) and Trifolium (Nagy et al., 2013). Draft genomes for such crops and cultivars are becoming increasingly common (Byrne et al., 2015; De Vega et al., 2015; VanBuren et al., 2015) as well as more evidence that model species like Brachypodium can direct research (Brkljacic et al., 2011; Rancour et al., 2012). Such research is providing clues to candidate genes which could be used for nutritional enhancement.
Candidate Genes for Nutritional Enhancement
Identification of potential candidate genes is usually through quantitative trait loci (QTL) analysis or marker-assisted selection (MAS) provided from the above completed genomes. Those identified are studied in relation to biomass and growth traits; in M. sativa QTL has been used for lodging resistance and vigor (McCord et al., 2014), plant height and regrowth following harvests in association with MsaciB (Robins et al., 2007), candidate gene analysis for flowering and stem height through CONSTANS-LIKE (Herrmann et al., 2010) and biochemical markers of ROS resistance genes for drought tolerance correlated to DM (Maghsoodi et al., 2017). The expression of other ROS associated genes of the Iron-Superoxide Dismutase family (Myouga et al., 2008) have also been linked to increases in DM in both the legume M. sativa (McKersie et al., 2000) and grass Lolium cultivars (Warnke et al., 2002).
In Lolium, transcriptomics showing differentially expressed genes between wild-type and a dwarf mutant enabled identification of three key genes associated with dwarfism (Li W. et al., 2017), which were subsequently used for forward screens. Markers are used to infer both phenotypic traits and to track inheritance to aid breeding. For instance chloroplast SSRs have been investigated in Lolium through a similar technique as above (Diekmann et al., 2012). A thorough re-annotation of the model forage M. truncatula genome has also identified hundreds of small, secreted peptides coded by both macronutrient-responsive and nodulation-responsive genes, which could aid reverse genetics for improving many forage crops, especially M. sativa (de Bang et al., 2017). Iterative mapping software such as BioMercator (Sosnowski et al., 2012) has been used in Lolium to perform meta-QTL analysis using readily available published data (Shinozuka et al., 2012), consequently providing new candidate genes from previous work including orthologs of rice amino acid biosynthesis genes and a marker for reproductive traits, showing how new algorithms can exploit old data. Moreover, BioMercator used to decipher flowering time and height in M. truncatula (Julier et al., 2007) directly implicated the above research into CONSTANS-LIKE in M. sativa (Herrmann et al., 2010; Julier et al., 2010). Such potential ease for transferring model plant knowledge to forage crop research is further discussed below.
Despite the need to ensure optimal nutritional content especially in the end-product feed, rapid vegetative biomass accumulation is the most desirable trait of a good forage crop, especially those which undergo extensive cutting throughout the growing season. Due to this phenomenon, candidate genes for improving the crops are associated with either photosynthesis or nitrogen use efficiency (NUE). More generally for resistance to biotic and abiotic stresses there is also a huge opportunity for improving traits in forage crops using our genetic knowledge from model plants (see Figure 3). Figure 3 summarizes some of the traits that can be considered for all forage crops.
FIGURE 3.
Targets for improved forage crops.
Such improvements in traits could be aided by achievements in transformation and genetic marker techniques. Reproducible and high efficiency transformation has been developed for temperate cultivars of Festuca (Wang and Ge, 2005; Zhang et al., 2006) and Lolium (Bajaj et al., 2006; Badenhorst et al., 2016); and more recently for some of the tropical grasses such as Pennisetum (Gondo et al., 2017) and Brachiaria (Cabral et al., 2015). Some examples of gene editing forage crops to confer stress tolerances have been successful in aiding both biomass increases but also nutritional quality. Transformation of M. sativa with the Arabidopsis Enhanced Drought Tolerance1 gene produced plants with not only increases in root length, shoot height and vegetative biomass, but also increases in proline, soluble sugar and chlorophyll content under drought stress when compared to wild-type (Zheng et al., 2017). Importantly these increases were shown both in the laboratory but also in field conditions. This study also identified the increased expression of many interesting genes, including M. sativa Heat Shock Protein23 (HSP23), a gene already shown to enhance abiotic stress tolerance in both Nicotiana tabacum and Festuca (Lee et al., 2012a,b) along with other members of the MsHSP family (Li et al., 2016; Li Z. et al., 2017). Similarly, for the Ethylene Response Factor (ERF) family studies have been shown that introducing the M. sativa gene into other plants can confer enhanced resistance to salinity; MsERF9 and MsERF11 in Nicotiana and Arabidopsis, respectively (Chen et al., 2012a,b).
Protein and N Budget
Forage NUE is a target for breeding, particularly as protein content of crops is so valuable. Protein accumulation is linked to N status and when the supply is supra-optimal greater storage occurs. When compared with grain crops that have been bred for high seed starch, forage crops often require N in greater amounts due to their increased growth, storage capacity and higher fiber content (Parsons et al., 1991). For forage crops, it is the leaf tissue biomass that is harvested rather than grains/roots/tubers. Principally NUE for forage crops can be based on N utilization efficiency (NUtE) as we are interested in the highest achievable biomass of the shoot which will form the content to be dried for feed production (Xu et al., 2012). Not only does this include biomass, but also the relative N levels in this tissue; it is not enough to only have a high yield of biomass in the shoots, it must also yield optimal amounts of N. Moreover, when looking at the effect of fertilizer use we are also interested in how both the biomass and N status change on application and thus also N uptake efficiency (NUpE). Forage crops offer challenges for NUE as there is a requirement for optimal yield of shoot biomass with a high N content (NUtE) while also optimizing N fertilizer acquisition (NUpE) throughout the growth season.
As NUE is an important criterion for biomass improvements, many genes relating to N acquisition or metabolism have been the subject of study in model systems. Additionally, genes important in carbon metabolism have also been the focus, due to the links between C:N ratios for plant growth (Jaradat et al., 2009). Despite the long evolutionary divergence between grasses and legumes, many key candidate genes have high genetic similarity, meaning one can use known genes which effect a trait in a forage crop from one species and investigate it within another. For example, a range of vegetative N storage proteins have been identified and the reviewed for leaves (Muntz, 1998) and roots (Bewley, 2002). To illustrate this further, the phylogeny in Figure 4 is the known and predicted coding sequences for rbcS including the model species Arabidopsis thaliana, many significant grass and legume crops. Many of the forage crops have high similarity in their coding sequence to more well-studied crop species. For example, Medicago and Trifolium rbcS sit closely to the legume species which have their genomes sequenced [Cajanus cajan, Cicer arietinum, Glycine max, Lotus japonicus, Medicago truncatula, and Phaseolus vulgaris (Jacob et al., 2016)]. Such sequences can provide a wealth of potential genes of interest for breeding programs (Araújo et al., 2015; Rauf et al., 2016). For example, investigation of Heat Shock Protein in M. truncatula, found a homologous HSP70 in M. sativa and had a substantial role in stress tolerance when conferred to A. thaliana (Li Z. et al., 2017).
FIGURE 4.
Phylogeny of temperate legumes and grasses for Ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit (rbcS), also including Arabidopsis thaliana and Marchantia polymorpha as outliers. Both known and predicted coding sequences for all species of interest were gathered from GenBank, PLAZA 3.0 Dicots and PLAZA 4.0 Monocots. Multiple sequence alignments (MSA) were performed using MAFFT v7 (Katoh and Standley, 2013) with a E-INS-I iterative refinement method and bootstrapping = 100. The phylogeny was built using Newick format in iTOL v3.4.3 (Letunic and Bork, 2007) and a radial phylogenetic tree produced. The tree was color coded to show model species, human food crops and forage crops for both legumes and grasses, although it should be noted that many can overlap in their uses.
In forage crop vegetative biomass, the most important nutrients for livestock are proteins and water-soluble carbohydrates (WSCs), and ideally the post-harvest quality of these should be maintained. There has been considerable interest in developing organ specific proteome reference maps for stems and leaves. The dominant proteins in these tissues are photosynthetic enzymes such as RuBisCO and RuBiCO small unit (rbsS), which for M. truncatula make up ∼28.9% of leaf tissue, or other carbon-fixation genes for example glyceraldehyde 3-phosphate dehydrogenase and triose phosphate isomerase, with structural protein such as lignin biosynthesis being more concentrated in stems (Watson et al., 2003). As the D-value of forage is mostly linked to cell wall concentration and a reduction of this can aid digestibility (Jung and Allen, 1995; Jung et al., 2012), some proteomes have looked even more specifically at such tissues (Gokulakannan and Niehaus, 2010).
Some research has focussed on transgenic approaches to increase and enhance amino acids and proteins. As many forages have low concentrations of the sulfur-containing amino acids of methionine and cysteine, both important in animal and human nutrition (Ball et al., 2006), some studies have specifically aimed at increasing these levels by over-expression. These have included using lupins (Lupinus albus) (Molvig et al., 1997; Tabe et al., 2010) and soybean (Dinkins et al., 2001; Tabe and Droux, 2002), used as forage sources.
Apart from cultivar differences which can be improved with breeding programs or specific transgenic approaches, the most significant changes in nutritional content is due to stresses (Araújo et al., 2015). Consequently, stress proteomes have also been used for vegetative tissue; lupin stem proteins have been analyzed under water stress to show increases in serine protease and cysteine protease required for remobilization of proteins (Pinheiro et al., 2005); in grasspea (Lathyrus sativus) seedlings under either salinity, low temperature or ABA stress gave rise to the identification of 48 stress-responsive proteins (SRPs) which include those important dominant proteins discussed above (Chattopadhyay et al., 2011); in M. sativa drought conditions showed remobilization of RuBisCO-derived N could compensate for the decreases in N assimilation (Aranjuelo et al., 2011). Moreover, through harvesting regimes, forage crops undergo extreme stress which has shown to cause the remobilization of vegetative storage proteins (VSPs) to boost new shoot regrowth in both Medicago and Trifolium as well as being important for cultivars with better cold tolerance (Avice et al., 2003), whereas Lolium has shown how defoliation increases the relative proportions of certain proteins, particularly asparagine and glutamine (Bigot et al., 1991).
Finally, the N consumed by livestock is recycled and increasing ruminant productivity is a major target for as the conversion of plant to microbial protein is inefficient. It was estimated that as much as 70% of the plant N eaten by animals for milk or meat production is excreted as ammonia or urea to the environment (MacRae and Ulyatt, 1974; Kingston-Smith et al., 2008; Kingston-Smith et al., 2010). Furthermore, the process of rumen fermentation is important for the generation of greenhouse gasses like methane (Bannink et al., 2008; Dijkstra et al., 2011).
Rhizosphere Microbiome
The impact of genomics extends beyond the crop plants to their environmental interface. For example, the rhizosphere microbiome is likely to be a future target for improving the nutritional quality of crops. Epiphytic bacteria living on and in the plant, may be important for crop health and nutrition, and some microorganisms can fix atmospheric N within legume root systems. Bacteria living with plants may be able to assist in digestion and absorption of forage eaten by livestock. These bacteria may improve the uptake of trace elements in the animal gut by the production of specific binding molecules and/or siderophores. In the soil, the rhizosphere microbiome is important for nutrient cycling and uptake, particularly in low input systems like those grown in the tropics. The inoculation of new forage crops with beneficial microorganisms is likely to be a target for research and use in future crops, coupled with rhizosphere microbiome research of root exudate composition.
