Abstract
Human apoE exhibits three major isoforms (apoE2, apoE3, and apoE4) corresponding to polymorphism in the APOE gene. Total plasma apoE concentrations are closely related to these isoforms, but the underlying mechanisms are unknown. We aimed to describe the kinetics of apoE individual isoforms to explore the mechanisms for variable total apoE plasma concentrations. We used LC-MS/MS to discriminate between isoforms by identifying specific peptide sequences in subjects (three E2/E3, three E3/E3, and three E3/E4 phenotypes) who received a primed constant infusion of 2H3-leucine for 14 h. apoE concentrations and leucine enrichments were measured hourly in plasma. Concentrations of apoE2 were higher than apoE3, and concentrations of apoE4 were lower than apoE3. There was no difference between apoE3 and apoE4 catabolic rates and between apoE2 and apoE3 production rates (PRs), but apoE2 catabolic rates and apoE4 PRs were lower. The mechanisms leading to the difference in total plasma apoE concentrations are therefore related to contrasted kinetics of the isoforms. Production or catabolic rates are differently affected according to the specific isoforms. On these grounds, studies on the regulation of the involved biochemical pathways and the impact of pathological environments are now warranted.
Keywords: peptide, liquid chromatography, tandem mass spectrometry, stable isotope tracers, lipoprotein/kinetics, lipoprotein/metabolism
apoE plays a key role in lipoprotein metabolism (especially triglyceride-rich lipoproteins) and as a ligand for the LDL receptor (LDLR), the LDLR-related protein (LRP), and heparan sulfate proteoglycans (HSPGs) (1–3). Human apoE is a 299 amino acid protein mostly expressed by the liver and the brain. The APOE gene is localized on chromosome 19 and exhibits three common alleles (ε2, ε3, and ε4) coding for three isoforms (apoE2, apoE3, and apoE4) that differ by single cysteine (C)-arginine (R) substitutions. apoE3 (C112, R158) is the most common isoform in 50–90% of the population. apoE2 (C112, C158) and apoE4 (R112, R158) are less frequent and found in 1–15% and 5–35% of the population, respectively (1, 2). Six phenotypes are found in humans: three homozygotes (E4/E4, E3/E3, and E2/E2) and three heterozygotes (E3/E4, E2/E3, and E2/E4) (2).
The LDLR, LRP, and HSPG binding functions of apoE2 are reduced compared with apoE3. This may lead, in the homozygote E2/E2 state, to type III combined hyperlipoproteinemia and increased CVD risk. In contrast, apoE4 and apoE3 show similar affinities for those receptors. apoE4 has been associated with increased CVD risk and also appears to be a strong genetic determinant for Alzheimer’s disease (1, 3–5). apoE plasma concentrations are closely related to APOE genotypes. Carriers of at least one ε2 or one ε4 allele respectively present with higher and lower plasma apoE levels than ε3/ε3 homozygotes (6–8). The mechanisms underlying these differences are still unknown.
Lipoprotein turnover can be assessed in vivo by measuring the incorporation of an injected tracer, usually 2H3-leucine, in apolipoproteins over time, allowing the determination of lipoprotein kinetic parameters such as their production rates (PRs) and fractional catabolic rates (FCRs) (9). This approach has been improved by new analytical techniques involving enzymatic proteolysis and LC-MS/MS (10, 11). LC-MS/MS is a powerful tool to simultaneously quantify several plasma proteins even at low concentrations (12, 13). This technique also allows the determination of protein polymorphisms (2, 14).
An LC-MS/MS method was recently developed to quantify apoE isoforms in both human plasma and cerebrospinal fluid (2). In this study, plasma apoE2 was more abundant than apoE3, and apoE3 more abundant than apoE4 in patients with ε2/ε3 and ε3/ε4 genotypes, respectively (7). To investigate why apoE isoform concentrations differ in vivo, we measured the isotopic enrichment of apoE isoforms in whole plasma and in the lipoproteins to determine their kinetics in a series of ε2/ε3, ε3/ε3, and ε3/ε4 patients who received a primed constant infusion of 2H3-leucine.
MATERIALS AND METHODS
Reagents and apparatus
UPLC/MS-grade acetonitrile, water, methanol, and 99% formic acid were purchased from Biosolve (Valkenswaard, The Netherlands). The 2H3-leucine was obtained from Cambridge Isotope Laboratories Inc. (Andover, MA). Ammonium bicarbonate was obtained from Sigma-Aldrich (Saint-Quentin Fallavier, France). Synthetic labeled and unlabeled peptides were purchased from Thermo Scientific Biopolymers (Darmstadt, Germany). Stock solutions of synthetic peptides were prepared at 1 mmol/l in 50% acetonitrile containing 0.1% formic acid and stored at −20°C until use. LC-MS/MS analyses were performed on a Xevo® TQD mass spectrometer with an ESI interface and an Acquity H-Class® UPLC™ device (Waters Corporation, Milford, MA). Data acquisition and analyses were performed with MassLynx® and TargetLynx® software, respectively (version 4.1; Waters Corporation).
Subjects and infusion protocol
Nine overweight male subjects (three ε2/ε3, three ε3/ε3, and three ε3/ε4; 49 ± 11 years old; BMI of 29 ± 3 kg/m2) with hypertriglyceridemia (plasma triglycerides: 248 ± 70 mg/dl) were included in this study. They did not receive any treatment. After an overnight fast, each subject received an intravenous bolus of 10 μmol/kg 2H3-leucine immediately followed by a constant intravenous infusion at 10 μmol/kg/h for 14 h. Blood samples were collected hourly in EDTA tubes (Venoject, Paris, France), and the plasma was separated by centrifugation at 4°C for 30 min and stored at −80°C until use. The Ethics Committee of Nantes University Hospital approved the clinical protocols, and written informed consent was obtained from each subject (trial numbers: NCT01216956 and V00002CA101).
Biochemical measurements
Cholesterol and triglyceride concentrations were measured using enzymatic kits from Boehringer Mannheim GmbH (Mannheim, Germany) according to the supplier’s instructions. Proprotein convertase subtilisin kexin type 9 (PCSK9) concentrations were measured in plasma by enzyme-linked immunosorbent assay according to the supplier’s instructions (R&D Systems, Lille, France).
Isolation of lipoproteins
Plasma lipoprotein fractions, including VLDL, IDL, LDL, and HDL, were separated by fast protein LC (FPLC) or by sequential ultracentrifugation methods (15, 16). Total cholesterol and triglyceride contents were measured in each FPLC fraction. FPLC fractions corresponding to the same lipoprotein class were pooled. Lipoprotein fractions (2 ml for FPLC, 800 μl for ultracentrifugation) were desalted and concentrated with 3 ml of 50 mmol/l ammonium bicarbonate buffer (pH 8) using a 5 kDa molecular mass cut-off filter for apolipoprotein enrichment measurements.
