Abstract
As adipose tissue is the major cholesterol storage organ and most of the intracellular cholesterol is distributed to lipid droplets (LDs), cholesterol homeostasis may have a role in the regulation of adipocyte size and function. ACATs catalyze the formation of cholesteryl ester (CE) from free cholesterol to modulate the cholesterol balance. Despite the well-documented role of ACATs in hypercholesterolemia, their role in LD development during adipogenesis remains elusive. Here, we identify ACATs as regulators of de novo lipogenesis and LD formation in murine 3T3-L1 adipocytes. Pharmacological inhibition of ACAT activity suppressed intracellular cholesterol and CE levels, and reduced expression of genes involved in cholesterol uptake and efflux. ACAT inhibition resulted in decreased de novo lipogenesis, as demonstrated by reduced maturation of SREBP1 and SREBP1-downstream lipogenic gene expression. Consistent with this observation, knockdown of either ACAT isoform reduced total adipocyte lipid content by approximately 40%. These results demonstrate that ACATs are required for storage ability of lipids and cholesterol in adipocytes.
Keywords: adipocytes, avasimibe, cholesterol metabolism, fatty acid synthesis, triglycerides, acyl-CoA:cholesterol acyltransferase
Adipose tissue is the primary depot for energy storage in a form of triglyceride (TG) within the body. Stored TG is then hydrolyzed to FAs in adipocytes by lipolysis during energy deficiency. Adipocytes are also the primary location for the deposition of unesterified free cholesterol (FC), mostly found in the cholesterol-rich endoplasmic reticulum (ER)-like surface layer of lipid droplets (LDs) as well as plasma membrane (1, 2). Several studies report a positive correlation between intracellular cholesterol level and TG content in adipocytes (1, 3–5). Indeed, cholesterol content in adipocytes appears to be associated with human obesity, as obese humans are reported to store 33–50% of body cholesterol in adipose tissue, while lean humans have about 25% (5). Conversely, altered cholesterol homeostasis results in impairment of systemic energy balance. For example, CD36-deficient mice with a defect in cholesterol uptake are resistant to high-fat diet-induced adipose tissue mass gain and ectopic hepatic lipid accumulation (6). In addition, mice with Niemann-Pick type C1 deficiency were susceptible to high-fat diet-induced weight gain (7).
The majority (>90%) of adipocyte cholesterol is found in the FC form and cholesteryl ester (CE) synthesis has been reported to be significantly low (4). Although intracellular FC is primarily derived from the dietary sources in adipocytes (4, 8), the synthesis of CE is catalyzed by ER resident ACATs/sterol O-acyltransferases (SOATs) (9). ACAT1 is ubiquitously expressed in different tissues to maintain cholesterol homeostasis, whereas ACAT2 is mainly expressed in the liver and intestine. The synthesized CE is then incorporated mostly into the lipid core of very low density lipoprotein, chylomicrons, and/or TG-rich LDs (10). Accumulated evidence suggests an important physiological function of ACATs in hypercholesterolemia and atherosclerosis. ACAT1-deficient mice or wild-type mice with transplanted ACAT1−/− bone marrow cells exhibit severe atherosclerosis (11, 12), while mice with myeloid-specific ACAT1 knockout are protected from atherosclerosis progression (13). Moreover, ACAT2-deficient mice are protected against diet-induced hypercholesterolemia due to a loss of cholesterol esterification activity in the intestine and liver (14). In addition, ACATs appear to play a key role in various diseases, such as cancer (15) and Alzheimer’s disease (16, 17). Despite this significant physiological importance of ACATs, their role in lipid metabolism in adipose tissue is still poorly understood.
Reportedly, the sterol synthesis pathway appears to play an important role in TG synthesis and LD formation in non-adipose tissue. For example, yeast mutants lacking both Lro1 (the yeast ortholog of mammalian LCAT) and Dga1 [the yeast ortholog of mammalian acyl-CoA:diacylglycerol acyltransferase (DGAT)2] are deficient in TG, but still maintain sterol ester-enriched LDs (18, 19). However, additional deletion of Are1 and Are2, the genes encoding sterol acyltransferases, in Lro1ΔDga1Δ yeast mutants abolishes LDs, indicating a key role of the sterol ester biosynthetic pathways in the maturation of LDs in yeast (19–21). Moreover, several studies demonstrate a potential role of cholesterol metabolism in adipocyte lipogenesis: i) genes involved in cholesterol metabolism and accumulation of FC and CE are differentially expressed during adipogenesis (22, 23); ii) altered de novo cholesterol biosynthesis by statin treatment suppresses lipogenesis in adipocytes (24, 25); and iii) perturbation of cholesterol export in adipocytes inhibits lipogenesis and adipocyte lipid storage, lowers adiposity, and increases systemic energy expenditure in vivo (26). Accordingly, we hypothesized that ACAT-regulated CE synthesis and accumulation modulates synthesis and storage of TG in LDs in adipocytes. Here, we show that ACATs are required for de novo lipogenesis and LD formation in adipocytes.
MATERIALS AND METHODS
Reagents
Insulin, dexamethasone, 3-isobutyl-1-methylxanthine, Oil Red O (ORO), Polybrene, free glycerol reagent (#F6428), glycerol standard solution (#G7793), TG reagent (#T2449), and avasimibe (AVA) (>98% of purity) were purchased from Sigma-Aldrich (St. Louis, MO). FCS and FBS were purchased from PAA (Dartmouth, MA). DMEM, penicillin/streptomycin, and sodium pyruvate were from VWR (Radnor, PA). TRIzol® reagent, SuperScriptII, and Lipofectamine 2000 were from Invitrogen (Carlsbad, CA). The 3-(4,5-dimethyl-thiazol-yl-2)-2,5-diphenyl tetrazolium bromide (MTT) was purchased from Alfa Aesar (Ward Hill, MA). The protein assay kit and iTaq™ Universal SYBR® Green Supermix were from Bio-Rad Laboratories (Hercules, CA). The 25-[N-[(7-nitro-2-1,3-benzoxadiazol-4-yl)methyl]amino]-27-norcholesterol (25-NBD-chol) (#810250p) was purchased from Avanti Polar Lipids (Alabaster, AL). Antibodies against SREBP1 (#sc8984) and β-actin and secondary HRP-conjugated mouse antibody were purchased from Santa Cruz Biotechnology (Dallas, TX). Secondary HRP-conjugated rabbit antibody was from the Jackson Laboratory (Bar Harbor, ME). Deuterium-glucose (1,2,3,4,5,6,6-D7, D-glucose, #DLM-2062) was from Cambridge Isotope Laboratories (Tewksbury, MA). The cholesterol assay kit (#K603-100) was from BioVision (Milpitas, CA).
Cell culture and treatment conditions
The 3T3-L1 murine preadipocytes purchased from American Type Culture Collection (Manassas, VA) were cultured and differentiated as described elsewhere (27, 28). The 3T3-L1 preadipocytes were maintained in 10% (v/v) FCS-DMEM with 100 U/ml penicillin/streptomycin and 0.11 g/l sodium pyruvate at 37°C with 5% CO2. After 2 days of postconfluency (designated as day 0), cells were differentiated with DMEM supplemented with 10% FBS and an adipogenic cocktail containing 5 μM dexamethasone, 0.5 mM 3-isobutyl-1-methyl-xanthine, and 167 nM insulin for 2 days (designated as days 0–2). Then, cells were maintained in 10% FBS-DMEM containing 167 nM insulin on days 2–4, followed by culturing in 10% FBS-DMEM. On day 6 or later, mature adipocytes were fixed with 3.7% paraformaldehyde and stained with ORO, as described previously (27). AVA was dissolved in DMSO and added to the cell culture medium such that the final DMSO concentration was less than or equal to 0.1% (v/v).
