Abstract
Lipid droplets (LDs) are ubiquitous organelles that contain neutral lipids and are surrounded by a phospholipid monolayer. How proteins specifically localize to the phospholipid monolayer of the LD surface has been a matter of extensive investigations. In the present study, we show that syntaxin 17 (Stx17), a soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) protein whose expression in the liver is regulated by diet, participates in LD biogenesis by regulating the distribution of acyl-CoA synthetase (ACSL)3, a key enzyme for LD biogenesis that redistributes from the endoplasmic reticulum (ER) to LDs during LD formation. Stx17 interacts with ACSL3, but not with LD formation-unrelated ACSL1 or ACSL4, through its SNARE domain. The interaction occurs at the ER-mitochondria interface and depends on the active site occupancy of ACSL3. Depletion of Stx17 impairs ACSL3 redistribution to nascent LDs. The defect in LD maturation due to Stx17 knockdown can be compensated for by ACSL3 overexpression, suggesting that Stx17 increases the efficiency of ACSL3 redistribution to LDs. Moreover, we show that the interaction between Stx17 and ACSL3 during LD maturation may be regulated by synaptosomal-associated protein of 23 kDa.
Keywords: endoplasmic reticulum, mitochondria-associated membrane, synaptosomal-associated protein of 23 kDa, soluble N-ethylmaleimide-sensitive factor attachment protein receptor, triacylglycerol
Lipid droplets (LDs) are ubiquitous organelles that store neutral lipids such as triacylglycerol (TAG) and sterol esters, and play central roles in energy and lipid metabolism (1). LDs are dynamic and diverse organelles, their size and number depending on cellular energy and the metabolic state, and their protein and lipid compositions varying with the cell type and the degree of LD maturation in individual cell types (2–4). Moreover, LDs are in contact with many organelles, including the endoplasmic reticulum (ER), mitochondria, and peroxisomes (5, 6). Recent studies revealed that LDs have additional functions, such as in ER stress responses, protein storage, protein degradation, and viral replication (7, 8).
LDs are unique among cellular organelles in that they are surrounded by a phospholipid monolayer. LD formation starts in the ER at predefined or random sites (9, 10) with the formation of lipid lenses of around 50 nm in the intermembrane space of the ER lipid bilayer (11). The formation of lenses and their enlargement as a consequence of lateral fusion and/or accumulation of more lipids generate curvature of the ER membrane. At this stage acyl-CoA synthetase (ACSL)3, an enzyme that provides acyl-CoA for LD formation, moves within the ER and becomes concentrated at emerging LD sites through its amphipathic α helices (9, 12). This enzyme is critical for LD expansion and maturation. Loss or overexpression of other ACSL family members does not affect LD biogenesis (9), highlighting the importance of ACSL3-mediated local synthesis of acyl-CoA for LD expansion. ACSL3 belongs to the class I LD proteins (13) that are characterized by their presence in the ER without LDs and translocation to the LD surface during LD formation or after reconnection of LDs to the ER via membrane bridges (14). Most class I proteins have hydrophobic regions with hairpin-like structures (13). Emerging LDs expand and then are recognized by proteins such as perilipins. Perilipins are class II proteins containing amphipathic α helices or other hydrophobic domains that move from the cytosol to the LD surface (13).
Syntaxin 17 (Stx17) was originally characterized as a t-soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) located in the smooth ER (15). Recent studies by us and other groups demonstrated that Stx17 localizes to the mitochondria-ER interface, including the mitochondria-associated ER membrane (MAM) (16), and plays roles in mitochondrial division (17), the fusion of stress-induced mitochondrial-derived vesicles with lysosomes (18), and autophagy in starved cells (19–22). In mitochondrial division and autophagosome formation, Stx17 functions not as a SNARE, but as a scaffold at the MAM (17, 20). For these functions and the MAM localization, the C-terminal hairpin-like hydrophobic domain (CHD) and the subsequent cytoplasmic tail of Stx17, but not its SNARE domain, are important (17). The MAM has versatile functions (16), including the synthesis of neutral lipids as well as phospholipids (23, 24).
In the present study, we demonstrated that Stx17 is required for LD biogenesis. We found that Stx17 interacts with ACSL3 and facilitates its translocation from the ER to the surface of LDs.
MATERIALS AND METHODS
Chemicals and antibodies
Triacsin C and digitonin were obtained from Enzo Life Sciences and Wako Chemicals, respectively. Oleic acid (OA), TAG, 1-oleoyl-2-acetyl-sn-glycerol (OAG), diacylglycerol acyltransferase (DGAT)1 inhibitor (PF-04620110), and DGAT2 inhibitor (PF-06424439) were obtained from Sigma-Aldrich. OA, TAG, and OAG were dissolved in DMSO, mixed with BSA, and used. Lipid Tox and BODIPY FL-C16 were obtained from Thermo Fisher Scientific. The following antibodies were purchased from Sigma-Aldrich: monoclonal FLAG (F3165) and polyclonal FLAG (F7425). The following antibodies were obtained from BD Bioscience Pharmingen: calnexin (CNX) (610523), Tom20 (612278), and Tim23 (611223). The following antibodies were purchased from Proteintech: Tip47 (10694-1-AP), synaptosomal-associated protein of 23 kDa (SNAP23) SNAP23 (10825-1-AP), and SNAP29 (12704-1-AP). The following antibodies were purchased from Abcam: polyclonal ACSL1 (ab76702) and α-tubulin (ab15246). Polyclonal and monoclonal antibodies against ACSL3 were obtained from GeneTeX (GTX112431) and Abnova (H00002181-B01P), respectively. The following antibodies were from the indicated sources: ACSL4 (Santa Cruz Biotechnology; H-53, sc-134507), HA (Sigma-Aldrich; H6908), and Alexa Fluor® 488 and 594 goat anti-mouse and -rabbit antibodies (Thermo Fisher Scientific; A-11001, A-11005, A-11008, and A-11012). Rabbit antibodies against Sec22b (25) and Stx5 (26) were produced in this laboratory and affinity purified. An antibody against Stx17 was prepared, as described previously (17), and used for immunofluorescence analysis. For immunoblotting, an anti-Stx17 antibody (Sigma-Aldrich; HPA001204) was used.
Cell culture
The 293T, HepG2, Huh7, and 3T3-L1 cells were grown in DMEM supplemented with 50 IU/ml penicillin, 50 μg/ml streptomycin, and 10% FCS. HeLa cells (RIKEN; RCB0007) were cultured in α-MEM supplemented with the same materials plus 2 mM l-glutamine. Stable transfectants were prepared as described previously (17). For the induction of LDs in HeLa cells, OA was added at a final concentration of 150 μM. Mouse primary hepatocytes were prepared using a collagenase liver perfusion technique, as described previously (27), with a minor modification. Female ddy mice (5 weeks) that had been fed or fasted for 24 h were used.
Plasmids and transfection
Plasmids encoding human Stx17 full-length and its derivatives were described previously (17). The plasmids for GFP-ACSL3 and GFP-HPos (9) were gifts from Dr. Albert Pol (IDIBAPS). The plasmid for FLAG-ACSL3 was constructed by inserting the cDNA for GFP-ACSL3 into the EcoRI/SmaI site of pFLAG-CMV-6a vector. The plasmids for FLAG-DGAT1/2, 3x-HA-lysophosphatidylcholine acyltransferase (LPCAT) 1/2, glycerol-3-phosphate O-acyltransferase (GPAT)4-GFP, and SU9-GFP were gifts from Dr. Robert V. Farese (Harvard University), Dr. Christoph Thiele (University of Bonn), Dr. Katsuko Tani (Tokyo University of Pharmacy and Life Sciences), and Dr. Naotada Ishihara (Kurume University), respectively. Deletion mutants of ACSL3 were constructed by inverse PCR. Transfection was carried out using LipofectAMINE Plus, LipofectAMINE2000 (Thermo Fisher Scientific), or polyethylenimine.
RNA interference
The following siRNAs were used: Stx17 (440), 5′-GGUAGUUCUCAGAGUUUGAUU-3′; Stx17 (194), 5′-CGAUCCAAUAUCCGAGAAAUU-3′; Stx17 (NC), 5′-GGAAAUUAAUGAUGUAAGA-3′; Stx17 (421), 5′-CACACUGGGGAGGCUGAAGCU-3′; Sec22b, 5′-CAGCATTGGATTCAAAGGCTA-3′; SNAP29, 5′-UAUCAUCCAGCUUUCUAAGGUUUGG-3′; ACSL1, 5′-AAGGAUGCUUUGCUUAUUCGA-3′; ACSL3, 5′-CCGAAGUGUGGGACUACAAUA-3′; and ACSL4, 5′-AUGCAUCAUAGCAAUUUGAUA-3′.
siRNAs were purchased from Japan Bio Services. HeLa, HepG2, Huh7, and 3T3-L1 cells were grown on 35 mm dishes, and siRNAs were transfected at a final concentration of 200 nM using Oligofectamine, LipofectAMINE2000, or LipofectAMINE2000 RNAi Max (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Immunoprecipitation
HeLa cells expressing FLAG-tagged proteins were lysed in lysis buffer (20 mM HEPES-KOH (pH 7.2), 150 mM KCl, 2 mM EDTA, 1 mM dithiothreitol, 1 μg/ml leupeptin, 1 μM pepstatin A, 2 μg/ml aprotinin, and 1 mM phenylmethylsulfonyl fluoride) containing 1% Triton X-100 or 10 mg/ml digitonin. After centrifugation, the supernatants were collected and immunoprecipitated with anti-FLAG M2 affinity beads (Sigma-Aldrich). The precipitated proteins were eluted with SDS sample buffer, and then analyzed by immunoblotting. Experiments were repeated two or three times with similar results.
TAG measurement
TAG measurement was performed using a triglyceride quantification colorimetric kit (BioVison; k662) according to the manufacturer’s protocol. The OD 570 nm was measured and the TAG amount was calculated using a standard curve.