The root is known to directly modify the rhizosphere population by altering the chemical constituents of root exudates. For example, the roots of the tropical grass Brachiaria specifically produce a chemical shown to inhibit nitrifying bacteria and to specifically block ammonia-oxidizing pathways in soil bacteria, the first step in the process of converting ammonium to nitrate (Byrnes et al., 2017). Soil ammonia-oxidizing bacteria quickly convert urea or NH4+ fertilizer to NO3-. Soil N form is fundamental for crop acquisition, as NO3- is mobile and readily leached while NH4+ binds. Nitrification inhibitors have been identified in root exudates from several legumes and grasses including sorghum and rice, but by far the largest activity was detected in the tropical grass Brachiaria humidicola (Subbarao et al., 2009). In rice, the ability of root exudates to inhibit nitrification varied between cultivars from 5 to 50%, but was not significantly higher in three ancestral lines (Tanaka et al., 2010). The biological nitrification inhibitor (BNI) activity of root exudates has been assayed using a recombinant luminescent reporter ammonia-oxidizing bacteria Nitrosomonas europaea (Subbarao et al., 2006). In Brachiaria, roots exudate the cyclic diterpene “brachialactone,” (Subbarao et al., 2009); brachialactone has a 5-8-5-membered ring system and a γ-lactone ring and contributed to 60–90% of the BNI activity released from the roots of this tropical grass. This exciting example offers the potential for transferring this trait to other forage crops to improve NUE. In the future synthetic pathways to produce plant nitrification inhibitors will be fully elucidated, providing the opportunity to capture this trait in forages and transfer to other crops to improve yield and nitrogen acquisition.
Digestibility
As protein digestion and uptake in livestock is directly related to energy availability (ME) (Nocek and Russell, 1988; McCarthy et al., 1989) it is important to increase WSC in many forage crops, especially grasses (Miller et al., 2001). In Lolium WSCs include fructans which are the most important storage polysaccharide and thus improved metabolism of fructan from sucrose can help improve the D-value (Chalmers et al., 2005). Use of distinct Fructan:Fructan 6G-fructosyltransferase sequence variants has shown to increase fructan levels at warmer temperatures in Lolium, thus hoping to aid development of high sugar-content grasses even at changing climates (Rasmussen et al., 2014). The amounts of WSCs are strongly associated with the N availability to the root (Roche et al., 2017) highlighting the importance of C:N balance in vegetative tissue (Louahlia et al., 2008). Furthermore, the amounts of WSCs varies between varieties as well as within the environment; Lolium cultivars AberMagic, AberDart, and AberElite all had highest growth rates correlated to highest WSC concentration during spring/summer, corresponding to high N availability from the roots alongside optimal photosynthesis conditions (Winters et al., 2010). Recent advances in the identification and manipulation of photosynthesis promoters for both Lolium perenne RBCS and Chlorophyll a/b Binding (CAB) (Panter et al., 2017) has provided transgenic lines for assessing increases in yield, fiber and, more importantly for digestibility, the fructan concentrations in both pseudostem and leaf blades in field trials (Badenhorst et al., 2018). Such work provides a platform for future studies to identify promotors important in other nutritional traits.
The amounts of resistant starch are important for the digestibility and nutritional content of forage crops. Resistant starch (RS) generally has lower digestibility until it reaches the large intestine (Englyst et al., 1999), where in ruminants more digestion can occur (Raigond et al., 2015). Research studies have shown that M. sativa has advantages as a feed source over cereals for enhanced D-value (Giuberti et al., 2018). One major difference between dietary RS is that it is seen to have advantages in the human diet by providing more fiber, but disadvantages in livestock feed for non-ruminants as it remains undigested. In general, lower RS will improve the digestibility of forage crops for both ruminant and non-ruminant livestock. As a crops D-value is closely linked with its starch, protein and lignin content, genomic studies have begun large-scale genome-wide association studies (GWAS) to confirm correlations across a range of traits, such as using three distinct alfalfa cultivars with a high-throughput genotyping-by-sequencing approach (Biazzi et al., 2017). However, this study did highlight that differences in SNPs associated in different tissue types (shoots and leaves) can vary in correlation with traits such as protein content, and so care must be taken when using GWAS to aid crop improvements.
Another substantial nutrient in forage crops is that of proanthocyanidins or condensed tannins (CTs). CTs bind to protein making it unavailable to digestion for ruminants until it reaches the rumen, and thusly an important trait in increasing the D-value of a crop (Min et al., 2003), although too high a CT content can be harmful restricting fermentation, especially in low leaf protein content species. A compromise is therefore desirable, with the moderate CT of 2–4% of the forage biomass giving the optimal D-value (Dixon et al., 2005). Whilst some species of legumes have optimum levels of CTs such as Lotus corniculatus, others such as Onobrychis viciifolia and Trifolium ambiguum are often poor choices for forage in many climates (Min et al., 2003; Baker, 2012); which means there is more scope to increase CTs concentrations in high yielding species where they are low such as M. sativa and Trifolium repens rather than increase growth traits aforementioned (Burggraaf et al., 2006; Salunkhe et al., 2017).
As the CT synthesis pathway has been well-characterized in Arabidopsis with the transcriptional regulators R2R3 MYB, bHLH, and WD40 protein identified as having a central role in final CT content (Lepiniec et al., 2006). Such knowledge can be used to manipulate forage crops. The R2R3 MYB homolog MtPAR in the M. truncatula seed coat has been characterized and hairy root transformation in alfalfa resulted in the accumulation of CTs to the level of ∼20 mg/g shoot biomass (Verdier et al., 2012), although this is still below the desirable concentration. A similar study showed that expression of the TaMYB14 transcription factor from a low-yielding forage activates CT biosynthesis in both Trifolium and Medicago (Hancock et al., 2012). Other approaches have involved characterizing early steps of CT biosynthesis in M. truncatula in the hope to later target crop relatives (Pang et al., 2007), whilst others have looked at how relative amounts of CT differ between leaves and higher concentration containing flowers to see if changing flowering in Trifolium could improve its D-value (Burggraaf et al., 2008). There has been an effort to engineer better digestibility in some forage cultivars (Wang and Brummer, 2012) and microbial pre-digestion after cutting and before feeding, including microbial supplements (Boyd et al., 2011; West and Bernard, 2011; Elghandour et al., 2015), can be used to enhance this.
Biomass Production
As biomass yield is the main target for forage crop improvement, more rapidly growing cultivars can be targeted for breeding. Studies have consequently focussed on heading date (Fe et al., 2015) and flowering time regulation (Skøt et al., 2011; Shinozuka et al., 2012) in Lolium by developing Genomic Prediction models and QTL mapping as described previously (Nuñez and Yamada, 2017). Manipulating genes involved in delayed senescence has been targeted for increasing biomass yields. The introduction of the 5′ flanking region of the Zea mays cysteine protease gene SEE1 in Lolium multiflorum has shown this promoter region to increase leaf lifespan by approximately 8–16 days (Li et al., 2004). A similar study using the Arabidopsis Senescence-Associated Gene12 (SAG12) promotor also delayed senescence in M. sativa with notable chlorophyll and yield increases even after 3 months of growth (Calderini et al., 2007). A final example is the expression of the Panicum virgatum NAC1 and NAC2 transcription factors in Arabidopsis atnap lines (mutants with defective senescence) to restore wild-type phenotype, predominantly measured using total chlorophyll concentrations (Yang et al., 2015).
However, fast growth must also be coupled with the ability of the plants to respond to cutting by providing rapid regrowth. Growth rates and recovery from cutting are traits that are relatively easy to select for in breeding trials, and have been of regular interest to researchers for many decades in both Lolium and Medicago (Wilman et al., 1977; Vance et al., 1979). In addition, cutting experiments using 13C and 15N in both Lolium and Medicago have shown how the soil is affected both for dissolved organic C and N, and microbial biomass, demonstrating that management schemes can be critical to subsequent soil health (Schmitt et al., 2013). An ability to rapidly regrow may increase the susceptibility of the plant to insect and pathogens and this is worthy of further investigation. The relationship between tissue wounding and plant immunity is a topic that is quickly developing and there is now good evidence that tissue growth rate is closely linked with immunity (Huot et al., 2014).
Thusly, management schemes for forage crops are very important for yield. For example, choosing when to cut or graze a crop is crucial for subsequent regrowth of the plant (Karn et al., 2006; Asaadi and Yazdi, 2011; Bumb et al., 2016). To assist in this choice there is scope for the use of molecular markers, with the future possibility of a PCR test for the optimal time harvest based on the expression of candidate genes like storage proteins. Such tissue testing of crops can also be used for decisions on the timing of fertilizer applications as the two evaluations are made at around the same time. There is scope to identify a suite of marker genes that can be used to help decide when these key decisions are made.
Mixed cropping schemes are already widely used for forage crops and there are clear advantages in growing legumes and grasses together. Legumes increase soil N through their N fixation symbiosis with Rhizobium, with their biological nitrogen fixation ranging from 32 to 115 kg ha-1 (Iannetta et al., 2016). This can in turn decrease subsequent fertilizer use for crops grown thereafter, a reduction between 23 and 31 kg N ha-1 (Preissel et al., 2015). Numerous intercropping regimes have been tested including modeling of various climatic and soil texture parameters (Bachinger and Reining, 2009). Transfer of N from legume to crop, including in grasslands, has been investigated (Pirhofer-Walzl et al., 2012). However, it is still unknown how this interaction affects N movement and leaching through the soil profile. Such an investigation is required to give evidence of environment changes as well as crop productivity. Mixed species cultivation also has advantages for disease and extreme weather resistance as the susceptibility of the plants to these stresses varies between cultivars and species. Forage breeding has focussed on monoculture selection regimes and there is scope for better mixed species crops that could be included in trials for new varieties. Some advantages and disadvantages of mixed forage crops are summarized below in Table 1.
Table 1.
The advantages and disadvantages of growing forage crops in mixed systems.
| Mixed cropping | |
|---|---|
| Advantages | Disadvantages |
| Soil nutrient availability-each species may have different strategies to mobilize nutrients | Grow rates and optimal harvest date can differ |
| Pathogen and pest susceptibility is different | Specialist equipment may be needed |
| Legume can supply N | Competition for resources |
| Stem support in canopy | One species may host pathogens |
| Root depth for water access and improved soil structure | Monitoring more than one species at a time to keep up with needs |
Growing forage crops for improved nutritional quality has not been a target for breeding programs, rather yield and climate tolerance have been the drivers. Future crops must be tolerant of climate changes and weather extremes. Unlike many crops where monocropping is most productive, forage crops have the advantage that they can be easily grown in combination without lowering productivity. Such a trend has been shown across multiple trials as well as increasing biodiversity (Tilman et al., 2001, 2006; Weigelt et al., 2009). As with any system that promotes biodiversity whilst still being productive, this can mean not only lowered costs to manage but also help cultural agriculture acceptability, with consumers becoming more aware of the effect the production of their food has environmentally (Scherr and McNeely, 2008).
Trace Elements
Plant research has focussed on the goal of biofortifying cereals, but there is also potential to improve the nutritional quality of forage crops. The economic importance of livestock production in the poorest parts of the world offers the opportunity to biofortify animal crops thereby improving the health of these animals and both directly and indirectly their owners. The knowledge base developed for grain biofortification (e.g., candidate plant metal transporters) has yet to be applied to forage crops. For example, transporter proteins for iron and zinc storage have been identified in cereals (Connorton et al., 2017; Menguer et al., 2017) and their equivalents in forages have yet to be identified.
Although very abundant in most soils, silicon is particularly required by grasses (Tubana et al., 2016) and is therefore likely to be important for the optimal growth of many forage crops. Silicon is important for cell wall structure and therefore resistance to pathogens and pests, however, it may have a negative impact on digestibility. The supply of this nutrient may become limiting for forage crops, particularly as the plant biomass is regularly removed from the field and silicon is not yet a routine addition to fertilizer.
Most species of forage crops can form mycorrhizal associations and this type of symbiosis is important for acquisition of trace elements. For natural grazing, these symbiotic associations are particularly important, but when fertilizer is added to cultivated forage crops mycorrhiza are suppressed (MacLean et al., 2017). Enhancing this symbiosis by inoculation of forage crops with mycorrhizal fungi has the potential to improve the mineral element composition of the feed. The fungal symbiosis has additional benefits for the plant by increasing the soil area mined for nutrients and water; this can be crucial during extreme weather events such as drought. Furthermore, a balanced and optimized root rhizosphere microbiome is essential for optimal root function and this applies to all crops including forage (Mommer et al., 2016).
