Abstract
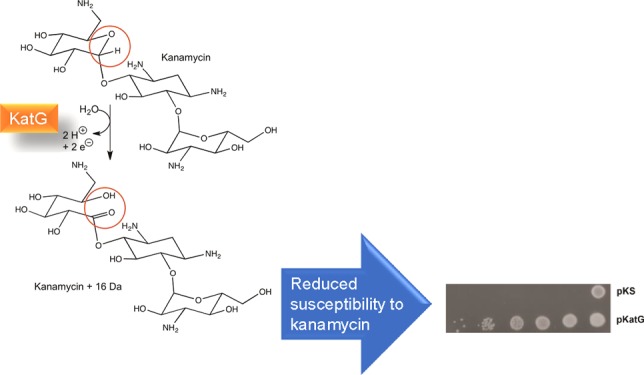
Resistance to antibiotics has become a serious problem for society, and there are increasing efforts to understand the reasons for and sources of resistance. Bacterial-encoded enzymes and transport systems, both innate and acquired, are the most frequent culprits for the development of resistance, although in Mycobacterium tuberculosis, the catalase-peroxidase, KatG, has been linked to the activation of the antitubercular drug isoniazid. While investigating a possible link between aminoglycoside antibiotics and the induction of oxidative bursts, we observed that KatG reduces susceptibility to aminoglycosides. Investigation revealed that kanamycin served as an electron donor for the peroxidase reaction, reducing the oxidized ferryl intermediates of KatG to the resting state. Loss of electrons from kanamycin was accompanied by the addition of a single oxygen atom to the aminoglycoside. The oxidized form of kanamycin proved to be less effective as an antibiotic. Kanamycin inhibited the crystallization of KatG, but the smaller, structurally related glycoside maltose did cocrystallize with KatG, providing a suggestion as to the possible binding site of kanamycin.
Introduction
Catalase-peroxidases found initially in bacteria and more recently in some fungi have been the focus of extensive study for over three decades but continue to provide puzzles and surprises. The first catalase-peroxidase was isolated from Escherichia coli as a broad-spectrum peroxidase with a significant catalase activity,1 and, following genetic characterization,2,3 named KatG. Subsequently, KatG became a focus of great interest when it was found to be the key determinant for the activation of isoniazid (INH) as an antitubercular drug.4 Compounding its apparent complexity is an oxidase activity that generates superoxide in the presence of electron donors such as isoniazid and reduced nicotinamide adenine dinucleotide.5,6
KatG is a homo-dimer with the 80 000 Da subunits having distinct N- and C-terminal domains that resemble each other and, at their core, also the core of plant peroxidases in both sequence and structure. The catalase and peroxidase activities of KatGs reside within the heme-containing N-terminal domain with the catalase activity requiring the cross-linked adduct of the side chains of a Met, a Tyr, and a Trp; a nearby mobile Arg; and a perhydroxy modification on the adduct Trp.7−9
The broad-spectrum peroxidase activity of KatG is typically assayed using 2,2′-azinobis(3-ethylbenzothiazolinesulfonic acid) (ABTS)9 or o-dianisidine,1 the oxidation products of which can be easily assayed colorimetrically. Small aromatic phenols and anilines are also substrates,10 albeit more difficult to assay. Isoniazid is cleaved to an isonicotinyl radical with coincident generation of superoxide, and the isonicotinyl radical can react with NAD+ to form an isonicotinyl-NAD radical, which is reduced by a superoxide to the active antitubercular drug isonicotinyl-NAD.11,12
A recent hypothesis suggests that bacterial killing caused by bactericidal aminoglycoside antibiotics is at least in part the result of a reactive oxygen response induced by the antibiotic.13,14 A corollary to this hypothesis is that KatG and other catalases might possibly ameliorate the effect of the antibiotic by reducing the level of reactive oxygen species, thereby enhancing antibiotic resistance. We report here that there is indeed a correlation between the presence of KatG and reduced susceptibility to the aminoglycoside antibiotics. However, the reduced susceptibility is a result of direct oxidation of the antibiotic by KatG as part of the peroxidatic cycle rather than KatG reacting with and removing reactive oxygen species.