Immunofluorescence microscopy and image analysis
For immunofluorescence microscopy, cells were fixed with 4% paraformaldehyde for 20 min at room temperature followed by permeabilization in 0.2% Triton X-100 in PBS for 10 min at room temperature or with ice-cold methanol for 1 min at room temperature, and blocked with 2% BSA in PBS for 10 min. The cells were incubated with a primary antibody [ACSL3 (1:50), Tip47 (1:50), Stx17 (1:50), FLAG (1:300), Tom20 (1:300), ACSL1 (1:50), or ACSL4 (1:20)] for 1 h at 37°C, followed by three washes in PBS and incubation with Alexa Fluor 488- or Alexa Fluor 594-conjugated anti-mouse or anti-rabbit IgG antibody (1:200) for 1 h at 37°C. When stained with LipidTox, the cells were incubated with LipidTox (dissolved in DMSO) for 30 min in PBS at room temperature. After washing in PBS, they were mounted with mounting medium (Dako) and observed using a 100× oil immersion objective lens (UPlan FI: NA = 1.3) under a laser scanning confocal microscope (OLYMPUS Fluoview FV300) with a pinhole of 3 AU. All images were single confocal sections.
ImageJ software (National Institutes of Health) was used to determine the size of LDs and the fluorescence intensity ratio between LipidTox and LD proteins. In each LD, the intensities of circular LipidTox fluorescence and surrounding FITC fluorescence were measured. This analysis was performed for randomly chosen 30 LDs in each cell, and 30 cells were analyzed in each experiment. Experiments were repeated three times.
Electron microscopy
Cells were fixed with a mixture of 2% formaldehyde and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) for 2 h, and then prepared for conventional observation as described previously (28). ER-LD contact sites were discerned when the distance between the ER membrane and the LD surface was 20 nm or less. The images of cytoplasmic LDs were randomly captured from five independent RNAi experiments (total number of analyzed LDs; siRNA control = 242, siRNA stx17 (440) = 252), and the frequency of ER-LD contact site formation was determined as the rational length of circular arch of an LD surface covered by contacted ER to the total perimeter of the LD. ER-LD contact length and the whole circumference of each LD were measured with ImageJ software.
Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching (FRAP) experiments were performed with an Olympus Fluoview 1000 laser scanning microscope equipped with a stage-top incubator (37°C, 5% CO2). To monitor LDs during FRAP experiments, cells expressing GFP constructs or labeled with BODIPY FL-C16 were incubated with LipidTox in Opti-MEM supplemented with 10% FCS before photobleaching. An arbitrary region containing several LDs and adjacent ER membranes (denoted as “LD”) or an ER region alone (denoted as “ER”) was photobleached using a 488 nm laser at 100% laser power for 2 s. After photobleaching, images of the individual LDs or the ER region were obtained at 0.5 s intervals. In each experiment, 30 cells were used, and more than three LDs in each cell were analyzed. Experiments were repeated three times. The recovered fluorescence intensity normalized to that before photobleaching was plotted.
Differentiation of 3T3-L1 cells and measurement of intensity of Oil Red O staining
Differentiation was induced using an AdipoInducer reagent (for animal cells) kit (Takara Bio, MK429) according to the manufacturer’s protocol. Briefly, 3T3-L1 cells were mock-transfected or transfected with siRNA Stx17 (421). At 72 h after transfection, cells were incubated with DMEM supplemented with 50 IU/ml penicillin, 50 μg/ml streptomycin, and 10% FCS, plus 10 μg/ml insulin solution, 2.5 μM dexamethasone, and 0.5 mM 3-isobutyl-1-methylxanthine, for 48 h. The medium was replaced with DMEM containing 10 μg/ml insulin, and then incubated for ∼9 days. For Oil Red O staining, cells were fixed, washed with 60% isopropanol for 1 min, and then incubated with a 60% Oil Red O solution (99% isopropanol/0.3% Oil Red O) for 15 min at 37°C. After washing with 60% isopropanol and drying, Oil Red O was dissolved in 100% isopropanol and the OD at 500 nm was measured.
Proximity ligation assay
A proximity ligation assay (PLA) was conducted using a PLA kit (Sigma-Aldrich) according to the manufacturer’s protocol. Thirty cells were analyzed in each assay. PLA dots were identified using the “analyze particle” program in the ImageJ software. Randomly, 30 cells were selected and the number of PLA dots was measured in each sample. The experiments were repeated three times.
Subcellular fractionation
Subcellular fractionation to isolate the MAM was performed as described previously (17). LD fractionation was performed as described previously (29). Cell were homogenized in buffer containing 225 mM mannitol and 75 mM sucrose and centrifuged at 8,000 g for 10 min to obtain mitochondrial pellets. The supernatant was overlaid with homogenization buffer omitting sucrose and centrifuged at 166,000 g for 5 h. The top of the supernatant was recovered as the LD fraction, and the rest of the supernatant and the pellet were recovered as the cytosolic and microsomal fractions, respectively. Experiments were repeated two or three times with similar results.
Statistical analyses
The results were averaged, expressed as the mean ± SD or SEM, and analyzed using a Student’s test. The P values are indicated by asterisks in the figures with the following notations: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
RESULTS
Stx17 is required for LD biogenesis
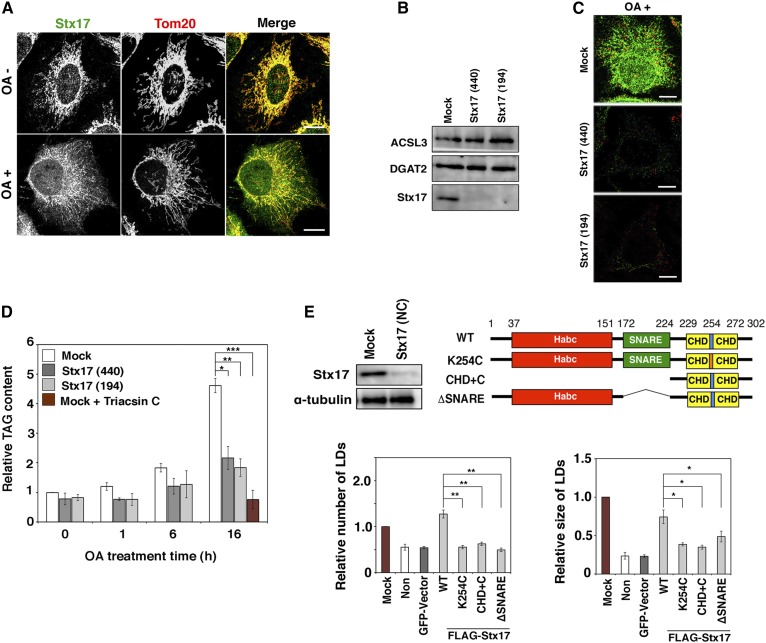
Although Stx17 is ubiquitously expressed, it is abundantly expressed in steroidogenic and hepatic cells (15), both of which have large numbers of LDs. This and the MAM localization of Stx17 prompted us to examine the role of Stx17 in LD biogenesis. To this end, we used HeLa cells that have only a few LDs under normal conditions. LD biogenesis can be induced by OA. At the level of immunofluorescence microscopy, Stx17 exhibited nearly perfect colocalization with mitochondria in OA-untreated cells, whereas OA treatment appeared to cause Stx17 to redistribute to a more diffuse pattern (Fig. 1A). We examined whether Stx17 is required for LD biogenesis by silencing the protein. We used two siRNAs (siRNA 440 and 194) (17) that were able to effectively knockdown Stx17 without affecting the expression levels of two important neutral lipid synthesizing enzymes, ACSL3 and DGAT2 (Fig. 1B). Stx17 silencing blocked OA-induced LD formation (Fig. 1C). In accordance with this, TAG synthesis was blocked in Stx17-silenced cells (Fig. 1D). The specific involvement of Stx17 in LD formation was demonstrated by the finding that depletion of SNAP29, a Stx17 partner in autophagy (19), or Sec22b, a partner in membrane trafficking (15), did not affect LD formation (supplemental Fig. S1A). Endogenous LDs were also diminished upon incubation of hepatic cells (HepG2 and Huh7 cells) with the siRNAs (supplemental Fig. S1B).
Fig. 1.
LD formation and TAG synthesis are impaired in Stx17-silenced cells. A: HeLa cells were incubated with or without 150 μM OA for 16 h, fixed, and then double immunostained for Stx17 and a mitochondrial marker, Tom20. Bars, 5 μm. B: HeLa cells were mock-transfected or transfected with siRNA Stx17 (440) or (194). After 72 h, the amounts of the indicated proteins were determined by immunoblotting. C: HeLa cells were mock-transfected or transfected with siRNA Stx17 (440) or (194). At 56 h after transfection, OA was added at a final concentration of 150 μM. After 16 h, the cells were fixed and stained with an anti-Stx17 antibody and LipidTox. Bars, 5 μm. D: HeLa cells were mock-transfected or transfected with siRNA Stx17 (440) or (194), treated with OA for the indicated times, and lysed, and then the amount of TAG was determined. As a negative control, mock-treated HeLa cells were incubated with OA in the presence of 10 μM triacsin C for 16 h, and then the amount of TAG was determined. The bar graph shows the means ± SD (n = 3). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. E: HeLa cells were mock-transfected or transfected with siRNA Stx17 (NC) targeting the 3′ noncoding region of Stx17, and the protein amounts of Stx17 and α-tubulin were determined by immunoblotting (upper left). Alternatively, HeLa cells or HeLa cells expressing the indicated FLAG-tagged constructs were transfected with siRNA Stx17 (NC), treated with OA for 16 h, fixed, and then stained with an anti-FLAG antibody and LipidTox. The bar graphs show the average number (lower left) and size (lower right) of LDs under each condition. Values are the mean ± SD (n = 3). *P ≤ 0.05; **P ≤ 0.01. “Non” denotes Stx17-silenced HeLa cells in which a vector was not transfected. Expression of an unrelated protein (GFP) had no effect on LD formation.
