Abstract
apoE is the primary lipid carrier within the CNS and the strongest genetic risk factor for late onset Alzheimer’s disease (AD). apoE is primarily lipidated via ABCA1, and both are under transcriptional regulation by the nuclear liver X receptor (LXR). Considerable evidence from genetic (using ABCA1 overexpression) and pharmacological (using synthetic LXR agonists) studies in AD mouse models suggests that increased levels of lipidated apoE can improve cognitive performance and, in some strains, can reduce amyloid burden. However, direct synthetic LXR ligands have hepatotoxic side effects that limit their clinical use. Here, we describe a set of small molecules, previously annotated as antagonists of the purinergic receptor, P2X7, which enhance ABCA1 expression and activity as well as apoE secretion, and are not direct LXR ligands. Furthermore, P2X7 is not required for these molecules to induce ABCA1 upregulation and apoE secretion, demonstrating that the ABCA1 and apoE effects are mechanistically independent of P2X7 inhibition. Hence, we have identified novel dual activity compounds that upregulate ABCA1 across multiple CNS cell types, including human astrocytes, pericytes, and microglia, through an indirect LXR mechanism and that also independently inhibit P2X7 receptor activity.
Keywords: adenosine 5′-triphosphate-binding cassette transporter A1, apolipoprotein E, nuclear receptors/liver X receptor, Alzheimer’s disease, brain, astrocyte, P2X7 receptor
Alzheimer’s disease (AD) is the most common form of dementia and results in severe memory impairment. The apoE gene, the primary lipid carrier in the CNS, is the most highly associated susceptibility locus for late onset AD and also plays major roles in the metabolism of Aβ peptides as well as in inflammation (1, 2). Previous work from our laboratory and others has established a key role for the ABCA1 protein, which effluxes cholesterol and phospholipids onto apolipoproteins, in regulating apoE levels and function in the CNS (3–7). ABCA1 deficiency leads to poorly lipidated and rapidly degraded apoE and can increase amyloid burden in AD mouse models (7, 8). Loss-of-function mutations in the human ABCA1 gene are reported to be associated with low plasma apoE and increased AD risk (9, 10). Conversely, in AD mice, selective overexpression of ABCA1 increases CNS apoE lipidation and markedly decreases amyloid deposition (6). Likewise, upregulation of ABCA1 through inhibition of microRNA-33 increases lipidation of apoE and decreases Aβ levels in the brain (11). Indeed, lipidated apoE has been proposed to promote the proteolytic degradation of Aβ peptides (12). Hence, upregulating ABCA1 expression or function may be of therapeutic interest for AD by increasing apoE lipidation, thereby contributing to decreased Aβ pathology.
Both ABCA1 and apoE are transcriptionally regulated by the nuclear receptors, liver X receptor (LXR), PPARγ, and retinoid X receptor (RXR) (13, 14). Direct LXR and RXR agonists stimulate ABCA1 activity and increase apoE lipidation. In animal models of AD, direct LXR agonists can reduce amyloid pathology and have shown considerable efficacy in maintaining cognitive function [reviewed in Hong and Tontonoz (15)]. However, their undesired side effects have limited advancement in clinical trials (16, 17) and there is considerable interest in developing new classes of ABCA1 and apoE modulators.
Here, we describe a set of novel compounds discovered in a phenotypic screen that included a counterscreen to eliminate direct LXR agonists that upregulate ABCA1 and apoE. Intriguingly, these compounds have dual activity, as they are also known antagonists of the purinergic receptor, P2X7, an ATP-gated cation-selective channel implicated in neurodegeneration and chronic inflammation (18), although the apoE modulating effects are independent of P2X7. As increasing ABCA1-mediated lipidation of apoE and reducing neuroinflammation are both desirable properties for potential AD therapeutics, these compounds may simultaneously allow the study of lipid metabolism and inflammation on AD pathogenesis due to their dual activity effects.
MATERIALS AND METHODS
Cell models and reagents
Human CCF-STTG1 astrocytoma cells, P2RX7−/− CCF-STTG1, and P2X7-overexpressing HEK293 cells (described below) were obtained from AstraZeneca (Sweden). WT HEK293, human HepG2 hepatoma, and human microglia clone 3 (HMC3) cell lines were purchased from ATCC (Manassas, VA). Immortalized LXR double knockout (LXRα−/LXRβ−) and LXR expressing (LXRα+/LXRβ−) mouse embryonic fibroblasts (MEFs) (19) were kindly provided by Dr. Peter Tontonoz. Primary human astrocytes, primary human brain vascular pericytes, and primary hepatocytes were purchased from ScienCell (Carlsbad, CA). Primary mouse mixed glia were cultured from postnatal days 0–2 WT or P2rx7−/− pups, as described (20). The compounds, AZ-1, AZ-2, and AZD9056, and the LXR antagonist, GSK2033, were provided by AstraZeneca. The P2X7 inhibitor, Brilliant Blue G (BBG), and the LXR agonists, T0901317 and GW3965, were purchased from Sigma-Aldrich. T0901317 was used as the positive control for most experiments, whereas GW3965 was used for experiments with primary cells due to the greater response of primary cells to GW3965 than T0901317. Commercial P2X7 antagonists, A740003, A804598, A839977, JNJ47965567, A438079, AZ11645373, and AZ10606120, were purchased from Tocris Bioscience (Bristol, UK). Stocks of all compounds were prepared in DMSO, except for BBG that was prepared in distilled deionized water.
Cell culture and treatment
Parental CCF-STTG1 cells were cultured in a mixed medium consisting of three parts of high-glucose DMEM with l-glutamine (Sigma, D6429) and one part of Ham’s F12 (Sigma, N6658) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin (P/S) (Gibco). P2RX7−/− CCF-STTG1 cells were cultured in RPMI (Gibco), 1640-GlutaMAX™ supplemented with 10% FBS, and 1% P/S. Primary mouse mixed glia, MEFs, and HEK293 cells were cultured in DMEM (Gibco) supplemented with 10% FBS, 2 mM l-glutamine, and 1% P/S; HepG2 cells were maintained in the same growth media further supplemented with 1 mM sodium pyruvate and 1× nonessential amino acids (Invitrogen). Primary human astrocytes, pericytes, and hepatocytes were cultured in their respective growth media (containing 2% FBS) provided by ScienCell. HMC3 cells were grown in Eagle’s minimum essential medium (ATCC) supplemented with 10% FBS.
For immunoblotting, mRNA, and select apoE ELISA assays, cells were seeded in either 12-well (CCF-STTG1: 300,000 cells/well; MEF: 100,000 cells/well; HepG2: 400,000 cells/well; primary mouse mixed glia: 250,000 cells/well; primary human astrocytes: 300,000 cells/well; primary human pericytes: 100,000 cells/well; primary human hepatocytes: 150,000 cells/well; HMC3: 100,000 cells/well) or 24-well (CCF-STTG1: 150,000 cells/well) plates in their respective standard growth media. After 24 h, cells were treated with treatment media [parental CCF-STTG1: 3:1 DMEM:F12 with 1% P/S and 1% FBS; P2RX7−/− CCF-STTG1: RPMI 1640-GlutaMAX™ with 1% P/S and 1% FBS; MEF and primary mouse mixed glia: 1:1 DMEM:F12 (Gibco, 11330) with 1% P/S and no serum; primary astrocytes, pericytes, and hepatocytes: their respective growth media; HepG2 and HMC3: 1:1 DMEM:F12 with 1% P/S and 1% FBS] containing DMSO, positive control T0901317 or GW3965, or test compounds for the indicated time intervals. For dose response experiments, compounds were serially diluted 1:3 to generate a range of concentrations. For all experiments, the final concentration of the vehicle DMSO was equalized for all treatment conditions.
Electrophoresis and immunoblotting
For native PAGE, medium samples were mixed with nondenaturing loading dye to a final concentration of 0.04% bromophenol blue, 4.0% glycerol, and 100 mM Tris (pH 6.8) and resolved on 6% nondenaturing Tris-HCl polyacrylamide gels. To visualize apoE, native gels were transferred as described below and probed with 1:1,000 anti-apoE antibody (Cell Signaling Technology, 13366S) overnight. For denaturing PAGE, cells were washed with 1× PBS and lysed in RIPA lysis buffer (20 mM Tris, 1% NP40 alternative, 5 mM EDTA, 50 mM NaCl, 10 mM Na pyrophosphate, 50 mM NaF, and complete protease inhibitor (Roche) (pH 7.4)]. Protein concentration was determined by BCA protein assay (Pierce). Cellular proteins (20–40 μg/well) were mixed with loading dye with a final concentration of 2% SDS and 1% β-mercaptoethanol, incubated for 5 min at 95°C, and resolved on 10% Tris-HCl polyacrylamide gels. Proteins were transferred onto PVDF membranes (Millipore) at 24 V overnight at 4°C. After blocking with 5% nonfat milk in PBS for 1 h, membranes were probed overnight at 4°C with 1:1,000 monoclonal mouse anti-ABCA1 (Neuromics, Edina, MN; MO13101), 1:2,000 rabbit anti-P2X7 (Alomone, Israel; APR-004), or 1:10,000 anti-GAPDH or anti-β-actin (Millipore) loading controls for 30 min. Membranes were washed with PBST (1× PBS with 0.05% Tween-20) and then incubated for 1 h with HRP-labeled anti-mouse (1:1,000 for ABCA1 detection, 1:5,000 for GAPDH or β-actin detection) or anti-rabbit (1:1,000 for P2X7 and native apoE) secondary antibodies (Jackson Immuno-Research). Results were visualized using ECL (Amersham) and blot images were captured with a Bio-Rad ChemiDoc MP imaging system (Bio-Rad). Band density was quantified using ImageJ software (version 1.47q, National Institutes of Health). Levels of ABCA1 were normalized to GAPDH (human) or β-actin (mouse).