Results
KatG Imparts Protection against Aminoglycoside Antibiotics
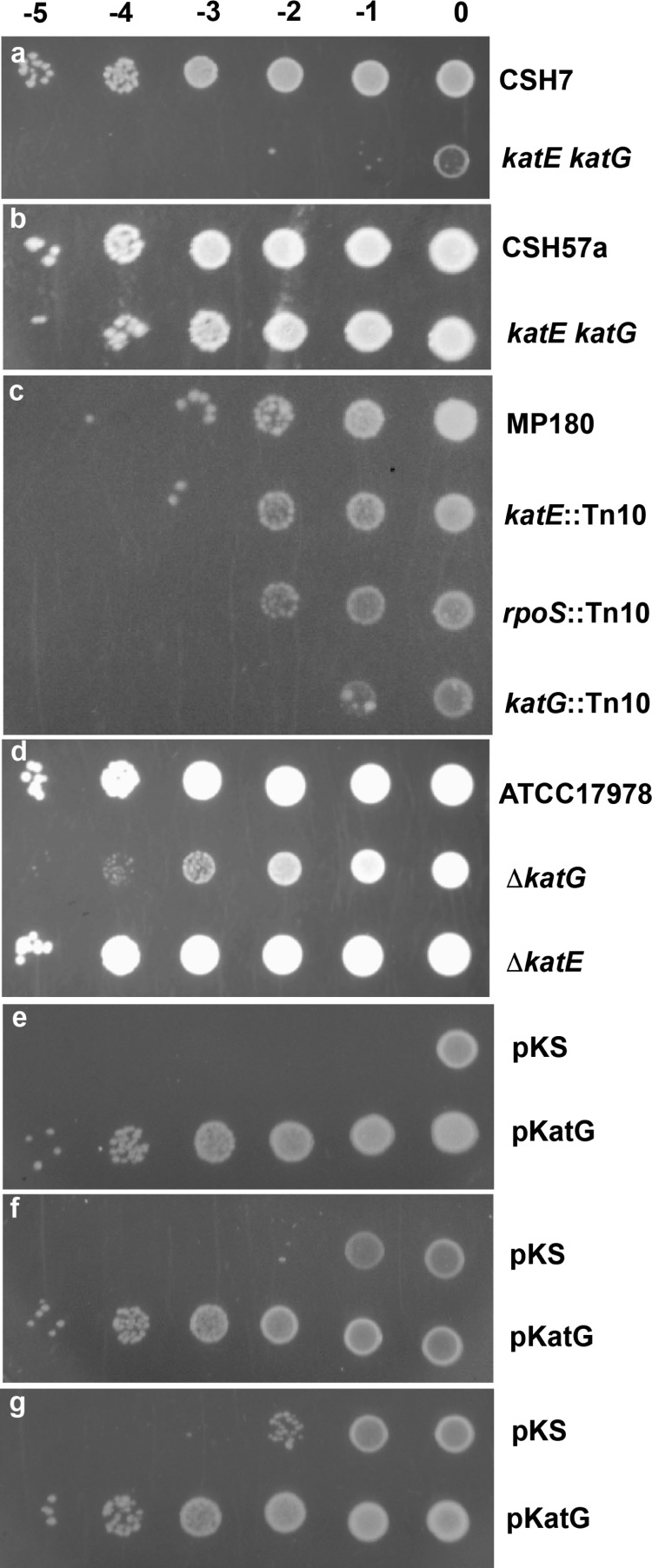
The initial rationale for investigating a possible role of KatG in aminoglycoside antibiotic resistance lay in the hypothesis that bactericidal antibiotics induce a reactive oxygen response that may be responsible for the bacterial killing.13,14 If one of the reactive oxygen species was hydrogen peroxide, the presence of catalase or peroxidase might reduce the effectiveness of the antibiotic. This was tested initially in a comparison of the sensitivity to kanamycin of the E. coli catalase mutants, UM1 and UM2 (both with katE katG genotype generated by nitrosoguanidine mutagenesis2 with their respective parental strains, CSH7 and CSH57a). Both mutants exhibited enhanced sensitivity to kanamycin compared to that of their parents, with UM1 (Figure 1a) exhibiting a much greater increase than that from UM2 (Figure 1b). The katG genes in UM1 and UM2 had both undergone a single base change, C to T in UM1, causing a Leu to Phe change at residue 138 close to the heme cavity, and G to A in UM2, causing a Gly to Asp change at residue 118 in the vicinity of mobile Arg426. Therefore, the large difference between UM1 and UM2 suggested that nitrosoguanidine had caused other mutations, and to assess more clearly the effect of unique catalase mutations, a series of isogenic strains was investigated. First, the isogenic series of MP180 (parent), UM120 (katE::Tn10), UM122 (rpoS::Tn10, originally katF::Tn10), and UM202 (katG::Tn10)15 was investigated, revealing enhanced sensitivity to kanamycin only in UM202 containing a disrupted katG gene (Figure 1c). A similar degree of enhanced sensitivity to kanamycin is observable in a strain of Acinetobacter baumannii with a deletion in katG, but not katE (Figure 1d).