Sample preparation and proteolytic digestion
Apolipoproteins (apoA-I, apoB100, apoC-II, apoC-III, and apoE) were analyzed in plasma, lipoprotein fractions, and concentrated lipoprotein fractions using a validated multiplexed assay involving trypsin proteolysis and the subsequent analysis of proteotypic peptides by LC-MS/MS (10). The method was updated for the quantification of apoE isoforms as described previously (2). A pool solution of unlabeled synthetic peptides (M0; Table 1) was constituted and serially diluted in water to obtain seven standard solutions ranging from 0.5 to 50 μmol/l (apoA-I), from 0.25 to 25 μmol/l (apoB100, apoC-II, and apoC-III), and from 0.1 to 10 μmol/l (apoE and isoforms). Plasma, lipoprotein, and standard samples (60 μl) were reduced [addition of 120 μl of ammonium bicarbonate (50 mmol/l) containing 7 mg/ml of RapidGest detergent (Waters Corporation) and incubation for 10 min at 80°C; then addition of dithiothreitol (100 mmol/l, 20 μl) and incubation for 20 min at 60°C], alkylated [addition of iodoacetamide (200 mmol/l, 20 μl) and incubation for 20 min at room temperature in the dark], and trypsin digested overnight [5 mg/ml in HCl (1 mmol/l, 30 μl) at 37°C] using the ready-to-use solutions of the ProteinWorks™ eXpress kit (Waters Corporation) according to the manufacturer’s instructions. Labeled proteotypic peptides (Table 1) were used as internal standards (ISs) and a mixed solution of standards was added to the digestion buffer to a final concentration of 0.5 μmol/l. After digestion, samples were cleaned using 30 mg Oasis HLB 1 cc cartridges (Waters Corporation). Cartridges were conditioned, equilibrated, loaded, washed, and eluted with methanol (1 ml), water (1 ml), samples (∼250 μl), 5% methanol (1 ml), and 80% methanol (500 μl), respectively. Eluates were dried under a nitrogen stream, reconstituted with 100 μl of 5% acetonitrile containing 0.1% formic acid, and 10 μl were injected into the LC-MS/MS system.
TABLE 1.
Analytical parameters used for each proteotypic peptide
| Protein | Name | Peptide | Fragment | Cone/Collision (V) | MRM (m/z) |
| apoA-I | M0 | ATEHLSTLSEK | y102+ | 25/15 | 406.2 → 573.2 |
| IS | ATEHLSTLSE-[13C615N4]R | y102+ | 25/15 | 408.9 → 577.2 | |
| apoB100 | M0 | ATGVLYDYVNK | y6+ | 34/23 | 622.4 → 915.6 |
| M3 | ATGVLYDYVNK | y6+ | 34/23 | 623.9 → 915.6 | |
| IS | ATGVLYDYVN-[13C615N2]K | y6+ | 34/23 | 626.4 → 923.6 | |
| apoC-II | M0 | TAAQNLYEK | y7+ | 35/20 | 519.7 → 865.7 |
| IS | TAAQNLYE-[13C615N2]K | y7+ | 35/20 | 523.7 → 873.7 | |
| apoC-III | M0 | GWVTDGFSSLK | y8+ | 40/35 | 598.2 → 854.1 |
| IS | GWVTDGFSSL-[13C615N2]K | y8+ | 40/35 | 602.2 → 862.1 | |
| apoE | M0 | LGPLVEQGR | y5+ | 25/30 | 484.8 → 588.3 |
| M3 | LGPLVEQGR | y5+ | 25/30 | 486.3 → 588.3 | |
| IS | LGPLVEQG-[13C615N4]R | y5+ | 25/30 | 489.8 → 598.3 | |
| apoE2 | M0 | [C]LAVYQAGAR | y8+ | 40/22 | 555.2 (527.7) → 835.7 |
| M3 | [C]LAVYQAGAR | y8+ | 40/22 | 556.7 (529.3) → 835.7 | |
| IS | [C]LAVYQAGA-[13C615N4]R | y8+ | 40/22 | 560.2 → 845.7 | |
| apoE4 | M0 | LGAD[M]EDVR | y8+ | 35/20 | 503.6 (511.6) → 892.6 (908.6) |
| M3 | LGADMEDVR | y8+ | 35/20 | 505.1 (513.1) → 892.6 (908.6) | |
| IS | LGADMEDV-[13C615N4]R | y8+ | 35/20 | 508.6 → 902.6 | |
| apoE2/E3 | M0 | LGADMEDV[C]GR | y6+ | 35/20 | 612.0 → 735.6 |
| M3 | LGADMEDV[C]GR | y6+ | 35/20 | 613.5 → 735.6 | |
| IS | LGADMEDV[C]G-[13C615N4]R | y6+ | 35/20 | 612.0 → 735.6 | |
| apoE3/E4 | M0 | LAVYQAGAR | y7+ | 40/22 | 475.0 → 764.7 |
| M3 | LAVYQAGAR | y7+ | 40/22 | 476.5 → 764.7 | |
| IS | LAVYQAGA-[13C615N4]R | y7+ | 40/22 | 480.0 → 774.7 |
Parentheses indicate secondary transitions that may occur. [C] indicates carbamydomethyl-cysteine (+57) and [M] indicates oxidized methionine. Underlined L indicates the putative incorporation site(s) of 2H3-leucine. M0, unlabeled peptide; M3, 2H3-leucine labeled peptide; MRM, multiple reaction monitoring.
Analytical parameters
Apolipoprotein analyses were carried out by LC-MS/MS. Proteotypic peptides were separated over 9 min on an Acquity® BEH C18 column (2.1 × 100 mm, 1.7 μm; Waters Corporation) held at 60°C with a linear gradient of mobile phase B (100% acetonitrile) in mobile phase A (5% acetonitrile), each containing 0.1% formic acid, at a flow rate of 600 μl/min. Mobile phase B was linearly increased from 1% to 50% for 5 min, kept constant for 1 min, returned to the initial condition over 1 min, and kept constant for 2 min before the next injection. Proteotypic peptides were then detected by the mass spectrometer with the ESI interface operating in the positive ion mode (capillary voltage, 3 kV; desolvatation gas (N2) flow and temperature, 900 l/h and 400°C; source temperature, 150°C). The multiple reaction monitoring mode was applied for MS/MS detection as detailed in Table 1.