Capillary Western blotting by Wes™ simple Western
ABCA1 levels in P2rx7−/− primary mouse mixed glia were assessed using an automated simple Western system (ProteinSimple) (21, 22). All procedures were performed with the manufacturer’s reagents, except for the primary antibodies described above, according to the user’s manual. Samples were loaded at a protein concentration of 0.5 μg/μl, incubated with primary antibodies (ABCA1, 1:5,000; β-actin, 1:500) for 60 min and secondary anti-mouse IgG for 30 min. Data were analyzed with Compass software (ProteinSimple). ABCA1 levels were normalized to β-actin.
Real-time quantitative PCR
Cells were lysed in TRIzol (Invitrogen). RNA was extracted and treated with DNase I according to the manufacturer’s protocol (Invitrogen). Real-time quantitative PCR (qPCR) was done with SYBR Green reagents (Roche) on a LightCycler96 system (Roche). The real-time qPCR primer sequences used in this study were previously described (23). Each sample was assayed at least in duplicate and normalized to GAPDH (human) or β-actin (mouse).
apoE ELISA
Secreted apoE levels in culture media were measured by an apoE ELISA protocol described previously (23). Fluorescence was read at 325Ex/420Em on an Infinite M200 Pro plate reader (Tecan Life Science, Switzerland).
Cholesterol efflux assay
CCF-STTG1 cells were seeded at 150,000 cells/well in 24-well plates and cultured for 24 h before labeling for 24 h with 1 μCi/ml of 3H-cholesterol (PerkinElmer Life Sciences) in growth media supplemented with DMSO, 1 μM T0901317, or 10 μM AZ-1 or AZ-2. Labeled cells were then washed and equilibrated in serum-free media for 60 min. Serum-free media containing the same drug treatments were then added to the cells in the absence [no acceptor (NA)] or presence of 10 μg/ml of exogenous lipid-free apoA-I (a kind gift from CSL Behring, Switzerland). After 24 h at 37°C, culture medium was collected and cells were lysed by addition of 0.1 M NaOH and 0.2% SDS, followed by incubation at room temperature for a minimum of 1 h. Radioactivity in media and cell lysate samples was quantified by scintillation counting (PerkinElmer). The percentage cholesterol efflux was calculated as the total counts per minute in the media divided by the sum of the counts per minute in the media plus in the cell lysate.
LXR-Gal4 reporter assay
To profile LXR agonist activity, we used a cell-based reporter gene assay based on expression of a chimeric receptor containing the yeast Gal4 DNA binding domain fused to the human LXRα or LXRβ ligand binding domain, as previously described for SHSY5Y cells (24). The assay was modified for performance in U2-OS cells, which have low endogenous steroid receptor activity (25). Briefly, U2-OS cells were transfected in bulk using Maxcyte technology (MaxCyte STX) with a pSGGAL human LXRα (amino acids 205–447) or pSGGAL human LXRβ (amino acids 216–461) expression plasmid and a 5xUAS Luc reporter plasmid. On the day of assay, cryopreserved Maxcyte transfected cells were thawed and seeded to 384-well plates in assay medium (DMEM, 1% FBS, 2 mM l-glutamine, 20 mM HEPES, 0.36% glucose, and 1% PEST). After at least 3 h recovery at 37°C and 5% CO2, compounds were added to the cell plates using Echo acoustic dispensing and incubated for an additional 40 h before measuring luciferase reporter gene activity using Steadylite plus substrate (PerkinElmer).
Electrophysiology on P2X7-HEK cells
HEK293 cells stably transfected with the human P2X7 receptor were grown in DMEM containing 10% FBS, 1% P/S, and 800 μg/ml geneticin (G418). Cells were plated on glass coverslips and grown for 48 h before experimentation. Cells were recorded at room temperature and at a holding potential of –50 mV. Patch pipettes were pulled from borosilicate glass, with resistances of 2–5 MΩ. Patch pipettes contained saline: 130 mM K-gluconate, 10 mM KCl, 0.1 mM CaCl2, 2 mM Mg-ATP, 1.1 mM EGTA, 10 mM HEPES, and pH 7.4 KOH. Bath saline contained: 140 mM NaCl, 2.5 mM KCl, 2.5 mM CaCl2, 1.2 mM MgCl2, 10 mM HEPES, 11 mM glucose, and pH 7.4 NaOH. Data were acquired at 10 kHz with an Axopatch 200B amplifier and analyzed off-line with pClamp 10 software (Molecular Devices). All cell culture materials were purchased from Invitrogen.
P2X7 CRISPR knockout cell line generation
CCF-STTG-1 cells were maintained in RPMI-1640 (Life Technologies) supplemented with 10% FBS (PAA Laboratories). Cells were transfected by Neon electroporation (two pulses at 1,200 V, 20 ms; Life Technologies), with a custom vector harboring a Cas9-T2A-GFP expression cassette and a U6-sgRNA expression cassette containing a guide sequence to P2X7 exon 3: 5′-AGAGCAGCGGTTGTGTCCC-3′. Following transfection, single GFP-positive cells were isolated by FACS after 48 h. Clones were screened for indels by PCR amplification of genomic DNA using the following primers: 5′-AGGTAGCGCCGGCCTCTGCT-3′ and 5′-AGCCCGAATCCCTGTGCCCTGG-3′. PCR products were Sanger sequenced. Sequence was also confirmed by next generation sequencing using the following primers: forward primer 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGAAAAGTGGAGAGGTTCGCC-3′; reverse primer 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGTGGGCTCAAATACACCCC-3′. P2X7 loss was confirmed by Western blotting using anti-P2X7 antibodies (Novus Biologicals, NB100-109), anti-GAPDH 14C10 (Cell Signaling Technology, 2118), and anti-rabbit HRP (Cell Signaling Technology, 7074) antibodies.
Statistics
To minimize inter-experimental noise, we performed statistical analysis using randomized block ANOVA on raw values with experimental runs as blocks (26). Because ANOVA is a linear model, raw data from immunoblot studies (target protein value over loading control protein value) were first log transformed and then analyzed by a blocked two-way (“Experiment” and “Drug” as the two factors) ANOVA model with Experiment being the blocking factor and with a Dunnett’s multiple comparison posttest (i.e., each drug condition compared with vehicle control). For real-time qPCR analysis, ΔCT values (target gene CT minus reference gene CT) were used in the same blocked two-way ANOVA model. For cholesterol efflux, GSK2033 inhibition, and MEF LXR-dependency experiments, a blocked three-way (Drug, Experiment, and “Additional condition” as the three factors) ANOVA with Experiment as the blocking factor with Sidak’s multiple-comparison tests was used to compare either the test compounds’ effect over vehicle control within each genotype/treatment condition or the effect of genotype/treatment themselves under each test compound condition.
For immunoblot and real-time qPCR results, data are plotted as mean fold-change over vehicle control ± 95% confidence interval (calculated from the aforementioned ANOVA analysis) of the indicated number of independent experiments. For ELISA and cholesterol efflux results, data are presented as mean measurement ± SD from the indicated number of experiments. All statistical analyses were performed using SPSS (version 23) and P values <0.05 were considered significant. Prism 5 (GraphPad Software) was used to graph all data.