Environmental Footprint of Forage Crops
As in all agriculture, improving water and nutrient use efficiency is a target for forage crops. The general fertilizer requirement of maize grown for forage and for grain are the same as that for a biomass crop. N requirements differ greatly for forage crops, and legumes and rhizome crops like Miscanthus have low N requirements (Dierking et al., 2017). Improving NUE using transporter marker genes as indicators of the crop status in the field could be valuable (Fan et al., 2017). Targeting particularly the NUpE component of NUE is important for minimizing the wasteful and environmentally damaging losses of excess N fertilizer additions.
As discussed above for protein content, biomass production and cutting/grazing decisions there is the potential to develop gene markers that can indicate the N status of each type of forage crop. Mixed plant communities tend to have better NUE, probably because each species has a different temporal pattern of N uptake, resulting from different growth rates and root architecture (Tilman et al., 2001; Weigelt et al., 2009). In more affluent countries the relatively low chemical fertilizer prices do not encourage more judicious use of fertilizer for forage crops, but the threat of legislation for overuse has provided a new incentive for better fertilizer use efficiency. There is plenty of scope for improving the NUE of forage crops particularly as breeding programs have not focussed on this trail. For water acquisition, the long tap roots of Medicago are ideal for penetrating deep for water and nutrients. Varietal differences in this important trait have long been known (McIntosh and Miller, 1980) and the choice of cultivar depends on the soil type, climate and cropping regime that is required.
Conclusions and Future Directions
Future Performance Improvements Using Genomics
The availability of genomics and bioinformatics has revolutionized all biology and as databases expand to include more species and cultivars this information can assist forage breeders to improve crop performance. The future possibilities for breeding of forage crops using Lolium as an example are shown in Figure 2. By comparing cultivar sequence information and using GWAS for traits such as high vegetative tissue concentrations of protein, NUpE or specific trace elements the nutritional quality and yield of forage crops can be improved. Some SNPs in key genes that have been identified in model plants can be the targets for gene editing techniques (Bonhomme et al., 2014; Slavov et al., 2014; Thorogood et al., 2017). TILLING lines are also being used in many forage crops to study gene function (Carelli et al., 2013; Dalmais et al., 2013; Manzanares et al., 2016). Furthermore, as shown with the rbcS example in Figure 4, sequence information can be used for the design of PCR primers which can be used for tissue testing. These tests can be used to rapidly identify general health and nutritional status of crops as well as specific pathogens. One bottleneck is likely to be the transfer of the new genetic information into forage crops. For example, GM forage crops may be more acceptable to the public, as if fed to animals their entry into the human food chain is indirect. The use of CRISPR/Cas9 technology may provide an acceptable route for such manipulations, and as with many crops such feasibility studies have begun in forage crops; the mutation of the Medicago sativa Squamosa Promoter Binding Protein Like9 (SPL9) has been attempted and validated (Gao et al., 2018), although poor genome editing efficiency is limiting advances at present. Many candidate genes have been identified which may be quickly transferred into forage crops, but the technology for transformation is limiting development of these improved plants. In the future genome editing may become more accepted, particularly perhaps for animal feed crops.
Focusing on Roots
As discussed above high-yield, low-input vegetative biomass is desired for forage crop production. This has meant aboveground phenotyping strategies are being widely developed using predominantly imaging and spectral data (Walter et al., 2012), although more research is needed to see how vegetative phenotyping will work across different species, especially in mixed-cropping systems. However, although the need for well-developed, established root systems is clearly important (Kell, 2011; Nacry et al., 2013), breeding for belowground traits has been largely disregarded. This is unsurprising as with all crops, root phenotyping is difficult, being hidden in the soil and therefore labor intensive and difficult to sample. Any current root system improvements have been the consequence of vegetative drought and salinity assays discussed previously.
Consequently, there has been a shift of focus toward breeding for underground traits in forage crops; across plant science this has been termed the next green revolution step (Lynch, 2007; Den Herder et al., 2010). Before phenotyping can even begin it is necessary to determine which kind of improvements are necessary, of which 2 main categories are found. The first is to improve root systems for the plant itself. This could include increasing fine root biomass, lateral root initiation, or in the case of legumes nodulation by Sinorhizobium, for increased nutrient uptake (Jackson et al., 1997; Ariel et al., 2010; Downie, 2010; Wang et al., 2010), or instead increasing root density or taproot length for either nutrient and/or water uptake, or resilience to stress such as defoliation (Dawson et al., 2004; Erice et al., 2007; Ghesquière et al., 2010; Kell, 2011).
The second category is the improvement of root systems to aid the environment. This target is to improve agricultural land not just for production but also in terms of the ecosystem services, and this is especially true in the case of forage crops (Marshall et al., 2016). Forages and grasslands can provide ecosystem services that are wide-ranging and highly linked to root function including soil C-sequestration important for climate change (Kell, 2011, 2012), or lowering run-off of land thus helping to lessen flooding and soil erosion (Macleod et al., 2013). The idea of using both non-leguminous and leguminous forage crops as cover crops to mitigate climate change is gaining appreciation, (Kaye and Quemada, 2017). Another point to note is that many perennial grasses including Miscanthus and Panicum can be used for biofuel production but the characteristics required for a forage crop do not always match with those of a biofuel (Yang and Udvardi, 2018), although efficient root function and structure is likely to be a characteristic desirable for both agricultural sectors.
Whether to improve plant performance or that of the environment, advances in phenotyping root systems will be crucial, including characterizing the plasticity of the system whilst the plant is growing. At present there are a plethora of root analysis software available (Paez-Garcia et al., 2015), but these require imaging roots either grown artificially such as on plates or already taken from the field and therefore evasive. There is therefore an increased interest in developing imaging techniques of plants grown in clear media to chart phenotypic changes throughout growth, or more promisingly the use of X-ray computed tomography (CT) scanning to give high resolution 3D models of the growing root system (Zhu et al., 2011).
Developing Management Systems
At present forage growers cannot easily and reliably determine the N status of their crops. For maximum biomass production, it is important to maintain the N status of the crop throughout the growing season and this requires an optimized soil N supply (Hofer et al., 2017). Application of too much N fertilizer results in wasteful run-off and sub-optimal supply results in decreased biomass production. Studies have already shown, through 15N labeling of Lolium, how deficiency caused by low N fertilizer application causes an increase in the protein substrate pool whereas the store pool decreased in size and turnover rate (Lehmeier et al., 2013). This highlights the importance of fertilizer studies for N composition of forage crop vegetative tissue. Maintaining N supply for maximal yield is limited by two factors: (1) unreliable and unreproducible tests for soil N levels (Knight, 2006) and (2) an easy reliable measure of the crop’s status.
Presently farmers take limited samples across their growing area in the hope that this is representative of the N in the whole plot through the growing season. Nevertheless, this does not indicate a plant’s N status or provide a measure of NUE. Some research has focused on the use of spectral data to evaluate crop efficiency (Foster et al., 2017), but such techniques require further investigation and can give false readings caused by pathogen attack. Sensors for N contents of soil are also being developed, however, these can be a costly solution (Shaw et al., 2016). Due to these problems, it may be better if the farmer could determine the crop N status directly and then make a more informed decision as to how they should subsequently fertilize the plot. This would enable more efficient fertilizer use, thus increasing forage biomass with lowered costs. Furthermore, for forage that includes legumes these N budget problems are complicated by the additional input of gaseous N-fixation. Other strategies of crop testing should be developed to reliably inform the grower of NUE efficiency.
Final Animal Product Studies
As forages are grown to rear livestock which in turn becomes food products for humans it is also important to view research in plant science from a livestock study prospective, of which has been touched upon above when discussing nutritional composition of crops. At present many countries adopt large-scale, concentrate-feeding led livestock production like that of the United States, with many potential human health risks due to bacteria, antibiotic-resistant bacteria, prion, and dioxin presence in end products (Sapkota et al., 2007). Despite a rise in concentrate-feeding, forage crops are still used widely as the main source of feed due to its high-yields of DM and energy for low costs (Reynolds, 2000), although usually studies focus on investigating a combination of both especially at various stages of development. For example, studies comparing growth of cattle fed a grass-diet instead of a linseed diet found the end product meat had a healthier fatty acid profile high in beneficial n-3 PUFAs, but the cattle were more slow-growing and thus the meat quality was poorer (Nuernberg et al., 2005). Similar outcomes have also been found for milk production from dairy cows in high-forage systems (Dewhurst et al., 2006). If improvements could be made in forage quality, especially more high-sugar varieties as outlined above, then potentially huge improvements in the animal production can be made.
In conclusion, utilizing the information obtained from the research effort to improve grain crops and the knowledge gathered from model systems like Arabidopsis, offers an excellent future perspective for improving the nutritional quality and yield for forage crops.
Author Contributions
NC and AM wrote the manuscript and conceived the perspective, read, and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This project was supported by grants BB/J004588/1 and BB/J004561/1 from the Biotechnology and Biological Sciences Research Council (BBSRC) and the John Innes Foundation. NC was supported by an iCASE studentship from the BBSRC, grant BB/M015203/1, with the support of The British Association of Green Crop Driers Ltd.