apoE genotype validation
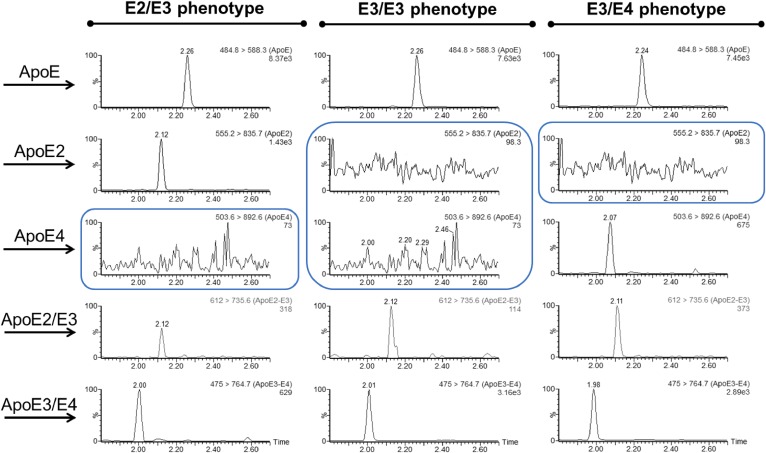
apoE genotypes were confirmed by LC-MS/MS in plasma samples according to the presence of different combinations of peptides (2) and illustrated in Fig. 1: E2/E2 phenotype (LGADMEDVCGR, CLAVYQAGAR), E2/E3 phenotype (LGADMEDVCGR, CLAVYQAGAR, LAVYQAGAR), E2/E4 phenotype (LGADMEDVCGR, CLAVYQAGAR, LGADMEDVR, LAVYQAGAR), E3/E3 phenotype (LGADMEDVCGR, LAVYQAGAR), E3/E4 phenotype (LGADMEDVCGR, LAVYQAGAR, LGADMEDVR), and E4/E4 phenotype (LGADMEDVR, LAVYQAGAR).
Fig. 1.
Selection of signature peptides from apoE sequences for isoform analysis. Phenotype identification and apoE isoform concentrations were assessed using different combinations of peptides. Both apoE2 and apoE4 carry a single specific peptide (CLAVYQAGAR and LGADMEDVR, respectively) unlike apoE3. For enrichment measurements in heterozygous patients, LAVYQAGAR and LGADMEDVCGR were used for apoE3 kinetics in E2/E3 and E3/E4 phenotypes, respectively. In homozygous E3/E3 patients, enrichments of LAVYQAGAR and LGADMEDVCGR were averaged. Blue indicates cysteine-arginine interchanges between isoforms.
Apolipoprotein quantification
Chromatographic peak area ratios between unlabeled peptides (M0) and their respective ISs constituted the detector responses. Standard solutions were used to plot calibration curves for peptide quantification. The linearity was expressed by the mean r2, which was greater than 0.985 for all peptides (linear regression, 1/x weighting, origin excluded). Each sample was assayed three times and the coefficients of variation did not exceed 11.3% for all peptides in all samples. Apolipoprotein concentrations were expressed in micromoles per liter, assuming 1 mol of peptide equivalent to 1 mol of protein. Concentrations were then converted to their standard unit (milligrams per deciliter) assuming a molecular mass of 28,079, 512,858, 8,204, 8,765, and 34,237 g/mol for apoA-I, apoB100, apoC-II, apoC-III, and apoE, respectively (www.uniprot.org). For the quantification of apoE isoforms, specific CLAVYQAGAR and LGADMEDVR peptides were used for apoE2 and apoE4, respectively. Unlike apoE2 and apoE4, the apoE3 isoform does not display any specific peptide. apoE3 concentration was therefore calculated by subtracting the concentrations measured for apoE2 (E2/E3 phenotype) or apoE4 (E3/E4 phenotype) from the total apoE (LGPLVEQGR) concentration. The common peptides of apoE2/E3 (LGADMEDVCGR) and apoE3/E4 (LAVYQAGAR) were used to confirm these apoE3 concentrations with acceptance criteria set at a maximum of 10% of variation between both approaches (2). Chemical modifications that might occur within some peptides were taken into account (secondary MRM transitions shown within parentheses, Table 1) and integrated to determine the exact concentrations of each apoE isoform (2).
Enrichments of apoE isoforms
2H3-leucine enrichments were assessed in apoE isoforms in plasma and concentrated lipoprotein fractions, as previously validated (10, 14). Enrichments were calculated as described previously from unlabeled (M0) and 2H3-leucine-labeled (M3) peptides (10, 17). Briefly, the isotope ratio (IR), corresponding to the M3/M0 percent ratio (%), was divided by the number of leucine residues in the peptide sequence. After baseline subtraction, the IR was converted to enrichment as follows: enrichment = (IR × 100) ÷ (100 + IR). Both apoE2 and apoE4 kinetics were investigated from their respective signature peptides (CLAVYQAGAR and LGADMEDVR, respectively). To minimize variability, two peptides located in the same areas were used for apoE3 kinetics, as illustrated in Fig. 1 (LAVYQAGAR for E2/E3 phenotype, LGADMEDVCGR for E3/E4 phenotype, and the average of both LAVYQAGAR and LGADMEDVCGR for the E3/E3 phenotype). Apolipoprotein enrichment measurements were performed on three replicates for all kinetic time points: coefficients of variation did not exceed 12.6%.
Enrichments of total apoE
Total apoE kinetics were investigated in plasma and concentrated lipoprotein fractions by the use of the common LGPLVEQGR peptide, as previously described and validated (10). Apolipoprotein enrichment measurements were performed on three replicates for all kinetic time points: coefficients of variation did not exceed 7.1%.
Precursor pool
The 2H3-leucine enrichments were investigated in VLDL apoB100 (10). Enrichment measurements were performed on three replicates for all kinetic time points and coefficients of variation did not exceed 5.1%.
Kinetic parameters
Kinetic analysis was achieved using the Simulation, Analysis, and Modeling II software (SAAM II; Epsilon Group, Charlottesville, VA). The labeling of apoE nearly reached the asymptotic maximal enrichment (precursor pool), which suggested a relatively rapid turnover over the time course of the study (17). Fractional synthetic rates (FSRs) were estimated using the following mono-exponential equation: protein labelingtime = protein labelingsteady-state × [1 − e−FSR×(time-delay)] (17). The protein labeling at steady state (precursor pool) was assumed to be close to that of a surrogate protein with fast turnover (i.e., VLDL apoB100, supplemental Fig. S1), and the delay parameter was set adjustable (0.01–1.00 h) for calculation. As expected (fasting state), apolipoprotein pool sizes were considered constant, as no significant variation was observed in apoE concentrations during the time course of the kinetic study. PRs were calculated as the product of the FSR and the average apoE concentration by assuming a plasma volume of 4.5% of body weight. At steady-state, the FCR is equal to the FSR.
Validation of kinetic parameters
Total apoE FCR was deduced from those of specific isoforms with the following equation: FCRTotal = [(Q1 × k1) + (Q2 × k2)] ÷ (Q1 + Q2). Pool sizes of isoforms 1 and 2 are expressed by Q1 and Q2, respectively, and the FCRs of isoforms 1 and 2 are expressed by k1 and k2, respectively. Kinetic parameters of total apoE were also investigated by the use of the common LGPLVEQGR peptide (10). Kinetic parameters obtained from both approaches (i.e., sum of isoforms vs. total apoE) were then compared.
apoE distribution within lipoproteins
Concentrations of apoE isoforms were measured in concentrated samples (60 μl) taking into account the concentration factor. Total apoE, apoA-I (HDL), and apoB100 (VLDL/IDL/LDL) contents were measured simultaneously.
Statistical analyses
Results are expressed as mean ± standard deviation. The nonparametric Spearman correlation test was carried out with GraphPad Prism software (version 6.0; GraphPad Software Inc., La Jolla, CA) and results were considered statistically significant at P < 0.05.