RESULTS
Compounds AZ-1 and AZ-2 enhance ABCA1 expression and activity, as well as apoE secretion, in human CCF-STTG1 astrocytoma cells
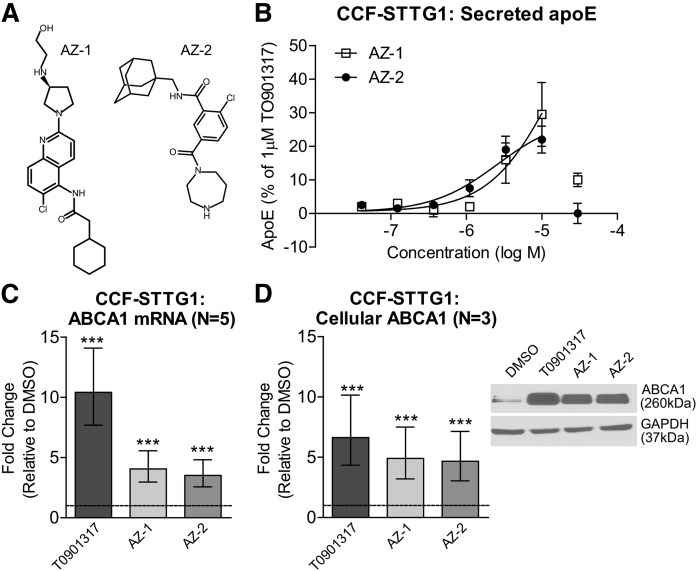
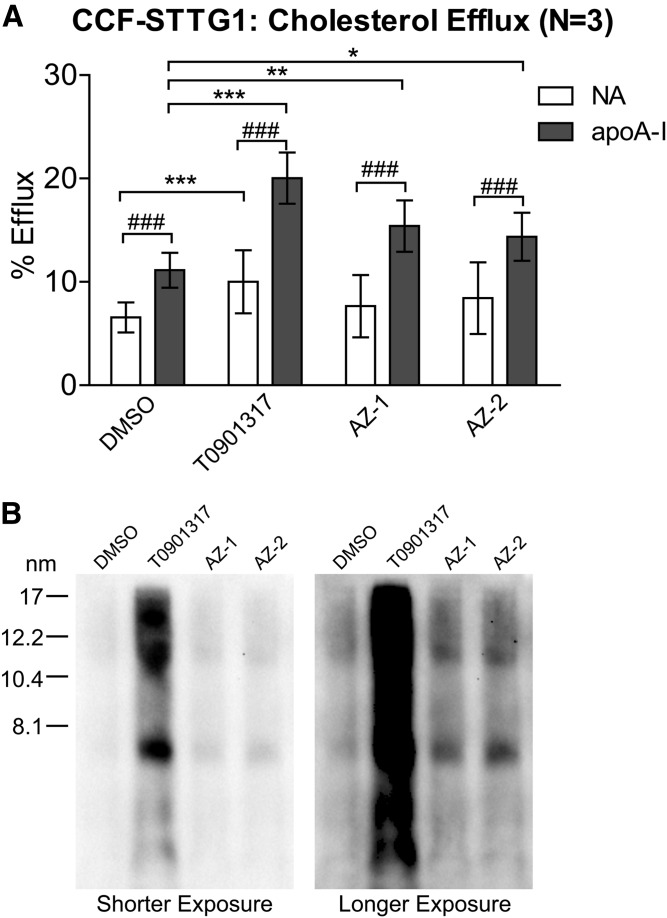
Compounds AZ-1 and AZ-2 (Fig. 1A) were identified through a phenotypic screen performed in human CCF-STTG1 astrocytoma cells for modulators of apoE secretion (Fig. 1B). Screening conditions were similar to those previously published in Fan et al. (23) and will be reported separately (unpublished observations). Both compounds were robust inducers of ABCA1; as at 10 μM, the maximum effective dose, both significantly upregulated ABCA1 mRNA expression (Fig. 1C), as well as cellular ABCA1 protein levels (Fig. 1D) in CCF-STTG1 cells after 72 h of treatment. To determine the effect of AZ-1 and AZ-2 on cholesterol efflux activity to apoA-I, the classical assay of ABCA1 activity, CCF-STTG1 cells were labeled with 3H-cholesterol in the presence of 1 μM T0901317 (a direct LXR agonist) or 10 μM of either AZ-1 or AZ-2 for 24 h. Cholesterol efflux was measured over a 24 h period in the continuous presence of the test compounds using unlipidated recombinant apoA-I as an exogenous lipid acceptor, as the low level of endogenous apoE secreted within 24 h has only a minor contribution to overall cholesterol efflux activity. As expected, the positive control compound, T0901317, which induces both ABCA1 and apoE expression, significantly increased cholesterol efflux both with and without apoA-I (Fig. 2A). By contrast, AZ-1 and AZ-2 significantly enhanced cholesterol efflux compared with the DMSO vehicle control only in the presence of exogenous apoA-I relative to T0901317 at the concentrations used (Fig. 2A). To determine whether overall apoE lipidation was affected by drug treatment, conditioned media were resolved on nondenaturing Tris gels to reveal the particle size distribution of the apoE-containing lipoproteins secreted by CCF-STTG1 cells. As expected and similar to T0901317, both AZ-1 and AZ-2 increased the amount of the HDL-like sized apoE particles ranging from ∼7 to 17 nm in diameter (Fig. 2B). These results demonstrate that both AZ-1 and AZ-2 increase ABCA1 expression and activity in CCF-STTG1 cells and elevate particles that resemble native lipidated apoE.
Fig. 1.
Compounds AZ-1 and AZ-2 modulate ABCA1 and apoE in CCF-STTG1 cells. A: Chemical structures of compounds AZ-1 and AZ-2. B: CCF-STTG1 cells were treated with AZ-1 and AZ-2 using a seven-point concentration response performed at half-log intervals (0.03–30 μM) for 72 h. Data are expressed as percent apoE secretion relative to DMSO (0%) and 1 μM of the positive control LXR agonist, T0901317 (100%). Error bars represent range of duplicate wells in one representative assay. ABCA1 mRNA levels were measured by real-time qPCR (C) and cellular ABCA1 protein levels were measured by immunoblot in CCF-STTG1 cells after 72 h treatment with vehicle control DMSO, 1 μM T0901317, or 10 μM AZ-1 or AZ-2 (D). Graphs represent fold-change over DMSO control (dashed line) and ±95% confidence intervals from N independent experiments indicated in brackets. ***P < 0.001 compared with vehicle control using blocked two-way ANOVA post hoc tests.
Fig. 2.
Compounds AZ-1 and AZ-2 enhance ABCA1 activity. A: CCF-STTG1 cells were labeled with 3H-cholesterol with cotreatment of DMSO alone, 1 μM T0901317, or 10 μM AZ-1 or AZ-2 for 24 h. Cholesterol efflux over 24 h in the absence (NA) or presence of 10 μg/ml of lipid-free apoA-I along with the above drug treatment was evaluated. Graphs represent mean percent efflux and SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 comparing drug effect over respective DMSO control; ###P < 0.001 comparing between NA versus apoA-I within each drug condition by blocked three-way ANOVA post hoc tests. B: Particle size distribution of apoE-containing lipoproteins in the unconcentrated 72 h-conditioned media from drug-treated CCF-STTG1 were assessed by 6% native PAGE followed by immunoblotting for apoE. The ladder on the left represents Stokes diameter. Due to the large signal intensity difference between T0901317-treated and other drug-treated samples, two exposures of the same blot are presented.
Compounds AZ-1 and AZ-2 are not LXR ligands, but activate the LXR pathway indirectly
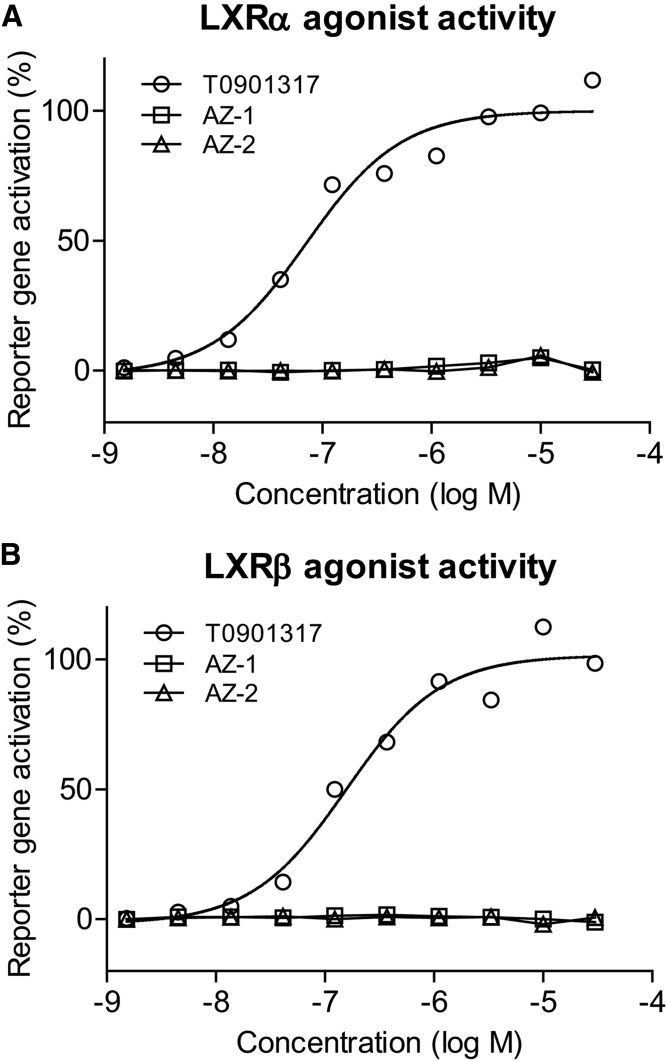
As both ABCA1 and apoE are LXR target genes, we next tested to determine whether AZ-1 and AZ-2 are direct LXR agonists using a U2-OS Gal4 chimeric cell-based reporter assay (25). Control and test compounds were added to U2-OS cells that were transfected with vectors containing a ligand-binding domain of LXRα or LXRβ receptor fused to the Gal4 DNA binding domain. Luciferase reporter gene activities were measured after 40 h of treatment. While the positive control compound, T0901317, exhibited strong agonist activity for both the LXRα and LXRβ receptors in a dose-dependent manner, neither AZ-1 nor AZ-2 showed activity at either LXR receptor, clearly demonstrating that these compounds do not directly bind LXR receptors (Fig. 3). Using the same chimeric reporter construct, lack of direct nuclear receptor activity was confirmed at 1 and 10 μM AZ-2 for several receptors, including LXRα, LXRβ, RXRα, RXRβ, RXRγ, PPARγ, and FXR, all of which were below the threshold of ≥2-fold activation (data not shown).
Fig. 3.
Compounds AZ-1 and AZ-2 are not direct LXR ligands. U2-OS LXR-Gal4 chimeric luciferase reporter assays were used to demonstrate that AZ-1 and AZ-2 (1.5 nM–30 μM) have no direct LXRα (A) or LXRβ (B) agonist activity, unlike the direct LXR agonist T091317, after 40 h of treatment.