References
- Alexandratos N., Bruinsma J., Bödeker G., Schmidhuber J., Broca S., Shetty P., et al. (2006). “World agriculture: towards 2030/2050. Interim report. Prospects for food, nutrition, agriculture and major commodity groups,” in ESA Working Paper No. 12-03 eds Bruinsma J., Alexandratos N. (Rome: FAO; ). [Google Scholar]
- American Dietetic Association and Dietitians of Canada (2003). Position of the American dietetic association and dietitians of Canada: vegetarian diets. J. Am. Diet. Assoc. 103 748–765. 10.1053/jada.2003.50142 [DOI] [PubMed] [Google Scholar]
- Aranjuelo I., Molero G., Erice G., Avice J. C., Nogués S. (2011). Plant physiology and proteomics reveals the leaf response to drought in alfalfa (Medicago sativa L.). J. Exp. Bot. 62 111–123. 10.1093/jxb/erq249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo S. S., Beebe S., Crespi M., Delbreil B., González E. M., Gruber V., et al. (2015). Abiotic stress responses in legumes: strategies used to cope with environmental challenges. Crit. Rev. Plant Sci. 34 237–280. 10.1080/07352689.2014.898450 [DOI] [Google Scholar]
- Ariel F., Diet A., Verdenaud M., Gruber V., Frugier F., Chan R., et al. (2010). environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell 22 2171–2183. 10.1105/tpc.110.074823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaadi A. M., Yazdi A. K. (2011). Phenological stage effects on forage quality of four forbs species. J. Food Agric. Environ. 9 380–384. [Google Scholar]
- Avice J. C., Dily F. L., Goulas E., Noquet C., Meuriot F., Volenec J. J., et al. (2003). Vegetative storage proteins in overwintering storage organs of forage legumes: roles and regulation. Can. J. Bot. 81 1198–1212. 10.1139/b03-122 [DOI] [Google Scholar]
- Bachinger J., Reining E. (2009). An empirical statistical model for predicting the yield of herbage from legume-grass swards within organic crop rotations based on cumulative water balances. Grass Forage Sci. 64 144–159. 10.1111/j.1365-2494.2009.00678.x [DOI] [Google Scholar]
- Badenhorst P., Smith K., Spangenberg G. (2016). Development of a molecular breeding strategy for the integration of transgenic traits in outcrossing perennial grasses. Agronomy 6:56 10.3390/agronomy6040056 [DOI] [Google Scholar]
- Badenhorst P. E., Panter S., Palanisamy R., Georges S., Smith K. F., Mouradov A., et al. (2018). Molecular breeding of transgenic perennial ryegrass (Lolium perenne L.) with altered fructan biosynthesis through the expression of fructosyltransferases. Mol. Breed. 38:21 10.1007/s11032-018-0776-3 [DOI] [Google Scholar]
- Bajaj S., Ran Y., Phillips J., Kularajathevan G., Pal S., Cohen D., et al. (2006). A high throughput Agrobacterium tumefaciens-mediated transformation method for functional genomics of perennial ryegrass (Lolium perenne L.). Plant Cell Rep. 25 651–659. 10.1007/s00299-005-0099-9 [DOI] [PubMed] [Google Scholar]
- Baker J. M. (2012). Vegetative propagation of kura clover: a field-scale test. Can. J. Plant Sci. 92 1245–1251. 10.4141/cjps2012-014 [DOI] [Google Scholar]
- Ball R. O., Courtney-Martin G., Pencharz P. B. (2006). The in vivo sparing of methionine by cysteine in sulfur amino acid requirements in animal models and adult humans. J. Nutr. 136 1682S–1693S. 10.1093/jn/136.6.1682S [DOI] [PubMed] [Google Scholar]
- Bannink A., France J., Lopez S., Gerrits W. J. J., Kebreab E., Tamminga S., et al. (2008). Modelling the implications of feeding strategy on rumen fermentation and functioning of the rumen wall. Anim. Feed Sci. Technol. 143 3–26. 10.1016/j.anifeedsci.2007.05.002 [DOI] [Google Scholar]
- Barrett B., Baird I., Woodfield D. (2009). “White clover seed yield: a case study in marker-assisted selection,” in Molecular Breeding of Forage and Turf eds Yamada T., Spangenberg G. (New York, NY: Springer; ), 241–250. [Google Scholar]
- Barry T. N., Manley T. R. (1984). The role of condensed tannins in the nutritional value of Lotus pedunculatus for sheep. 2. Quantitative digestion of carbohydrates and proteins. Br. J. Nutr. 51 493–504. 10.1079/BJN19840055 [DOI] [PubMed] [Google Scholar]
- Bélanger G., Castonguay Y., Bertrand A., Dhont C., Rochette P., Couture L., et al. (2006). Winter damage to perennial forage crops in eastern Canada: causes, mitigation, and prediction. Can. J. Plant Sci. 86 33–47. 10.4141/P04-171 [DOI] [Google Scholar]
- Bergman E. N. (1990). Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70 567–590. 10.1152/physrev.1990.70.2.567 [DOI] [PubMed] [Google Scholar]
- Bewley J. D. (2002). Root storage proteins, with particular reference to taproots. Can. J. Bot. 80 321–329. 10.1139/b02-025 [DOI] [Google Scholar]
- Biazzi E., Nazzicari N., Pecetti L., Brummer E. C., Palmonari A., Tava A., et al. (2017). Genome-wide association mapping and genomic selection for alfalfa (Medicago sativa) forage quality traits. PLoS One 12:e0169234. 10.1371/journal.pone.0169234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot J., Lefevre J., Boucaud J. (1991). Changes in the amide and amino acid composition of xylem exudate from perennial ryegrass (Lolium perenne L.) during regrowth after defoliation. Plant Soil 136 59–64. 10.1007/bf02465220 [DOI] [Google Scholar]
- Blackmore T., Thorogood D., Skøt L., Mcmahon R., Powell W., Hegarty M. (2016). Germplasm dynamics: the role of ecotypic diversity in shaping the patterns of genetic variation in Lolium perenne. Sci. Rep. 6:22603. 10.1038/srep22603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller B., Green S. L. (2010). “Genetic resources,” in Fodder Crops and Amenity Grasses. Handbook of Plant Breeding eds Boller B., Posselt U., Veronesi F. (New York, NY: Springer; ), 13–38. 10.1007/978-1-4419-0760-8_2 [DOI] [Google Scholar]
- Bonhomme M., Andre O., Badis Y., Ronfort J., Burgarella C., Chantret N., et al. (2014). High-density genome-wide association mapping implicates an F-box encoding gene in Medicago truncatula resistance to Aphanomyces euteiches. New Phytol. 201 1328–1342. 10.1111/nph.12611 [DOI] [PubMed] [Google Scholar]
- Boufaïed H., Chouinard P. Y., Tremblay G. F., Petit H. V., Michaud R., Bélanger G. (2003). Fatty acids in forages. I. Factors affecting concentrations. Can. J. Anim. Sci. 83 501–511. 10.4141/A02-098 [DOI] [Google Scholar]
- Boyd J., West J. W., Bernard J. K. (2011). Effects of the addition of direct-fed microbials and glycerol to the diet of lactating dairy cows on milk yield and apparent efficiency of yield. J. Dairy Sci. 94 4616–4622. 10.3168/jds.2010-3984 [DOI] [PubMed] [Google Scholar]
- Brkljacic J., Grotewold E., Scholl R., Mockler T., Garvin D. F., Vain P., et al. (2011). Brachypodium as a model for the grasses: today and the future. Plant Physiol. 157 3–13. 10.1104/pp.111.179531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugger D., Windisch W. M. (2015). Environmental responsibilities of livestock feeding using trace mineral supplements. Anim. Nutr. 1 113–118. 10.1016/j.aninu.2015.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruins M. (2016). Breeding Targets for Ryegrass in Europe 3. Available at: http://european-seed.com/breeding-targets-ryegrass-europe/ [accessed December 04 2017]. [Google Scholar]
- Bumb I., Garnier E., Bastianelli D., Richarte J., Bonnal L., Kazakou E. (2016). Influence of management regime and harvest date on the forage quality of rangelands plants: the importance of dry matter content. AoB Plants 8:plw045. 10.1093/aobpla/plw045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggraaf V. T., Waghorn G. C., Woodward S. L., Thom E. R. (2008). Effects of condensed tannins in white clover flowers on their digestion in vitro. Anim. Feed Sci. Technol. 142 44–58. 10.1016/j.anifeedsci.2007.07.001 [DOI] [Google Scholar]
- Burggraaf V. T., Woodward S. L., Woodfield D. R., Thom E. R., Waghorn G. C., Kemp P. D. (2006). Morphology and agronomic performance of white clover with increased flowering and condensed tannin concentration. N. Z. J. Agric. Res. 49 147–155. 10.1080/00288233.2006.9513704 [DOI] [Google Scholar]
- Byrne S. L., Nagy I., Pfeifer M., Armstead I., Swain S., Studer B., et al. (2015). A synteny-based draft genome sequence of the forage grass Lolium perenne. Plant J. 84 816–826. 10.1111/tpj.13037 [DOI] [PubMed] [Google Scholar]
- Byrnes R. C., Nùñez J., Arenas L., Rao I., Trujillo C., Alvarez C., et al. (2017). Biological nitrification inhibition by Brachiaria grasses mitigates soil nitrous oxide emissions from bovine urine patches. Soil Biol. Biochem. 107 156–163. 10.1016/j.soilbio.2016.12.029 [DOI] [Google Scholar]
- CAST (2013). Animal Feed vs. Human Food: Challenges and Opportunities in Sustaining Animal Agriculture Toward 2050. Ames, IA: CAST. [Google Scholar]
- CIAT and CGIAR (2015). Improved Forages to Enhancing Farm Productivity, Climate Change Resilience, and Environmental Sustainability in Eastern and Southern Africa [Online]. Nairobi: International Centre for Tropical Agriculture (CIAT). [Google Scholar]
- Cabral G. B., Carneiro V. T. C., Rossi M. L., Da Silva J. P., Martinelli A. P., Dusi D. M. A. (2015). Plant regeneration from embryogenic callus and cell suspensions of Brachiaria brizantha. In Vitro Cell. Dev. Biol. Plant 51 369–377. 10.1007/s11627-015-9690-0 [DOI] [Google Scholar]
- Calderini O., Bovone T., Scotti C., Pupilli F., Piano E., Arcioni S. (2007). Delay of leaf senescence in Medicago sativa transformed with the ipt gene controlled by the senescence-specific promoter SAG12. Plant Cell Rep. 26 611–615. 10.1007/s00299-006-0262-y [DOI] [PubMed] [Google Scholar]
- Carelli M., Calderini O., Panara F., Porceddu A., Losini I., Piffanelli P., et al. (2013). Reverse genetics in Medicago truncatula using a TILLING mutant collection. Methods Mol. Biol. 1069 101–118. 10.1007/978-1-62703-613-9_9 [DOI] [PubMed] [Google Scholar]
- Chalmers J., Lidgett A., Cummings N., Cao Y., Forster J., Spangenberg G. (2005). Molecular genetics of fructan metabolism in perennial ryegrass. Plant Biotechnol. J. 3 459–474. 10.1111/j.1467-7652.2005.00148.x [DOI] [PubMed] [Google Scholar]
- Chattopadhyay A., Subba P., Pandey A., Bhushan D., Kumar R., Datta A., et al. (2011). Analysis of the grasspea proteome and identification of stress-responsive proteins upon exposure to high salinity, low temperature, and abscisic acid treatment. Phytochemistry 72 1293–1307. 10.1016/j.phytochem.2011.01.024 [DOI] [PubMed] [Google Scholar]
- Chen T., Yang Q., Gruber M., Kang J., Sun Y., Ding W., et al. (2012a). Expression of an alfalfa (Medicago sativa L.) ethylene response factor gene MsERF8 in tobacco plants enhances resistance to salinity. Mol. Biol. Rep. 39 6067–6075. 10.1007/s11033-011-1421-y [DOI] [PubMed] [Google Scholar]
- Chen T., Yang Q., Zhang X., Ding W., Gruber M. (2012b). An alfalfa (Medicago sativa L.) ethylene response factor gene, MsERF11, enhances salt tolerance in transgenic Arabidopsis. Plant Cell Rep. 31 1737–1746. 10.1007/s00299-012-1287-z [DOI] [PubMed] [Google Scholar]
- Chilliard Y., Glasser F., Ferlay A., Bernard L., Rouel J., Doreau M. (2007). Diet, rumen biohydrogenation and nutritional quality of cow and goat milk fat. Eur. J. Lipid Sci. Technol. 109 828–855. 10.1002/ejlt.200700080 [DOI] [Google Scholar]