Figure 1.
Susceptibility of E. coli and A. baumannii strains to aminoglycoside antibiotics. Cultures were grown to an absorbance of 1.0–1.5 at 600 nm and diluted to an absorbance of 1.0 at 600 nm, followed by dilution by 10–5, 10–4, 10–3, 10–2, 10–1, and 0, from left to right, and spotting on Luria broth (LB) plates containing 1.0 μg/mL kanamycin (a–e), 1.0 μg/mL gentamycin (f), and 1.0 μg/mL tobramycin (g). (a) E. coli CSH1 (parent) and nitrosoguanidine-generated UM1 (katE katG). (b) E. coli CSH57a (parent) and nitrosoguanidine-generated UM2 (katE katG). (c) Isogenic E. coli group of MP180 (parent), UM120 (katE::Tn10), UM122 (rpoS::Tn10), and UM202 (katG::Tn10). (d) A. baumannii series of ATCC17978 (WT) and its ΔkatG and ΔkatE derivatives. (e–g) UM262 (katG::Tn10 katE) transformed with pKS and pBpKatG (8).
The fact that a single gene dosage of katG elicits an increased tolerance to kanamycin suggested that overexpression of katG from a multicopy plasmid might possibly elicit greater tolerance, and this is indeed the case. Elevated KatG levels significantly enhance tolerance to kanamycin (Figure 1e) and only slightly less so against gentamycin (Figure 1f) and tobramycin (Figure 1g). The control strain contained the parent plasmid to correct for the effects of the multicopy plasmid. We would like to point out that the difference in susceptibility was observed only when cells were spotted on the agar plate and not using two-fold serial dilution method (data not shown). We suspect that this is because spotting cells directly on the agar plate from an overnight culture (as described in Materials and Methods) ensured that there was high enough expression of KatG to mediate tolerance to kanamycin.
Kanamycin as an Electron Source for the Reduction of KatG Compound I*
The reduced susceptibility to kanamycin induced by KatG is consistent with the hypothesis that KatG ameliorates an oxidative burst involving H2O2, but the fact that the more reactive catalase, KatE or HPII, provides no such protection suggested a different mechanism, most likely involving either the oxidase or the peroxidase activity. Involvement of the oxidase activity was ruled out by the absence of superoxide generation and by the lack of oxygen depletion in a mixture of KatG and kanamycin (data not shown). This led to a focus on the peroxidatic activity of KatG as the mediator of enhanced tolerance to kanamycin.
KatG was initially characterized as having a broad-spectrum peroxidase activity with substrates ranging from ABTS and o-dianisidine to small aromatic amines, phenols, and INH. To determine if the substrate range extended to aminoglycoside antibiotics, the compound I* ferryl intermediate mixture16 of KatG was challenged with kanamycin and the rate of return of the hypochromic Soret peak from 413 nm to the resting state at 407 was monitored spectrophotometrically (Figure 2). The oxoferryl form of KatG naturally undergoes a slow autocatalytic reduction with electrons drawn from oxidizable side chains in the protein (Figure 2a),17 but kanamycin (Figure 2b) substantially enhanced the rate of reduction (Figure 2c).
Figure 2.