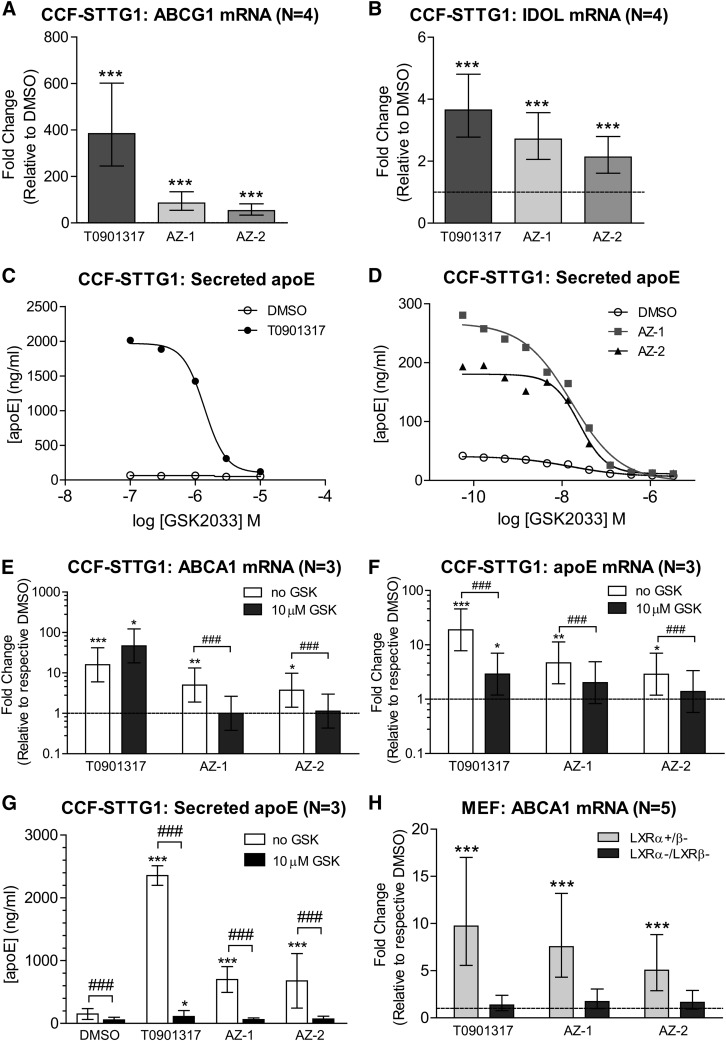
That AZ-1 and AZ-2 upregulate ABCA1 and apoE without being direct LXR agonists raised the possibility that these compounds indirectly activate LXR signaling. To test this hypothesis, we tested to determine whether AZ-1 and AZ-2 could also stimulate other LXR target genes. Treatment of CCF-STTG1 cells with AZ-1 and AZ-2 significantly increased mRNA levels of ABCG1 (Fig. 4A) and inducible degrader of the LDL receptor (IDOL) (Fig. 4B), suggesting that AZ-1 and AZ-2 can modulate expression of ABCA1 and other LXR genes to various degrees. We then used two methods to test whether ABCA1 and apoE upregulation required activation of the LXR pathway. First, CCF-STTG1 cells were cotreated with a known LXR antagonist, GSK2033, to pharmacologically inhibit LXR activity. Concentration-response curves of GSK2033 were established using half-log increments to measure suppression of T0901317-, AZ-1-, or AZ-2-induced apoE secretion (Fig. 4C, D). While more than 10 μM of GSK2033 was needed to completely inhibit T0901317-induced upregulation of apoE secretion (Fig. 4C), 300 nM of GSK2033 was sufficient to completely block AZ-1- or AZ-2-induced apoE secretion (Fig. 4D), demonstrating that both AZ-1 and AZ-2 require LXR activity to increase apoE secretion. Notably, GSK2033 itself displayed a modest inhibitory activity on baseline apoE secretion, which is not surprising as LXR activity likely contributes to endogenous apoE regulation in astrocytes (27). Furthermore, 10 μM GSK2033 completely abolished the ability of 10 μM AZ-1 or AZ-2 to induce ABCA1 mRNA expression, apoE mRNA expression, and secreted apoE levels (Fig. 4E–G). While 10 μM of GSK2033 was not sufficient to inhibit the ability of 1 μM of T0901317 to induce ABCA1 mRNA levels (Fig. 4E), this dose of GSK2033 reduced T0901317-induced apoE expression (Fig. 4F) and abolished apoE secretion (Fig. 4G). Of note, as GSK2033 reduced baseline ABCA1 mRNA levels by more than 90%, cellular ABCA1 protein levels were undetectable by immunoblotting in any GSK2033-treated condition (data not shown).
Fig. 4.
AZ-1 and AZ-2 activate endogenous LXR signaling. ABCG1 (A) and IDOL (B) mRNA levels were measured by real-time qPCR in CCF-STTG1 cells after 72 h treatment with vehicle control DMSO, 1 μM T0901317, or 10 μM AZ-1 or AZ-2. Graphs represent fold-change over DMSO control (dashed line) and ±95% confidence intervals from four independent experiments. ***P < 0.001 compared with vehicle control using blocked two-way ANOVA post hoc tests. C: Log dose response curves for a known LXR antagonist, GSK2033 (0.1–10 μM), on apoE secretion by CCF-STTG1 in cotreatment with DMSO or 1 μM T0901317 for 72 h. D: Log dose response curves for GSK2033 (0.05 nM to 30 μM) on apoE secretion in cotreatment with DMSO or 10 μM AZ-1 or AZ-2 for 72 h. ABCA1 mRNA (E), apoE mRNA (F), and media apoE (G) levels were measured following 72 h treatment of 1 μM T0901317 or 10 μM AZ-1 or AZ-2 without or with 10 μM GSK2033 (GSK). Graphs are presented either as fold-change over respective DMSO control (dashed line) and ±95% confidence interval for C and D, or as mean concentration and SD for G from three experiments. *P < 0.05, **P < 0.01, ***P < 0.001 comparing drug effect over respective DMSO control; ###P < 0.001 comparing between no GSK2033 versus GSK2033 inhibition within each drug condition by blocked three-way ANOVA post hoc tests. H: LXR-knockout (LXRα−/β−) and LXRα-expressing (LXRα+/β−) MEF cells were treated with DMSO or drugs for 48 h. ABCA1 mRNA levels were measured by real-time qPCR. The graph represents fold-change over respective DMSO control (dashed line) ±95% confidence interval from five experiments.
Second, immortalized MEFs deficient for both LXRα and LXRβ and isogenic MEFs that were reconstituted with LXRα were used to confirm whether AZ-1- or AZ-2-induced upregulation of ABCA1 expression requires LXR activity. T0901317-, AZ-1-, and AZ-2-induced ABCA1 upregulation was completely abolished in LXRα and LXRβ double knockout MEFs (Fig. 4H). These data confirm that LXR activity is required for AZ-1- and AZ-2-induced ABCA1 expression. Taken together, as neither compound is a direct LXR ligand, these data show that AZ-1 and AZ-2 lead to indirect activation of the endogenous LXR pathway.
Compounds AZ-1 and AZ-2 have confirmed purinergic receptor P2X7 antagonist activity
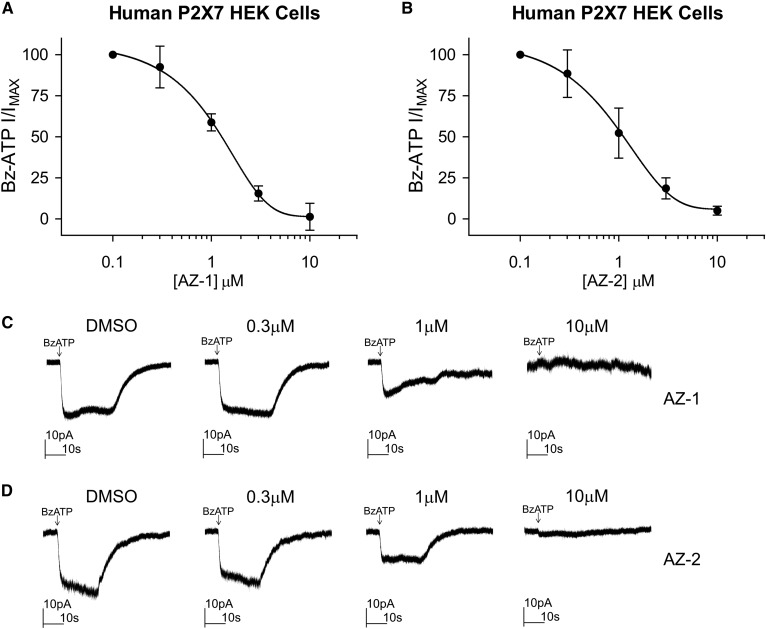
The novel compounds AZ-1 and AZ-2 have been described in patent applications US20010003121 and US6492355, respectively (28, 29). AZ-2 shares an N-(1-adamantylmethyl)-2-chloro-benzamide core with AZD9056 (PubChem CID 10161381), a well-tolerated drug developed by AstraZeneca that has been previously evaluated for treatment of rheumatoid arthritis, osteoarthritis, chronic obstructive pulmonary disease, and Crohn’s disease (30, 31). AZ-1 belongs to a different chemical class of P2X7 antagonists (i.e., quinolyl amide). To confirm the activity of these compounds as P2X7 antagonists, HEK293 cells stably expressing full-length human P2X7 were cotreated with Bz-ATP, a known P2X7 agonist, in the presence of increasing concentrations of AZ-1 and AZ-2 from 0.1 to 10 μM. Inhibition curves for Bz-ATP membrane currents demonstrated that both compounds inhibited ligand-induced receptor activity with an IC50 of 1.1 μM for AZ-1 and an IC50 of 1.0 μM for AZ-2 (Fig. 5A, B). At 10 μM, both AZ-1 and AZ-2 blocked Bz-ATP-induced whole-cell currents (Fig. 5C, D).