- Clapham W. M., Foster J. G., Neel J. P. S., Fedders J. M. (2005). Fatty acid composition of traditional and novel forages. J. Agric. Food Chem. 53 10068–10073. 10.1021/jf0517039 [DOI] [PubMed] [Google Scholar]
- Collins R. P., Helgadóttir Á, Frankow-Lindberg B. E., Skøt L., Jones C., Skøt K. P. (2012). Temporal changes in population genetic diversity and structure in red and white clover grown in three contrasting environments in northern Europe. Ann. Bot. 110 1341–1350. 10.1093/aob/mcs058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connorton J. M., Jones E. R., Rodriguez-Ramiro I., Fairweather-Tait S., Uauy C., Balk J. (2017). Wheat vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiol. 174 2434–2444. 10.1104/pp.17.00672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer H., Kessler C. S., Sundberg T., Leach M. J., Schumann D., Adams J., et al. (2017). Characteristics of americans choosing vegetarian and vegan diets for health reasons. J. Nutr. Educ. Behav. 49 561–567. 10.1016/j.jneb.2017.04.011 [DOI] [PubMed] [Google Scholar]
- Dalmais M., Antelme S., Ho-Yue-Kuang S., Wang Y., Darracq O., d’Yvoire M. B., et al. (2013). A TILLING platform for functional genomics in Brachypodium distachyon. PLoS One 8:e65503. 10.1371/journal.pone.0065503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson L. A., Thornton B., Pratt S. M., Paterson E. (2004). Morphological and topological responses of roots to defoliation and nitrogen supply in Lolium perenne and Festuca ovina. New Phytol. 161 811–818. 10.1111/j.1469-8137.2004.00979.x [DOI] [PubMed] [Google Scholar]
- de Araüjo M. R. A., Coulman B. E., Rakow G. (2002). Genetic variation, heritability and progeny testing in meadow bromegrass. Plant Breed. 121 417–424. 10.1046/j.1439-0523.2002.739124.x [DOI] [Google Scholar]
- de Bang T., Lundquist P. K., Dai X., Boschiero C., Zhuang Z., Pant P., et al. (2017). Genome-wide Identification of Medicago peptides involved in macronutrient responses and nodulation. Plant Physiol. 175 17.01096. 10.1104/pp.17.01096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vega J. J., Ayling S., Hegarty M., Kudrna D., Goicoechea J. L., Ergon Å, et al. (2015). Red clover (Trifolium pratense L.) draft genome provides a platform for trait improvement. Sci. Rep. 5:17394. 10.1038/srep17394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Herder G., Van Isterdael G., Beeckman T., De Smet I. (2010). The roots of a new green revolution. Trends Plant Sci. 15 600–607. 10.1016/j.tplants.2010.08.009 [DOI] [PubMed] [Google Scholar]
- Derner J. D., Hunt L., Filho K. E., Ritten J., Capper J., Han G. (2017). “Livestock production systems,” in Rangeland Systems: Processes, Management and Challenges ed. Briske D. D. (Cham: Springer; ), 347–372. 10.1007/978-3-319-46709-2_10 [DOI] [Google Scholar]
- Dewhurst R. J., Scollan N. D., Youell S. J., Tweed J. K. S., Humphreys M. O. (2001). Influence of species, cutting date and cutting interval on the fatty acid composition of grasses. Grass Forage Sci. 56 68–74. 10.1046/j.1365-2494.2001.00247.x [DOI] [Google Scholar]
- Dewhurst R. J., Shingfield K. J., Lee M. R. F., Scollan N. D. (2006). Increasing the concentrations of beneficial polyunsaturated fatty acids in milk produced by dairy cows in high-forage systems. Anim. Feed Sci. Technol. 131 168–206. 10.1016/j.anifeedsci.2006.04.016 [DOI] [Google Scholar]
- Di Paola A., Rulli M. C., Santini M. (2017). Human food vs. animal feed debate. A thorough analysis of environmental footprints. Land Use Policy 67 652–659. 10.1016/j.landusepol.2017.06.017 [DOI] [Google Scholar]
- Diekmann K., Hodkinson T. R., Barth S. (2012). New chloroplast microsatellite markers suitable for assessing genetic diversity of Lolium perenne and other related grass species. Ann. Bot. 110 1327–1339. 10.1093/aob/mcs044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierking R. M., Allen D. J., Cunningham S. M., Brouder S. M., Volenec J. J. (2017). Nitrogen reserve pools in two Miscanthus × giganteus genotypes under contrasting N managements. Front. Plant Sci. 8:1618. 10.3389/fpls.2017.01618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra J., Oenema O., Bannink A. (2011). Dietary strategies to reducing N excretion from cattle: implications for methane emissions. Curr. Opin. Environ. Sustain. 3 414–422. 10.1016/j.cosust.2011.07.008 [DOI] [Google Scholar]
- Dinkins R. D., Srinivasa Reddy M. S., Meurer C. A., Yan B., Trick H., Thibaud-Nissen F., et al. (2001). Increased sulfur amino acids in soybean plants overexpressing the maize 15 kDa zein protein. In Vitro Cell. Dev. Biol. Plant 37 742–747. 10.1007/s11627-001-0123-x [DOI] [Google Scholar]
- Dixon R. A., Xie D.-Y., Sharma S. B. (2005). Proanthocyanidins – a final frontier in flavonoid research? New Phytol. 165 9–28. 10.1111/j.1469-8137.2004.01217.x [DOI] [PubMed] [Google Scholar]
- Downie J. A. (2010). The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 34 150–170. 10.1111/j.1574-6976.2009.00205.x [DOI] [PubMed] [Google Scholar]
- Elgersma A., Ellen G., Van Der Horst H., Muuse B. G., Boer H., Tamminga S. (2003). Influence of cultivar and cutting date on the fatty acid composition of perennial ryegrass (Lolium perenne L.). Grass Forage Sci. 58 323–331. 10.1046/j.1365-2494.2003.00384.x [DOI] [Google Scholar]
- Elgersma A., Tamminga S., Ellen G. (2006). Modifying milk composition through forage. Anim. Feed Sci. Technol. 131 207–225. 10.1016/j.anifeedsci.2006.06.012 20105538 [DOI] [Google Scholar]
- Elghandour M. M. Y., Salem A. Z. M., Castañeda J. S. M., Camacho L. M., Kholif A. E., Chagoyán J. C. V. (2015). Direct-fed microbes: A tool for improving the utilization of low quality roughages in ruminants. J. Integr. Agric. 14 526–533. 10.1016/S2095-3119(14)60834-0 [DOI] [Google Scholar]
- Englyst K. N., Englyst H. N., Hudson G. J., Cole T. J., Cummings J. H. (1999). Rapidly available glucose in foods: an in vitro measurement that reflects the glycemic response. Am. J. Clin. Nutr. 69 448–454. 10.1093/ajcn/69.3.448 [DOI] [PubMed] [Google Scholar]
- Erb K.-H., Mayer A., Kastner T., Sallet K.-E., Haberl H. (2012). The Impact of Industrial Grain Fed Livestock Production on Food Security: An Extended Literature Review. Vienna: Institute of Soil Ecology. [Google Scholar]
- Erice G., Irigoyen J. J., Sánchez-Díaz M., Avice J.-C., Ourry A. (2007). Effect of drought, elevated CO2 and temperature on accumulation of N and vegetative storage proteins (VSP) in taproot of nodulated alfalfa before and after cutting. Plant Sci. 172 903–912. 10.1016/j.plantsci.2006.12.013 [DOI] [Google Scholar]
- FAO (2010). Challenges and Opportunities for Carbon Sequestration in Grassland Systems: A Technical Report on Grassland Management and Climate Mitigation. (Rome: Food and Agriculture Organization of the United Nations; ). [Google Scholar]
- Fan X., Naz M., Fan X., Xuan W., Miller A. J., Xu G. (2017). Plant nitrate transporters: from gene function to application. J. Exp. Bot. 68 2463–2475. 10.1093/jxb/erx011 [DOI] [PubMed] [Google Scholar]
- Fe D., Cericola F., Byrne S., Lenk I., Ashraf B. H., Pedersen M. G., et al. (2015). Genomic dissection and prediction of heading date in perennial ryegrass. BMC Genomics 16:921. 10.1186/s12864-015-2163-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster A. J., Kakani V. G., Mosali J. (2017). Estimation of bioenergy crop yield and N status by hyperspectral canopy reflectance and partial least square regression. Precis. Agric. 18 192–209. 10.1007/s11119-016-9455-8 [DOI] [Google Scholar]
- Foyer C. H., Lam H. M., Nguyen H. T., Siddique K. H., Varshney R. K., Colmer T. D., et al. (2016). Neglecting legumes has compromised human health and sustainable food production. Nat. Plants 2:16112. 10.1038/nplants.2016.112 [DOI] [PubMed] [Google Scholar]
- Freeman K. W., Girma K., Arnall D. B., Mullen R. W., Kent K. L., Teal R. K., et al. (2007). By-Plant prediction of corn forage biomass and nitrogen uptake at various growth stages using remote sensing and plant height. Agron. J. 99 530–536. 10.2134/agronj2006.0135 [DOI] [Google Scholar]
- Gao R., Feyissa B. A., Croft M., Hannoufa A. (2018). Gene editing by CRISPR/Cas9 in the obligatory outcrossing Medicago sativa. Planta 247 1043–1050. 10.1007/s00425-018-2866-1 [DOI] [PubMed] [Google Scholar]
- Ghesquière M., Humphreys M. W., Zwierzykowski Z. (2010). “Festulolium,” in Fodder Crops and Amenity Grasses eds Boller B., Posselt U. K., Veronesi F. (New York, NY: Springer; ), 288–311. [Google Scholar]
- Giuberti G., Rocchetti G., Sigolo S., Fortunati P., Lucini L., Gallo A. (2018). Exploitation of alfalfa seed (Medicago sativa L.) flour into gluten-free rice cookies: nutritional, antioxidant and quality characteristics. Food Chem. 239 679–687. 10.1016/j.foodchem.2017.07.004 [DOI] [PubMed] [Google Scholar]
- Gokulakannan G. G., Niehaus K. (2010). Characterization of the Medicago truncatula cell wall proteome in cell suspension culture upon elicitation and suppression of plant defense. J. Plant Physiol. 167 1533–1541. 10.1016/j.jplph.2010.06.023 [DOI] [PubMed] [Google Scholar]
- Gondo T., Umami N., Muguerza M., Akashi R. (2017). Plant regeneration from embryogenic callus derived from shoot apices and production of transgenic plants by particle inflow gun in dwarf napier grass (Pennisetum purpureum Schumach.). Plant Biotechnol. 34 143–150. 10.5511/plantbiotechnology.17.0623a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber F. (2002). The synthesis of ammonia from its elements Nobel Lecture. Resonance 7 86–94. 10.1007/BF02836189 [DOI] [Google Scholar]
- Hancock K. R., Collette V., Fraser K., Greig M., Xue H., Richardson K., et al. (2012). Expression of the R2R3-MYB transcription factor TaMYB14 from Trifolium arvense activates proanthocyanidin biosynthesis in the legumes Trifolium repens and Medicago sativa. Plant Physiol. 159 1204–1220. 10.1104/pp.112.195420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfoot C. G., Hazlewood G. P. (1988). “Lipid metabolism in the rumen,” in The Rumen Microbial Ecosystem ed. Hobson P. M. (London: Elsevier; ), 285–322. [Google Scholar]
- Hatfield R. D., Jung H. J. G., Broderick G., Jenkins T. C. (2007). “Nutritional chemistry of forages,” in Forages: The Science of Grassland Agriculture 6th Edn, eds Barnes R. F., Nelson C. J., Moore K. J., Collins M. (Ames, IA: Blackwell Publishing; ), 467–485. [Google Scholar]
- Helminger W., Martins C., Henderyckx E., Scuvée B. (2016). Agriculture , Forestry and Fishery Statistics, eds Forti R., Brussels M. Henrard.: European Union. [Google Scholar]
- Herrero M., Grace D., Njuki J., Johnson N., Enahoro D., Silvestri S., et al. (2013). The roles of livestock in developing countries. Animal 7 3–18. 10.1017/S1751731112001954 [DOI] [PubMed] [Google Scholar]
- Herrmann D., Barre P., Santoni S., Julier B. (2010). Association of a CONSTANS-LIKE gene to flowering and height in autotetraploid alfalfa. Theor. Appl. Genet. 121 865–876. 10.1007/s00122-010-1356-z [DOI] [PubMed] [Google Scholar]