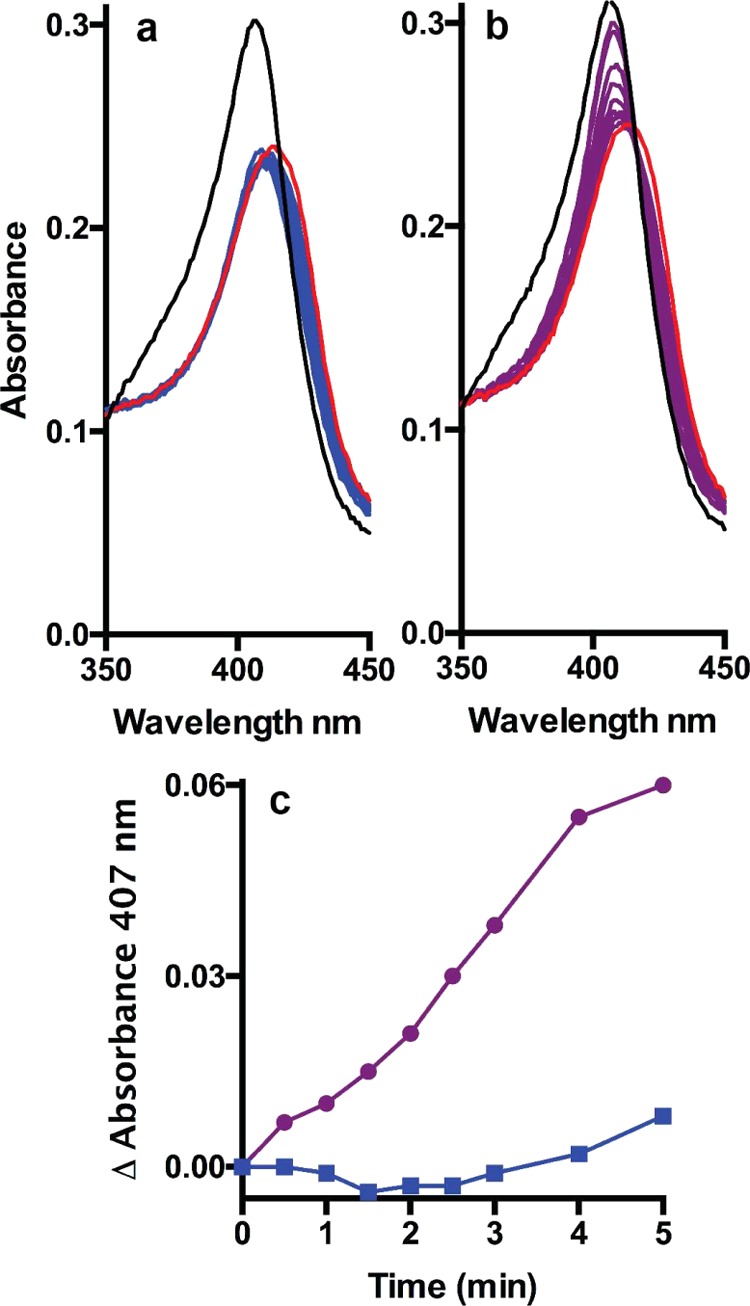
Reduction of compound I* ferryl intermediate mixture of KatG by kanamycin. (a, b) Resting-state spectrum (3.1 μM) is shown in black, and the spectrum of the oxoferryl species after 1 min reaction with a 10× excess of peracetic acid is shown in red. The mixtures were then incubated at 20 °C without (a) and with (b) a 100× excess of kanamycin, and spectra were collected at 0.5, 1, 1.5, 2, 2.5, 3, 4, and 5 min (blue in (a) and purple in (b)). (c) Changes in absorbance at 407 nm from the spectra in (a) (blue) and (b) (purple).
Kanamycin as an Electron Source for Reactivation of the Catalatic Process in KatG
The catalase reaction of KatGs undergoes a gradual inactivation during catalatic turnover, attributed to the accumulation of a partially oxidized17 and catalatically inactive state,18 and the rate of inactivation is enhanced in the presence of carbon monoxide (CO).19 Originally, inactivation in the presence of CO was attributed to the accumulation of an inactive ferrous–CO [Fe2+–CO] complex (19), but because (1) the peroxidase activity is not inactivated coincidentally (Figure S1a), (2) the inactivation can be reversed by electron donation from ABTS (Figures 3 and S1), and (3) there is no electron density associated with the heme iron in a crystal soaked with H2O2 in the presence of CO (Figure S2 and Table S3, 5KT9), it is more likely that a partially oxidized state of KatG is simply accumulating more rapidly in the presence of CO. Like ABTS, which donates electrons to reduce the partially oxidized state of KatG back to the resting state, kanamycin is also effective in reversing the inhibition of KatG during catalatic turnover in the presence of CO, consistent with it being an effective electron donor for KatGs (Figure 3).