RESULTS
apoE genotype validation
apoE genotypes were confirmed by LC-MS/MS in the nine subjects according to the presence or the absence of proteotypic peptides (Fig. 2). Although we used a limited number of patients, precluding adequate statistical analyses, the lipid/lipoprotein/apolipoprotein levels (Table 2) between E2/E3, E3/E3, and E3/E4 groups were similar, including the parameters PCSK9, apoB100, LDL cholesterol, triglyceride, apoC-II, and apoC-III. In contrast, apoE plasma concentrations appeared quite different between groups: 7.2 ± 1.1 mg/dl for E2/E3, 3.8 ± 0.7 mg/dl for E3/E3, and 3.1 ± 0.4 mg/dl for E3/E4. As shown in Fig. 3, E3/E3 patients displayed nearly twice as much apoE3 (3.8 ± 0.7 mg/dl) as E2/E3 (2.1 ± 0.8 mg/dl) and E3/E4 (2.1 ± 0.5 mg/dl) patients. In E2/E3 individuals, the apoE2 plasma concentration was higher than apoE3 (5.1 ± 0.3 mg/dl vs. 2.1 ± 0.8 mg/dl, respectively). In E3/E4 individuals, apoE4 plasma concentrations were lower than apoE3 (1.0 ± 0.1 mg/dl vs. 2.1 ± 0.5 mg/dl, respectively).
Fig. 2.
Identification of apoE phenotypes by selective combination of proteotypic peptides. LC-MS/MS chromatograms obtained in plasma from representative subjects.
TABLE 2.
Clinical and biochemical characteristics of patients
| Parameters | E2/E3 | E3/E3 | E3/E4 |
| Number | 3 | 3 | 3 |
| Age (years) | 51 ± 5 | 45 ± 15 | 44 ± 9 |
| BMI (kg/m2) | 31 ± 3 | 27 ± 3 | 28 ± 4 |
| TC (mg/dl) | 193 ± 38 | 209 ± 28 | 231 ± 9 |
| TG (mg/dl) | 207 ± 43 | 290 ± 80 | 228 ± 47 |
| HDL-C (mg/dl) | 41 ± 5 | 41 ± 12 | 40 ± 5 |
| LDL-C (mg/dl) | 114 ± 28 | 116 ± 28 | 150 ± 8 |
| apoA-I (mg/dl) | 128 ± 27 | 139 ± 28 | 138 ± 14 |
| apoB100 (mg/dl) | 105 ± 18 | 119 ± 17 | 116 ± 4 |
| apoC-II (mg/dl) | 4 ± 1 | 4 ± 2 | 4 ± 2 |
| apoC-III (mg/dl) | 19 ± 5 | 27 ± 7 | 22 ± 4 |
| apoE (mg/dl) | 7.2 ± 1.1 | 3.8 ± 0.7 | 3.1 ± 0.4 |
| PCSK9 (ng/ml) | 300 ± 136 | 369 ± 170 | 298 ± 65 |
Values are expressed as mean ± standard deviation. TC, total cholesterol; TG, triglycerides; HDL-C, HDL cholesterol; LDL-C, LDL cholesterol.
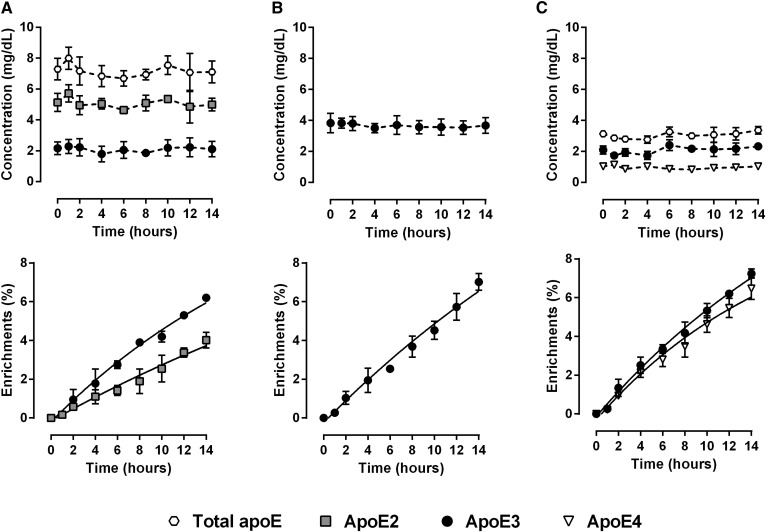
Fig. 3.
Kinetics of plasma apoE isoforms. Mean plasma concentrations and mean changes in 2H3-leucine incorporation over the course of the tracer infusion in subjects with E2/E3 phenotype (A), E3/E3 phenotype (B), and E3/E4 phenotype (C). Values are presented as mean ± standard deviation (n = 3). Total apoE concentrations are shown as indicative in heterozygote patients.
Kinetics of whole plasma apoE isoforms
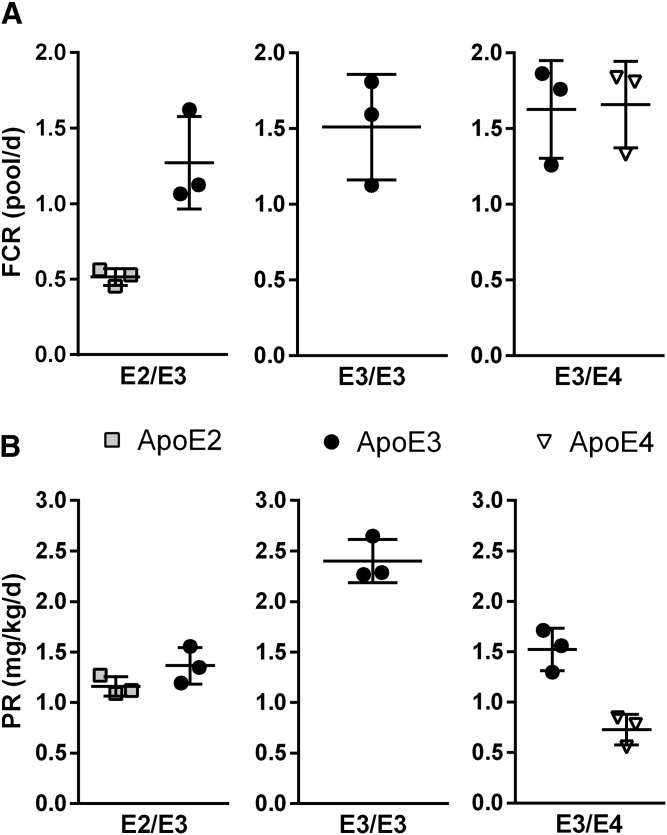
Whole plasma enrichment curves of apoE3 in tracer over time were similar in E2/E3, E3/E3, and E3/E4 patients (Fig. 3). Tracer enrichments of apoE2 were nearly half of those of apoE3 measured in E2/E3 patients (Fig. 3A). Enrichment of apoE4 in tracer was slightly less than that of apoE3 measured in E3/E4 patients (Fig. 3C). Of note, we did not observe any marked analytical biases for enrichment measurements obtained from both LAVYQAGAR and LGADMEDVCGR peptides (supplemental Fig. S2). The FCRs of apoE3 were similar in E2/E3 (1.27 ± 0.31 pool/day), E3/E3 (1.51 ± 0.35 pool/day), and E3/E4 (1.63 ± 0.32 pool/day) subjects (Fig. 4). In E3/E4 patients, the apoE4 FCR (1.67 ± 0.29 pool/day) was in the same range as that of apoE3 (1.63 ± 0.32 pool/day). In E2/E3 individuals, the apoE2 FCR (0.51 ± 0.05 pool/day) was lower than apoE3 (1.27 ± 0.31 pool/day). The PR of apoE3 was 2.40 ± 0.21 mg/kg/day in E3/E3 subjects, and about half of that in E2/E3 and E3/E4 individuals (1.36 ± 0.18 mg/kg/day and 1.52 ± 0.21 mg/kg/day, respectively), which is consistent with the presence of only one ε3 allele in heterozygotes. In E2/E3 individuals, the PR of apoE2 was found to be similar to that of apoE3 (1.16 ± 0.10 mg/kg/day vs. 1.36 ± 0.18 mg/kg/day). In sharp contrast, in E3/E4 individuals, the PR of apoE4 was twice lower than that of apoE3 (0.73 ± 0.15 mg/kg/day vs. 1.52 ± 0.21 mg/kg/day).