Cell viability assay
An MTT assay was performed to evaluate the effect of AVA (0–20 μM) on cell viability. The 3T3-L1 adipocytes differentiated in a 24-well plate for 6 days were treated with the indicated AVA concentrations for 48 h. The MTT solution (0.5 mg/ml) was then applied to the cells for 1 h at 37°C with 5% CO2. Precipitated formazan was dissolved in DMSO and quantified at 595 nm using a microplate reader (Beckman-Coulter, Brea, CA).
Cholesterol visualization and quantification
TLC was employed to analyze the lipid profile in adipocytes. Briefly, mature adipocytes (day 8) differentiated with or without AVA (20 μM) during days 4–8 were collected. Lipids were extracted via the Folch method. The extracted lipids were dissolved in chloroform/methanol (2:1 by volume), and 750 μg protein-related lipids were loaded in a small spot on a TLC silica plate (MilliporeSigma, Burlington, MA). The mobile phase was hexane:ether:acetic acid (80:20:1). Lipids were detected with iodine (29). The intensity of the iodine-stained lipids was quantified using ImageJ software (The National Institutes of Health).
The intracellular cholesterol level was quantified by a cholesterol assay kit according to the manufacturer’s protocol. CE was then calculated by subtracting the value of FC from the value of total cholesterol. To visualize the cholesterol in adipocytes, 25-NBD-chol was employed. During adipogenesis (days 0–6), 3T3-L1 cells grown in a 96-well plate were subjected to 10% (v/v) FBS-DMEM containing 25-NBD-chol (1 μg/ml in DMSO) or DMSO control in the presence or absence of AVA (20 μM) for the indicated period of time. After aspirating the medium and rinsing the cells with 1× PBS, the fluorescence intensity in the cells was quantified using a microplate reader (SpectraMax Gemini EM, Molecular Devices, San Jose, CA) at an excitation wavelength of 497 nm and an emission wavelength of 551 nm. Additionally, 25-NBD-chol (1 μg/ml) was added to adipocytes (day 6) in the presence of AVA (20 μM) or DMSO control for 2 h. DNA-binding AT-specific fluorochrome DAPI was used for nuclear staining. Fluorescent cell images were obtained under confocal microscopy (Nikon A1R_MP, Nikon Instruments, Inc., Melville, NY).
Multimodal coherent anti-Stokes Raman scattering microscopy and stimulated Raman scattering imaging
Intracellular LDs in 3T3-L1 adipocytes were visualized by coherent anti-Stokes Raman scattering (CARS)-2-photon excitation fluorescence analysis, as described previously (28). Briefly, the CARS signal of TG in adipocytes was detected with the pump laser and Stokes laser set at 2,840 cm−1. The CARS signal was then processed for visualization via an air condenser (numerical aperture, 0.55), a 600/65 nm band-pass filter, and a photomultiplier tube (H7422-40; Hamamatsu, Japan). Deuterium-labeled glucose was used to visualize and quantify de novo lipogenesis by stimulated Raman scattering (SRS) imaging. The 3T3-L1 preadipocytes were cultured in glass-bottomed dishes (#D35-10-1.5-N; Cellvis Inc.). D-glucose (25 mM) in glucose-free DMEM supplemented with 10% FBS-DMEM and 167 nM insulin was used to differentiate cells during days 2–4 with the indicated treatment. On day 4, cells were washed with 1× PBS and fixed with 10% neutral buffered formalin for 30 min, and then proceeded to SRS imaging, as described previously (30). Briefly, two femtosecond lasers produced from a Ti:Sapphire laser (Chameleon Vision, Coherent) and an optical parametric oscillator were used as pump and Stokes, respectively. The pump wavelength was tuned to 830 nm. The Stokes beam was tuned to 1,090 nm for imaging at a C-H vibration mode around 2,850 cm−1, and 1,005 nm for imaging at C-D vibration mode around 2,120 cm−1. A 40× water-immersion objective lens (UplanSApo, Olympus) was used to focus the laser on the sample. The acquisition time for an image with 512 × 512 pixels was 1.12 s. SRS images were analyzed using ImageJ software. The lipid amount was quantified using the threshold method in ImageJ.
Lentiviral shRNA-mediated knockdown of ACAT1 and ACAT2
Plasmids encoding shRNA for mouse ACAT1 (TRCN0000197536; Open Biosystems) and for mouse ACAT2 (SHCLNG-NM_146064, TRCN0000246787; Sigma-Aldrich) were extracted with QIAGEN HiSpeed Plasmid Midi kit. Lentivirus production was modified from the standard protocol (31). The 293T cells and the third generation of lentivirus packaging plasmids were gifts from Dr. Timothy Ratliff’s laboratory, Purdue University. Briefly, 293T cells were transfected with 10 μg of the pLKO.1-target (pLKO.1-ACAT1, pLKO.1-ACAT2, or pLKO.1-CTRL), 7.5 μg of pMLDg/pRRE, 7.5 μg of pRSV-rev, and 5 μg of pVSV-g using Lipofectamine 2000. After 16 h, fresh 10% FBS-DMEM was applied to the transfected cells and the virus medium was then collected 48 h later. Virus particles were harvested by filtration (0.45 μm pore size) following centrifugation (40,000 g for 2 h). The harvested viruses were employed to infect 3T3-L1 preadipocytes in the presence of 10 μg/ml Polybrene to express ACAT1 or ACAT2 shRNA and silence their target gene expression via RNA interference. Finally, successfully transfected cells were selected by puromycin (1 μg/ml) and differentiated to adipocytes for various analyses. Gene knockdown efficiency was determined by real-time PCR assay.
Immunoblotting
The 3T3-L1 cells cultured in 6-well plates were harvested in cell lysis buffer [50 mM Tris-HCl (pH 7.4), 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl, 1 mM EDTA, protease inhibitor cocktail, 1 mM sodium orthovanadate, and 10 mM sodium fluoride]. These samples were incubated in ice for 30 min and vortexed periodically. After centrifugation at 18,000 g for 1 min, the supernatants were transferred to new tubes for further analysis. The protein concentration was determined by Bradford assay (32) using the Bio-Rad protein assay. Fifty micrograms of protein with loading buffer were loaded in each lane of a 7.5% SDS-PAGE gel and transferred to a polyvinylidene difluoride membrane. Immunoblot was performed with SREBP1 and β-actin antibodies overnight at 4°C followed by secondary HRP-conjugated rabbit and mouse antibodies, respectively. Pierce ECL Plus Western blotting reagents were applied to develop the protein bands and ImageJ software was used to quantify the band intensity. Band intensity of SREBP1 was normalized to the band intensity of β-actin in the same sample lane.
Real-time PCR analysis
Total RNA was extracted from various tissues or cells by TriZol reagent. cDNAs were synthesized using SuperScript II reverse transcriptase and random primers according to the manufacturer’s protocol. Real-time PCR with iTaq™ Universal SYBR® Green Supermix was used to quantify gene expression by StepOne real-time PCR system (Applied Biosystems). The primer sequences are listed in Table 1. Data was normalized to β-actin or ribosomal protein L27 (RPL27, an 18S ribosomal protein), as indicated, and analyzed using the ΔΔCt method.
TABLE 1.