Figure 3.
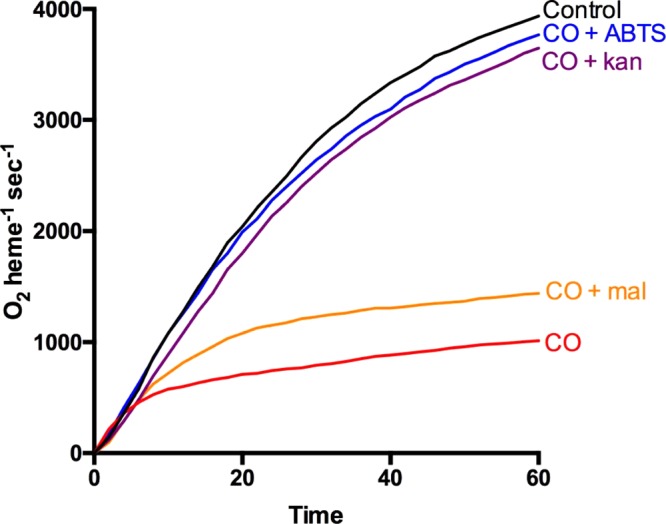
Kanamycin prevents the inactivation of catalase activity in the presence of CO. Oxygen evolution in a solution of 1.74 pmol/mL BpKatG in pH 7.0 potassium phosphate buffer (black) and in the same buffer saturated with CO (red), supplemented with 0.4 mM ABTS (blue), 2.0 mM kanamycin (purple), and 3 mM maltose (orange).
Mass Spectrometric Identification of the Kanamycin Oxidation Product
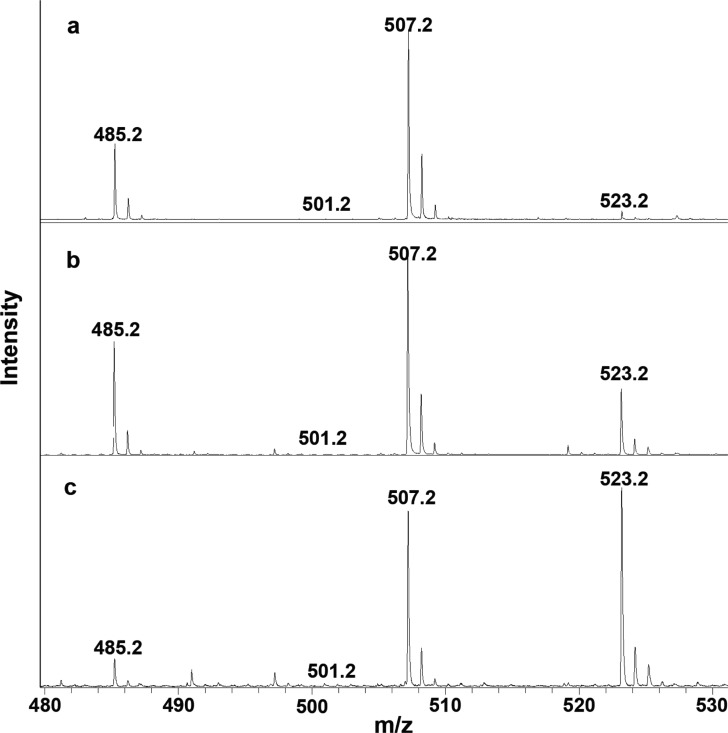
Peroxidatic substrates by definition donate electrons to reduce the compound I* or ferryl intermediate mixture16 generated following oxidation of the peroxidase heme, but the final substrate oxidation product varies with the substrate ranging from radical formation in the case of ABTS to quinone formation in the case of some phenols. To confirm that kanamycin was indeed being oxidized in the peroxidatic reaction, samples of kanamycin before and after treatment were subjected to mass spectrometry analysis. The matrix-assisted laser desorption ionization (MALDI) conditions produce kanamycin ions at the expected m/z 485.2 and also an associated sodium ion at m/z 507.2 (485.2 + 22.0). In addition, samples of untreated kanamycin contain barely detectable ions at m/z 501.2 (485.2 + 16.0) and m/z 523.2 (507.2 + 16.0) (Figure 4). Incubation of kanamycin with KatG increases the ion at m/z 523.2 (Figure 4b) to 10–20% of the total, and treatment with KatG and glucose oxidase and glucose (the latter to provide H2O2 for KatG oxidation and probably chemical oxidation of kanamycin) increases the m/z 523.2 ion to >50% of the total (Figure 4c). The m/z 501.2 ion was never intense enough under any conditions to be measured accurately. The conclusion to be drawn from the increment of 16 Da is that a single oxygen atom is added to kanamycin, a somewhat unusual reaction in the absence of a loss of hydrogen atoms.