Fig. 4.
Kinetic parameters of plasma apoE isoforms. FCRs (A) and PRs (B) estimated with a mono-exponential equation in subjects with E2/E3 phenotype, E3/E3 phenotype, and E3/E4 phenotype.
Validation of kinetic data
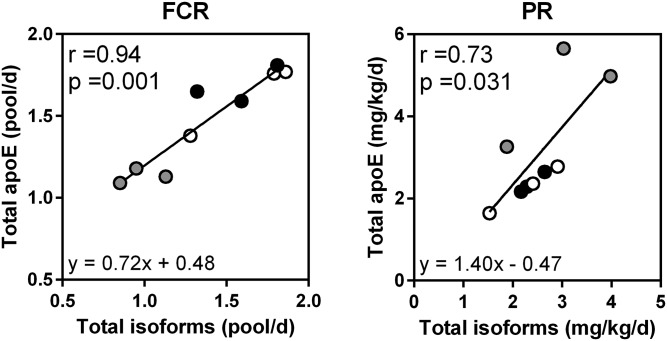
The enrichment curves of total apoE in tracer over time were also investigated with the common LGPLVEQGR peptide (supplemental Fig. S3). In all patients, the total plasma apoE FCR and PR were, on average, 1.48 ± 0.29 pool/day and 3.09 ± 1.35 mg/kg/day, respectively. The total apoE FCR and PR were also calculated in all patients from kinetic data obtained specifically for each isoform and related peptides. From these data, FCR and PR were, on average, 1.40 ± 0.38 pool/day and 2.54 ± 0.72 mg/kg/day, respectively. We did not find any marked difference in total apoE kinetic parameters using both approaches. As shown in Fig. 5, there was a significant correlation between FCRs measured using both approaches (r = 0.94, P = 0.001), as well as a significant correlation between PRs (r = 0.73, P = 0.031) despite the heterogeneity in the apoE2 PR.
Fig. 5.
Validation of plasma apoE kinetic parameters. Total FCRs and PRs were calculated for each subject from kinetic data of apoE isoforms, and then compared (Spearman correlation test) with those obtained directly from the LGPLVEQGR peptide used for total apoE detection. Gray, black, and white circles indicate E2/E3, E3/E3, and E3/E4 phenotypes, respectively.
Distribution of apoE isoforms within lipoproteins
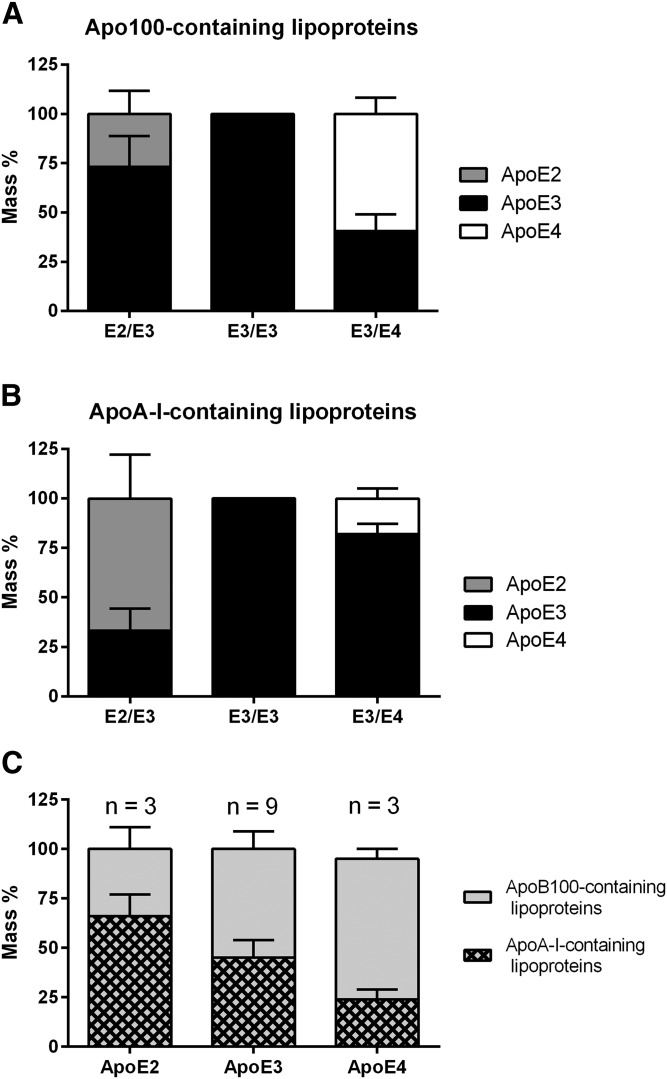
Compared with the whole plasma concentrations, FPLC and sequential ultracentrifugations gave similar and satisfactory recovery rates for structural apolipoproteins, such as apoA-I for HDL (94% and 84%, respectively) and apoB100 for VLDL, IDL, and LDL (79% and 88%, respectively). A poor recovery was obtained for apoE (48% and 51%, respectively) (supplemental Table S1), likely because apoE sheds off surface lipoprotein particles easily. It is noteworthy that separation of IDLs that were rich in apoE was not optimal (supplemental Fig. S4) and led to a ∼130-fold dilution of the original sample by FPLC. In addition, apoE2 and apoE4 peptides displayed ∼10-fold lower ionization yields than the common apoE peptide, precluding accurate detection of both isoforms in FPLC fractions despite a concentration procedure. The distribution of apoE isoforms within lipoprotein classes was therefore investigated in the nondiluted fractions obtained after ultracentrifugation. As shown in Fig. 6A, the major apoE isoform found in apoB100-containing lipoproteins from E2/E3 individuals was apoE3 (73.2 ± 15.6%), whereas the major apoE isoform in apoA-I-containing lipoproteins was apoE2 (66.7 ± 22.3%). In contrast, the major apoE isoform present in apoB100-containing lipoproteins from E3/E4 individuals was apoE4 (59.3 ± 8.3%), whereas the major apoE isoform in apoA-I-containing lipoproteins was apoE3 (82.0 ± 5.6%) (Fig. 6B), indicating a higher affinity of apoE2 for HDL and of apoE4 for apoB100-containing lipoproteins, while apoE3 distributed homogeneously between lipoprotein classes (Fig. 6C) in those hypertriglyceridemic patients.