Primer sequences used for real-time PCR
| Genes | Primer Sequence (5′-3′) | |
| Forward | Reverse | |
| ACAT1 | TCGCACTCCTCATCCTATTCG | GTACCAGCCTTCCTTCATCGAT |
| ACAT2 | AGACTTGGTGCAATGGACTCGAC | CATAGGGCCCGATCCAACAG |
| UCP1 | TCATCATCAATTGTACAGAGCTGGTA | CGTCATCTGCCAGTATTTTGTTG |
| Leptin | CACACACGCAGTCGGTATCC | AGCCCAGGAATGAAGTCCAA |
| SREBP1 | GGCACTGAAGCAAAGCTGAAT | GCAAGAAGCGGATGTAGTCGAT |
| PPARr | CCCAATGGTTGCTGATTACAAAT | CTACTTTGATCGCACTTTGGTATTCT |
| FAS | ACCACTGCATTGACGGCCGG | GGGTCAGGCGGGAGACCGAT |
| SCD1 | GGTGATGTTCCAGAGGAGGTACTAC | AGCGTGGGCAGGATGAAG |
| MGAT1 | CTGGTTCTGTTTCCCGTTGT | TGGGTCAAGGCCATCTTAAC |
| DGAT1 | GTGCACAAGTGGTGCATCAG | CAGTGGGATCTGAGCCATCA |
| DGAT2 | GCCGTGTGGCGCTACTTC | GTGGTCAGCAGGTTGTGTGTCT |
| Adipoq | GGAACTTGTGCAGGTTGGAT | GCTTCTCCAGGCTCTCCTTT |
| Resistin | TGCCAGTGTGCAAGGATAGACT | CGCTCACTTCCCCGACAT |
| SR-BI | GGCTGCTGTTTGCTGCG | GCTGCTTGATGAGGGAGGG |
| CD36 | GTTCTTCCAGCCAATGCCTTT | ATGTCTAGCACACCATAAGATGTACAGTT |
| ABCA1 | GGTTTGGAGATGGTTATACAATAGTTGT | TTCCCGGAAACGCAAGTC |
| ABCG1 | AGGTCTCAGCCTTCTAAAGTTCCTC | TCTCTCGAAGTGAATGAAATTTATCG |
| SREBP1a | AGATGTGCGAACTGGACACA | CATCTTTAAAGCAGCGGGTG |
| SREBP1c | GGAGCCATGGATTGCACATT | CATCTTTAAAGCAGCGGGTG |
| SREBP2 | TGTGAACCTGGCCGAGTGT | CGCTGTCAGGTGGATCTCAA |
| β-Actin | AGATGACCCAGATCATGTTTGAGA | CACAGCCTGGATGGCTACGT |
| RPL27 | CCTGGCCGGACGCTACT | AGGTGCCATCGTCAATGTTCT |
UCP1, uncoupling protein 1.
Statistical analysis
Data are presented as mean ± SEM. Statistical analysis was performed using the Student’s two-tailed t-test to compare between groups. One-way ANOVA with Bonferroni post hoc test was performed to analyze the data generated in Figs. 1D, 1E, 3A, and supplemental Fig. S1 with SAS 9.2 (SAS® Institute Inc., Cary, NC). P < 0.05 was considered statistically significant.
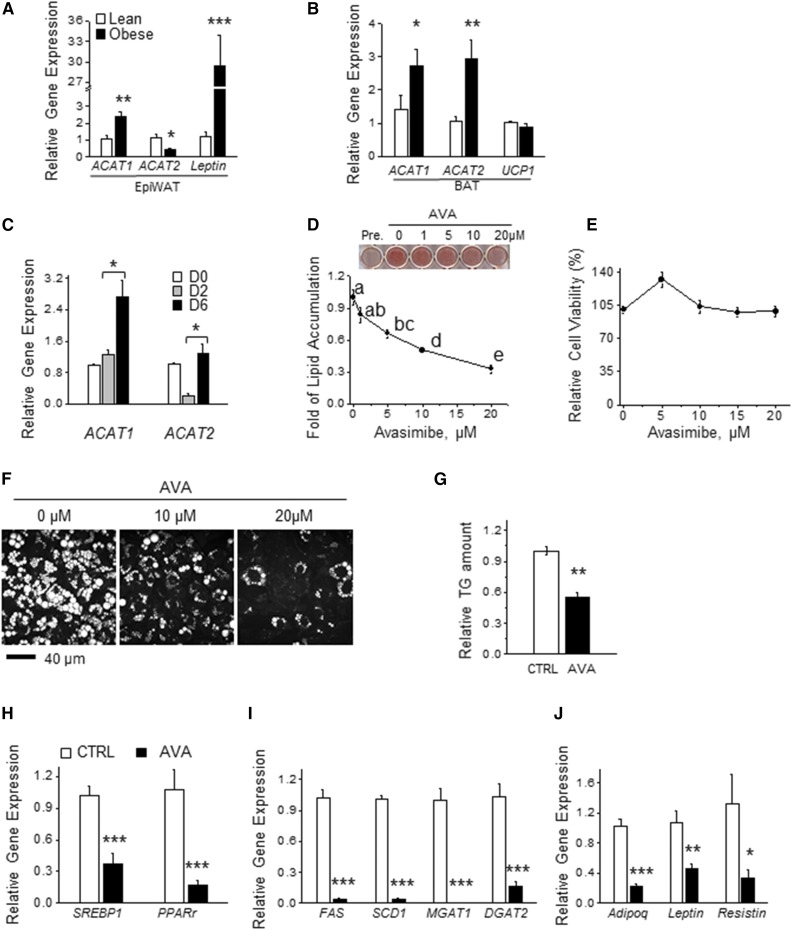
Fig. 1.
Inhibiting ACATs suppresses lipid accumulation in adipocytes during adipogenesis. A, B: mRNA levels of ACAT1 and ACAT2 determined by real-time PCR in epiWAT (A) and BAT (B) from chow diet-fed mice and age-matched DIO mice (male, 13 weeks old, C57BL/6J, Jackson Laboratory). Signals were normalized to RPL27 (n = 4). C: mRNA levels of ACAT1 and ACAT2 in 3T3-L1 cells during adipogenesis as determined by real-time PCR and normalized to β-actin (n = 3, repeated three times). D: ORO staining from adipocytes (day 6) that were differentiated with or without AVA at various concentrations (1–20 μM) for 6 days (n = 3). E: Adipocyte viability upon AVA treatment (0–20 μM) for 48 h was determined by MTT assay (n = 3). F: CARS image analysis of intracellular LDs in adipocytes that were differentiated in the presence or absence of AVA (10 or 20 μM) for 9 days (n = 3). G: Mature adipocytes differentiated with or without AVA (20 μM) during days 4–8 were subjected to TLC. The intensity of the TG spots in TLC was quantified by ImageJ and normalized to control (CTRL) (n = 3). mRNA levels of genes involved in adipogenic transcription program (H), lipid synthesis (I), and adipokine production (J) were determined by real-time PCR with signals normalized β-actin (n = 3). All the data were normalized to control. Data presented are expressed as mean ± SEM. Student’s two-tailed t-test was applied to A–C and G–J. *P < 0.05; **P < 0.01; ***P < 0.001. One-way ANOVA with Bonferroni post hoc test was applied in D and E, and different lowercase letters indicate significant different (P < 0.05).