- Hirata M. (2011). “Forage crop production,” in The Role of Food, Agriculture, Forestry and Fisheries in Human Nutrition ed. Squires V. R. (Oxford: Encyclopedia of Life Support Systems; ), 181–202. [Google Scholar]
- Hofer D., Suter M., Buchmann N., Luscher A. (2017). Nitrogen status of functionally different forage species explains resistance to severe drought and post-drought overcompensation. Agric. Ecosyst. Environ. 236 312–322. 10.1016/j.agee.2016.11.022 [DOI] [Google Scholar]
- Humphreys M. W., Canter P. J., Thomas H. M. (2003). Advances in introgression technologies for precision breeding within the Lolium - Festuca complex. Ann. Appl. Biol. 143 1–10. 10.1111/j.1744-7348.2003.tb00263.x [DOI] [Google Scholar]
- Humphreys M. W., O’donovan S. A., Farrell M. S., Gay A. P., Kingston-Smith A. H. (2014). The potential of novel Festulolium (2n = 4x = 28) hybrids as productive, nutrient-use-efficient fodder for ruminants. Food Energy Secur. 3 98–110. 10.1002/fes3.50 [DOI] [Google Scholar]
- Huot B., Yao J., Montgomery B. L., He S. Y. (2014). Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant 7 1267–1287. 10.1093/mp/ssu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannetta P. P., Young M., Bachinger J., Bergkvist G., Doltra J., Lopez-Bellido R. J., et al. (2016). A comparative nitrogen balance and productivity analysis of legume and non-legume supported cropping systems: the potential role of biological nitrogen fixation. Front. Plant Sci. 7:1700. 10.3389/fpls.2016.01700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y., Hamano K., Kang D.-J., Idota S., Nishiwaki A. (2015). Cadmium phytoremediation potential of napiergrass cultivated in Kyushu, Japan. Appl. Environ. Soil Sci. 2015:6 10.1155/2015/756270 [DOI] [Google Scholar]
- Jackson R. B., Mooney H. A., Schulze E.-D. (1997). A global budget for fine root biomass, surface area, and nutrient?contents. Proc. Natl. Acad. Sci. U.S.A. 94 7362–7366. 10.1073/pnas.94.14.7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob C., Carrasco B., Schwember A. R. (2016). Advances in breeding and biotechnology of legume crops. Plant Cell Tissue Organ Cult. 127 561–584. 10.1007/s11240-016-1106-2 [DOI] [Google Scholar]
- Jahufer M. Z. Z., Cooper M., Ayres J. F., Bray R. A. (2002). Identification of research to improve the efficiency of breeding strategies for white clover in Australia - a review. Aust. J. Agric. Res. 53 239–257. 10.1071/AR01110 [DOI] [Google Scholar]
- Jaradat A. A., Johnson J. M. F., Weyers S. L., Barbour N. W. (2009). Determinants and prediction of carbon/nitrogen ratio in five diverse crop plants. Commun. Soil Sci. Plant Anal. 40 2688–2711. 10.1080/00103620903173756 [DOI] [Google Scholar]
- Julier B., Herrmann D., Barre P., Pierre J.-B. (2010). “From a model to a crop species: constans is involved in aerial morphogenesis of lucerne,” in Sustainable use of Genetic Diversity in Forage and Turf Breeding ed. Huyghe C. (Dordrecht: Springer; ). [Google Scholar]
- Julier B., Huguet T., Chardon F., Ayadi R., Pierre J.-B., Prosperi J.-M., et al. (2007). Identification of quantitative trait loci influencing aerial morphogenesis in the model legume Medicago truncatula. Theor. Appl. Genet. 114 1391–1406. 10.1007/s00122-007-0525-1 [DOI] [PubMed] [Google Scholar]
- Jung H. G., Allen M. S. (1995). Characteristics of plant cell walls affecting intake and digestibility of forages by ruminants. J. Anim. Sci. 73 2774–2790. 10.2527/1995.7392774x [DOI] [PubMed] [Google Scholar]
- Jung H.-J. G., Samac D. A., Sarath G. (2012). Modifying crops to increase cell wall digestibility. Plant Sci. 185 65–77. 10.1016/j.plantsci.2011.10.014 [DOI] [PubMed] [Google Scholar]
- Karn J. F., Berdahl J. D., Frank A. B. (2006). Nutritive quality of four perennial grasses as affected by species, cultivar, maturity, and plant tissue. Agron. J. 98 1400–1409. 10.2134/agronj2005.0293 [DOI] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J. P., Quemada M. (2017). Using cover crops to mitigate and adapt to climate change. A review. Agron. Sustain. Dev. 37:4 10.1007/s13593-016-0410-x [DOI] [Google Scholar]
- Kell D. B. (2011). Breeding crop plants with deep roots: their role in sustainable carbon, nutrient and water sequestration. Ann. Bot. 108 407–418. 10.1093/aob/mcr175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell D. B. (2012). Large-scale sequestration of atmospheric carbon via plant roots in natural and agricultural ecosystems: why and how. Philos. Trans. R. Soc. B Biol. Sci. 367 1589–1597. 10.1098/rstb.2011.0244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston-Smith A. H., Davies T. E., Edwards J. E., Theodorou M. K. (2008). From plants to animals; the role of plant cell death in ruminant herbivores. J. Exp. Bot. 59 521–532. 10.1093/jxb/erm326 [DOI] [PubMed] [Google Scholar]
- Kingston-Smith A. H., Edwards J. E., Huws S. A., Kim E. J., Abberton M. (2010). Plant-based strategies towards minimising ’livestock’s long shadow’. Proc. Nutr. Soc. 69 613–620. 10.1017/s0029665110001953 [DOI] [PubMed] [Google Scholar]
- Knight M. (2006). Soil Mineral Nitrogen Testing : Practice and Interpretation. Wymondham: The Arable Group. [Google Scholar]
- Krausmann F., Erb K.-H., Gingrich S., Lauk C., Haberl H. (2008). Global patterns of socioeconomic biomass flows in the year 2000: a comprehensive assessment of supply, consumption and constraints. Ecol. Econ. 65 471–487. 10.1016/j.ecolecon.2007.07.012 [DOI] [Google Scholar]
- Lee K.-W., Cha J.-Y., Kim K.-H., Kim Y.-G., Lee B.-H., Lee S.-H. (2012a). Overexpression of alfalfa mitochondrial HSP23 in prokaryotic and eukaryotic model systems confers enhanced tolerance to salinity and arsenic stress. Biotechnol. Lett. 34 167–174. 10.1007/s10529-011-0750-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.-W., Choi G. J., Kim K.-Y., Ji H. J., Park H. S., Kim Y.-G., et al. (2012b). Transgenic expression of MsHsp23 confers enhanced tolerance to abiotic stresses in tall fescue. Asian Aust. J. Anim. Sci. 25 818–823. 10.5713/ajas.2012.12034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees G. L. (1992). Condensed tannins in some forage legumes: their role in the prevention of ruminant pasture bloat. Basic Life Sci. 59 915–934. 10.1007/978-1-4615-3476-1_55 [DOI] [PubMed] [Google Scholar]
- Lehmeier C. A., Wild M., Schnyder H. (2013). Nitrogen stress affects the turnover and size of nitrogen pools supplying leaf growth in a grass. Plant Physiol. 162 2095–2105. 10.1104/pp.113.219311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepiniec L., Debeaujon I., Routaboul J. M., Baudry A., Pourcel L., Nesi N., et al. (2006). Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57 405–430. 10.1146/annurev.arplant.57.032905.105252 [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2007). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23 127–128. 10.1093/bioinformatics/btl529 [DOI] [PubMed] [Google Scholar]
- Li Q., Robson P. R. H., Bettany A. J. E., Donnison I. S., Thomas H., Scott I. M. (2004). Modification of senescence in ryegrass transformed with IPT under the control of a monocot senescence-enhanced promoter. Plant Cell Rep. 22 816–821. 10.1007/s00299-004-0762-6 [DOI] [PubMed] [Google Scholar]
- Li W., Katin-Grazzini L., Gu X., Wang X., El-Tanbouly R., Yer H., et al. (2017). Transcriptome analysis reveals differential gene expression and a possible role of gibberellins in a shade-tolerant mutant of perennial ryegrass. Front. Plant Sci. 8:868. 10.3389/fpls.2017.00868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Long R., Zhang T., Wang Z., Zhang F., Yang Q., et al. (2017). Molecular cloning and functional analysis of the drought tolerance gene MsHSP70 from alfalfa (Medicago sativa L.). J. Plant Res. 130 387–396. 10.1007/s10265-017-0905-9 [DOI] [PubMed] [Google Scholar]
- Li Z.-Y., Long R.-C., Zhang T.-J., Yang Q.-C., Kang J.-M. (2016). Molecular cloning and characterization of the MsHSP17.7 gene from Medicago sativa L. Mol. Biol. Rep. 43 815–826. 10.1007/s11033-016-4008-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louahlia S., Laine P., Macduff J. H., Ourry A., Humphreys M., Boucaud J. (2008). Interactions between reserve mobilization and regulation of nitrate uptake during regrowth of Lolium perenne L.: putative roles of amino acids and carbohydrates. Botany 86 1101–1110. 10.1139/B08-066 [DOI] [Google Scholar]
- Luo K., Jahufer M. Z. Z., Wu F., Di H., Zhang D., Meng X., et al. (2016). Genotypic variation in a breeding population of yellow sweet clover (Melilotus officinalis). Front. Plant Sci. 7:972. 10.3389/fpls.2016.00972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. P. (2007). Roots of the second green revolution. Aust. J. Bot. 55 493–512. 10.1071/BT06118 [DOI] [Google Scholar]
- MacLean A. M., Bravo A., Harrison M. J. (2017). Plant signaling and metabolic pathways enabling arbuscular mycorrhizal symbiosis. Plant Cell. 29 2319–2335. 10.1105/tpc.17.00555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macleod C. J. A., Humphreys M. W., Whalley W. R., Turner L., Binley A., Watts C. W., et al. (2013). A novel grass hybrid to reduce flood generation in temperate regions. Sci. Rep. 3:1683. 10.1038/srep01683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae J. C., Ulyatt M. J. (1974). Quantitative digestion of fresh herbage by sheep: II. The sites of digestion of some nitrogenous constituents. J. Agric. Sci. 82 309–319. 10.1017/S0021859600059207 [DOI] [Google Scholar]
- Maghsoodi M., Razmjoo J., Gheysari M. (2017). Application of biochemical markers for the assessment of drought tolerance in alfalfa (Medicago sativa L.) cultivars. Grassland Sci. 63 3–14. 10.1111/grs.12144 [DOI] [Google Scholar]
- Manzanares C., Yates S., Ruckle M., Nay M., Studer B. (2016). TILLING in forage grasses for gene discovery and breeding improvement. New Biotechnol. 33 594–603. 10.1016/j.nbt.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Marshall A. H., Collins R. P., Humphreys M. W., Scullion J. (2016). A new emphasis on root traits for perennial grass and legume varieties with environmental and ecological benefits. Food Energy Secur. 5 26–39. 10.1002/fes3.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy R. D., Klusmeyer T. H., Vicini J. L., Clark J. H., Nelson D. R. (1989). Effects of source of protein and carbohydrate on ruminal fermentation and passage of nutrients to the small intestine of lactating cows. J. Dairy Sci. 72 2002–2016. 10.3168/jds.S0022-0302(89)79324-3 [DOI] [PubMed] [Google Scholar]
- McCord P., Gordon V., Saha G., Hellinga J., Vandemark G., Larsen R., et al. (2014). Detection of QTL for forage yield, lodging resistance and spring vigor traits in alfalfa (Medicago sativa L.). Euphytica 200 269–279. 10.1007/s10681-014-1160-y [DOI] [Google Scholar]
- Mcevoy C., Woodside J. V. (2010). “Vegetarian and vegan diets: weighing the claims,” in Nutrition Guide for Physicians eds Wilson T., Bray G. A., Temple N. J., Struble M. B. (Totowa, NJ: Humana Press; ), 81–93. [Google Scholar]
- McIntosh M. S., Miller D. A. (1980). Development of root-branching in three alfalfa cultivars. Crop Sci. 20 807–809. 10.2135/cropsci1980.0011183X002000060033x [DOI] [Google Scholar]