To gain insight into the mechanism by which Stx17 participates in LD biogenesis, we examined which domains of Stx17 are responsible for LD biogenesis. To address this, we performed rescue experiments using siRNA [Stx17 (NC)] that targets the 3′ noncoding region of Stx17 (Fig. 1E). In Stx17-silenced cells, FLAG-tagged Stx17 wild-type showed restored size and number of LDs, excluding the possibility of an off-target effect of the siRNAs used (Fig. 1E, bar graphs). We examined the ability of several Stx17 mutants to compensate for Stx17 depletion. Although the expression levels of the mutants were similar to that of wild-type Stx17 (data not shown), no rescue was observed for Stx17 K254C in which Lys254 in the middle of the CHD was replaced by Cys, the CHD+C mutant, or the ΔSNARE mutant (Fig. 1E, bar graphs), suggesting that both the SNARE domain and the CHD with the C-terminal cytoplasmic region, the latter of which is required for the MAM localization (17), are involved in LD biogenesis.
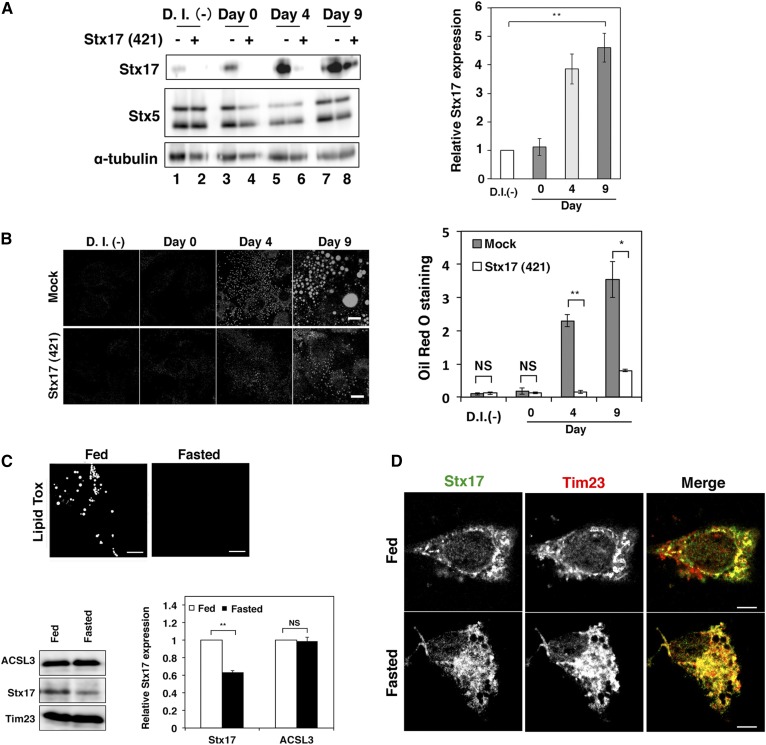
To verify the physiological importance of Stx17 in LD biogenesis, we examined the effect of Stx17 silencing on the differentiation of 3T3-L1 preadipocytes into adipocytes. As differentiation progressed, the expression level of Stx17 markedly increased (Fig. 2A). Silencing of Stx17 inhibited LD formation, as demonstrated by a reduced increase in Oil Red O staining (Fig. 2B).
Fig. 2.
Expression of Stx17 is regulated during 3T3-L1 cell differentiation and by diet. A: The 3T3-L1 cells were mock-transfected or transfected with siRNA targeting mouse Stx17 siRNA (421), and then differentiation was induced with an AdipoInducer reagent. Protein expression levels were determined by immunoblotting using the indicated antibodies. The bar graph on the right shows the relative level of the Stx17 protein normalized to that of α-tubulin. D.I. (-) denotes no differentiation induction. The bar graph shows the means ± SD (n = 3). **P ≤ 0.01. B: Oil Red O fluorescence was observed. Bars, 5 μm. The bar graph on the right shows the quantitation of Oil Red O staining. Values are the means ± SD (n = 3). *P ≤ 0.05; **P ≤ 0.01; NS, not significant. C: Mouse primary hepatocytes were stained with LipidTox (upper panels) or analyzed by immunoblotting (lower panels). The bar graph shows the relative level of the Stx17 protein normalized to that of Tim23. Values are the means ± SD (n = 3). Bars, 5 μm. **P ≤ 0.01; NS, not significant. D: Mouse primary hepatocytes were double immunostained with antibodies against Stx17 and a mitochondrial marker, Tim 23. Bars, 5 μm.
We next examined the localization and expression of Stx17 in primary hepatocytes. When primary hepatocytes were prepared from fed and 24 h-fasted juvenile mice, the expression level of Stx17 in fed hepatocytes that had abundant LDs was found to be substantially higher than that in fasted hepatocytes with only a few LDs (Fig. 2C). Immunofluorescence microscopy showed that Stx17 displayed less colocalization with mitochondria in fed hepatocytes than in fasted hepatocytes (Fig. 2D), which is reminiscent of Stx17 localization in OA-treated HeLa cells (Fig. 1A). Consistent with a previous study (30), we observed that primary hepatocytes prepared from 14 h-fasted adult mice contained more LDs than fed mice (data not shown). The difference in the accumulation of LDs in primary hepatocytes between juvenile and adult mice in response to starvation may reflect a difference in the level of accumulated fat in the adipose tissue, which is degraded during starvation to provide free fatty acids in blood that are subsequently taken up by the liver and transiently stored as TAG in LDs.
Depletion of Stx17 causes aberrant distribution of ACSL3 on the LD surface
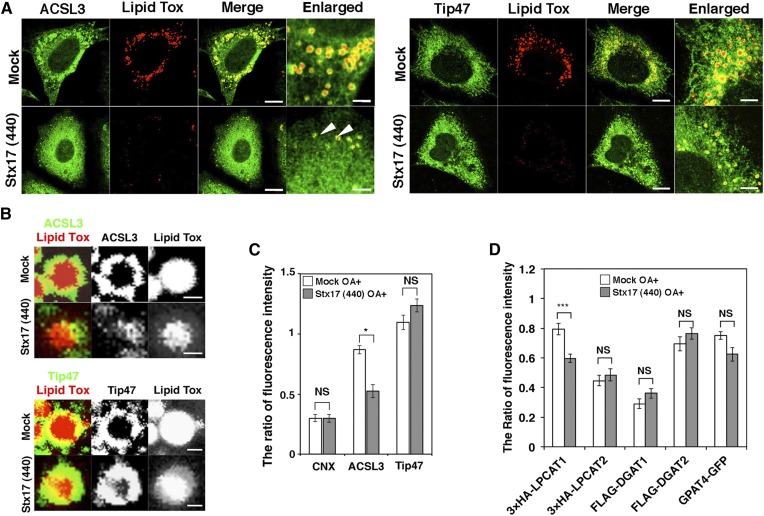
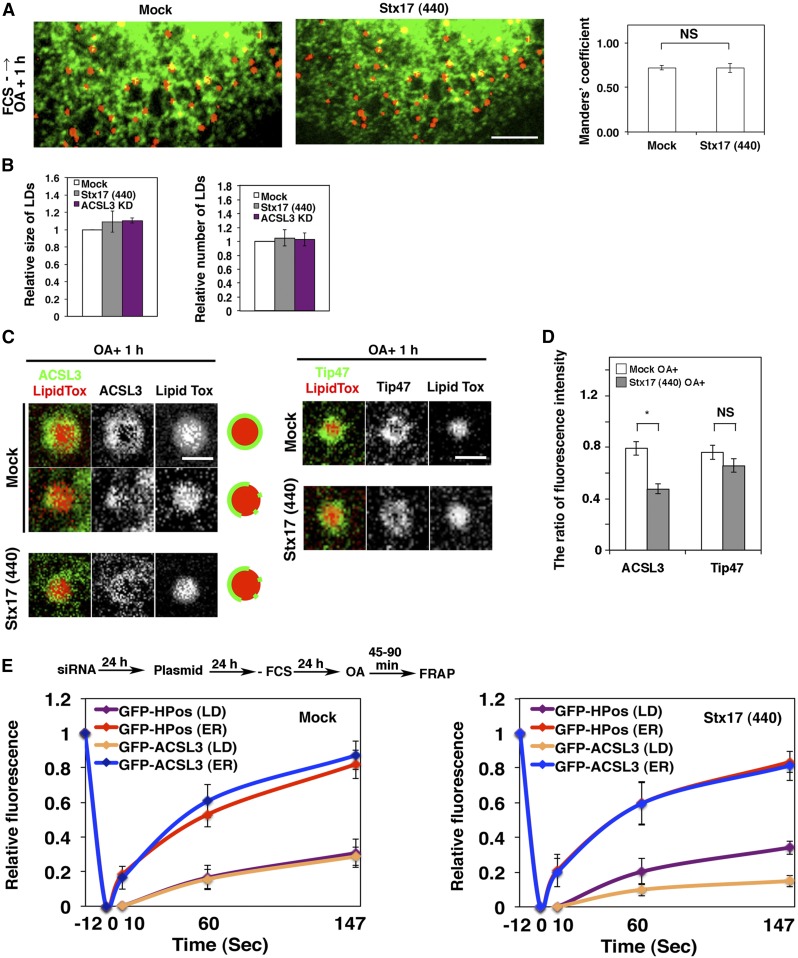
ACSL3, an abundant ACSL family member on LDs in certain cells (31), redistributes from a microdomain of the ER to the LD surface during LD formation, and the enzymatic activity of ACSL3 plays a crucial role in LD expansion (9, 32). Because TAG synthesis was markedly compromised in Stx17-silenced cells, we reasoned that Stx17 depletion might have caused ACSL3 dysfunction. Therefore, we examined the localization of ACSL3 in Stx17-silenced cells. As reported previously (9), ACSL3 was detected on the surface of LDs exhibiting a circular distribution in mock-treated cells (Fig. 3A, B; left upper row and top row, respectively). In Stx17-silenced cells, on the other hand, ACSL3 exhibited a crescent-like distribution on the surface of LDs (Fig. 3A, B; left lower row and second row, respectively). Importantly, there was no difference in the circular distribution of Tip47/PLIN3, a LD-localized protein that redistributes from the cytosol, between mock- and Stx17-silenced cells (Fig. 3A, B; right two rows and third and bottom rows, respectively). Quantification revealed that the recruitment of ACSL3 to the surface of LDs was specifically suppressed by Stx17 silencing (Fig. 3C).