Figure 4.
Mass spectrometry analysis of the oxidation products of kanamycin. (a) Spectrum of kanamycin before treatment with KatG or glucose oxidase. The predominant ion has an associated sodium ion, 485.2 + 22.0 = 507.2. The sample was incubated with (b) 1 μg/mL KatG for 15 min and (c) 1 μg/mL KatG supplemented with 5 mg/mL glucose oxidase and 5 mM glucose for 15 min. Sodium ion adducts of carbohydrates are a well-documented product of MALDI.20
Oxidation of Kanamycin Reduces Its Effectiveness as an Antibiotic
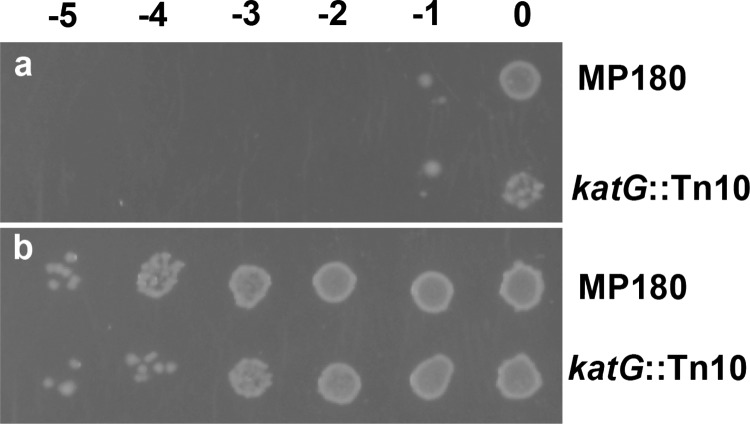
The correlation of reduced susceptibility to kanamycin with apparent oxidation of kanamycin suggested that the oxidized form of kanamycin is a less effective antibiotic than kanamycin itself. This hypothesis was tested by treating kanamycin with a mixture of glucose oxidase and glucose with and without KatG and spotting bacteria on plates containing either treated or untreated kanamycin (1.0 μg/mL). As predicted, growth of both MP180 and UM202 is much less susceptible to oxidized kanamycin as compared with untreated kanamycin (Figure 5). Somewhat surprisingly, KatG did not enhance the change in susceptibility involving glucose oxidase plus glucose, and KatG oxidized with peracetic acid elicits a similar, albeit less striking, response (not shown). Clearly, chemical oxidation by H2O2 is the main pathway for kanamycin oxidation in this experiment.
Figure 5.
Susceptibility of MP180 and UM202 to untreated (a) and oxidized kanamycin (b). Oxidized kanamycin (1.0 μg/mL) was generated in a mixture with 5 mg/mL glucose oxidase and 5 mM glucose for 2.5 h at room temperature. The presence of KatG in the mixture did not change the resulting pattern of growth. Untreated kanamycin (1.0 μg/mL) was incubated for the same time with glucose but no oxidase. The solutions were then added to the agar as the plates were poured. Dilutions of MP180 and UM202 were plated as in Figure 1.
Binding of Maltose to KatG
Implicit in the observation of oxoferryl reduction by glycosides is the existence, however transient, of an interaction or association of the glycosides with KatG. A binding site for INH on KatG was identified by cocrystallization,12 and the same strategy was applied to kanamycin with 100 mM kanamycin included in the crystallization solution with BpKatG. Despite repeated attempts, crystals did not form in the presence of kanamycin, and formed only in its absence, suggesting that kanamycin was interfering with the crystallization process. With two glycosidic bonds and three six-membered rings, kanamycin is a large glycoside, and cocrystallization was also attempted with the smaller disaccharide, maltose. Crystals of KatG formed in the presence of 100 mM maltose and X-ray diffraction data were collected to 1.80 Å and refined to Rcryst and Rfree values of 13.6 and 16.2%, respectively (Table S3). Inspection of the resulting electron density maps revealed a region of strong Fo–Fcdensity near Arg506 that was not present in a crystal grown in the absence of maltose. Inclusion of a single molecule of glucose in the model satisfied the density (Figure 6), but there was no density corresponding to the second glucose unit of the maltose, which was presumably present in multiple conformations.