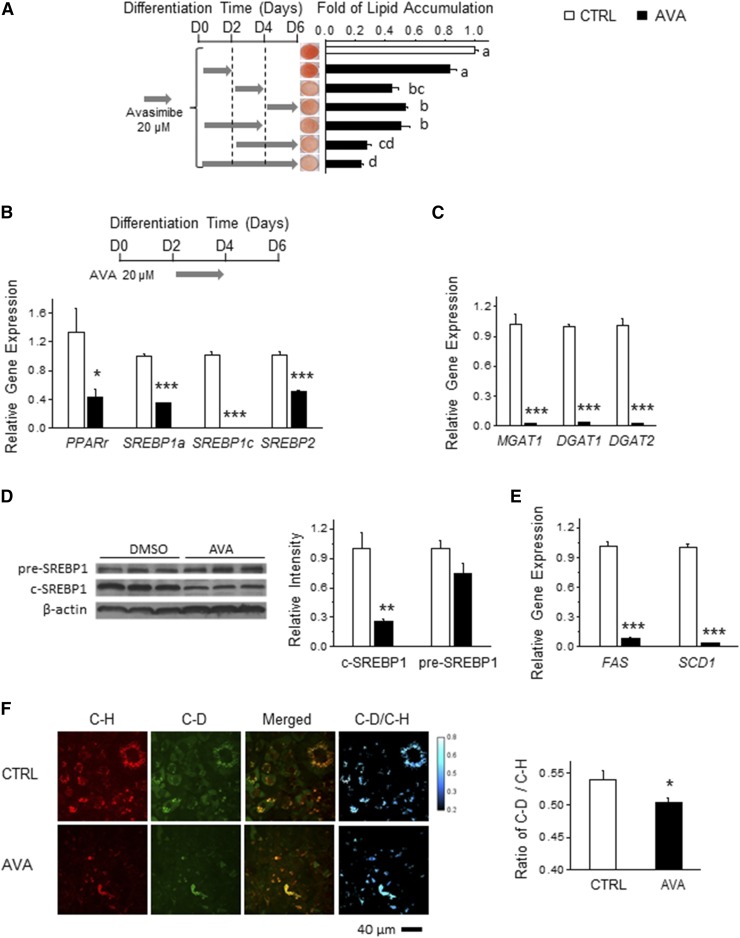
Fig. 3.
ACAT inhibition reduces lipogenic gene expression in adipogenesis through inhibition of SREBP1 processing. A: The 3T3-L1 preadipocytes were differentiated with or without AVA (20 μM) for the indicated period of time. After 6 days of differentiation, ORO staining was performed and the corresponding quantification was calculated to determine the intracellular lipid content. Representative images are presented (n = 3). The 3T3-L1 cells were cocultured with or without AVA (20 μM) for 48 h from day 2 and were harvested to determine mRNA levels of PPARγ, SREBP1a, SREBP1c, and SREBP2 (B) and MGAT1, DGAT1, and DGAT2 (C) (n = 3). Adipocytes differentiated in the presence or absence of AVA (10 μM, during days 2–6) were subjected to immunoblotting to determine SREBP1 level (D) and real-time PCR to determine the mRNA levels of SREBP1 downstream genes (E) (n = 3). F: During adipogenesis, 3T3-L1 cells (day 2) were cultured in medium supplemented with deuterium-glucose (D-Glucose) (25 mM) in the presence or absence of AVA (10 μM) for 2 days. SRS imaging was taken at the carbon-deuterium (C-D) vibration (∼2,120 cm−1) and the carbon-hydrogen (C-H) vibration (∼2,850 cm−1), indicating total lipids and deuterium-incorporated lipids, respectively. ImageJ was used for quantification (n = 3). Data are presented are mean ± SEM. One-way ANOVA with Bonferroni post hoc test was applied to A, and different lowercase letters indicate significant different (P < 0.05). Student’s two-tailed t-test was applied to B–F. *P < 0.05; **P < 0.01; ***P < 0.001.
RESULTS
ACAT inhibition suppresses lipid accumulation in adipocytes
We first examined the expression patterns of ACAT1 and ACAT2 in adipose tissue from lean and diet-induced obese (DIO) mice. ACAT1 mRNA expression was higher (∼2.2-fold) while ACAT2 was lower (∼0.4-fold) in epididymal white adipose tissue (epiWAT) isolated from DIO mice compared with that in age-matched lean mice (Fig. 1A). On the other hand, brown adipose tissue (BAT) from DIO mice had higher mRNA levels of both ACAT1 (∼1.9-fold) and ACAT2 (∼2.8-fold) compared with those in age-matched lean mice (Fig. 1B). These results imply a positive correlation between adipose ACAT1 expression and adiposity in vivo. Next, to determine the role of ACATs in adipocyte function, we examined the mRNA levels of ACAT1 and ACAT2 during adipogenesis of 3T3-L1 murine preadipocytes. While the mRNA level of ACAT1 increased ∼2.2-fold during adipogenesis (Fig. 1C), the ACAT2 mRNA level markedly decreased during the early stage of adipogenesis (Fig. 1C), indicating a positive correlation between ACAT1 expression and adipocyte differentiation in vitro. To further understand the role of ACAT in adipogenesis, we next examined the effect of ACAT inhibition on adipogenesis using AVA (CI-1011), a clinically proven ACAT inhibitor (33). AVA suppressed lipid accumulation in differentiating 3T3-L1 preadipocytes in a dose-dependent manner, with a maximum decrease of 70% at 20 μM AVA, as judged by ORO staining (Fig. 1D). AVA showed little effect on cell viability (Fig. 1E). Consistently, another ACAT inhibitor, CI-976 (34), also blocked lipid accumulation in 3T3-L1 adipocytes (supplemental Fig. S1A) with an undetectable effect on cell viability (supplemental Fig. S1B). Supporting this result, CARS microscopy revealed AVA-induced reduction of LD size and number during adipogenesis (Fig. 1F).
As the majority of lipids found in LDs in adipocytes are TG (3), we tested to determine whether AVA altered adipocyte TG content. Lipids extracted from AVA-treated adipocytes contained about 45% less TG than controls (Fig. 1G), as assessed by TLC. These results suggest that AVA-inhibited lipid accumulation was largely attributed to reduced TG content in adipocyte LDs. AVA treatment also resulted in an approximately 57–96% reduction in mRNA levels of genes involved in the adipogenic transcription program (e.g., PPARγ and SREBP1) (Fig. 1H), lipid synthesis [e.g., FAS, stearoyl-CoA desaturase-1 (SCD1), acyl-CoA:monoacylglycerol acyltransferase (MGAT)1, and DGAT2] (Fig. 1I), and adipokine production (e.g., adiponectin, leptin, and resistin) (Fig. 1J) compared with those in the control group. Taken together, our results indicate that inhibiting ACAT activity suppresses LD formation and adipogenesis in vitro.
ACAT inhibition alters intracellular cholesterol balance in adipocytes
To understand the impact of ACATs on intracellular adipocyte cholesterol levels, we examined the effect of ACAT inhibition on FC and CE levels during adipogenesis. Consistent with a previous study (23), we observed that adipogenesis was associated with an increase in intracellular FC level in 3T3-L1 adipocytes (Fig. 2A). Notably, AVA treatment during adipogenesis significantly suppressed the intracellular levels of FC (Fig. 2A) and CE (supplemental Fig. S2) in adipocytes.