Fig. 6.
Distribution of apoE isoforms within lipoproteins in hypertriglyceridemic patients. ApoE isoforms were assayed in apoB100-containing lipoproteins (i.e., VLDL + IDL + LDL) (A) and in apoA-I-containing lipoproteins (i.e., HDL) (B) obtained by ultracentrifugation. C: Summary of apoE isoform distribution within lipoproteins. Concentrations were normalized to the total content of apoE recovered in each lipoprotein subclass. Values are expressed as mean ± standard deviation.
Enrichments of apoE within lipoproteins
We were not able to detect 2H3-leucine enrichments of apoE isoforms in lipoprotein fractions because of insufficient sensitivity. Kinetic enrichments of total apoE were therefore investigated within lipoproteins by using the common LGPLVEQGR peptide and we did not observe any pronounced difference on total apoE kinetics between groups (supplemental Fig. S3). In all patients, total apoE FCRs were 0.49 ± 0.08 pool/day and 2.95 ± 0.65 pool/day and PRs were 0.47 ± 0.12 mg/kg/day and 2.59 ± 0.91 mg/kg/day in HDL and VLDL, respectively.
DISCUSSION
We investigated the plasma concentrations, lipoprotein distribution, and kinetic parameters of apoE2, apoE3, and apoE4 in human plasma by LC-MS/MS. Total circulating apoE concentrations and kinetics were different according to apoE isoforms with different repartitions within lipoproteins. We showed that the differences in the whole plasma apoE isoform concentrations stemmed from a reduced clearance rate of the apoE2 isoform and from a reduced PR of the apoE4 isoform, compared with apoE3.
One limitation of the study was the small number of subjects and the lack of ε2/ε2, ε2/ε4, or ε4/ε4 patients. This was related to the very low frequencies of these genotypes in our medical environment. A second limitation of our study was that we used a simple mathematical approach. Because of the small number of subjects per group, we did not develop compartment models including a delay, which might have provided a better fit to the experimental data (9). This compartmental analysis will be mandatory when more subjects are analyzed. But a specific study is required because some patients, especially with E2 isoforms, are few and difficult to recruit. Finally, we did not use calibration solutions with known enrichments for each proteotypic peptide. This was a third limitation and we cannot totally rule out any analytical bias in assessing apoE2 and apoE4 enrichments.
LC-MS/MS is not only reliable to simultaneously quantify several proteins (12, 13, 18) but also to study their polymorphisms (2, 14) and to measure their kinetics (10, 11). This approach involves a trypsin proteolysis before analysis of signature peptides carefully selected to maximize sensitivity, specificity, and stability. Peptide candidate selection is a crucial step that is unfortunately limited when considering polymorphic modifications. Here, we have optimized our previous protocol (10, 12, 14) to quantify and study total apoE and each isoform in human plasma. Despite our efforts to set up optimal proteolysis conditions (2, 19), some of our peptides displayed 10- to 15-fold reduced sensitivities by MS compared with the peptide selected for total apoE measurement, likely because these peptides either contain a methionine or a cysteine residue responsible for side chain reactions and poor stability (2, 10, 20, 21). This reduced sensitivity and stability did not allow the measurement of 2H3-leucine enrichments in apoE isoforms in lipoprotein fractions. We were able to get only total apoE kinetics within lipoproteins with the common and more sensitive LGPLVEQGR peptide. Although total apoE kinetic parameters in both HDL and VLDL were in agreement with previous reports (10, 22, 23), we did not observe any marked difference between patients with heterozygous phenotypes. To assess the kinetics of apoE isoforms, the common LGPLVEQGR peptide, therefore, appears to be limited to homozygous phenotypes.
Another hurdle, unrelated to the MS technology, is the exchange of apoE between lipoproteins and their shedding off of the lipoprotein surface by ultracentrifugation, a clear bias for accurate determination of apoE pool sizes (10, 22). In that respect, immuno-affinity separations or softer ultracentrifugation techniques could yield better recovery rates (12, 22, 24). The exchangeability of apolipoproteins could also be a limitation to the determination of apoC’s kinetic enrichment curves. However, apoC-II enrichment curves in VLDL and HDL are similar, and those of apoC-III much closer than those observed for apoE. Furthermore, apoE enrichment curves in VLDL and HDL parallel those observed for VLDL-apoB100 and HDL-apoA-I (supplemental Figs. S1, S3). While apoC displayed similar kinetics in VLDL and HDL, apoE enrichment curves were sharply different between VLDL and HDL, and relatively close to those of VLDL-apoB100 and HDL-apoA-I, respectively (10, 22), indicating that the differences observed in apoE enrichment between lipoprotein subclasses is genuine and that apoE exchange is limited.
We observed a preferential association of apoE4 with apoB100-containing lipoproteins, in agreement with previous reports (25–27). The presence of a positive charge in the arginine residue at position 112 of apoE4 enhances its affinity for lipids compared with apoE3 (25, 28), and further strengthens its association with VLDL. In addition, the absence of cysteines at positions 112 and 158 reduces apoE4’s ability to establish disulfide bonds with HDL-apoA-II (25, 29). In contrast, we observed a preferential association of apoE2 with HDL, in line with a previous study (30). In contrast with apoE4, the cysteine residue at position 112 in apoE3 and at positions 112 and 158 in apoE2 allows the formation of apoE/apoA-II heterodimers and could explain their preferential association within HDL compared with apoE4 (25, 29, 31, 32). The cysteine residue at position 158 in apoE2 alters its conformation and its ability to bind to the LDLR (28).Whether this might reduce apoE2’s ability to associate with apoB100-containing lipoproteins is not established (25, 30).
As anticipated, the catabolic rate of apoE2 was slower than that of apoE3 or apoE4, as previously shown with radio-isotopes (29) or with 13C6-leucine in a pilot study conducted in humans (one subject from each genotype, E3/E3, E3/E4, E4/E4 and E2/E4) (33). As mentioned above, the presence of a cysteine instead of an arginine at position 158 reduces the affinity of apoE2 for the LDLR by ∼98% (8, 28), and its affinity for the LRP or HSPG by ∼50%, compared with apoE3 (30), consistent with reduced catabolism. Another mechanism has been proposed (34). Because the turnover of VLDL is much faster than that of HDL, the preferential distribution of apoE2 within HDL could also explain why apoE2 is cleared more slowly than apoE3 or apoE4. In addition, the reduced apoE2 FCR could also be explained by its association with apoA-II in HDL, as detailed above. It has been suggested that both apo(E2/A-II) and apo(A-II/E2/A-II) complexes could prevent LDLR binding by masking the apoE2 component (31, 32).