Figure 6.
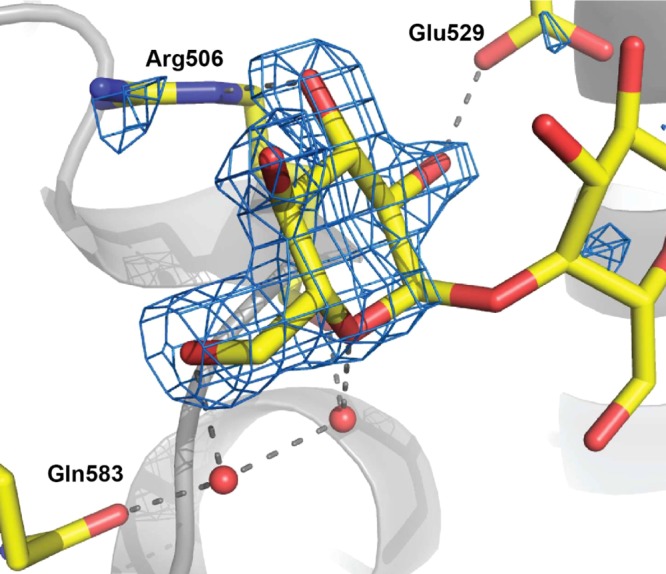
Maltose binding to KatG. The Fo–Fc omit electron density map at 7.0σ was calculated without maltose in the model.
The maltose–KatG interactions include two direct hydrogen-bond interactions, C2–OH with OE2 of E529 (2.5 Å) and C3–OH with NE of R506 (2.8 Å) as well as two indirect interactions, one with C6–OH through water with OE1 of Q583 and one with the hexose ring O5 through water with C=O of R506 (Figure 6). The occupancy is clearly greater in the B subunit where the site falls in a shallow pocket just at the edge of the crystal interface, although there are no direct contacts with the symmetry-related subunit. KatG was also crystallized in the presence of glucose, and the resulting electron density maps contained a region of density in the same location as that for maltose, albeit much weaker, suggesting much lower occupancy. The binding site at the crystal interface may explain why crystals did not form in the presence of kanamycin, but there is otherwise no indication of where kanamycin might be binding to KatG.
The interaction of maltose suggested that it too might be a peroxidatic electron donor. However, unlike kanamycin, maltose did not substantially enhance the rate of autocatalytic reduction of the compound I* ferryl intermediates (data not shown) and only weakly prevented the inactivation of KatG in the presence of CO (Figure 3). Maltose was oxidized to a limited extent by KatG and effectively by glucose oxidase (Figure S3).
Discussion
In less than a century since the first clinical use of antibiotics, it is feared that the preantibiotic era might be returning as a result of resistance of bacterial pathogens to almost all classes of antibiotics. Consequently, new and effective antibiotics are urgently needed. Understanding the molecular mechanisms leading to reduced susceptibility to antibiotics is key to finding new and effective antibiotics that bypass resistance mechanisms. In this work, we show that catalase-peroxidase KatG can lead to the modification leading to inactivation of aminoglycoside activity and therefore contribute toward a bacterial cell’s reduced susceptibility to these antibiotics.
Catalase-peroxidases have garnered considerable notoriety from the role of KatG in the activation of the pro-drug INH into its antitubercular form, isonicotinyl-NAD. Mutation of the katG gene in Mycobacterium tuberculosis at a number of different locations slows IN-NAD synthesis, leading to INH resistance.12,21 Our data show that the opposite is true in the case of aminoglycosides; it is the presence of KatG that leads to reduced susceptibility or tolerance to aminoglycoside antibiotics. One of the three common mechanisms giving rise to aminoglycoside resistance is enzymatic modification involving acetylation, adenylylation, or phosphorylation.22 We show for the first time that the oxidation of kanamycin caused by KatG therefore presents a fourth mechanism of aminoglycoside modification leading to inactivation. As shown in Figure 1, the effect was also observed, albeit less pronounce than that for kanamycin, for other members of aminoglycosides, i.e., gentamycin and tobramycin. How broad-based the effect of KatG-mediated aminoglycoside tolerance might be in Gram-negative species remains to be determined, but clearly it cannot be a factor in organisms such as Pseudomonas aeruginosa that do not produce KatG. However, it should also be noted that an oxidative burst in bacteria may lead to chemical oxidation of kanamycin and reduced susceptibility.