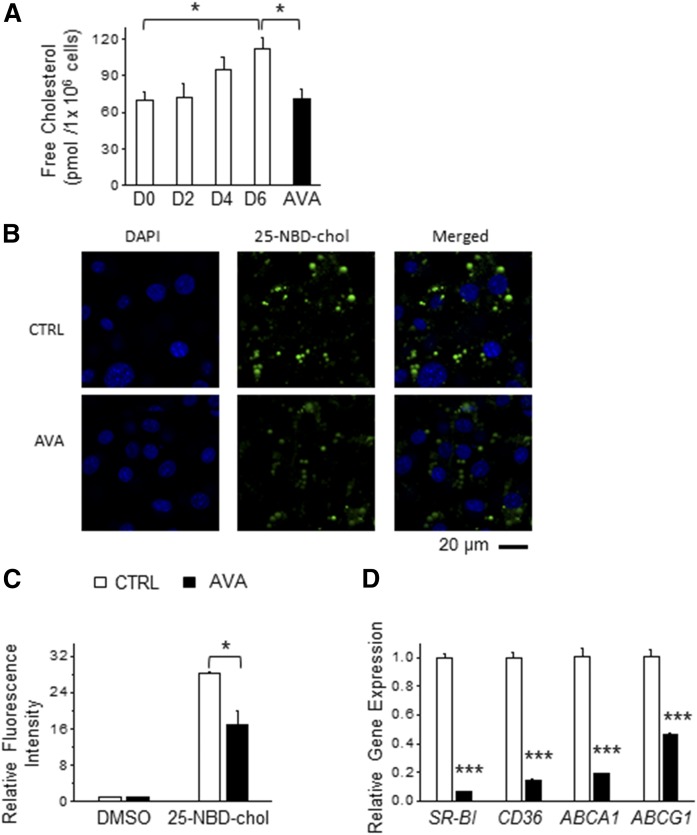
Fig. 2.
ACAT inhibition alters the intracellular cholesterol balance in adipocytes. A: The 3T3-L1 preadipocytes were differentiated in the presence or absence of 20 μM AVA for 6 days and the cells were harvested at various stages of adipogenesis as indicated [day (D)0, D2, D4, and D6] for measurement of FC level (n = 3). B: The 3T3-L1 adipocytes (D6) treated with a fluorescent cholesterol analog (25-NBD-chol, 1 μg/ml) and DAPI in the presence or absence of AVA (20 μM) for 2 h. Representative images by confocal microscopy are presented (n = 3). C: The 3T3-L1 cells (day 2) were cultured in medium supplemented with or without AVA (20 μM) or 25-NBD-chol (1 μg/ml) for 48 h, and then the intracellular fluorescence intensity was quantified (n = 5). Data were normalized to DMSO-treated control. D: mRNA levels of genes involved in cholesterol uptake (SR-BI and CD36) and cholesterol efflux (ABCA1 and ABCG1) were determined by real-time PCR and normalized to β-actin (n = 3). Data presented are the mean ± SEM; *P < 0.05; **P < 0.01; ***P < 0.001.
As cholesterol in adipocytes is largely associated with cellular membranes and LDs and ACATs play a role in cholesterol absorption and intracellular cholesterol homeostasis in nonadipocytes (35, 36), we examined the effect of AVA treatment on incorporation of 25-NBD-chol, a fluorescent cholesterol analog (37), in adipocytes. We found that both 2 h and 48 h of AVA treatment in mature adipocytes suppressed incorporation of 25-NBD-chol in adipocytes (Fig. 2B, C). However, AVA treatment showed no effect on 25-NBD-chol incorporation in differentiating adipocytes when treated during the early stage of adipogenesis (i.e., days 0–2) (data not shown). Moreover, AVA treatment in adipocytes during the late stage of adipogenesis resulted in a marked reduction of mRNA levels of genes involved in cholesterol uptake [scavenger receptor class BI (SR-BI) and CD36] as well as genes in cholesterol efflux (ABCA1 and ABCG1) by 53–94% (Fig. 2D). Taken together, we demonstrate that ACAT inhibition effectively lowered intracellular cholesterol levels in differentiated adipocytes by altering both cholesterol uptake and possibly cholesterol efflux.
ACAT inhibition reduced lipogenic gene expression in adipogenesis through inhibition of SREBP1 processing
To understand the molecular basis underlying the inhibitory effect of ACAT inhibition on LD formation in adipocytes, we first attempted to identify the critical stage of adipogenesis that is specifically targeted by ACAT inhibition in adipocytes. Differentiating 3T3-L1 cells exposed to the adipogenic cocktail were treated with 20 μM AVA at various time points, as illustrated in Fig. 3A. As shown in Fig. 3A, adipocytes incubated with AVA during days 0–2 had similar levels of lipid accumulation as control adipocytes. However, adipocytes treated with AVA during days 2–4, days 2–6, or days 4–6 exhibited more than 50% reduction in intracellular lipid content compared with control adipocytes (Fig. 3A). As expected, adipocytes treated with AVA during days 2–4 displayed reduced mRNA levels of genes involved in lipogenesis (PPARγ, SREBP1a, SREBP1c, and SREBP2) by 57–99% and TG synthesis (MGAT1, DGAT1, and DGAT2) by more than 95% (Fig. 3B, C).
Given the role of SREBPs in the regulation of genes in cholesterol synthesis and uptake, FA synthesis, and TG homeostasis (38), we hypothesized that ACAT inhibition would impair SREBP-regulated de novo lipogenesis. To test this hypothesis, we first investigated the effect of AVA on SREBP1 processing in adipocytes. Compared with nontreated adipocytes, AVA reduced SREBP1 cleavage by ∼70% (Fig. 3D). Consequently, AVA treatment reduced mRNA expression of SREBP1 downstream genes, such as FAS and SCD1, by approximately 96% (Fig. 3E). Additionally, we employed noninvasive SRS microscopy coupled with deuterium-labeled glucose to trace the impact of AVA on de novo lipogenesis in adipocytes (30). This method allowed us to visualize and quantify de novo lipogenesis in LDs as indicated by the C-H bond vibration from existing lipids, and the C-D bond vibration from newly synthesized deuterium-containing lipids. Accordingly, the ratio of the C-D signal over C-H signal indicates the level of de novo lipogenesis. AVA treatment suppressed both LD formation and deuterium-labeled FA production, which effectively lowered de novo lipogenesis by 7.4% (Fig. 3F). Taken together, we found that ACAT inhibition suppressed TG accumulation in adipocytes mainly through reducing de novo lipogenesis in vitro, and in part by abrogating SREBP1 maturation and expression of its downstream lipogenic genes.