Fig. 3.
Aberrant distribution of ACSL3 on LDs in Stx17-silenced cells. A: HeLa cells were mock-transfected or transfected with siRNA Stx17 (440), treated with OA for 16 h, fixed, and then stained with LipidTox and an antibody against ACSL3 or Tip47. Bars in normal images, 5 μm, and in enlarged images, 1 μm. Arrows indicate LDs with aberrant ACSL3 localization. B: Enlarged images of A. Bars, 200 nm. C: Quantitation of data in B. The fluorescence intensities of CNX, ACSL3, and Tip47 surrounding LDs relative to that of LipidTox were plotted. The bar graph shows the means ± SEM (n = 3). *P ≤ 0.05; NS, not significant. D: HeLa cells were mock-transfected or transfected with siRNA Stx17 (440). After 48 h, cells were transfected with one of the indicated 3x-HA-, FLAG-, or GFP-tagged constructs, and incubated for 8 h, and then OA was added. After 16 h, cells were fixed and stained with LipidTox and an antibody against HA or FLAG. The staining intensity of HA, FLAG, or GFP relative to that of LipidTox was plotted. Values are the means ± SEM (n = 3). ***P ≤ 0.001; NS, not significant.
As ACSL1 and ACSL4 also localize to LDs in certain cells (3), we examined the effect of silencing of these ACSLs. ACSL1 was not colocalized with LDs, whereas ACSL4 showed some colocalization in HeLa cells (supplemental Fig. S2A). Silencing of either protein did not significantly affect LD formation, whereas ACSL3 depletion blocked LD formation (supplemental Fig. S2B).
Next, we examined whether Stx17 regulates the localization of other class I LD proteins. LPCAT1 and -2 are lipid synthesis enzymes that are related to the Lands cycle and relocate from the ER to LDs during LD maturation to regulate the size of LDs (33, 34). GPAT4, which catalyzes the first step of TAG synthesis, has also been reported to redistribute from the ER to LDs during LD formation (14). DGAT1 and -2 catalyze the last step of TAG synthesis, and DGAT2 is known to localize to LDs as well as MAMs (14, 24, 35). Among these enzymes, the localization of only LPCAT1 was significantly affected upon Stx17 depletion (Fig. 3D). It should be noted that no significant translocation defect was observed for LPCAT2, the closest homolog of LPCAT1 among the LPCAT family proteins (36).
Stx17 regulates the redistribution of ACSL3 to LDs
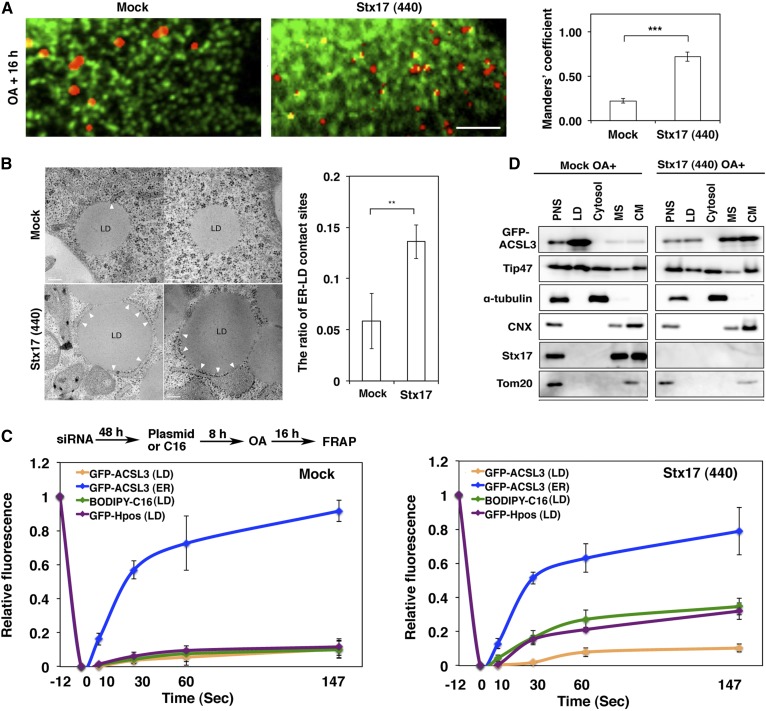
When LD position was compared with that of the ER by immunofluorescence microscopy, many large LDs in mock-treated cells appeared not to be fully in contact with the ER, whereas small LDs in Stx17-silenced cells might be adjacent to the ER (Fig. 4A). Because the resolution of microscopy is limited, we performed electron microscopy to determine the position of LDs relative to the ER. The data showed that the area of LDs that is in contact with the ER in Stx17-silenced cells was significantly larger than that in control cells (Fig. 4B). This may suggest that LDs may remain immature and connected to or close to the ER due to loss of Stx17. To test this, we performed FRAP experiments. In mock-treated cells, the signals of LD-localized BODIPY FL-C16, a membrane-permeable fatty acid that is efficiently incorporated into the TAG pool (37, 38), as well as those of GFP-HPos, which redistributes from the ER to LDs during LD formation (9), were not recovered after photobleaching (Fig. 4C, left; supplemental Fig. S3A), consistent with the view that mature LDs become detached from and/or not fully associated with the ER. On the other hand, significant recovery of the signals derived from these probes was observed after photobleaching of Stx17-silenced cells (Fig. 4C, right; supplemental Fig. S3B), implying that at least some LDs remained associated with the ER. Nevertheless, little recovery was observed for LD-localized GFP-ACSL3 in Stx17-silenced cells. In contrast to LD-localized GFP-ACSL3, an ER-localized GFP-ACSL3 fraction was rapidly recovered regardless of whether Stx17 was present or not (Fig. 4C). Subcellular fractionation analysis showed that, in the case of mock-treated cells, both GFP-ACSL3 and Tip47 were recovered in the LD fraction (Fig. 4D, left), consistent with the view that at least some LDs had pinched off from and/or not fully associated with the ER. In the case of Stx17-silenced cells, on the other hand, Tip47 was also recovered in the LD fraction, but the amount of GFP-ACSL3 recovered in the LD fraction was substantially reduced, along with its appearance in the microsomal and crude mitochondrial fractions (Fig. 4D, right), the latter of which contained the MAM (16). Obviously, Tip47 in the LD fraction was not due to contamination of ER membranes because the ER/MAM marker, CNX, and the ER/Golgi marker, Sec22b, were not recovered in this fraction (Fig. 4D, right). We do not exclude the possibility that some LDs recovered in the LD fraction had been loosely attached to the ER, and dissociated as a consequence of mechanical disruption of cells.
Fig. 4.
Stx17 regulates the redistribution of ACSL3 to LDs. A: HeLa cells transiently expressing GFP-KDEL were mock-transfected or transfected with siRNA Stx17 (440), treated with OA for 16 h, fixed, and then stained with LipidTox. Bar, 5 μm. The degree of colocalization between LipidTox and GFP-KDEL was measured with Manders’ coefficients. The bar graphs show the means ± SD (n = 3). ***P ≤ 0.001. B: Electron micrographs of mock-treated (upper row) and Stx17-silenced cells (lower row) after 16 h OA treatment. Bars, 200 nm. The graph on the right shows the quantitation of EM data. The ratio of the area of ER-LD contact sites relative to the circumference of the LD surface was plotted. Values are the means ± SD (n = 5). **P ≤ 0.01. More than 30 LDs were analyzed in each experiment. C: HeLa cells were treated as depicted in the figure, and then FRAP experiments were performed. The graphs show the relative signal intensity after photobleaching. Values are the means ± SD (n = 3). D: Mock-treated and Stx17-silenced HeLa cells transiently expressing GFP-ACSL3 were incubated with OA for 16 h, and then subjected to LD fractionation analysis. PNS, postnuclear supernatant; MS, microsomes; CM, crude mitochondria.
Next, we performed FRAP experiments at an early stage of LD formation. At this stage, nascent LDs are expected to be fully associated with the ER. To this end, HeLa cells were incubated in the absence of FCS for 24 h to allow the consumption of preformed LDs, and then incubated with OA for 1 h to induce the formation of new LDs. The position relative to the ER, size, and number of LDs in Stx17-depleted cells were indistinguishable from those in mock-treated cells (Fig. 5A, B), suggesting that Stx17 is not required for the early stage of LD formation. It should be noted that similar results were obtained in cells depleted of ACSL3 (Fig. 5B), consistent with the notion that ACSL3 is required for LD maturation and expansion (9, 32). At this time point, Tip47 fully decorated LDs, whereas ACSL3 exhibited a disrupted distribution around the surface of LDs in Stx17-silenced cells (Fig. 5C, D), as observed in cells incubated with OA for 16 h (Fig. 3B, C). FRAP analysis showed that the signals for GFP-ACSL3, as well as those of GFP-HPos, were recovered after photobleaching in mock-treated cells (Fig. 5E, left). Similarly, the signals of GFP-HPos were recovered in Stx17-silenced cells (Fig. 5E, right). However, little recovery of the signal was observed for GFP-ACSL3, verifying that GFP-ACSL3 redistribution to LDs is impaired in Stx17-depleted cells.
Fig. 5.