Fig. 5.
AZ-1 and AZ-2 exhibit P2X7 receptor antagonist activity. A: Concentration-dependent inhibition of Bz-ATP (100 μM) evoked responses by compound AZ-1 in HEK293 cells that stably overexpress the full-length human P2X7 receptor. The graph represents mean and SD from four cells. B: Concentration-dependent inhibition of Bz-ATP evoked responses by compound AZ-2. The graph represents mean and SD from five cells. C, D: Sample traces of whole-cell currents evoked by Bz-ATP (indicated by the arrow) in the absence or presence of a range of concentrations of AZ-1 or AZ-2.
Other structurally distinct P2X7 antagonists do not induce ABCA1 or apoE
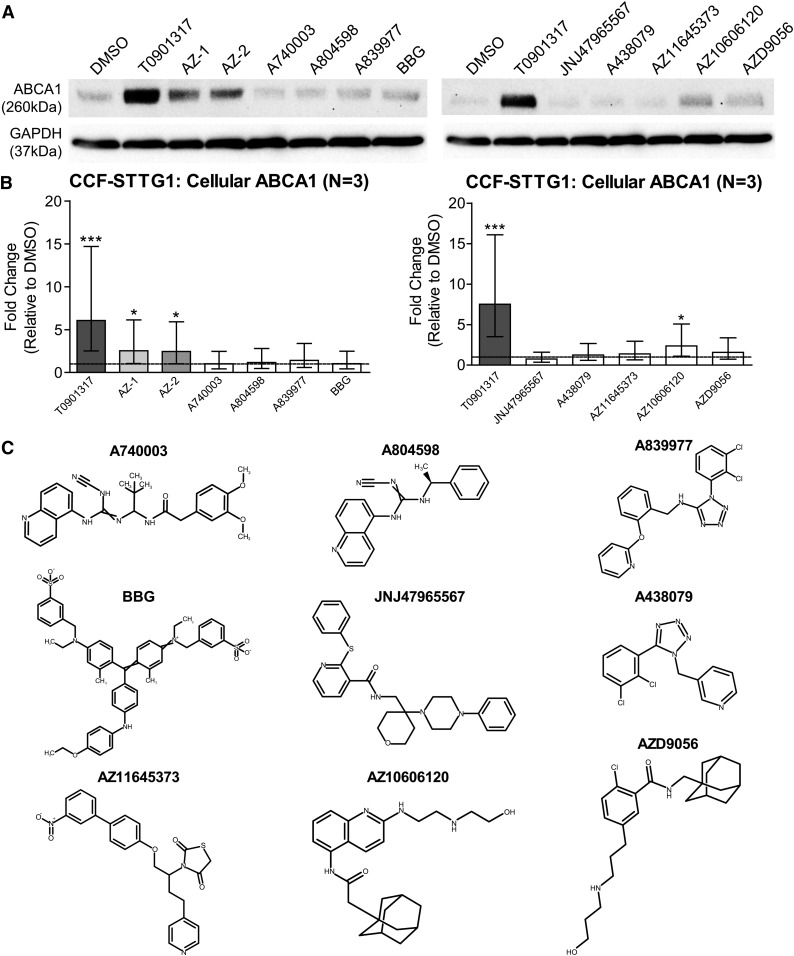
Nine additional P2X7 antagonists, including AZD9056, A740003, A804598, A839977, BBG, JNJ47965567, A438079, AZ11645373, and AZ10606120, were then tested for their ability to increase ABCA1 protein levels at 10 μM in CCF-STTG1 cells. Eight of the nine P2X7 antagonists screened did not induce ABCA1 protein levels, demonstrating that ABCA1 upregulation is not a general activity of known P2X7 antagonists. Only one compound, AZ10606120, modestly but significantly increased cellular ABCA1 protein levels (Fig. 6A, B). When this compound panel was examined for its effect on apoE secretion from CCF-STTG1, only AZ10606120 and AZD9056 exhibited trends toward increasing secreted apoE in a dose-dependent manner (data not shown). Similar to compounds AZ-2 and AZD9056, AZ10606120 also contains an adamantyl moiety, but has a quinolyl acetamide instead of a benzamide group and the amide moiety is reversed between AZ-2/AZD9056 and AZ10606120 (Fig. 6C). Given that several additional structurally distinct P2X7 antagonists do not exhibit ABCA1 or apoE activities, the ability of AZ-1 and AZ-2 to induce ABCA1 is unlikely to depend upon P2X7 antagonism.
Fig. 6.
Structurally distinct P2X7 antagonists do not induce ABCA1 or apoE. CCF-STTG1 cells were treated with 10 μM of published P2X7 antagonists A740003, A804598, A839977, BBG, JNJ47965567, A438079, AZ11645373, AZ10606120, and AZD9056, along with AZ-1, AZ-2, and controls for 72 h. A: Representative immunoblots of cellular ABCA1. B: Quantification of ABCA1 protein levels. The graphs represent fold changes over DMSO control (dashed line) and ±95% confidence interval from three independent experiments. *P < 0.05, ***P < 0.001 compared with vehicle control using blocked two-way ANOVA post hoc tests. C: Chemical structures of the tested P2X7 antagonists.
Inactivation of the P2X7 receptor does not abolish the ability of AZ-1 and AZ-2 to induce ABCA1
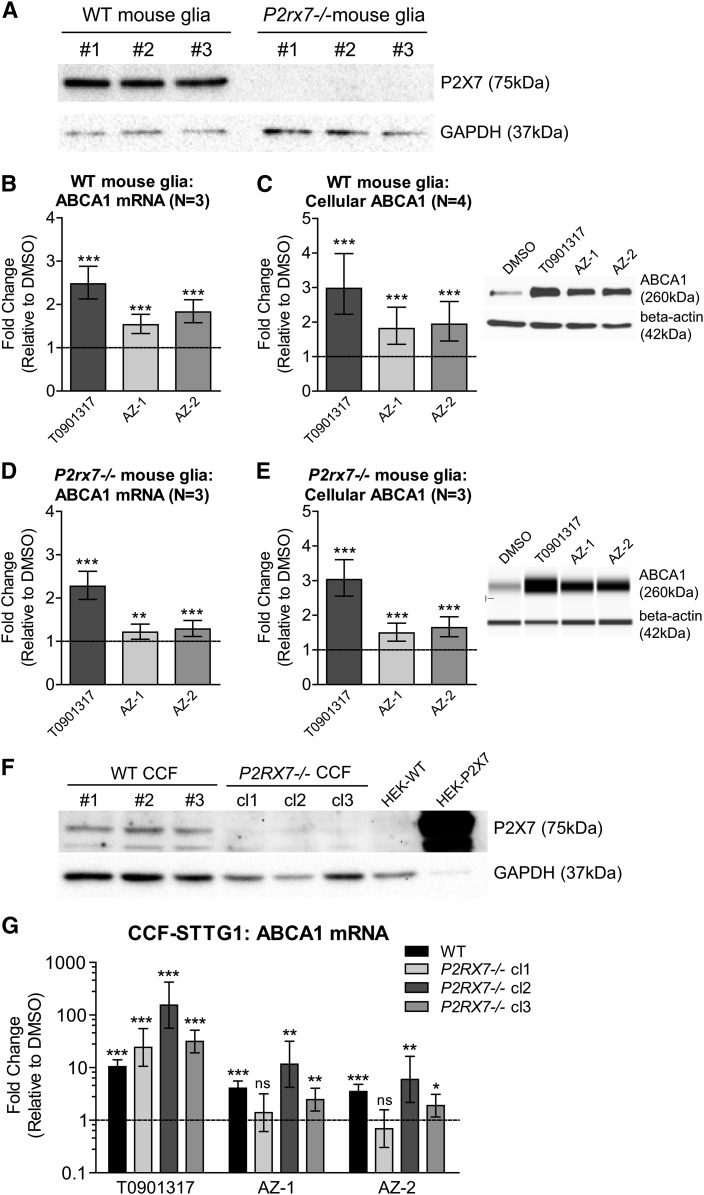
To determine whether antagonism of the P2X7 receptor is required for induction of ABCA1 expression, we cultured primary mixed glia derived from either WT or P2X7-deficient (P2rx7−/−) mice (Fig. 7A). Consistent with the CCF-STTG1 results, primary murine glia also showed significantly increased ABCA1 mRNA (Fig. 7B) and protein levels (Fig. 7C) upon treatment with compounds AZ-1 and AZ-2 for 72 h. As both compounds retained the ability to induce ABCA1 in P2rx7−/− murine glia (Fig. 7D, E), ABCA1 induction and P2X7 antagonism appear to be separable activities of AZ-1 and AZ-2.
Fig. 7.