ACATs are required for lipid accumulation in adipocytes
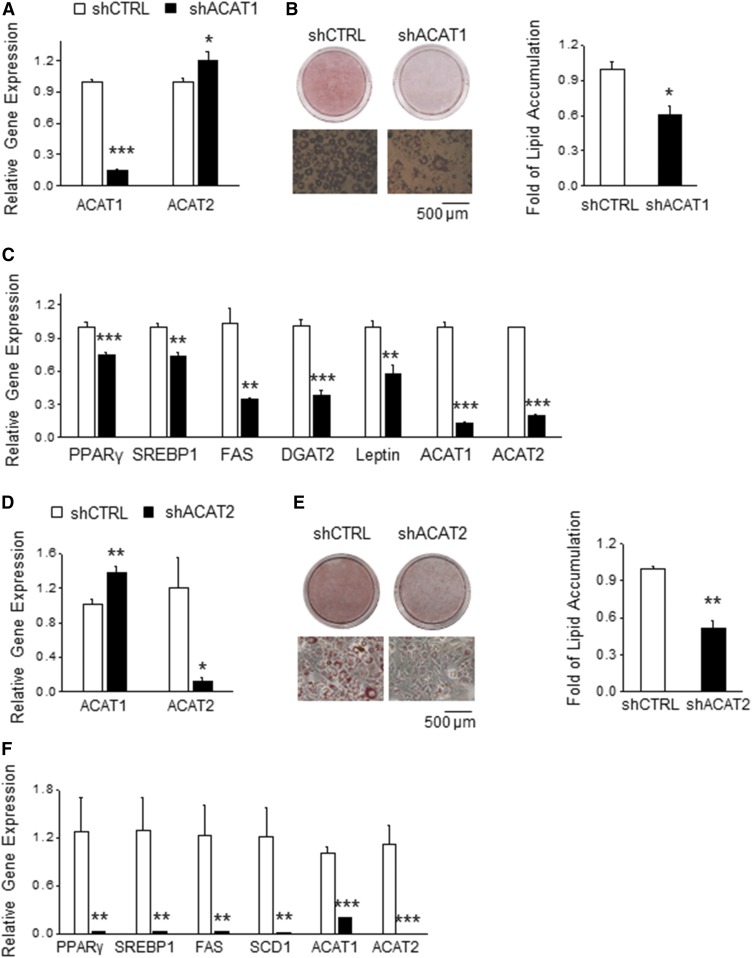
In order to verify the role of ACAT in LD development in adipocytes, we stably knocked down ACAT1 in 3T3-L1 preadipocytes. Knockdown efficiency was 80%, as determined by real-time PCR (Fig. 4A). ACAT1 knockdown slightly increased ACAT2 expression (Fig. 4A), with no effect on DGAT1 and DGAT2 levels (supplemental Fig. S3). After 6 days of differentiation, shACAT adipocytes accumulated 40% less lipids that shCTRL adipocytes (Fig. 4B) and displayed reduced expression of PPARγ and SREBP1 and its downstream genes, FAS and DGAT2 (Fig. 4C). To test the requirement of ACAT2 for LD generation in adipocytes, we stably silenced ACAT2 in 3T3-L1 preadipocytes. This resulted in 80% suppression of ACAT2 expression (Fig. 4D). ACAT2 knockdown slightly increased ACAT1 level (Fig. 4D), but showed no effect on DGAT1 and DGAT2 levels (supplemental Fig. S3). After 6 days of differentiation, shACAT2 adipocytes contained 55% less lipids than the shCTRL adipocytes (Fig. 4E). Consistently, ACAT2 knockdown dramatically reduced mRNA levels of genes involved in TG synthesis (PPARγ, SREBP1, FAS, and SCD1) (Fig. 4F). We also observed a decrease of ACAT1 mRNA level in differentiated shACAT2 adipocytes (Fig. 4F). Collectively, these results implicate that both ACAT genes are required for LD development in adipocytes in vitro.
Fig. 4.
ACAT1 or ACAT2 are required for lipid accumulation. A: Preadipocytes were infected with lentiviral particles carrying either an ACAT1-targeted shRNA (shACAT1) or a scrambled shRNA (shCTRL). mRNA levels of ACAT1 and ACAT2 in ACAT1 knockdown (shACAT1) or control (shCTRL) preadipocytes were measured by real-time PCR (n = 3) to determine the knockdown specificity and efficacy. B: shCTRL and shACAT1 preadipocytes were differentiated to mature adipocytes for 6 days and then stained with ORO to quantify lipid content (n = 3). Representative images are shown. C: Mature adipocytes differentiated from shCTRL and shACAT1 preadipocytes were subjected to real-time PCR to determine the mRNA levels of genes involved in adipogenesis, lipid synthesis, and adipokine production (n = 3). D: mRNA levels of ACAT1 and ACAT2 in ACAT2 knockdown (shACAT2) or control (shCTRL) preadipocytes were measured by real-time PCR (n = 3) to determine the knockdown specificity and efficacy. E: shCTRL and shACAT2 preadipocytes were differentiated to mature adipocytes for 6 days and then stained with ORO to quantify lipid content (n = 3). Representative images are shown. F: Mature adipocytes differentiated from shCTRL and shACAT2 preadipocytes were subjected to real-time PCR to determine mRNA levels of genes involved in adipogenesis and lipid synthesis (n = 3). Data presented are the mean ± SEM and analyzed by Student’s two-tailed t-test. *P < 0.05; **P < 0.01; ***P < 0.001.
DISCUSSION
Adipose tissue is a major cholesterol storage organ (1), and its intracellular cholesterol level is positively correlated with an increase in fat cell size and TG level in adipocytes (3–5). Although FC and CE appear to play an important role in TG synthesis and LD formation, at least in hepatocytes (39) and Saccharomyces cerevisiae (19), the role of ACAT in LD formation in adipocytes remains poorly understood.
Herein, we demonstrated that ACATs are required to maintain intracellular TG and cholesterol levels during adipogenesis. This inhibitory effect of ACATs was, at least in part, mediated by suppressing SREBP1-dependent de novo lipogenesis. Our finding is in agreement with the previous studies of a role of ACAT in TG synthesis in other cell types. In HepG2 cells, pharmacological inhibition of ACAT lowered the synthesis of TG, cholesterol, and CE (40). In obese patients, higher ACAT activity correlated with increased lipid levels in the liver (41). Conversely, silencing ARE1 (the yeast homolog of mammalian ACAT1) in yeast lowered the TG level without changing cholesterol levels (42). We speculate that the effect of ACAT on TG level is partly through modulation of the SREBP1 pathway, as evidenced by AVA-inhibited SREBP1 maturation and expression of SREBP1 target genes. It has been well-established that SREBP1-regulated lipogenesis is largely controlled by the intracellular cholesterol pool of FC and CE (43), reflecting a balance between uptake, efflux, and de novo synthesis. Our study shows that ACAT inhibition resulted in a decrease in intracellular FC level, and FC uptake and efflux in differentiating adipocytes. We also found a decreased intracellular CE level in differentiating adipocytes when treated with AVA (supplemental Fig. S2). These results indicate that ACAT inhibition suppresses the overall intracellular cholesterol pool of FC and CE in adipocytes. Our study is supported by the previously reported role of ACAT in modulating the intracellular cholesterol pool in various cell types: In macrophages (44) and in bone metastasis-derived PC-3 cells (15), ACAT inhibition resulted in increased intracellular FC levels and decreased intracellular CE levels. In neurons, however, blocking ACAT1 activity suppressed CE synthesis without changing intracellular FC levels (16). Locally, CE is formed on the ER membrane where it modulates the function of ER resident ACATs and SREBP1 (38). Thus, it is plausible that our finding of AVA-inhibited SREBP1 function and TG synthesis is likely to be attributed to both an altered local cholesterol balance on the ER membrane and a decreased intracellular cholesterol pool in adipocytes.
There is increasing evidence that cholesterol uptake and efflux are associated with changes in energy metabolism and function of adipocytes. Enhanced cholesterol uptake by upregulating oxidized low density lipoprotein receptor 1 improved FA uptake in adipocytes (45). Additionally, blocking cholesterol transport by NPC2 knockdown impaired autophagy-related mitochondrial function and blunted lipopolysaccharide-stimulated inflammation in adipocytes (46). Moreover, adipocyte-specific ABCA1-deficient mice with impaired cholesterol export displayed reduced lipogenesis, adipocyte lipid storage and adiposity, and increased systemic energy expenditure, thereby impairing diet-induced obesity in vivo (26). In contrast, alteration of cholesterol efflux in epiWAT by administration of ABCG1 shRNA resulted in lowering cellular cholesterol, inflammation, and fat storage (47). Our study identified ACAT as an important regulator in the existing paradigm concerning the role of the intracellular cholesterol pool in the lipogenic ability of adipocytes. Additional work is needed to determine the physiological consequence of the ACAT-regulated intracellular cholesterol pool in adipose tissue to systemic energy balance and metabolism in vivo.