Stx17 regulates the redistribution of ACSL3 to nascent LDs. A: HeLa cells transiently expressing GFP-KDEL were mock-transfected or transfected with siRNA Stx17 (440), starved for 24 h in the absence of FCS followed by incubation with OA for 1 h, fixed, and then stained with LipidTox. Bar, 5 μm. The degree of colocalization between LipidTox and GFP-KDEL was measured with Manders’ coefficients. NS, not significant. B: HeLa cells were mock-transfected or transfected with siRNA Stx17 (440) or siRNA targeting ACSL3, and incubated for 72 h without FCS for the last 24 h. OA was added, and cells were incubated for 1 h, fixed, stained with LipidTox and an antibody against ACSL3, and quantified. Values are the means ± SD (n = 3). C: HeLa cells were treated as described in A and stained with LipidTox and an antibody against ACSL3 or Tip47. Bars, 200 nm. D: Quantitation of data in C. *P ≤ 0.05; NS, not significant. The fluorescence intensities of ACSL3 and Tip47 surrounding LDs relative to that of LipidTox were plotted. Values are the means ± SEM (n = 3). E: Nascent LDs were analyzed by FRAP using a protocol depicted in the figure.
We performed similar knockdown experiments using Huh7 cells that abundantly contain LDs without OA treatment. As observed in HeLa cells, Stx17 depletion impaired the translocation of ACSL3 to LDs (not shown). Subcellular fractionation demonstrated the prominent presence of ACSL3 in LDs in mock-treated Huh7 cells (supplemental Fig. S4A, left), whereas Stx17 depletion caused ACSL3 redistribution to the microsomal and crude mitochondria membrane fractions (supplemental Fig. S4A, right), as seen in the case of GFP-ACSL3 in HeLa cells (Fig. 4D). The results of FRAP experiments were essentially the same as those obtained using HeLa cells. In mock-treated cells, little FRAP was observed for both GFP-HPos and GFP-ACSL3 (supplemental Fig. S4B, upper panel), whereas GFP-HPos fluorescence, but not GFP-ACSL3 fluorescence, was considerably recovered in Stx17-depleted cells (supplemental Fig. S4B, lower panel). We then examined the early stage of LD formation. To this end, Huh7 cells were fasted for 24 h followed by incubation with FCS for 1 h. The position of LDs relative to the ER was similar between mock-treated and Stx17-silenced cells, but ACSL3 redistribution to LDs was impaired in Stx17-silenced cells (not shown). Subcellular fractionation demonstrated that Stx17 depletion inhibits the redistribution of ACSL3 from the microsomal and crude mitochondrial fractions to LDs (supplemental Fig. S4C, right). As in the case of HeLa cells (Fig. 4C), the signals of GFP-HPos and GFP-ACSL3 were similarly recovered in mock-treated cells (supplemental Fig. S4D, upper panel), whereas the recovery of GFP-ACSL3 fluorescence was much lower than that of GFP-HPos in Stx17-silenced cells (supplemental Fig. S4D, lower panel), consistent with the subcellular fractionation data (supplemental Fig. S4C).
Binding between Stx17 and ACSL3
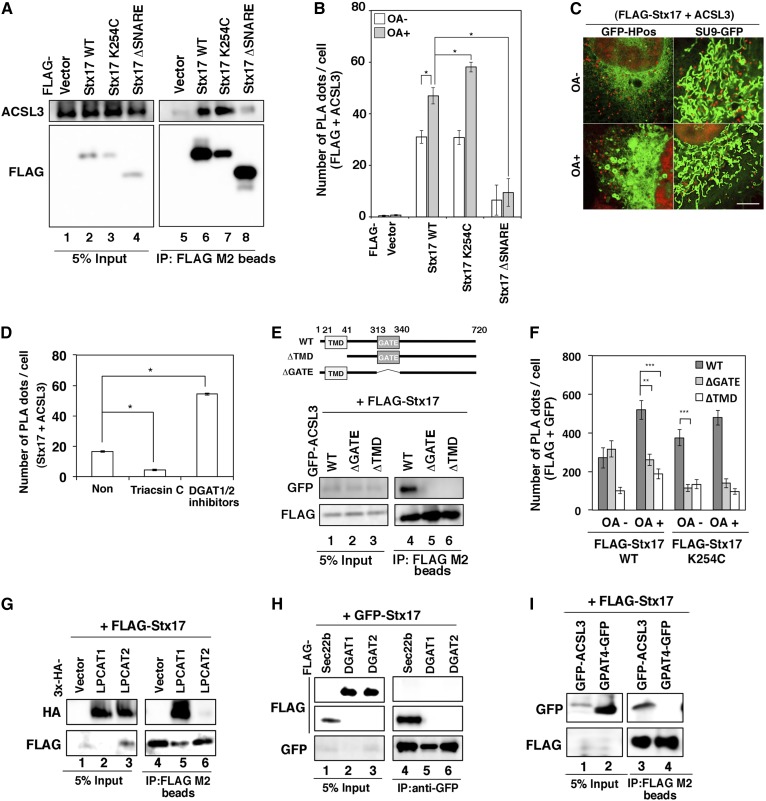
Because the redistribution of ACSL3 to LDs was suppressed in Stx17-silenced cells, Stx17 might regulate the localization of ACSL3 through protein-protein interaction. To test this possibility, we performed immunoprecipitation and PLA. Significant amounts of endogenous ACSL3 were coprecipitated with FLAG-Stx17 wild-type and the K254C mutant, but not with the ΔSNARE mutant (Fig. 6A). Similar results were obtained for PLA (Fig. 6B), suggesting that Stx17 interacts with ACSL3 via its SNARE domain. PLA combined with the ER-LD and mitochondrial markers (GFP-HPos and Su9-GFP) revealed that the interaction between Stx17 and ACSL3 mainly occurs close to mitochondria, rather than to HPos-positive ER membranes, regardless of whether OA is present or not (Fig. 6C).
Fig. 6.
The SNARE domain of Stx17 and GATE domain of ACSL3 are indispensable for the interaction between Stx17 and ACSL3. A: HeLa cells were transfected with one of the indicated FLAG-Stx17 constructs. At 24 h after transfection, cell lysates were prepared and immunoprecipitated (IP) using anti-FLAG M2 beads. The 5% input and the precipitated proteins were analyzed with the indicated antibodies. B: HeLa cells were transfected with one of the indicated FLAG-Stx17 constructs and then incubated for 24 h. Cells were incubated without or with OA for 16 h and fixed, and PLA was performed using antibodies against FLAG and ACSL3. The bar graph shows the means ± SEM (n = 3). *P ≤ 0.05. C: HeLa cells stably expressing FLAG-Stx17 were transfected with GFP-HPos or Su9-GFP. At 24 h after transfection, cells were incubated without or with OA for 16 h, and then PLA was performed using antibodies against FLAG and ACSL3. Bar, 5 μm. D: HeLa cells were incubated with OA for 16 h in the absence (Non) or presence of 10 μM triacsin C or DGAT1/2 inhibitors (80 μM DPF-04620110 and 40 μM PF-06424439). PLA was performed using antibodies against Stx17 and ACSL3. The bar graph shows the means ± SEM (n = 3). *P ≤ 0.05. E: The immunoprecipitation experiments were conducted as described in A using HeLa cells transiently expressing FLAG-Stx17 and the indicated GFP-ACSL3 constructs. F: HeLa cells stably expressing FLAG-Stx17 wild-type or the K254C mutant were transfected with GFP-ACSL3 wild-type, the ΔGATE mutant, or the ΔTMD mutant, and treated as described in B. PLA was performed using antibodies against FLAG and GFP. The bar graph shows the means ± SEM (n = 3). **P ≤ 0.01; ***P ≤ 0.001. G–I: HeLa cells were transfected with GFP- or FLAG-Stx17 and one of the indicated constructs. At 24 h after transfection, cell lysates were prepared and immunoprecipitated using an anti-GFP or -FLAG antibody. The 5% input and the precipitated proteins were analyzed with the indicated antibodies.
Taking advantage of the in situ nature of PLA, we examined whether inhibitor/substrate binding to ACSL3 affects the interaction between ACSL3 and Stx17. The proximity between Stx17 and ACSL3 was suppressed by triacsin C (Fig. 6D), a competitive inhibitor for ACSL (39). To allow accumulation of OA and/or acyl-CoA in cells, we blocked OA-induced TAG synthesis by DGAT inhibitors. We reasoned that inhibition of DGAT 1 and -2 in the presence of OA would allow the accumulation of OA and/or acyl-CoA within cells, although we could not exclude the possibility that some OA and acyl-CoA are consumed by oxidation in mitochondria or secreted into medium (37). DGAT1/2 inhibitors enhanced the proximity between Stx17 and ACSL3 (Fig. 6D). These results suggest that the interaction of ACSL3 with Stx17 is dependent on the inhibitor/substrate in the active site of ACSL3.
We next examined which domains of ACSL3 are required for the interaction with Stx17. ACSL3 has a transmembrane domain (TMD) and a GATE domain (Fig. 6E), the latter of which is implicated in the control of access of the fatty acid substrate to the catalytic site of each of the two subunits (40, 41). Binding experiments revealed that both the TMD and GATE domains of ACSL3 are required for the interaction with Stx17 (Fig. 6E), although the ΔGATE mutant as well as the wild-type protein may be close to Stx17 in the absence of OA (Fig. 6F). These findings combined with the fact that the SNARE domain of Stx17 is required for LD formation (Fig. 1E) suggest that the interaction of ACSL3 with Stx17 is important for the redistribution of ACSL3 to LDs.
It should be noted that ACSL1 or ACSL4, neither of which is responsible for LD formation (supplemental Fig. S2B), did not show proximity to FLAG-Stx17 (supplemental Fig. S2C). Moreover, LPCAT1, whose LD translocation was reduced upon Stx17 depletion (Fig. 3D), was found to bind to FLAG-Stx17, whereas none of the other class I LD proteins (LPCAT2, DGAT1, DGAT2, and GPAT4), whose redistribution to LDs was not affected upon Stx17 depletion (Fig. 3D), bound to FLAG/GFP-Stx17 (Fig. 6G–I), manifesting a correlation between the translocation defect and the binding to Stx17.