Induction of ABCA1 by AZ-1 and AZ-2 is P2X7 receptor independent. A: Representative immunoblot confirming lack of P2X7 receptor protein in primary mixed glia derived from P2rx7−/− mice. Untreated cell lysates of each genotype from three experiments are presented. ABCA1 mRNA levels (B) and cellular ABCA1 protein levels(C) in WT mixed glia treated with DMSO, 1 μM T0901317, or 10 μM AZ-1 or AZ-2 for 72 h. ABCA1 mRNA levels (D) and cellular ABCA1 protein levels (E) in P2rx7−/− mixed glia treated with DMSO, 1 μM T0901317, or 10 μM AZ-1 or AZ-2 for 72 h. Graphs represent fold change over DMSO control (dashed line) and ±95% confidence interval from N independent experiments indicated in brackets. **P < 0.01, ***P < 0.001 compared with vehicle control by blocked two-way ANOVA post hoc tests. F: Representative immunoblot showing cellular P2X7 protein levels in WT and P2RX7−/− CCF-STTG1 (CCF) cells. G: ABCA1 mRNA levels in WT CCF-STTG1 (WT), P2RX7−/− clone 1 (P2RX7−/− cl1), P2RX7−/− clone 2 (P2RX7−/− cl2), and P2RX7−/− clone 3 (P2RX7−/− cl3) CCF-STTG1 cells treated with DMSO, 1 μM T0901317, or 10 μM AZ-1 or AZ-2 for 72 h. The graphs represent fold changes over respective DMSO control (dashed line) and ±95% confidence interval from four experiments for WT and three experiments for each P2RX7−/− clone. *P < 0.05, **P < 0.01, ***P < 0.001 compared with respective DMSO by blocked two-way ANOVA post hoc tests. ns, not significant.
To provide further confirmation that the ability of AZ-1 and AZ-2 to induce ABCA1 is independent of P2X7, we assessed P2RX7 knockout CCF-STTG1 cells generated by the CRISPR/Cas9 method. Three clones cultured from the same P2RX7−/− CCT-STTG1 line (clones 1–3) that were confirmed to have no detectable P2X7 protein (Fig. 7F) were compared with parental WT CCF-STTG1 cells. Two of the three clones evaluated, clones 2 and 3, retained AZ-1- and AZ-2-mediated ABCA1 upregulation in the absence of P2X7 protein (Fig. 7G). By contrast, clone 1 showed no ABCA1 response to AZ-1 and AZ-2, although direct induction by T0901317 was intact (Fig. 7G). As we noted that each CRISPR/Cas9 knockout clone exhibited distinct cell morphology, we assessed three available clones and found the majority to retain significant ABCA1 induction in the absence of P2RX7. Finally, although parental CCF-STTG1 cells express P2X7 mRNA, they were unresponsive to Bz-ATP, a classical P2X7 agonist (data not shown).
Compounds AZ-1 and AZ-2 induce ABCA1 in other CNS cells that lack detectable P2X7
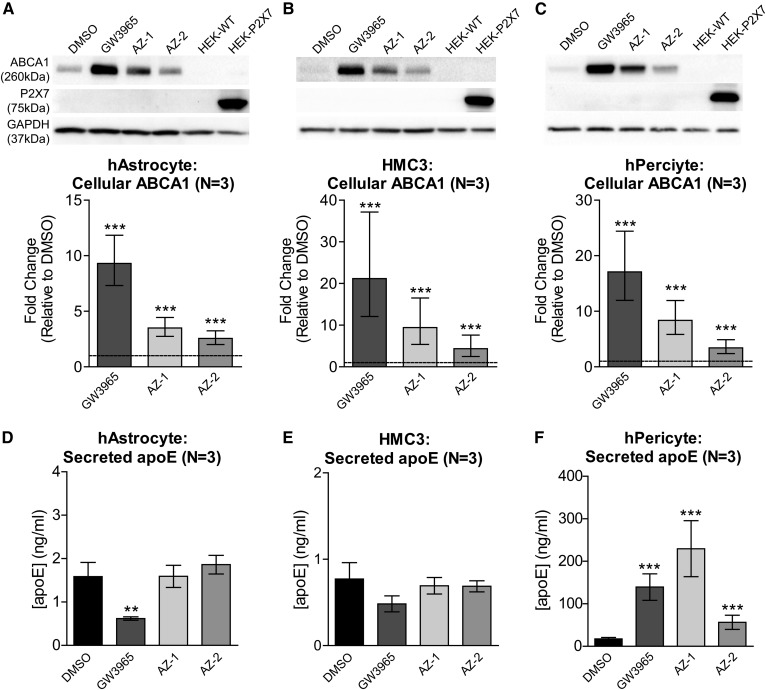
We further explored the responses to compounds AZ-1 and AZ-2 in CNS cells, including primary human astrocytes, a human microglia cell line, HMC3, and primary human brain vascular pericytes. Interestingly, ABCA1 protein levels were significantly elevated by AZ-1 and AZ-2 in all three cell types, whereas the P2X7 receptor was completely undetectable by immunoblotting in any of these cell types (Fig. 8A–C). However, we did detect P2X7 real-time qPCR amplification products of the expected size on an agarose gel for all three cell types, suggesting that these cells still express P2X7 transcript (data not shown). These data provide additional evidence that the ability of AZ-1 and AZ-2 to modulate ABCA1 expression is independent of P2X7. Furthermore, neither compound showed apoE activity in either primary human astrocytes (Fig. 8D) or HMC3 microglia (Fig. 8E), while AZ-1 and AZ-2 significantly increased apoE secretion from primary human pericytes (Fig. 8F), indicating that compounds AZ-1 and AZ-2 consistently induce ABCA1, but differentially modulate apoE in a cell type-specific manner.
Fig. 8.
AZ-1 and AZ-2 induce ABCA1 in other CNS cells that lack detectable P2X7. Primary human astrocytes (hAstrocyte) (A, D), human microglia cell line HMC3 (B, E), and primary human brain vascular pericytes (hPericyte) (C, F) were treated with DMSO, 1 μM GW3965, or 10 μM AZ-1 or AZ-2 for 72 h. A–C: Top panels: Representative immunoblots of cellular ABCA1 and P2X7 protein levels. WT HEK293 (HEK-WT) and transfected HEK293 overexpressing human P2X7 receptors (HEK-P2X7) were used as negative and positive controls for P2X7 detection. GAPDH serves as the loading control. Bottom panels: Quantification of ABCA1 protein levels. The graphs represent fold change over DMSO (dashed line) and ±95% confidence interval from three independent experiments. ***P < 0.001 compared with DMSO by blocked two-way ANOVA post hoc tests. D–F: Secreted apoE levels in conditioned media were measured by apoE ELISA. The graph represents mean concentration and SD from three experiments. **P < 0.01, ***P < 0.001 compared with DMSO by blocked two-way ANOVA post hoc tests.
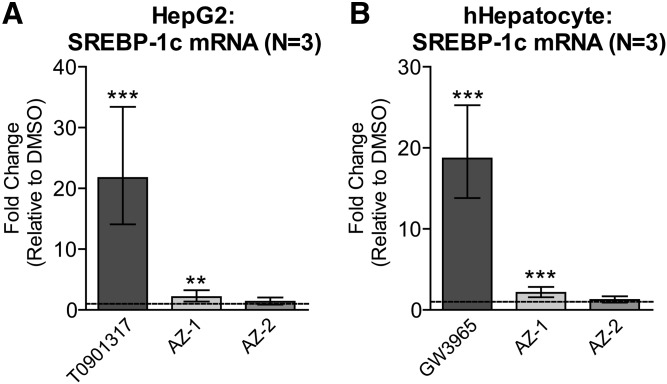
AZ-1 and AZ-2 lead to minimal or no SREBP-1c induction in human liver cells
Induction of hepatic SREBP-1c, an LXR target that induces lipogenesis and leads to hepatic steatosis and hypertriglyceridemia (15), is a major liability of current direct LXR agonists. Importantly, and in contrast to T0901317, AZ-1 caused minimal SREBP-1c induction, whereas AZ-2 showed no effect on SREBP-1c expression in either HepG2 hepatoma cells (Fig. 9A) or primary human hepatocytes (Fig. 9B), suggesting that these compounds may avoid the hepatotoxicity of LXR agonists.
Fig. 9.
Minimal or no SREBP-1c induction by P2X7 antagonists in liver cells. HepG2 cells (A) and primary human hepatocyte (hHepatocyte) cells (B) were treated with DMSO, 1 μM T0901317 or GW3965, or 10 μM AZ-1 or AZ-2 for 48 h. SREBP-1c mRNA levels were measured by real-time qPCR. Data are expressed as fold change (mean ± 95% confidence interval from three experiments) relative to control treatment (dashed line). **P < 0.01, ***P < 0.001 compared with DMSO by blocked two-way ANOVA post hoc tests.
DISCUSSION
ABCA1 is a desirable therapeutic target for AD, as it mediates lipidation of apoE in the CNS and consistently demonstrates beneficial effects on cognition and neuropathology in multiple AD models (32). In people, ABCA1 functional capacity may be impaired with cognitive decline, as ABCA1-mediated cholesterol efflux capacity was reported to be reduced by 30% in the cerebrospinal fluid (CSF) of patients with mild cognitive impairment and AD patients compared with cognitively healthy controls using a BHK ABCA1-expressing cell line cholesterol efflux assay and human CSF (33). Therapies that specifically boost ABCA1 activity could be promising and, perhaps, more important than CSF apoE levels, as the concentrations of apoE and apoA-I in CSF do not clearly correlate with efflux capacity and will also be influenced by lipoprotein uptake. Indeed, a recently described apoE-derived peptide (CS-6253) was reported to elevate ABCA1 levels and increase apoE4 lipidation leading to improved brain pathology and cognitive defects in apoE4-targeted replacement mice (34). As the known hepatic steatosis side effects of direct LXR and RXR agonists pose significant safety challenges to chronic and long-term use, new compound classes that upregulate ABCA1 expression and activity independent of direct LXR transactivation are highly desirable.