- McKersie B. D., Murnaghan J., Jones K. S., Bowley S. R. (2000). Iron-superoxide dismutase expression in transgenic alfalfa increases winter survival without a detectable increase in photosynthetic oxidative stress tolerance. Plant Physiol. 122 1427–1437. 10.1104/pp.122.4.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menguer P. K., Vincent T., Miller A. J., Brown J. K. M., Vincze E., Borg S., et al. (2017). Improving zinc accumulation in cereal endosperm using HvMTP1, a transition metal transporter. Plant Biotechnol. J. 16 63–71. 10.1111/pbi.12749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuriot F., Avice J. C., Simon J. C., Laine P., Decau M. L., Ourry A. (2004). Influence of initial organic N reserves and residual leaf area on growth, N uptake, N partitioning and N storage in alfalfa (Medicago sativa) during post-cutting regrowth. Ann. Bot. 94 311–321. 10.1093/aob/mch144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuriot F., Decau M.-L., Morvan-Bertrand A., Prud’homme M.-P., Gastal F., Simon J.-C., et al. (2005). Contribution of initial C and N reserves in Medicago sativa recovering from defoliation: impact of cutting height and residual leaf area. Funct. Plant Biol. 32 321–334. 10.1071/FP04151 [DOI] [PubMed] [Google Scholar]
- Miller L. A., Moorby J. M., Davies D. R., Humphreys M. O., Scollan N. D., Macrae J. C., et al. (2001). Increased concentration of water-soluble carbohydrate in perennial ryegrass (Lolium perenne L.): milk production from late-lactation dairy cows. Grass Forage Sci. 56 383–394. 10.1046/j.1365-2494.2001.00288.x [DOI] [Google Scholar]
- Min B. R., Barry T. N., Attwood G. T., Mcnabb W. C. (2003). The effect of condensed tannins on the nutrition and health of ruminants fed fresh temperate forages: a review. Anim. Feed Sci. Technol. 106 3–19. 10.1016/S0377-8401(03)00041-5 [DOI] [Google Scholar]
- Molvig L., Tabe L. M., Eggum B. O., Moore A. E., Craig S., Spencer D., et al. (1997). Enhanced methionine levels and increased nutritive value of seeds of transgenic lupins (Lupinus angustifolius L.) expressing a sunflower seed albumin gene. Proc. Natl. Acad. Sci. 94 8393–8398. 10.1073/pnas.94.16.8393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommer L., Hinsinger P., Prigent-Combaret C., Visser E. J. W. (2016). Advances in the rhizosphere: stretching the interface of life. Plant Soil 407 1–8. 10.1007/s11104-016-3040-9 [DOI] [Google Scholar]
- Mueller-Harvey I. (2006). Unravelling the conundrum of tannins in animal nutrition and health. J. Sci. Food Agric. 86 2010–2037. 10.1002/jsfa.2577 [DOI] [Google Scholar]
- Muir J. P., Pitman W. D., Foster J. L. (2011). Sustainable, low-input, warm-season, grass–legume grassland mixtures: mission (nearly) impossible? Grass Forage Sci. 66 301–315. 10.1111/j.1365-2494.2011.00806.x [DOI] [Google Scholar]
- Muntz K. (1998). Deposition of storage proteins. Plant Mol. Biol. 38 77–99. 10.1023/A:1006020208380 [DOI] [PubMed] [Google Scholar]
- Myouga F., Hosoda C., Umezawa T., Iizumi H., Kuromori T., Motohashi R., et al. (2008). A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. Plant Cell 20 3148–3162. 10.1105/tpc.108.061341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIMSS (2017). NE1010: Breeding and Genetics of Forage Crops to Improve Productivity, Quality, and Industrial Uses [Online]. Available at: https://www.nimss.org/projects/1314 [accessed November 30 2017] [Google Scholar]
- Nacry P., Bouguyon E., Gojon A. (2013). Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 370 1–29. 10.1007/s11104-013-1645-9 [DOI] [Google Scholar]
- Nagy I., Barth S., Mehenni-Ciz J., Abberton M. T., Milbourne D. (2013). A hybrid next generation transcript sequencing-based approach to identify allelic and homeolog-specific single nucleotide polymorphisms in allotetraploid white clover. BMC Genomics 14:100. 10.1186/1471-2164-14-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Njuki J., Sanginga P. C. (2013). “Gender and livestock: key issues, challenges and opportunities,” in Women, Livestock Ownership and Markets: Bridging the gender gap in Eastern and Southern Africa eds Njuki J., Sanginga P. C. (New York, NY: Routledge; ). [Google Scholar]
- Nocek J. E., Russell J. B. (1988). Protein and energy as an integrated system. Relationship of ruminal protein and carbohydrate availability to microbial synthesis and milk production. J. Dairy Sci. 71 2070–2107. 10.3168/jds.S0022-0302(88)79782-9 [DOI] [Google Scholar]
- Nuernberg K., Dannenberger D., Nuernberg G., Ender K., Voigt J., Scollan N. D., et al. (2005). Effect of a grass-based and a concentrate feeding system on meat quality characteristics and fatty acid composition of longissimus muscle in different cattle breeds. Livest. Prod. Sci. 94 137–147. 10.1016/j.livprodsci.2004.11.036 [DOI] [Google Scholar]
- Nuñez F., Yamada T. (2017). Molecular regulation of flowering time in grasses. Agronomy 7:17 10.3390/agronomy7010017 [DOI] [Google Scholar]
- Osbourn D. F. (1980). “The feeding value of grass and grass products,” in Grass: Its Production and Utilization ed. Holmes W. (Oxford: Blackwell Scientific Publications; ), 70–124. [Google Scholar]
- Paez-Garcia A., Motes C., Scheible W.-R., Chen R., Blancaflor E., Monteros M. (2015). Root traits and phenotyping strategies for plant improvement. Plants 4 334–355. 10.3390/plants4020334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y., Peel G. J., Wright E., Wang Z., Dixon R. A. (2007). Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol. 145 601–615. 10.1104/pp.107.107326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panter S., Mouradov A., Badenhorst P., Martelotto L., Griffith M., Smith K., et al. (2017). Re-programming photosynthetic cells of perennial ryegrass (Lolium perenne L) for fructan biosynthesis through transgenic expression of fructan biosynthetic genes under the control of photosynthetic promoters. Agronomy 7:36 10.3390/agronomy7020036 [DOI] [Google Scholar]
- Parsons A. J., Harvey A., Woledge J. (1991). Plant-animal interactions in a continuously grazed mixture. I. Differences in the physiology of leaf expansion and the fate of leaves of grass and clover. J. Appl. Ecol. 28 619–634. 10.2307/2404572 [DOI] [Google Scholar]
- Paterson E., Sim A. (1999). Rhizodeposition and C-partitioning of Lolium perenne in axenic culture affected by nitrogen supply and defoliation. Plant Soil 216 155–164. 10.1023/A:1004789407065 [DOI] [Google Scholar]
- Pfeifer M., Martis M., Asp T., Mayer K. F. X., Lübberstedt T., Byrne S., et al. (2013). The perennial ryegrass GenomeZipper: targeted use of genome resources for comparative grass genomics. Plant Physiol. 161 571–582. 10.1104/pp.112.207282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano E., Annicchiarico P., Romani M., Pecetti L. (2007). Effect of the number of parents and their combining ability on the performance of synthetic varieties in tall fescue. Aust. J. Agric. Res. 58 1100–1105. 10.1071/AR06424 [DOI] [Google Scholar]
- Piluzza G., Sulas L., Bullitta S. (2014). Tannins in forage plants and their role in animal husbandry and environmental sustainability: a review. Grass Forage Sci. 69 32–48. 10.1111/gfs.12053 [DOI] [Google Scholar]
- Pinheiro C., Kehr J., Ricardo C. P. (2005). Effect of water stress on lupin stem protein analysed by two-dimensional gel electrophoresis. Planta 221 716–728. 10.1007/s00425-004-1478-0 [DOI] [PubMed] [Google Scholar]
- Pirhofer-Walzl K., Rasmussen J., Høgh-Jensen H., Eriksen J., Søegaard K., Rasmussen J. (2012). Nitrogen transfer from forage legumes to nine neighbouring plants in a multi-species grassland. Plant Soil 350 71–84. 10.1007/s11104-011-0882-z [DOI] [Google Scholar]
- Poehlman J. M. (ed.) (1987). “Breeding forage crops,” in Breeding Field Crops. (Dordrecht: Springer), 625–679. 10.1007/978-94-015-7271-2_22 [DOI] [Google Scholar]
- Preissel S., Reckling M., Schläfke N., Zander P. (2015). Magnitude and farm-economic value of grain legume pre-crop benefits in Europe: a review. Field Crops Res. 175 64–79. 10.1016/j.fcr.2015.01.012 [DOI] [Google Scholar]
- Raigond P., Ezekiel R., Raigond B. (2015). Resistant starch in food: a review. J. Sci. Food Agric. 95 1968–1978. 10.1002/jsfa.6966 [DOI] [PubMed] [Google Scholar]
- Rancour D. M., Marita J. M., Hatfield R. D. (2012). Cell wall composition throughout development for the model grass Brachypodium distachyon. Front. Plant Sci. 3:266. 10.3389/fpls.2012.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S., Parsons A. J., Xue H., Liu Q., Jones C. S., Ryan G. D., et al. (2014). Transcript profiling of fructan biosynthetic pathway genes reveals association of a specific fructosyltransferase isoform with the high sugar trait in Lolium perenne. J. Plant Physiol. 171 475–485. 10.1016/j.jplph.2013.12.008 [DOI] [PubMed] [Google Scholar]
- Rauf S., Sienkiewicz-Paderewska D., Malinowski D. P., Hussain M. M., Niazi I. A. K., Kausar M. (2016). “Forages: ecology, breeding objectives and procedures,” in Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits eds Al-Khayri J. M., Jain S. M., Johnson D. V. (Cham: Springer; ), 149–201. 10.1007/978-3-319-22518-0_5 [DOI] [Google Scholar]
- Reynolds C. K. (2000). “Forage evaluation using measurements of energy metabolism,” in Forage Evaluation in Ruminant Nutrition, eds Givens D. I., Owen E., Axford R. F. E., Omed H. M. (Wallingford: CABI Publishing; ). [Google Scholar]
- Riday H., Brummer E. C. (2007). Narrow sense heritability and additive genetic correlations in alfalfa subsp. falcata. J. Iowa Acad. Sci. 114 28–34. [Google Scholar]
- Robins J. G., Bauchan G. R., Brummer E. C. (2007). Genetic mapping forage yield, plant height, and regrowth at multiple harvests in tetraploid alfalfa (Medicago sativa L.). Crop Sci. 47 11–18. 10.2135/cropsci2006.07.0447 [DOI] [Google Scholar]
- Roche J., Turnbull M. H., Guo Q., Novák O., Späth J., Gieseg S. P., et al. (2017). Coordinated nitrogen and carbon remobilization for nitrate assimilation in leaf, sheath and root and associated cytokinin signals during early regrowth of Lolium perenne. Ann. Bot. 119 1353–1364. 10.1093/aob/mcx014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckle M., Meier M., Frey L., Eicke S., Kölliker R., Zeeman S., et al. (2017). Diurnal leaf starch content: an orphan trait in forage legumes. Agronomy 7:16 10.3390/agronomy7010016 [DOI] [Google Scholar]
- Salunkhe D. K., Chavan J. K., Kadam S. S. (2017). “Occurrence, nature, and composition,” in Dietary Tannins : Consequences and Remedies ed. Salunkhe D. K. (Boca Raton, FL: CRC Press; ), 54–60. [Google Scholar]
- Sapkota A. R., Lefferts L. Y., Mckenzie S., Walker P. (2007). What do we feed to food-production animals? A review of animal feed ingredients and their potential impacts on human health. Environ. Health Perspect. 115 663–670. 10.1289/ehp.9760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherr S. J., McNeely J. A. (2008). Biodiversity conservation and agricultural sustainability: towards a new paradigm of ‘ecoagriculture’ landscapes. Philos. Trans. R. Soc. B Biol. Sci. 363 477–494. 10.1098/rstb.2007.2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A., Pausch J., Kuzyakov Y. (2013). C and N allocation in soil under ryegrass and alfalfa estimated by 13C and 15N labelling. Plant Soil 368 581–590. 10.1007/s11104-012-1536-5 [DOI] [Google Scholar]