Transient overexpression of ACSL3 can compensate for Stx17 depletion
Why is Stx17 required for ACSL3 to redistribute to nascent LDs? A clue came from the observation that, upon OA treatment, endogenous old ACSL3 protein was less efficiently redistributed to LDs than newly transiently synthesized GFP-ACSL3 (compare Fig. 7A with Fig. 4D). We reasoned that Stx17 might prevent the interaction of ACSL3 with ER proteins that prohibit redistribution of ACSL3 to LDs. If so, transient overexpression of ACSL3 may allow this protein to reach LDs even in the absence of Stx17. When transiently overexpressed, FLAG-tagged ACSL3 was found to translocate LDs and restore LD formation in Stx17-silenced cells (Fig. 7B), implying that Stx17 is dispensable for LD biogenesis if ACSL3 is abundantly expressed. It should be noted that the expression level of GFP-ACSL3 was comparable to that of endogenous ACSL3 (Fig. 7B, lane 1), thus explaining at least partly why GFP-ACSL3 failed to translocate to LDs in the absence of Stx17.
Fig. 7.
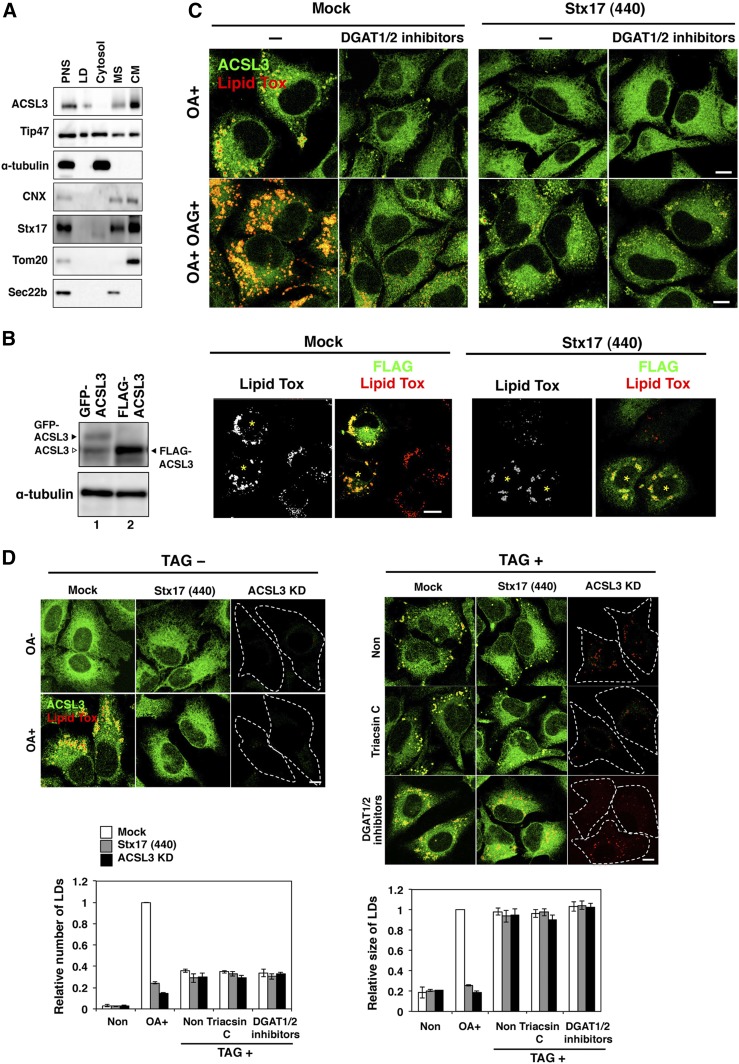
Transient overexpression of ACSL3 can compensate for an Stx17 depletion effect on LD formation. A: HeLa cells were incubated with OA for 16 h, and then subjected to LD fractionation analysis. PNS, postnuclear supernatant; MS, microsomes; CM, crude mitochondria. B: HeLa cells were mock-transfected or transfected with siRNA Stx17 (440). At 48 h, cells were transfected with the plasmid for FLAG-ACSL3 and incubated for 24 h, and then analyzed by immunoblotting using antibodies against ACSL3 and α-tubulin (left, lane 2). As a control, cells transiently expressing GFP-ACSL3 were analyzed (left, lane 1). Of note is that GFP-ACSL3 expression was comparable to that of endogenous ACSL3, whereas FLAG-ACSL3 was expressed in approximately 4-fold excess over endogenous ACSL3. Alternatively, cells overexpressing FLAG-ACSL3 were stained with LipidTox and an antibody against FLAG (right). Bar, 5 μm. Asterisks indicate cells expressing FLAG-ACSL3. C: Mock-treated and Stx17-silenced HeLa cells were incubated with 150 μM OA with or without 150 μM OAG for 16 h in the absence or presence of DGAT inhibitors, and then stained with LipidTox and an antibody against ACSL3. Bars, 5 μm. D: Mock-treated, Stx17-silenced, and ACSL3-silenced HeLa cells were incubated without or with 150 μM OA or 150 μM TAG in the absence or presence of triacsin C or DGAT1/2 inhibitors for 16 h, and then stained with LipidTox and an antibody against ACSL3. Bars, 5 μm. The bar graph shows the means ± SD (n = 3).
The ACSL3 overexpression experiments also suggest that the Stx17 depletion-induced defect in LD formation is not due to secondary effects, such as the shortage of diacylglycerol, substrate for DGAT, or ER structure disruption. To exclude the possibility that diacylglycerol is not adequately supplied in Stx17-silenced cells due to metabolic disturbance, we used membrane-permeable OAG. In mock-treated cells, addition of OAG stimulated OA-induced LD formation, and this LD formation was blocked by DGAT1/2 inhibitors, implying that OAGs incorporated into cells were converted to TAG to produce LDs (Fig. 7C, left). In contrast, LD formation was not stimulated upon the addition of OAG in Stx17-depleted cells (Fig. 7C, right). These results exclude the possibility that impaired LD formation in Stx17-silenced cells is attributed to the lack of diacylglycerol and are consistent with the view that the supply of acyl-CoA on site of LD biogenesis is blocked in Stx17-silenced cells.
Finally, we examined the ability of Stx17-silenced cells to form and/or retain LDs when TAG is fully present within cells. To this end, we incubated Stx17-silenced cells with TAG complexed with BSA. A previous study showed that TAG and other lipids and fatty acids, when complexed with BSA or low density lipoprotein, are taken up by a variety of cells (42). When HeLa cells were incubated with TAG, LDs were formed (Fig. 7D, right). This LD formation was not inhibited by triacsin C or DGAT1/2 inhibitors, suggesting that TAG incorporated into cells directly, not through degradation and resynthesis, induced LD formation. The number and size of LDs formed were indistinguishable between mock-treated and Stx17-silenced cells. These results suggest that Stx17-silenced cells retain the capacity to form LDs.
SNAP23 can outcompete the ACSL3 -Stx17 interaction
Given that Stx17 regulates the redistribution of ACSL3 from the ER to LDs likely through protein-protein interaction, we reasoned that other protein(s) might modulate this interaction. We focused on SNAP23 because previous studies revealed the involvement of this protein in LD formation and localization (43, 44), and our interactome analysis identified SNAP23 as one of the Stx17-interacting proteins (not shown).
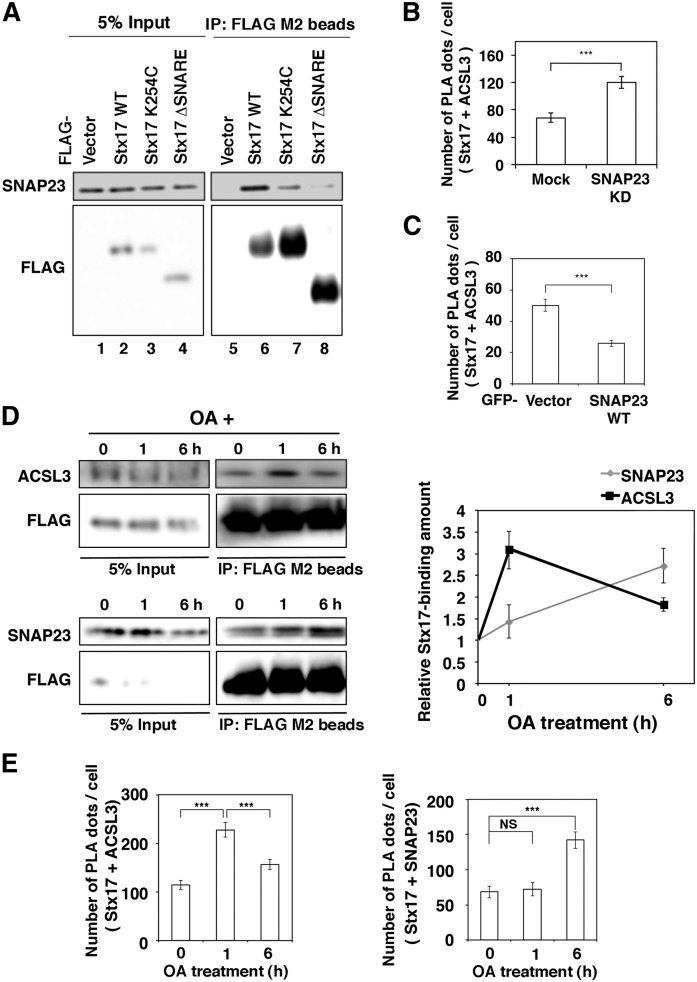
We determined the region of Stx17 responsible for the interaction with SNAP23 by immunoprecipitation. As expected, the interaction of Stx17 with SNAP23 was abolished on deletion of the SNARE domain of Stx17 (Fig. 8A, lane 8). Interestingly, the interaction was drastically reduced on mutation at Lys254 located in the middle of the CHD, even though the resultant mutant retained the SNARE domain (Fig. 8A, lane 7). Because both ACSL3 and SNAP23 interact with the SNARE domain of Stx17, we sought to determine whether they bind to Stx17 in a synergic or competitive manner. When SNAP23 was knocked down, the number of PLA dots representing the proximity between Stx17 and ACSL3 at the endogenous level was significantly increased (Fig. 8B), whereas ectopic expression of SNAP23 reduced the number of the PLA dots (Fig. 8C). These findings suggest that SNAP23 and ACSL3 compete for Stx17 binding.