We also showed that reduced apoE4 concentrations were associated with a 2-fold reduction in apoE4 PR compared with apoE3 and apoE2. This is not due to different gene expression of the three isoforms (35). However, the secretion of apoE2 and apoE4 by macrophages appears to be significantly reduced, compared with that of apoE3 (35), indicating that posttranslational mechanisms governing apoE secretion could be related to its isoforms. Another mechanism could involve the recycling of apoE within the hepatocytes. After the initial secretion, a part of apoE is submitted to a reuptake and is immediately recycled to contribute to the overall production (36). Compared with apoE3, the intracellular hepatocyte recycling of apoE4 derived from VLDL appeared to be lower (37). apoE4 from VLDL is also apparently recycled via distinct cellular pathways (38). The precise cellular mechanisms underpinning the reduced secretion rate of apoE4 clearly remains to be elucidated.
In this study, we have evaluated a novel approach to assess the kinetic parameters of plasma apoE isoforms. We showed that the variations of total apoE plasma concentrations (E2/E3 > E3/E3 > E3/E4) associated with these phenotypes can be explained by reduced catabolic rates for apoE2 and reduced PRs for apoE4. Improvements in the sensitivity of our techniques and in our modeling approach are warranted to assess the kinetics of each apoE isoform within lipoprotein subclasses.
Supplementary Material
Acknowledgments
The authors thank the Therassay core facility for the provision of FPLC AKTA© and the staff of the Clinical Investigation Center of the University Hospital in Nantes, especially Eliane Hivernaud for her invaluable help with patients and blood collection.
Footnotes
Abbreviations:
- FCR
- fractional catabolic rate
- FPLC
- fast protein LC
- FSR
- fractional synthetic rate
- HSPG
- heparan sulfate proteoglycan
- IR
- isotope ratio
- IS
- internal standard
- LDLR
- LDL receptor
- LRP
- LDL receptor-related protein
- PCSK9
- proprotein convertase subtilisin kexin type 9
- PR
- production rate
- TRL
- triglyceride-rich lipoprotein
This work was supported by an Allocation de Recherche Chaire Mixte (Inserm-Université) and Agence Nationale de la Recherche Program Grant ANR-16-RHUS-0007 CHOPIN to G.L. Additional financial support was provided by the Biogenouest CORSAIRE core facility. The authors declare no competing financial interest.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Mahley R. W., and Rall S. C. Jr. 2000. Apolipoprotein E: far more than a lipid transport protein. Annu. Rev. Genomics Hum. Genet. 1: 507–537. [DOI] [PubMed] [Google Scholar]
- 2.Martínez-Morillo E., Nielsen H. M., Batruch I., Drabovich A. P., Begcevic I., Lopez M. F., Minthon L., Bu G., Mattsson N., Portelius E., et al. 2014. Assessment of peptide chemical modifications on the development of an accurate and precise multiplex selected reaction monitoring assay for apolipoprotein e isoforms. J. Proteome Res. 13: 1077–1087. [DOI] [PubMed] [Google Scholar]
- 3.Dominiczak M. H., and Caslake M. J.. 2011. Apolipoproteins: metabolic role and clinical biochemistry applications. Ann. Clin. Biochem. 48: 498–515. [DOI] [PubMed] [Google Scholar]
- 4.Mahley R. W., Huang Y., and Rall S. C. Jr. 1999. Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia): questions, quandaries and paradoxes. J. Lipid Res. 40: 1933–1949. [PubMed] [Google Scholar]
- 5.Mahley R. W. 2016. Apolipoprotein E: from cardiovascular disease to neurodegenerative disorders. J. Mol. Med. (Berl.). 94: 739–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moriarty P. M., Varvel S. A., Gordts P. L., McConnell J. P., and Tsimikas S.. 2017. Lipoprotein (a) mass levels increase significantly according to APOE genotype. Arterioscler. Thromb. Vasc. Biol. 37: 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martínez-Morillo E., Hansson O., Atagi Y., Bu G., Minthon L., Diamandis E. P., and Nielsen H. M.. 2014. Total apolipoprotein E levels and specific isoform composition in cerebrospinal fluid and plasma from Alzheimer’s disease patients and controls. Acta Neuropathol. 127: 633–643. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen K. L. 2016. Plasma levels of apolipoprotein E, APOE genotype and risk of dementia and ischemic heart disease: a review. Atherosclerosis. 255: 145–155. [DOI] [PubMed] [Google Scholar]
- 9.Barrett P. H., Chan D. C., and Watts G. F.. 2006. Design and analysis of lipoprotein tracer kinetics studies in humans. J. Lipid Res. 47: 1607–1619. [DOI] [PubMed] [Google Scholar]
- 10.Croyal M., Fall F., Ferchaud-Roucher V., Chétiveaux M., Zaïr Y., Ouguerram K., Krempf M., and Nobécourt E.. 2016. Multiplexed peptide analysis for kinetic measurements of major human apolipoproteins by LC/MS/MS. J. Lipid Res. 57: 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan Y., Zhou H., Mahsut A., Rohm R. J., Berejnaia O., Price O., Chen Y., Castro-Perez J., Lassman M. E., McLaren D., et al. 2014. Static and turnover kinetic measurement of protein biomarkers involved in triglyceride metabolism including apoB48 and apoA5 by LC/MS/MS. J. Lipid Res. 55: 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tavori H., Christian D., Minnier J., Plubell D., Shapiro M. D., Yeang C., Giunzioni I., Croyal M., Duell P. B., Lambert G., et al. 2016. PCSK9 association with lipoprotein(a). Circ. Res. 119: 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ceglarek U., Dittrich J., Becker S., Baumann F., Kortz L., and Thiery J.. 2013. Quantification of seven apolipoproteins in human plasma by proteotypic peptides using fast LC-MS/MS. Proteomics Clin. Appl. 7: 794–801. [DOI] [PubMed] [Google Scholar]
- 14.Croyal M., Ouguerram K., Passard M., Ferchaud-Roucher V., Chétiveaux M., Billon-Crossouard S., de Gouville A. C., Lambert G., Krempf M., and Nobécourt E.. 2015. Effects of extended-release nicotinic acid on apolipoprotein (a) kinetics in hypertriglyceridemic patients. Arterioscler. Thromb. Vasc. Biol. 35: 2042–2047. [DOI] [PubMed] [Google Scholar]