In summary, our study demonstrates that ACAT plays a critical role in regulating the cholesterol and TG storage ability of adipocytes in that ACAT inhibition and ACAT deficiency resulted in suppression of lipogenesis and the intracellular cholesterol pool. These results highlight an important role of ACAT in linking cholesterol metabolism to TG synthesis in adipocytes.
Supplementary Material
Acknowledgments
The authors thank Drs. Ta-Yuan Chang and Catherine C. Y. Chang for their comments on the study design; they appreciate Bindley Bioscience Center for the use of imaging facility; they thank Jonathan Kershaw for editorial assistance; and they appreciate the insightful comments and technical assistance from Kimberly K. Buhman, Scott A. Crist, and Renee Vickman.
Footnotes
Abbreviations:
- AVA
- avasimibe
- BAT
- brown adipose tissue
- CARS
- coherent anti-Stokes Raman scattering
- CE
- cholesteryl ester
- DGAT
- acyl-CoA:diacylglycerol acyltransferase
- DIO
- diet-induced obese
- epiWAT
- epididymal white adipose tissue
- ER
- endoplasmic reticulum
- FC
- free cholesterol
- LD
- lipid droplet
- MGAT
- acyl-CoA:monoacylglycerol acyltransferase
- MTT
- 3-(4,5-dimethyl-thiazol-yl-2)-2, 5-diphenyl tetrazolium bromide
- 25-NBD-chol
- 25-[N-[(7-nitro-2-1,3-benzoxadiazol-4-yl)methyl]amino]-27-norcholesterol
- ORO
- Oil Red O
- RPL27
- ribosomal protein L27
- SCD1
- stearoyl-CoA desaturase-1
- SR-BI
- scavenger receptor class BI
- SRS
- stimulated Raman scattering
- TG
- triglyceride
This work was supported in part by the Purdue Research Foundation (K-H.K.) and a study abroad scholarship from the Chinese Scholar Council (Y.Z.). K-H.K. is a co-founder of EFIL Pharmaceuticals Corporation. The other authors declare no conflicts of interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Farkas J., Angel A., and Avigan M. I.. 1973. Studies on the compartmentation of lipid in adipose cells. II. Cholesterol accumulation and distribution in adipose tissue components. J. Lipid Res. 14: 344–356. [PubMed] [Google Scholar]
- 2.Prattes S., Horl G., Hammer A., Blaschitz A., Graier W. F., Sattler W., Zechner R., and Steyrer E.. 2000. Intracellular distribution and mobilization of unesterified cholesterol in adipocytes: triglyceride droplets are surrounded by cholesterol-rich ER-like surface layer structures. J. Cell Sci. 113: 2977–2989. [DOI] [PubMed] [Google Scholar]
- 3.Le Lay S., Krief S., Farnier C., Lefrere I., Le Liepvre X., Bazin R., Ferre P., and Dugail I.. 2001. Cholesterol, a cell size-dependent signal that regulates glucose metabolism and gene expression in adipocytes. J. Biol. Chem. 276: 16904–16910. [DOI] [PubMed] [Google Scholar]
- 4.Kovanen P. T., Nikkila E. A., and Miettinen T. A.. 1975. Regulation of cholesterol synthesis and storage in fat cells. J. Lipid Res. 16: 211–223. [PubMed] [Google Scholar]
- 5.Schreibman P. H., and Dell R. B.. 1975. Human adipocyte cholesterol. Concentration, localization, synthesis, and turnover. J. Clin. Invest. 55: 986–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vroegrijk I. O., van Klinken J. B., van Diepen J. A., van den Berg S. A., Febbraio M., Steinbusch L. K., Glatz J. F., Havekes L. M., Voshol P. J., Rensen P. C., et al. . 2013. CD36 is important for adipocyte recruitment and affects lipolysis. Obesity (Silver Spring). 21: 2037–2045. [DOI] [PubMed] [Google Scholar]
- 7.Jelinek D., Castillo J. J., Heidenreich R. A., and Garver W. S.. 2015. The C57BL/6J Niemann-Pick C1 mouse model with decreased gene dosage is susceptible to increased weight gain when fed a high-fat diet: Confirmation of a gene-diet interaction. Gene. 568: 112–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tilvis R. S., Kovanen P. T., and Miettinen T. A.. 1978. Release of newly synthesized squalene, methyl sterols and cholesterol from human adipocytes in the presence of lipoproteins. Scand. J. Clin. Lab. Invest. 38: 83–87. [DOI] [PubMed] [Google Scholar]
- 9.Chang C. C. Y., Lee C-Y. G., Chang E. T., Cruz J. C., Levesque M. C., and Chang T-Y.. 1998. Recombinant acyl-CoA:cholesterol acyltransferase-1 (ACAT-1) purified to essential homogeneity utilizes cholesterol in mixed micelles or in vesicles in a highly cooperative manner. J. Biol. Chem. 273: 35132–35141. [DOI] [PubMed] [Google Scholar]
- 10.Chang T. Y., Chang C. C., Lin S., Yu C., Li B. L., and Miyazaki A.. 2001. Roles of acyl-coenzyme A:cholesterol acyltransferase-1 and -2. Curr. Opin. Lipidol. 12: 289–296. [DOI] [PubMed] [Google Scholar]
- 11.Fazio S., Major A. S., Swift L. L., Gleaves L. A., Accad M., Linton M. F., and Farese R. V. Jr. 2001. Increased atherosclerosis in LDL receptor-null mice lacking ACAT1 in macrophages. J. Clin. Invest. 107: 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su Y. R., Dove D. E., Major A. S., Hasty A. H., Boone B., Linton M. F., and Fazio S.. 2005. Reduced ABCA1-mediated cholesterol efflux and accelerated atherosclerosis in apolipoprotein E-deficient mice lacking macrophage-derived ACAT1. Circulation. 111: 2373–2381. [DOI] [PubMed] [Google Scholar]
- 13.Huang L. H., Melton E. M., Li H., Sohn P., Rogers M. A., Mulligan-Kehoe M. J., Fiering S. N., Hickey W. F., Chang C. C., and Chang T. Y.. 2016. Myeloid acyl-CoA:cholesterol acyltransferase 1 deficiency reduces lesion macrophage content and Suppresses atherosclerosis progression. J. Biol. Chem. 291: 6232–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buhman K. K., Accad M., Novak S., Choi R. S., Wong J. S., Hamilton R. L., Turley S., and Farese R. V. Jr. 2000. Resistance to diet-induced hypercholesterolemia and gallstone formation in ACAT2-deficient mice. Nat. Med. 6: 1341–1347. [DOI] [PubMed] [Google Scholar]
- 15.Yue S., Li J., Lee S. Y., Lee H. J., Shao T., Song B., Cheng L., Masterson T. A., Liu X., Ratliff T. L., et al. . 2014. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 19: 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shibuya Y., Chang C. C., and Chang T. Y.. 2015. ACAT1/SOAT1 as a therapeutic target for Alzheimer’s disease. Future Med. Chem. 7: 2451–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhattacharyya R., and Kovacs D. M.. 2010. ACAT inhibition and amyloid beta reduction. Biochim. Biophys. Acta. 1801: 960–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C. F., Natter K., and Kohlwein S. D.. 2009. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 284: 30981–30993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandager L., Gustavsson M. H., Stahl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., and Stymne S.. 2002. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277: 6478–6482. [DOI] [PubMed] [Google Scholar]
- 20.Oelkers P., Cromley D., Padamsee M., Billheimer J. T., and Sturley S. L.. 2002. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 277: 8877–8881. [DOI] [PubMed] [Google Scholar]