Fig. 8.
Stx17 changes the binding partner from ACSL3 to SNAP23 during LD maturation. A: HeLa cells were transfected with one of the indicated FLAG-tagged constructs and then incubated for 24 h. Cell lysates were prepared and immunoprecipitated (IP) using anti-FLAG M2 beads. The 5% input and the precipitated proteins were analyzed with antibodies against SNAP23 and FLAG. B, C: HeLa cells were mock-transfected or transfected with siRNA targeting SNAP23 (B), or transfected with the GFP vector or GFP-SNAP23 wild-type (C). PLA was performed using antibodies against Stx17 and ACSL3. The bar graph shows the means ± SEM (n = 3). ***P ≤ 0.001. D: HeLa cells stably expressing FLAG-Stx17 were incubated with OA for the indicated times. Cell lysates were prepared and immunoprecipitated using anti-FLAG M2 beads. The 5% input and the precipitated proteins were analyzed with the indicated antibodies. The graph on the right shows the relative band intensity. Values are the means ± SD (n = 3). E: HeLa cells were treated with OA for the indicated times, and PLA was performed using antibodies against Stx17 and ACSL3 or SNAP23. The bar graphs show the means ± SEM (n = 3). **P ≤ 0.001; NS, not significant.
Given that SNAP23 and ACSL3 compete for Stx17 binding, we were interested in whether the interactions of Stx17 with ACSL3 and SNAP23 change during LD biogenesis. Immunoprecipitation (Fig. 8D) and PLA (Fig. 8E) revealed that the binding of ACSL3 to Stx17 was augmented at a 1 h incubation with OA and then decreased, whereas the binding of Stx17 to SNAP23 increased up to 6 h.
DISCUSSION
The protein and lipid compositions of LDs vary not only with the cell type but also in the degrees of LD maturation and consumption in individual cell types (2–4). The importance of the recruitment of proteins to LDs in LD biogenesis is highlighted by the finding that depletion of LD-localized ACSL3 impaired LD formation, whereas depletion of non-LD-localized members of the ACSL family, ACSL1 and ACSL4, had no impact on LD biogenesis (Ref. 9 and this study), suggesting that on-site supply of acyl-CoA is necessary for the growth of LDs. Therefore, how proteins selectively bind to the LD surface, which is surrounded by a phospholipid monolayer, in a maturation stage-dependent manner is one of the most key questions to be addressed in LD biogenesis. In this study, we demonstrated that Stx17, a SNARE protein localized in the MAM, regulates the translocation of ACSL3 and perhaps LPCAT1 from the ER to nascent LDs. On the other hand, the distribution of some other class I LD proteins, such as DGAT2 and GPAT4, and class II LD proteins, such as Tip47/PLIN3, which translocate from the ER and cytosol to LDs, respectively, was found to be independent of Stx17. These findings suggest that Stx17 selectively interacts with and regulates the localization of a subset of proteins for LD biogenesis.
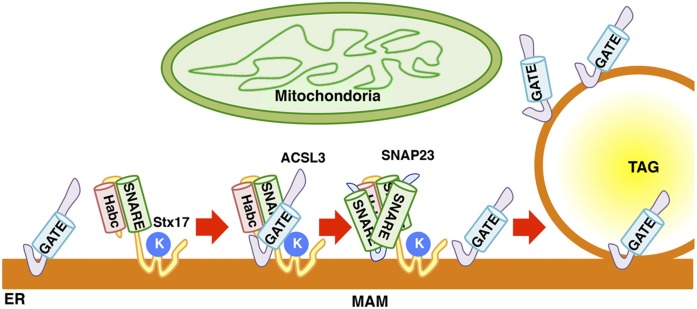
The SNARE domain of Stx17 was found to be responsible for the interaction with ACSL3 (Fig. 6A, B) and SNAP23 (Fig. 8A). For binding to SNAP23, not only the SNARE domain of Stx17, but also Lys254, which is a critical residue for MAM localization (17), is required. The requirement of both the SNARE domain and Lys254 of Stx17 for SNAP23 binding matches the requirement of Stx17 domains for LD biogenesis (Fig. 1E). Moreover, the Stx17-ACSL3 interaction is dependent on the level of SNAP23 in cells (Fig. 8B, C). As LDs mature, the binding partner of Stx17 is changed from ACSL3 to SNAP23 (Fig. 8D, E). Given these observations, it is tempting to speculate that SNAP23 is a factor that regulates the interaction of Stx17 and ACSL3 and facilitates ACSL3 redistribution to nascent LDs. Based on the present observation, we envisage that the interaction of Stx17 and ACSL3 begins at the MAM, a site distant from LD biogenesis sites, under conditions in which acyl-CoA is produced by ACSL3, possibly as a consequence of the supply of fatty acids (Fig. 9). Subsequently, ACSL3 is released from Stx17 due to the binding of SNAP23 to Stx17. ACSL3 may translocate to LD biogenesis sites and finally enters the phospholipid monolayer surrounding LDs. Of course, we do not exclude the possibility that Stx17 facilitates ACSL3 redistribution to LDs by different mechanisms.
Fig. 9.
Working model of the function of Stx17 in LD maturation. For details, see the Discussion. K denotes Lys254.
Overexpression of ACSL3 in Stx17-silenced cells allows ACSL3 redistribution to LDs (Fig. 7B). This suggests that Stx17 is not essential for LD biogenesis, but increases the efficiency of ACSL3 redistribution to assist LD maturation. The dispensability of Stx17 is consistent with the finding that Stx17 is not present in many organisms, including yeast, that retain LDs, although Stx17 is one of the last eukaryotic common ancestors (17). Perhaps, during expansion in number of ER proteins and ER complexity in evolution, many proteins might have been produced that prohibit the redistribution of ACSL3 from the ER to LDs. At the onset of LD formation, Stx17 may interact with ACSL3 to compete out proteins that function as an obstacle for ACSL3 redistribution to LDs. This role of Stx17 may be compatible with the observation that transiently expressed ACSL3 can reach the LD surface without Stx17. Newly synthesized ACSL3s may be abundant enough to reach the LD surface, some are even trapped by obstacle proteins en route to LDs. Moreover, this role of Stx17 predicts that the expression level of Stx17 relative to those of ACSL3 and ACSL3-binding proteins is important for LD biogenesis. Indeed, Stx17 is abundantly expressed in steroidogenic and hepatic cells (15), both of which have large numbers of LDs, and Stx17 expression in the liver is dependent on diet (Fig. 2C). Furthermore, Stx17 expression is induced during differentiation of preadipocytes (Fig. 2A).
In conclusion, our present results disclose that Stx17 facilitates the redistribution of ACSL3 to LDs. A recent study revealed that seipin/BSCL2 supports ACSL3 delivery to LDs by regulating the ER-LD contact (45). This role is different from that of Stx17 in that Stx17 is localized distant from the ER-LD contact and regulates ACSL3 redistribution. Future studies should reveal the detailed mechanism of how Stx17 confers redistribution competency on ACSL3 for LD biogenesis and of how this novel function of Stx17 relates to its previously assigned function in cholesterol and steroid hormone metabolism (15, 46).
Supplementary Material
Acknowledgments
The authors thank Dr. Albert Pol, Dr. Christoph Thiele, Dr. Robert V. Farese and the Gladstone Institutes, UCSF, Dr. Katsuko Tani, and Dr. Naotada Ishihara for the generous gifts of plasmids. The authors are indebted to Ms. Risako Yoshida, Mr. Toshiki Amemiya, and Ms. Miwa Yokoyama for their technical assistance.