- 15.Chétiveaux M., Nazih H., Ferchaud-Roucher V., Lambert G., Zaïr Y., Masson M., Ouguerram K., Bouhours D., and Krempf M.. 2002. The differential apoA-I enrichment of prebeta1 and alphaHDL is detectable by gel filtration separation. J. Lipid Res. 43: 1986–1993. [DOI] [PubMed] [Google Scholar]
- 16.Ouguerram K., Chetiveaux M., Zair Y., Costet P., Abifadel M., Varret M., Boileau C., Magot T., and Krempf M.. 2004. Apolipoprotein B100 metabolism in autosomal-dominant hypercholesterolemia related to mutations in PCSK9. Arterioscler. Thromb. Vasc. Biol. 24: 1448–1453. [DOI] [PubMed] [Google Scholar]
- 17.Zhou H., Castro-Perez J., Lassman M. E., Thomas T., Li W., McLaughlin T., Dan X., Jumes P., Wagner J. A., Gutstein D. E., et al. 2013. Measurement of apo(a) kinetics in human subjects using a microfluidic device with tandem mass spectrometry. Rapid Commun. Mass Spectrom. 27: 1294–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Percy A. J., Chambers A. G., Yang J., Hardie D. B., and Borchers C. H.. 2014. Advances in multiplexed MRM-based protein biomarker quantitation toward clinical utility. Biochim. Biophys. Acta. 1844: 917–926. [DOI] [PubMed] [Google Scholar]
- 19.Hoofnagle A. N. 2010. Quantitative clinical proteomics by liquid chromatography-tandem mass spectrometry: assessing the platform. Clin. Chem. 56: 161–164. [DOI] [PubMed] [Google Scholar]
- 20.Wildsmith K. R., Han B., and Bateman R. J.. 2009. Method for the simultaneous quantitation of apolipoprotein E isoforms using tandem mass spectrometry. Anal. Biochem. 395: 116–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krastins B., Prakash A., Sarracino D. A., Nedelkov D., Niederkofler E. E., Kiernan U. A., Nelson R., Vogelsang M. S., Vadali G., Garces A., et al. 2013. Rapid development of sensitive, highthroughput, quantitative and highly selective mass spectrometric targeted immunoassays for clinically important proteins in human plasma and serum. Clin. Biochem. 46: 399–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Batal R., Tremblay M., Barrett P. H., Jacques H., Fredenrich A., Mamer O., Davignon J., and Cohn J. S.. 2000. Plasma kinetics of apoC-III and apoE in normolipidemic and hypertriglyceridemic subjects. J. Lipid Res. 41: 706–718. [PubMed] [Google Scholar]
- 23.Bach-Ngohou K., Ouguerram K., Nazih H., Maugère P., Ripolles-Piquer B., Zaïr Y., Frénais R., Krempf M., and Bard J. M.. 2002. Apolipoprotein E kinetics: influence of insulin resistance and type 2 diabetes. Int. J. Obes. Relat. Metab. Disord. 26: 1451–1458. [DOI] [PubMed] [Google Scholar]
- 24.Agnani G., Bard J. M., Candelier L., Delattre S., Fruchart J. C., and Clavey V.. 1991. Interaction of LpB, LpB:E, LpB:C-III, and LpB:C-III:E lipoproteins with the low density lipoprotein receptor of HeLa cells. Arterioscler. Thromb. 11: 1021–1029. [DOI] [PubMed] [Google Scholar]
- 25.Weisgraber K. H. 1990. Apolipoprotein E distribution among human plasma lipoproteins: role of the cysteine-arginine interchange at residue 112. J. Lipid Res. 31: 1503–1511. [PubMed] [Google Scholar]
- 26.Steinmetz A., Jakobs S., Motzny S., and Kaffarnik H.. 1989. Differential distribution of apolipoprotein E isoforms in human plasma lipoproteins. Arteriosclerosis. 9: 405–411. [DOI] [PubMed] [Google Scholar]
- 27.Gregg R. E., Zech L. A., Schaefer E. J., Stark D., Wilson D., and Jr. Brewer H. B.. 1986. Abnormal in vivo metabolism of apolipoprotein E4 in humans. J. Clin. Invest. 78: 815–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phillips M. C. 2014. Apolipoprotein E isoforms and lipoprotein metabolism. IUBMB Life. 66: 616–623. [DOI] [PubMed] [Google Scholar]
- 29.Ikewaki K., Zech L. A., Jr. Brewer H. B., and Rader D. J.. 2002. Comparative in vivo metabolism of apolipoproteins E2 and E4 heterozygous apoE2/E4 subjects. J. Lab. Clin. Med. 140: 369–374. [DOI] [PubMed] [Google Scholar]
- 30.Mahley R. W., and Ji Z. S.. 1999. Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J. Lipid Res. 40: 1–16. [PubMed] [Google Scholar]
- 31.Innerarity T. L., Mahley R. W., Weisgraber K. H., and Bersot T. P.. 1978. Apoprotein (E–A-II) complex of human plasma lipoproteins. II. Receptor binding activity of a high density lipoprotein subfraction modulated by the apo(E–A-II) complex. J. Biol. Chem. 253: 6289–6295. [PubMed] [Google Scholar]
- 32.Tozuka M., Hidaka H., Miyachi M., Furihata K., Katsuyama T., and Kanai M.. 1992. Identification and characterization of apolipoprotein(AII-E2-AII) complex in human plasma lipoprotein. Biochim. Biophys. Acta. 1165: 61–67. [DOI] [PubMed] [Google Scholar]
- 33.Wildsmith K. R., Basak J. M., Patterson B. W., Pyatkivskyy Y., Kim J., Yarasheski K. E., Wang J. X., Mawuenyega K. G., Jiang H., Parsadanian M., et al. 2012. In vivo human apolipoprotein E isoform fractional turnover rates in the CNS. PLoS One. 7: e38013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown M. S., Kovanen P. T., and Goldstein J. L.. 1981. Regulation of plasma cholesterol by lipoprotein receptors. Science. 212: 628–635. [DOI] [PubMed] [Google Scholar]
- 35.Cullen P., Cignarella A., Brennhausen B., Mohr S., Assmann G., and Von Echardstein A.. 1998. Phenotype-dependent differences in apolipoprotein E metabolism and in cholesterol homeostasis in human monocyte-derived macrophages. J. Clin. Invest. 101: 1670–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swift L. L., Farkas M. H., Major A. S., Vayi-Nagy K., Linton M. F., and Fazio S. A.. 2001. Recycling pathway for resecretion of internalized apolipoprotein E in liver cells. J. Biol. Chem. 276: 22965–22970. [DOI] [PubMed] [Google Scholar]
- 37.Heeren J., Grewal T., Laatsch A., Becker N., Rinninger F., Rye K. A., and Beisiegel U.. 2004. Impaired recycling of apolipoprotein E4 is associated with intracellular cholesterol accumulation. J. Biol. Chem. 279: 55483–55492. [DOI] [PubMed] [Google Scholar]
- 38.Kockx M., Jessup W., and Kritharides L.. 2008. Regulation of endogenous apolipoprotein E secretion by macrophages. Arterioscler. Thromb. Vasc. Biol. 28: 1060–1067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.