- 21.Zweytick D., Leitner E., Kohlwein S. D., Yu C., Rothblatt J., and Daum G.. 2000. Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 267: 1075–1082. [DOI] [PubMed] [Google Scholar]
- 22.Kawamura M., Jensen D. F., Wancewicz E. V., Joy L. L., Khoo J. C., and Steinberg D.. 1981. Hormone-sensitive lipase in differentiated 3T3-L1 cells and its activation by cyclic AMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA. 78: 732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keay S., and Grossberg S. E.. 1980. Interferon inhibits the conversion of 3T3-L1 mouse fibroblasts into adipocytes. Proc. Natl. Acad. Sci. USA. 77: 4099–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan T., Hamilton M. P., Mundy D. I., Chua S. C., and Scherer P. E.. 2009. Impact of simvastatin on adipose tissue: pleiotropic effects in vivo. Endocrinology. 150: 5262–5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeda T., and Horiuchi N.. 2009. Simvastatin suppresses leptin expression in 3T3-L1 adipocytes via activation of the cyclic AMP-PKA pathway induced by inhibition of protein prenylation. J. Biochem. 145: 771–781. [DOI] [PubMed] [Google Scholar]
- 26.Cuffe H., Liu M., Key C. C., Boudyguina E., Sawyer J. K., Weckerle A., Bashore A., Fried S. K., Chung S., and Parks J. S.. Targeted deletion of adipocyte Abca1 (ATP-binding cassette transporter A1) impairs diet-induced obesity. Arterioscler. Thromb. Vasc. Biol. Epub ahead of print. January 18, 2018; doi:10.1161/ATVBAHA.117.309880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim C. Y., Le T. T., Chen C., Cheng J. X., and Kim K. H.. 2011. Curcumin inhibits adipocyte differentiation through modulation of mitotic clonal expansion. J. Nutr. Biochem. 22: 910–920. [DOI] [PubMed] [Google Scholar]
- 28.Kwon J. Y., Seo S. G., Yue S., Cheng J. X., Lee K. W., and Kim K. H.. 2012. An inhibitory effect of resveratrol in the mitotic clonal expansion and insulin signaling pathway in the early phase of adipogenesis. Nutr. Res. 32: 607–616. [DOI] [PubMed] [Google Scholar]
- 29.Uchida A., Slipchenko M. N., Cheng J. X., and Buhman K. K.. 2011. Fenofibrate, a peroxisome proliferator-activated receptor alpha agonist, alters triglyceride metabolism in enterocytes of mice. Biochim. Biophys. Acta. 1811: 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J., and Cheng J. X.. 2014. Direct visualization of de novo lipogenesis in single living cells. Sci. Rep. 4: 6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tiscornia G., Singer O., and Verma I. M.. 2006. Production and purification of lentiviral vectors. Nat. Protoc. 1: 241–245. [DOI] [PubMed] [Google Scholar]
- 32.Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 33.Lee H. T., Sliskovic D. R., Picard J. A., Roth B. D., Wierenga W., Hicks J. L., Bousley R. F., Hamelehle K. L., Homan R., Speyer C., et al. . 1996. Inhibitors of acyl-CoA: cholesterol O-acyl transferase (ACAT) as hypocholesterolemic agents. CI-1011: an acyl sulfamate with unique cholesterol-lowering activity in animals fed noncholesterol-supplemented diets. J. Med. Chem. 39: 5031–5034. [DOI] [PubMed] [Google Scholar]
- 34.Bocan T. M., Mueller S. B., Uhlendorf P. D., Newton R. S., and Krause B. R.. 1991. Comparison of CI-976, an ACAT inhibitor, and selected lipid-lowering agents for antiatherosclerotic activity in iliac-femoral and thoracic aortic lesions. A biochemical, morphological, and morphometric evaluation. Arterioscler. Thromb. 11: 1830–1843. [DOI] [PubMed] [Google Scholar]
- 35.Repa J. J., Buhman K. K., Farese R. V. Jr., Dietschy J. M., and Turley S. D.. 2004. ACAT2 deficiency limits cholesterol absorption in the cholesterol-fed mouse: impact on hepatic cholesterol homeostasis. Hepatology. 40: 1088–1097. [DOI] [PubMed] [Google Scholar]
- 36.Miyazaki A., Kanome T., and Watanabe T.. 2005. Inhibitors of acyl-coenzyme a: cholesterol acyltransferase. Curr. Drug Targets Cardiovasc. Haematol. Disord. 5: 463–469. [DOI] [PubMed] [Google Scholar]
- 37.Mukherjee S., Zha X., Tabas I., and Maxfield F. R.. 1998. Cholesterol distribution in living cells: fluorescence imaging using dehydroergosterol as a fluorescent cholesterol analog. Biophys. J. 75: 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein J. L., DeBose-Boyd R. A., and Brown M. S.. 2006. Protein sensors for membrane sterols. Cell. 124: 35–46. [DOI] [PubMed] [Google Scholar]
- 39.Borradaile N. M., de Dreu L. E., Wilcox L. J., Edwards J. Y., and Huff M. W.. 2002. Soya phytoestrogens, genistein and daidzein, decrease apolipoprotein B secretion from HepG2 cells through multiple mechanisms. Biochem. J. 366: 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taghibiglou C., Van Iderstine S. C., Kulinski A., Rudy D., and Adeli K.. 2002. Intracellular mechanisms mediating the inhibition of apoB-containing lipoprotein synthesis and secretion in HepG2 cells by avasimibe (CI-1011), a novel acyl-coenzyme A: cholesterol acyltransferase (ACAT) inhibitor. Biochem. Pharmacol. 63: 349–360. [DOI] [PubMed] [Google Scholar]
- 41.Sahlin S., Granstrom L., Gustafsson U., Stahlberg D., Backman L., and Einarsson K.. 1994. Hepatic esterification rate of cholesterol and biliary lipids in human obesity. J. Lipid Res. 35: 484–490. [PubMed] [Google Scholar]
- 42.Sandager L., Dahlqvist A., Banas A., Stahl U., Lenman M., Gustavsson M., and Stymne S.. 2000. An acyl-CoA:cholesterol acyltransferase (ACAT)-related gene is involved in the accumulation of triacylglycerols in Saccharomyces cerevisiae. Biochem. Soc. Trans. 28: 700–702. [PubMed] [Google Scholar]
- 43.Brown M. S., and Goldstein J. L.. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 89: 331–340. [DOI] [PubMed] [Google Scholar]
- 44.An S., Jang Y. S., Park J. S., Kwon B. M., Paik Y. K., and Jeong T. S.. 2008. Inhibition of acyl-coenzyme A:cholesterol acyltransferase stimulates cholesterol efflux from macrophages and stimulates farnesoid X receptor in hepatocytes. Exp. Mol. Med. 40: 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chui P. C., Guan H. P., Lehrke M., and Lazar M. A.. 2005. PPARgamma regulates adipocyte cholesterol metabolism via oxidized LDL receptor 1. J. Clin. Invest. 115: 2244–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo H., Zhao M., Qiu X., Deis J. A., Huang H., Tang Q. Q., and Chen X.. 2016. Niemann-Pick type C2 deficiency impairs autophagy-lysosomal activity, mitochondrial function, and TLR signaling in adipocytes. J. Lipid Res. 57: 1644–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frisdal E., Le Lay S., Hooton H., Poupel L., Olivier M., Alili R., Plengpanich W., Villard E. F., Gilibert S., Lhomme M., et al. . 2015. Adipocyte ATP-binding cassette G1 promotes triglyceride storage, fat mass growth, and human obesity. Diabetes. 64: 840–855. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.