Footnotes
Abbreviations:
- ACSL
- acyl-CoA synthetase
- CHD
- C-terminal hairpin-like hydrophobic domain
- CNX
- calnexin
- DGAT
- diacylglycerol acyltransferase
- ER
- endoplasmic reticulum
- FRAP
- fluorescence recovery after photobleaching
- GPAT
- glycerol-3-phosphate O-acyltransferase
- LD
- lipid droplet
- LPCAT
- lysophosphatidylcholine acyltransferase
- MAM
- mitochondria-associated endoplasmic reticulum membrane
- OA
- oleic acid
- OAG
- 1-oleoyl-2-acetyl-sn-glycerol
- PLA
- proximity ligation assay
- SNARE
- soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- SNAP23
- synaptosomal-associated protein of 23 kDa
- Stx17
- syntaxin 17
- TAG
- triacylglycerol
- TMD
- transmembrane domain
This work was supported in part by Ministry of Education, Culture, Sports, Science and Technology Grants-in-Aid for Scientific Research 25291029 and 26650066 (to M.T.), and 26111520 and 26713016 (to K.A.), and the Ministry of Education, Culture, Sports, Science and Technology-supported program for the Strategic Research Foundation at Private Universities (to M.T. and K.A.). The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Walther T. C., and Farese R. V. Jr. 2012. Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81: 687–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohsaki Y., Suzuki M., and Fujimoto T.. 2014. Open questions in lipid droplet biology. Chem. Biol. 21: 86–96. [DOI] [PubMed] [Google Scholar]
- 3.Pol A., Gross S. P., and Parton R. G.. 2014. Biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J. Cell Biol. 204: 635–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiam A. R., and Beller M.. 2017. The why, when and how of lipid droplet diversity. J. Cell Sci. 130: 315–324. [DOI] [PubMed] [Google Scholar]
- 5.Gao Q., and Goodman J. M.. 2015. The lipid droplet-a well-connected organelle. Front. Cell Dev. Biol. 3: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohsaki Y., Suzuki M., and Fujimoto T.. 2017. The lipid droplet and the endoplasmic reticulum. Adv. Exp. Med. Biol. 997: 111–120. [DOI] [PubMed] [Google Scholar]
- 7.Welte M. A. 2015. Expanding roles for lipid droplets. Curr. Biol. 25: R470–R481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stordeur C., Puth K., Sáenz J. P., and Ernst R.. 2014. Crosstalk of lipid and protein homeostasis to maintain membrane function. Biol. Chem. 395: 313–326. [DOI] [PubMed] [Google Scholar]
- 9.Kassan A., Herms A., Fernández-Vidal A., Bosch M., Schieber N. L., Reddy B. J., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. . 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203: 985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiam A. R., and Forêt L.. 2016. The physics of lipid droplet nucleation, growth and budding. Biochim. Biophys. Acta. 1861: 715–722. [DOI] [PubMed] [Google Scholar]
- 11.Choudhary V., Ojha N., Golden A., and Prinz W. A.. 2015. A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cell Biol. 211: 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poppelreuther M., Rudolph B., Du C., Großmann R., Becker M., Thiele C., Ehehalt R., and Füllekrug J.. 2012. The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J. Lipid Res. 53: 888–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kory N., Farese R. V. Jr., and Walther T. C.. 2016. Targeting fat: mechanisms of protein localization to lipid droplets. Trends Cell Biol. 26: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilfling F., Wang H., Haas J. T., Krahmer N., Gould T. J., Uchida A., Cheng J. X., Graham M., Christiano R., Fröhlich F., et al. . 2013. Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets. Dev. Cell. 24: 384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steegmaier M., Oorschot V., Klumperman J., and Scheller R. H.. 2000. Syntaxin 17 is abundant in steroidogenic cells and implicated in smooth endoplasmic reticulum membrane dynamics. Mol. Biol. Cell. 11: 2719–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vance J. E. 2014. MAM (mitochondria-associated membranes) in mammalian cells: lipids and beyond. Biochim. Biophys. Acta. 1841: 595–609. [DOI] [PubMed] [Google Scholar]
- 17.Arasaki K., Shimizu H., Mogari H., Nishida N., Hirota N., Furuno A., Kudo Y., Baba M., Baba N., Cheng J., et al. . 2015. A role for the ancient SNARE syntaxin 17 in regulating mitochondrial division. Dev. Cell. 32: 304–317. [DOI] [PubMed] [Google Scholar]
- 18.McLelland G. L., Lee S. A., McBride H. M., and Fon E. A.. 2016. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J. Cell Biol. 214: 275–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itakura E., Kishi-Itakura C., and Mizushima N.. 2012. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151: 1256–1269. [DOI] [PubMed] [Google Scholar]
- 20.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. . 2013. Autophagosomes form at ER-mitochondria contact sites. Nature. 495: 389–393. [DOI] [PubMed] [Google Scholar]
- 21.Takáts S., Nagy P., Varga Á., Pircs K., Kárpáti M., Varga K., Kovács A. L., Hegedűs K., and Juhász G.. 2013. Autophagosomal syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201: 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diao J., Liu R., Rong Y., Zhao M., Zhang J., Lai Y., Zhou Q., Wilz L. M., Li J., Vivona S., et al. . 2015. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 520: 563–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusiñol A. E., Cui Z., Chen M. H., and Vance J. E.. 1994. A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 269: 27494–27502. [PubMed] [Google Scholar]
- 24.Stone S. J., Levin M. C., Zhou P., Han J., Walther T. C., and Farese R. V. Jr. 2009. The endoplasmic reticulum enzyme DGAT2 is found in mitochondria-associated membranes and has a mitochondrial targeting signal that promotes its association with mitochondria. J. Biol. Chem. 284: 5352–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirose H., Arasaki K., Dohmae N., Takio K., Hatsuzawa K., Nagahama M., Tani K., Yamamoto A., Tohyama M., and Tagaya M.. 2004. Implication of ZW10 in membrane trafficking between the endoplasmic reticulum and Golgi. EMBO J. 23: 1267–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizoguchi T., Nakajima K., Hatsuzawa K., Nagahama M., Hauri H. P., Tagaya M., and Tani K.. 2000. Determination of functional regions of p125, a novel mammalian Sec23p-interacting protein. Biochem. Biophys. Res. Commun. 279: 144–149. [DOI] [PubMed] [Google Scholar]
- 27.Miyazaki M. 1977. Primary culture of adult rat liver cells. I. Preparation of isolated cells from trypsin-perfused liver of adult rat. Acta Med. Okayama. 31: 351–360. [PubMed] [Google Scholar]
- 28.Ohsaki Y., Kawai T., Yoshikawa Y., Cheng J., Jokitalo E., and Fujimoto T.. 2016. PML isoform II plays a critical role in nuclear lipid droplet formation. J. Cell Biol. 212: 29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki M., Murakami T., Cheng J., Kano H., Fukata M., and Fujimoto T.. 2015. ELMOD2 is anchored to lipid droplets by palmitoylation and regulates adipocyte triglyceride lipase recruitment. Mol. Biol. Cell. 26: 2333–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chitraju C., Trötzmüller M., Hartler J., Wolinski H., Thallinger G. G., Lass A., Zechner R., Zimmermann R., Köfeler H. C., and Spener F.. 2012. Lipidomic analysis of lipid droplets from murine hepatocytes reveals distinct signatures for nutritional stress. J. Lipid Res. 53: 2141–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujimoto Y., Itabe H., Sakai J., Makita M., Noda J., Mori M., Higashi Y., Kojima S., and Takano T.. 2004. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim. Biophys. Acta. 1644: 47–59. [DOI] [PubMed] [Google Scholar]
- 32.Fujimoto Y., Itabe H., Kinoshita T., Homma K. J., Onoduka J., Mori M., Yamaguchi S., Makita M., Higashi Y., Yamashita A., et al. . 2007. Involvement of ACSL in local synthesis of neutral lipids in cytoplasmic lipid droplets in human hepatocyte HuH7. J. Lipid Res. 48: 1280–1292. [DOI] [PubMed] [Google Scholar]
- 33.Moessinger C., Kuerschner L., Spandl J., Shevchenko A., and Thiele C.. 2011. Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J. Biol. Chem. 286: 21330–21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moessinger C., Klizaite K., Steinhagen A., Philippou-Massier J., Shevchenko A., Hoch M., Ejsing C. S., and Thiele C.. 2014. Two different pathways of phosphatidylcholine synthesis, the Kennedy Pathway and the Lands Cycle, differentially regulate cellular triacylglycerol storage. BMC Cell Biol. 15: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuerschner L., Moessinger C., and Thiele C.. 2008. Imaging of lipid biosynthesis: how a neutral lipid enters lipid droplets. Traffic. 9: 338–352. [DOI] [PubMed] [Google Scholar]
- 36.Shindou H., and Shimizu T.. 2009. Acyl-CoA:lysophospholipid acyltransferases. J. Biol. Chem. 284: 1–5. [DOI] [PubMed] [Google Scholar]
- 37.Rambold A. S., Cohen S., and Lippincott-Schwartz J.. 2015. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Dev. Cell. 32: 678–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somwar R., Roberts C. T. Jr., and Varlamov O.. 2011. Live-cell imaging demonstrates rapid cargo exchange between lipid droplets in adipocytes. FEBS Lett. 585: 1946–1950. [DOI] [PubMed] [Google Scholar]
- 39.Omura S., Tomoda H., Xu Q. M., Takahashi Y., and Iwai Y.. 1986. Triacsins, new inhibitors of acyl-CoA synthetase produced by Streptomyces sp. J. Antibiot. (Tokyo). 39: 1211–1218. [DOI] [PubMed] [Google Scholar]
- 40.Soupene E., Dinh N. P., Siliakus M., and Kuypers F. A.. 2010. Activity of the acyl-CoA synthetase ACSL6 isoforms: role of the fatty acid Gate-domains. BMC Biochem. 11: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hisanaga Y., Ago H., Nakagawa N., Hamada K., Ida K., Yamamoto M., Hori T., Arii Y., Sugahara M., Kuramitsu S., et al. . 2004. Structural basis of the substrate specific two-step catalysis of long chain fatty acyl-CoA synthetase dimer. J. Biol. Chem. 279: 31717–31726. [DOI] [PubMed] [Google Scholar]
- 42.Hameed R. 2013. Analysis of lipid uptake and processing in cultured cells. PhD Dissertation. Universität Bonn, Bonn, Germany. http://hss.ulb.uni-bonn.de/2013/3112/3112.pdf.
- 43.Boström P., Andersson L., Rutberg M., Perman J., Lidberg U., Johansson B. R., Fernandez-Rodriguez J., Ericson J., Nilsson T., Borén J., et al. . 2007. SNARE proteins mediate fusion between cytosolic lipid droplets and are implicated in insulin sensitivity. Nat. Cell Biol. 9: 1286–1293. [DOI] [PubMed] [Google Scholar]
- 44.Jägerström S., Polesie S., Wickström Y., Johansson B. R., Schröder H. D., Højlund K., and Boström P.. 2009. Lipid droplets interact with mitochondria using SNAP23. Cell Biol. Int. 33: 934–940. [DOI] [PubMed] [Google Scholar]
- 45.Salo V. T., Belevich I., Li S., Karhinen L., Vihinen H., Vigouroux C., Magré J., Thiele C., Hölttä-Vuori M., Jokitalo E., et al. . 2016. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35: 2699–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin Y. Hou X., Shen W. J., Hanssen R., Khor V. K., Cortez Y., Roseman A. N., Azhar S., and Kraemer F. B.. 2016. SNARE-mediated cholesterol movement to mitochondria supports steroidogenesis in rodent cells. Mol. Endocrinol. 30: 234–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.