This study reports that two compounds, AZ-1 and AZ-2, induce ABCA1 expression across multiple CNS cell types, including astrocytes, pericytes, and microglia. Importantly, AZ-1 and AZ-2 are also selective antagonists of P2X7, yet their effects on ABCA1 and apoE induction are independent of P2X7, demonstrating that AZ-1 and AZ-2 have dual functions, both of which are desirable for AD. The increase in ABCA1 expression is associated with enhanced ABCA1 cholesterol efflux activity resulting in apoE particles that resemble native lipidated apoE and, in some cases, significantly upregulated apoE secretion. A unique feature of AZ-1 and AZ-2 is that they are not direct LXR ligands. For AZ-2, we further confirmed lack of direct activity at RXR and PPARγ, which, together with LXR, comprise the principal known nuclear receptors that transcriptionally regulate ABCA1 and apoE (13, 14).
While AZ-1 and AZ-2 are not direct LXR ligands, they do require LXR activity to induce ABCA1 and apoE, and we hypothesize that they converge indirectly on endogenous LXR signaling pathways, as expression of other target genes, including ABCG1 and IDOL, are also affected. AZ-1 belongs to a family of quinolyl amide P2X7 antagonists, whereas AZ-2 is part of an adamantane series. AZ-2 is structurally similar to AZD9056, which is also able to induce apoE. AZD9056 has been evaluated in phase 2 clinical trials for the treatment of rheumatoid arthritis, osteoarthritis, chronic obstructive pulmonary disease, and Crohn’s disease, and was well-tolerated in all studies, but efficacious only for Crohn’s disease (30, 31). Finally, AZ10606120, a selective high-affinity antagonist that functions by a mode of allosteric inhibition of P2X7, was also able to upregulate both ABCA1 and apoE (35).
All the P2X7 antagonists we tested are well-characterized inhibitors of the P2X7 receptor. The structure activity relationships of the adamantane series of P2X7 antagonists have been well explored and compound 21, which has high activity at the rat P2X7 receptor, shares substantial similarity, namely the N-(1-adamantylmethyl)-2-chloro-benzamide core, with AZ-2 (36, 37). We further confirmed P2X7 antagonist activity of compounds AZ-1 and AZ-2 to block ligand-induced whole-cell currents in HEK-P2X7 cells. Additionally, AZ-1 and AZ-2 have IC50s of 56 and 6.9 nM, respectively, in Bz-ATP-stimulated THP-1 cells when measuring inhibition of pore-mediated ethidium bromide uptake as described (28, 29).
There is mounting evidence that P2X receptor signaling pathways may play a substantial role in neurodegenerative diseases. P2X2 has recently been reported to mediate synaptic loss and dyshomeostasis in primary rat hippocampal neurons exposed to oligomeric Aβ (38) and P2X7 has previously been suggested as an AD target (18). P2X7 is reported to be elevated around amyloid plaques in humans, and upregulation of P2X7 in mice is age dependent, parallels Aβ accumulation, precedes plaque development, and correlates with synaptotoxicity (39). BBG, a blood-brain barrier-permeable rodent P2X7 antagonist, has been reported to attenuate toxicity of injected Aβ42 in these models (40, 41). The high concentration of ATP required to activate the P2X7 receptor usually occurs following a “danger signal,” such as injury, damage, and inflammation, and P2X7 is thought to have negligible activity under normal conditions (42). Indeed, we observed that ABCA1 and apoE activity conferred by AZ-1 and AZ-2 is P2X7-independent, supported by the findings that: 1) genetic deletion of P2RX7 in mouse and human glia does not abolish the ability of AZ-1 and AZ-2 to upregulate ABCA1; and 2) these compounds induce ABCA1 in primary CNS cells that lack detectable P2X7. To explore whether compounds AZ-1 and AZ-2 have off target inhibition at other P2X or P2Y receptors, we performed a concentration response of suramin, a broad-spectrum antagonist that inhibits various P2X and P2Y receptors, but has little activity against P2X7 at low micromolar concentrations. Suramin-treated CCF-STTF1 cells had no upregulation of apoE, even up to concentrations of 100 μM (43) (not shown). In addition, we used FLIPR intracellular calcium assays to specifically measure selectivity at P2X4, which shares a close physical and functional association with P2X7 as well as P2X3 (44). Neither AZ-1 nor AZ-2 functionally inhibited these channels (not shown). These findings imply that inhibition of related P2X or P2Y family members is likely not responsible for modulation of lipid metabolism in response to AZ-1 or AZ-2.
That P2X7 is not the target by which AZ-1 and AZ-2 induce ABCA1 activity raises the obvious question about what this target may be and how it intersects with endogenous LXR signaling pathways. The mechanisms that enable LXR target gene expression in certain cellular contexts in response to these compounds likely depend on the levels and composition of transcription factors and coregulators, as well as epigenetic modifications. AZ-1 and AZ-2 are structurally distinct, but share P2X7 antagonism and modulation of lipid signaling through an unknown target mechanism that results in induction of ABCA1. AZ-1 and AZ-2 compounds can now be used as unique dual-activity tool compounds to further investigate interactions between endogenous LXR signaling, inflammatory pathways, and lipoprotein metabolism.
Acknowledgments
The authors would like to thank Dean Brown (AstraZeneca) for reviewing the manuscript.
Footnotes
Abbreviations:
- AD
- Alzheimer’s disease
- BBG
- Brilliant Blue G
- CSF
- cerebrospinal fluid
- HMC3
- human microglia clone 3
- IDOL
- inducible degrader of the LDL receptor
- LDLR
- low-density lipoprotein receptor
- LXR
- liver X receptor
- MEF
- mouse embryonic fibroblast
- P/S
- penicillin/streptomycin
- RXR
- retinoid X receptor
This work was supported by operating funding provided by AstraZeneca through the AstraZeneca ApoE Alzheimer Disease Academic Alliance to C.L.W. and Weston Brain Institute Rapid Response Grant RR150031 to I.K. The authors declare no conflict of interest. C.R., S.B., J.M., O.E., M.M., R.H., N.J.B. and M.W.W. were employees of AstraZeneca at the time the studies were conducted. C.L.W. receives research support from Pfizer on an unrelated project.
REFERENCES
- 1.Holtzman D. M., Herz J., and Bu G.. 2012. Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2: a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolf A. B., Valla J., Bu G., Kim J., LaDu M. J., Reiman E. M., and Caselli R. J.. 2013. Apolipoprotein E as a beta-amyloid-independent factor in Alzheimer’s disease. Alzheimers Res. Ther. 5: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirsch-Reinshagen V., Zhou S., Burgess B. L., Bernier L., McIsaac S. A., Chan J. Y., Tansley G. H., Cohn J. S., Hayden M. R., and Wellington C. L.. 2004. Deficiency of ABCA1 impairs apolipoprotein E metabolism in brain. J. Biol. Chem. 279: 41197–41207. [DOI] [PubMed] [Google Scholar]
- 4.Donkin J. J., Stukas S., Hirsch-Reinshagen V., Namjoshi D., Wilkinson A., May S., Chan J., Fan J., Collins J., and Wellington C. L.. 2010. ATP-binding cassette transporter A1 mediates the beneficial effects of the liver X receptor agonist GW3965 on object recognition memory and amyloid burden in amyloid precursor protein/presenilin 1 mice. J. Biol. Chem. 285: 34144–34154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirsch-Reinshagen V., Maia L. F., Burgess B. L., Blain J. F., Naus K. E., McIsaac S. A., Parkinson P. F., Chan J. Y., Tansley G. H., Hayden M. R., et al. . 2005. The absence of ABCA1 decreases soluble ApoE levels but does not diminish amyloid deposition in two murine models of Alzheimer disease. J. Biol. Chem. 280: 43243–43256. [DOI] [PubMed] [Google Scholar]
- 6.Wahrle S. E., Jiang H., Parsadanian M., Kim J., Li A., Knoten A., Jain S., Hirsch-Reinshagen V., Wellington C. L., Bales K. R., et al. . 2008. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J. Clin. Invest. 118: 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koldamova R., Staufenbiel M., and Lefterov I.. 2005. Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice. J. Biol. Chem. 280: 43224–43235. [DOI] [PubMed] [Google Scholar]
- 8.Wahrle S. E., Jiang H., Parsadanian M., Hartman R. E., Bales K. R., Paul S. M., and Holtzman D. M.. 2005. Deletion of Abca1 increases Abeta deposition in the PDAPP transgenic mouse model of Alzheimer disease. J. Biol. Chem. 280: 43236–43242. [DOI] [PubMed] [Google Scholar]
- 9.Reynolds C. A., Hong M. G., Eriksson U. K., Blennow K., Bennet A. M., Johansson B., Malmberg B., Berg S., Wiklund F., Gatz M., et al. . 2009. A survey of ABCA1 sequence variation confirms association with dementia. Hum. Mutat. 30: 1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordestgaard L. T., Tybjaerg-Hansen A., Nordestgaard B. G., and Frikke-Schmidt R.. 2015. Loss-of-function mutation in ABCA1 and risk of Alzheimer’s disease and cerebrovascular disease. Alzheimers Dement. 11: 1430–1438. [DOI] [PubMed] [Google Scholar]