- Shaw R., Lark R. M., Williams A. P., Chadwick D. R., Jones D. L. (2016). Characterising the within-field scale spatial variation of nitrogen in a grassland soil to inform the efficient design of in-situ nitrogen sensor networks for precision agriculture. Agric. Ecosyst. Environ. 230 294–306. 10.1016/j.agee.2016.06.004 [DOI] [Google Scholar]
- Shinozuka H., Cogan N. O., Spangenberg G. C., Forster J. W. (2012). Quantitative Trait Locus (QTL) meta-analysis and comparative genomics for candidate gene prediction in perennial ryegrass (Lolium perenne L.). BMC Genet. 13:101. 10.1186/1471-2156-13-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skøt L., Sanderson R., Thomas A., Skøt K., Thorogood D., Latypova G., et al. (2011). Allelic variation in the perennial ryegrass FLOWERING LOCUS T gene is associated with changes in flowering time across a range of populations. Plant Physiol. 155 1013–1022. 10.1104/pp.110.169870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavov G. T., Nipper R., Robson P., Farrar K., Allison G. G., Bosch M., et al. (2014). Genome-wide association studies and prediction of 17 traits related to phenology, biomass and cell wall composition in the energy grass Miscanthus sinensis. New Phytol. 201 1227–1239. 10.1111/nph.12621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnowski O., Charcosset A., Joets J. (2012). BioMercator V3: an upgrade of genetic map compilation and quantitative trait loci meta-analysis algorithms. Bioinformatics 28 2082–2083. 10.1093/bioinformatics/bts313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao G. V., Ishikawa T., Ito O., Nakahara K., Wang H. Y., Berry W. L. (2006). A bioluminescence assay to detect nitrification inhibitors released from plant roots: a case study with Brachiaria humidicola. Plant Soil 288 101–112. 10.1007/s11104-006-9094-3 [DOI] [Google Scholar]
- Subbarao G. V., Nakahara K., Hurtado M. P., Ono H., Moreta D. E., Salcedo A. F., et al. (2009). Evidence for biological nitrification inhibition in Brachiaria pastures. Proc. Natl. Acad. Sci. U.S.A. 106 17302–17307. 10.1073/pnas.0903694106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabe L., Wirtz M., Molvig L., Droux M., Hell R. (2010). Overexpression of serine acetlytransferase produced large increases in O-acetylserine and free cysteine in developing seeds of a grain legume. J. Exp. Bot. 61 721–733. 10.1093/jxb/erp338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabe L. M., Droux M. (2002). Limits to sulfur accumulation in transgenic lupin seeds expressing a foreign sulfur-rich protein. Plant Physiol. 128 1137–1148. 10.1104/pp.010935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J. P., Nardi P., Wissuwa M. (2010). Nitrification inhibition activity, a novel trait in root exudates of rice. AoB Plants 2010:plq014. 10.1093/aobpla/plq014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas H., Humphreys M. O. (1991). Progress and potential of interspecific hybrids of Lolium and Festuca. J. Agric. Sci. 117 1–8. 10.1017/S0021859600078916 [DOI] [Google Scholar]
- Thornton P. K. (2010). Livestock production: recent trends, future prospects. Philos. Trans. R. Soc. B Biol. Sci. 365 2853–2867. 10.1098/rstb.2010.0134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorogood D., Yates S., Manzanares C., Skot L., Hegarty M., Blackmore T., et al. (2017). A novel multivariate approach to phenotyping and association mapping of multi-locus gametophytic self-incompatibility reveals S, Z, and other loci in a perennial ryegrass (Poaceae) population. Front. Plant Sci. 8:1331. 10.3389/fpls.2017.01331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D., Reich P. B., Knops J., Wedin D., Mielke T., Lehman C. (2001). Diversity and productivity in a long-term grassland experiment. Science 294 843–845. 10.1126/science.1060391 [DOI] [PubMed] [Google Scholar]
- Tilman D., Reich P. B., Knops J. M. H. (2006). Biodiversity and ecosystem stability in a decade-long grassland experiment. Nature 441 629–632. 10.1038/nature04742 [DOI] [PubMed] [Google Scholar]
- Tubana B. S., Babu T., Datnoff L. E. (2016). A review of silicon in soils and plants and its role in US agriculture: history and future perspectives. Soil Sci. 181 393–411. 10.1097/ss.0000000000000179 [DOI] [Google Scholar]
- Van Elswyk M. E., McNeill S. H. (2014). Impact of grass/forage feeding versus grain finishing on beef nutrients and sensory quality: the U.S. experience. Meat Sci. 96 535–540. 10.1016/j.meatsci.2013.08.010 [DOI] [PubMed] [Google Scholar]
- VanBuren R., Bryant D., Edger P. P., Tang H., Burgess D., Challabathula D., et al. (2015). Single-molecule sequencing of the desiccation-tolerant grass Oropetium thomaeum. Nature 527 508–511. 10.1038/nature15714 [DOI] [PubMed] [Google Scholar]
- Vance C. P., Heichel G. H., Barnes D. K., Bryan J. W., Johnson L. E. (1979). Nitrogen fixation, nodule development, and vegetative regrowth of alfalfa (Medicago sativa L.) following Harvest. Plant Physiol. 64 1–8. 10.1104/pp.64.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdier J., Zhao J., Torres-Jerez I., Ge S., Liu C., He X., et al. (2012). MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula. Proc. Natl. Acad. Sci. U.S.A. 109 1766–1771. 10.1073/pnas.1120916109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel K. P., Pedersen J. F. (1993). “Breeding systems for cross-pollinated perennial grasses,” in Plant Breeding Reviews ed. Janick J. (New York, NY: John Wiley & Sons, Inc; ), 251–274. 10.1002/9780470650035.ch7 [DOI] [Google Scholar]
- Wallace R. J., Onodera R., Cotta M. A. (1997). “Metabolism of nitrogen-containing compounds,” in The Rumen Microbial Ecosystem eds Hobson P. N., Stewart C. S. (Dordrecht: Springer; ), 283–328. 10.1007/978-94-009-1453-7_7 [DOI] [Google Scholar]
- Walter A., Studer B., Kölliker R. (2012). Advanced phenotyping offers opportunities for improved breeding of forage and turf species. Ann. Bot. 110 1271–1279. 10.1093/aob/mcs026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Suo H., Zheng Y., Liu K., Zhuang C., Kahle K. T., et al. (2010). The soybean root-specific protein kinase GmWNK1 regulates stress-responsive ABA signaling on the root system architecture. Plant J. 64 230–242. 10.1111/j.1365-313X.2010.04320.x [DOI] [PubMed] [Google Scholar]
- Wang Z.-Y., Brummer E. C. (2012). Is genetic engineering ever going to take off in forage, turf and bioenergy crop breeding? Ann. Bot. 110 1317–1325. 10.1093/aob/mcs027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. Y., Ge Y. (2005). Agrobacterium-mediated high efficiency transformation of tall fescue (Festuca arundinacea). J. Plant Physiol. 162 103–113. 10.1016/j.jplph.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Warnke E., Barker E., Brilman A., Young C., Cook L. (2002). Inheritance of superoxide dismutase (Sod-1) in a perennial x annual ryegrass cross and its allelic distribution among cultivars. Theor. Appl. Genet. 105 1146–1150. 10.1007/s00122-002-1007-0 [DOI] [PubMed] [Google Scholar]
- Watson B. S., Asirvatham V. S., Wang L., Sumner L. W. (2003). Mapping the proteome of barrel medic Medicago truncatula. Plant Physiol. 131 1104–1123. 10.1104/pp.102.019034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigelt A., Weisser W. W., Buchmann N., Scherer-Lorenzen M. (2009). Biodiversity for multifunctional grasslands: equal production in high-diversity low-input and low-diversity high-input systems. Biogeosciences 6 1675–1706. 10.5194/bg-6-1695-2009 [DOI] [Google Scholar]
- Weimer P. J. (1996). Why don’t ruminal bacteria digest cellulose faster? J. Dairy Sci. 79 1496–1502. 10.3168/jds.S0022-0302(96)76509-8 [DOI] [PubMed] [Google Scholar]
- West J. W., Bernard J. K. (2011). Effects of addition of bacterial inoculants to the diets of lactating dairy cows on feed intake, milk yield, and milk composition. Prof. Anim. Sci. 27 122–126. 10.15232/S1080-7446(15)30458-7 [DOI] [Google Scholar]
- Wilman D., Droushiotis D., Mzamane M. N., Shim J. S. (1977). The effect of interval between harvests and nitrogen application on initiation, emergence and longevity of leaves, longevity of tillers and dimensions and weights of leaves and ‘stems’ in Lolium. J. Agric. Sci. 89 65–79. 10.1017/S0021859600027209 [DOI] [Google Scholar]
- Winters A., Leemans D., Morris S. M., Pippel J., Lovatt A. J., Charlton A., et al. (2010). “High-sugar perennial ryegrass as a feed-stock for bioconversion to platform chemicals,” in Aspects of Applied Biology, Non Food Uses of Crops eds Smith L., Stafford A., Weightman R. (Wellesbourne: Association of Applied Biologists; ), 79–86. [Google Scholar]
- Wood J. D., Richardson R. I., Nute G. R., Fisher A. V., Campo M. M., Kasapidou E., et al. (2004). Effects of fatty acids on meat quality: a review. Meat Sci. 66 21–32. 10.1016/S0309-1740(03)00022-6 [DOI] [PubMed] [Google Scholar]
- Xu G., Fan X., Miller A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63 153–182. 10.1146/annurev-arplant-042811-105532 [DOI] [PubMed] [Google Scholar]
- Yang J., Udvardi M. (2018). Senescence and nitrogen use efficiency in perennial grasses for forage and biofuel production. J. Exp. Bot. 69 855–865. 10.1093/jxb/erx241 [DOI] [PubMed] [Google Scholar]
- Yang J., Worley E., Torres-Jerez I., Miller R., Wang M., Fu C., et al. (2015). PvNAC1 and PvNAC2 are associated with leaf senescence and nitrogen use efficiency in switchgrass. BioEnergy Res. 8 868–880. 10.1007/s12155-014-9566-x 26935010 [DOI] [Google Scholar]
- Yates S. A., Swain M. T., Hegarty M. J., Chernukin I., Lowe M., Allison G. G., et al. (2014). De novo assembly of red clover transcriptome based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genomics 15:453. 10.1186/1471-2164-15-453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.-J., Dong J.-L., Liang B.-G., Jin Y.-S., Wang T. (2006). Highly efficient embryogenesis and plant regeneration of tall fescue (Festuca arundinacea Schreb.) from mature seed-derived calli. In Vitro Cell. Dev. Biol. Plant 42 114–118. 10.1079/ivp2005733 [DOI] [Google Scholar]
- Zheng G., Fan C., Di S., Wang X., Xiang C., Pang Y. (2017). Over-expression of Arabidopsis EDT1 gene confers drought tolerance in alfalfa (Medicago sativa L.). Front. Plant Sci. 8:2125. 10.3389/fpls.2017.02125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Ingram P. A., Benfey P. N., Elich T. (2011). From lab to field, new approaches to phenotyping root system architecture. Curr. Opin. Plant Biol. 14 310–317. 10.1016/j.pbi.2011.03.020 [DOI] [PubMed] [Google Scholar]
- Zhu Y.-G., Pilon-Smits E. A., Zhao F.-J., Williams P. N., Meharg A. A. (2009). Selenium in higher plants: understanding mechanisms for biofortification and phytoremediation. Trends Plant Sci. 14 436–442. 10.1016/j.tplants.2009.06.006 [DOI] [PubMed] [Google Scholar]