- 11.Kim J., Yoon H., Horie T., Burchett J. M., Restivo J. L., Rotllan N., Ramirez C. M., Verghese P. B., Ihara M., Hoe H. S., et al. . 2015. microRNA-33 regulates ApoE lipidation and amyloid-beta metabolism in the brain. J. Neurosci. 35: 14717–14726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang Q., Lee C. Y., Mandrekar S., Wilkinson B., Cramer P., Zelcer N., Mann K., Lamb B., Willson T. M., Collins J. L., et al. . 2008. ApoE promotes the proteolytic degradation of Abeta. Neuron. 58: 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wildsmith K. R., Holley M., Savage J. C., Skerrett R., and Landreth G. E.. 2013. Evidence for impaired amyloid beta clearance in Alzheimer’s disease. Alzheimers Res. Ther. 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pérez E., Bourguet W., Gronemeyer H., and de Lera A. R.. 2012. Modulation of RXR function through ligand design. Biochim. Biophys. Acta. 1821: 57–69. [DOI] [PubMed] [Google Scholar]
- 15.Hong C., and Tontonoz P.. 2014. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat. Rev. Drug Discov. 13: 433–444. [DOI] [PubMed] [Google Scholar]
- 16.Chisholm J. W., Hong J., Mills S. A., and Lawn R. M.. 2003. The LXR ligand T0901317 induces severe lipogenesis in the db/db diabetic mouse. J. Lipid Res. 44: 2039–2048. [DOI] [PubMed] [Google Scholar]
- 17.Gao M., and Liu D.. 2013. The liver X receptor agonist T0901317 protects mice from high fat diet-induced obesity and insulin resistance. AAPS J. 15: 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skaper S. D., Debetto P., and Giusti P.. 2010. The P2X7 purinergic receptor: from physiology to neurological disorders. FASEB J. 24: 337–345. [DOI] [PubMed] [Google Scholar]
- 19.Kim W. K., Meliton V., Park K. W., Hong C., Tontonoz P., Niewiadomski P., Waschek J. A., Tetradis S., and Parhami F.. 2009. Negative regulation of Hedgehog signaling by liver X receptors. Mol. Endocrinol. 23: 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan J., Stukas S., Wong C., Chan J., May S., DeValle N., Hirsch-Reinshagen V., Wilkinson A., Oda M. N., and Wellington C. L.. 2011. An ABCA1-independent pathway for recycling a poorly lipidated 8.1 nm apolipoprotein E particle from glia. J. Lipid Res. 52: 1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Neill R. A., Bhamidipati A., Bi X., Deb-Basu D., Cahill L., Ferrante J., Gentalen E., Glazer M., Gossett J., Hacker K., et al. . 2006. Isoelectric focusing technology quantifies protein signaling in 25 cells. Proc. Natl. Acad. Sci. USA. 103: 16153–16158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen U., Squaglia N., Boge A., and Fung P. A.. 2011. The Simple Western™: a gel-free, blot-free, hands-free Western blotting reinvention. Nat. Methods. 8: . [Google Scholar]
- 23.Fan J., Zareyan S., Zhao W., Shimizu Y., Pfeifer T. A., Tak J. H., Isman M. B., Van den Hoven B., Duggan M. E., Wood M. W., et al. . 2016. Identification of a chrysanthemic ester as an apolipoprotein E inducer in astrocytes. PLoS One. 11: e0162384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swahn B. M., Macsari I., Viklund J., Ohberg L., Sjodin J., Neelissen J., and Lindquist J.. 2009. Liver X receptor agonists with selectivity for LXRbeta; N-aryl-3,3,3-trifluoro-2-hydroxy-2-methylpropionamides. Bioorg. Med. Chem. Lett. 19: 2009–2012. [DOI] [PubMed] [Google Scholar]
- 25.Sedlák D., Paguio A., and Bartůněk P.. 2011. Two panels of steroid receptor luciferase reporter cell lines for compound profiling. Comb. Chem. High Throughput Screen. 14: 248–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lew M. 2007. Good statistical practice in pharmacology. Problem 2. Br. J. Pharmacol. 152: 299–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuercher W. J., Buckholz R. G., Campobasso N., Collins J. L., Galardi C. M., Gampe R. T., Hyatt S. M., Merrihew S. L., Moore J. T., Oplinger J. A., et al. . 2010. Discovery of tertiary sulfonamides as potent liver X receptor antagonists. J. Med. Chem. 53: 3412–3416. [DOI] [PubMed] [Google Scholar]
- 28.Baxter, A., McInallyT. , MortimoreM. , and CladingboelD. , inventors; AstraZeneca UK Limited, assignee. Novel compounds. United States patent US 20,010,003,121. June 7, 2001..
- 29.Alcaraz, L., FurberM. , and MortimoreM. , inventors; AstraZeneca AB, assignee. Adamantane derivatives. United States patent US 6,492,355. December 10, 2002..
- 30.Keystone E. C., Wang M. M., Layton M., Hollis S., McInnes I. B., and Team D. C. S.. 2012. Clinical evaluation of the efficacy of the P2X7 purinergic receptor antagonist AZD9056 on the signs and symptoms of rheumatoid arthritis in patients with active disease despite treatment with methotrexate or sulphasalazine. Ann. Rheum. Dis. 71: 1630–1635. [DOI] [PubMed] [Google Scholar]
- 31.Eser A., Colombel J. F., Rutgeerts P., Vermeire S., Vogelsang H., Braddock M., Persson T., and Reinisch W.. 2015. Safety and efficacy of an oral inhibitor of the purinergic receptor P2X7 in adult patients with moderately to severely active Crohn’s disease: a randomized placebo-controlled, double-blind, phase IIa study. Inflamm. Bowel Dis. 21: 2247–2253. [DOI] [PubMed] [Google Scholar]
- 32.Koldamova R., Fitz N. F., Lefterov I.. 2014. ATP-binding cassette transporter A1: from metabolism to neurodegeneration. Neurobiol. Dis. 72: 13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yassine H. N., Feng Q., Chiang J., Petrosspour L. M., Fonteh A. N., Chui H. C., and Harrington M. G.. 2016. ABCA1-mediated cholesterol efflux capacity to cerebrospinal fluid is reduced in patients with mild cognitive impairment and Alzheimer’s disease. J. Am. Heart Assoc. 5: e002886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boehm-Cagan A., Bar R., Liraz O., Bielicki J. K., Johansson J. O., and Michaelson D. M.. 2016. ABCA1 agonist reverses the ApoE4-driven cognitive and brain pathologies. J. Alzheimers Dis. 54: 1219–1233. [DOI] [PubMed] [Google Scholar]
- 35.Allsopp R. C., Dayl S., Schmid R., and Evans R. J.. 2017. Unique residues in the ATP gated human P2X7 receptor define a novel allosteric binding pocket for the selective antagonist AZ10606120. Sci. Rep. 7: 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guile S. D., Alcaraz L., Birkinshaw T. N., Bowers K. C., Ebden M. R., Furber M., and Stocks M. J.. 2009. Antagonists of the P2X(7) receptor. From lead identification to drug development. J. Med. Chem. 52: 3123–3141. [DOI] [PubMed] [Google Scholar]
- 37.Furber M., Alcaraz L., Bent J. E., Beyerbach A., Bowers K., Braddock M., Caffrey M. V., Cladingboel D., Collington J., Donald D. K., et al. . 2007. Discovery of potent and selective adamantane-based small-molecule P2X(7) receptor antagonists/interleukin-1beta inhibitors. J. Med. Chem. 50: 5882–5885. [DOI] [PubMed] [Google Scholar]
- 38.Sáez-Orellana F., Fuentes-Fuentes M. C., Godoy P. A., Silva-Grecchi T., Panes J. D., Guzman L., Yevenes G., Gavilan J., Egan T. M., Aguayo L. G., et al. . 2018. P2X receptor overexpression induced by soluble oligomers of amyloid beta peptide potentiates synaptic failure and neuronal dyshomeostasis in cellular models of Alzheimer’s disease. Neuropharmacology. 128: 366–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee H. G., Won S. M., Gwag B. J., and Lee Y. B.. 2011. Microglial P2X(7) receptor expression is accompanied by neuronal damage in the cerebral cortex of the APPswe/PS1dE9 mouse model of Alzheimer’s disease. Exp. Mol. Med. 43: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Diaz-Hernandez J. I., Gomez-Villafuertes R., Leon-Otegui M., Hontecillas-Prieto L., Del Puerto A., Trejo J. L., Lucas J. J., Garrido J. J., Gualix J., Miras-Portugal M. T., et al. . 2012. In vivo P2X7 inhibition reduces amyloid plaques in Alzheimer’s disease through GSK3beta and secretases. Neurobiol. Aging. 33: 1816–1828. [DOI] [PubMed] [Google Scholar]
- 41.Ryu J. K., and McLarnon J. G.. 2008. Block of purinergic P2X(7) receptor is neuroprotective in an animal model of Alzheimer’s disease. Neuroreport. 19: 1715–1719. [DOI] [PubMed] [Google Scholar]
- 42.Kaczmarek-Hájek K., Lörinczi E., Hausmann R., and Nicke A.. 2012. Molecular and functional properties of P2X receptors–recent progress and persisting challenges. Purinergic Signal. 8: 375–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.North R. A. 2002. Molecular physiology of P2X receptors. Physiol. Rev. 82: 1013–1067. [DOI] [PubMed] [Google Scholar]
- 44.Stokes L., Jiang L. H., Alcaraz L., Bent J., Bowers K., Fagura M., Furber M., Mortimore M., Lawson M., Theaker J., et al. . 2006. Characterization of a selective and potent antagonist of human P2X(7) receptors, AZ11645373. Br. J. Pharmacol. 149: 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]