The cocrystallization of maltose with KatG near the crystal interface and absence of crystal formation with kanamycin suggest that the two glycosides may bind at the same site but that the larger kanamycin molecule interferes with crystal formation. In addition, the orientation of the single glucose unit in the crystal provides insight into the probable orientation of kanamycin. Both hexose units of kanamycin can potentially fit into the same site, but the C3″–NH2 hexose unit would place the NH2 group adjacent to Arg506, whereas the C6′–NH2 hexose unit would have the NH2 group more favorably adjacent to C=O of Gln583. Therefore, it is the cleavage of the C1′–O–C4 glycosidic linkage that is shown in Figure S4.
The oxidation reaction is somewhat unusual in that a single oxygen atom is added without the loss of hydrogen atoms, most likely a result of cleavage of the hexose C5′–O–C1′ bonds with coincident C1′ oxidation (Figure S4). Overall, two electrons are lost in the process, sufficient to reduce the compound I* ferryl intermediate mixture back to the resting state in KatG. However, it is impossible to conclude a direct stoichiometry especially in light of the very slow turnover rate of 0.5–1.0 per minute in KatG, similar to that of INH activation.
In summary, using a combination of phenotypic, structural, biochemical, and genetic approaches, we show that overexpression of KatG can result in a reduced susceptibility to the aminoglycoside kanamycin. Although the presence of KatG may not confer clinical resistance, it can result in reduced intracellular concentrations of the active kanamycin molecule, which in turn can potentially result in the accumulation of other resistance mechanisms such as efflux and target-site mutation(s).
Materials and Methods
Bacterial Strains and Chemicals
A list of bacterial strains and plasmids is provided in Table S1. All chemicals were obtained from Sigma-Aldrich or Fisher unless otherwise stated. All media components were obtained from Becton-Dickinson, and all restriction enzymes were obtained from Invitrogen.
Creation of Gene Deletions of katG and katE in A. baumannii
Both katG (A1S_0412) and katE (A1S_1386) deletions in A. baumannii ATCC17978 were carried out using a homologous recombination-based method as described previously23 with slight modifications. Briefly, an upstream portion and a downstream portion of the target gene were amplified using the primers described in Table S1, which contained added FRT regions flanking 3′ and 5′ ends, respectively, and a gentamicin resistance (GmR) marker containing complementary FRT regions cloned between the upstream and the downstream portions of the target gene using sequence overlap extension polymerase chain reaction (PCR)24 to generate the gene knockout cassette. This knockout cassette was then cloned into suicide vector pMO13024 digested with SmaI, and the resulting plasmid was transformed into E. coli SM10 cells for conjugation into A. baumannii ATCC17978. Conjugants were grown in 10% sucrose to induce the second recombination event using the counter-selectable sacB marker in pMO130, and ultimately the GmR marker was excised using the Flp-FRT recombination to yield the unmarked deletions in both katG and katE genes. katG was deleted leaving 3 bp in the 5′-end and 9 bp in the 3′-end of the gene, resulting in a deletion of 2139 bp from 2154 bp long gene. In the case of katE, a total of 1869 bp were deleted from the full gene (2139 bp) leaving 247 bp in the 5′-end and 25 bp in the 3′-end. Both gene deletions were confirmed initially with PCR. The oligonucleotides used are listed in Table S2.
Enzyme Assays
Catalase activity was determined by the method of Rørth and Jensen25 in a Gilson oxygraph equipped with a Clark electrode. One unit of catalase is defined as the amount that decomposes 1 μmol of H2O2 in 1 min in a 60 mM H2O2 solution at pH 7.0 and 37 °C. Peroxidase activity was determined using 2,2′-azinobis(3-ethylbenzothiazolinesulfonic acid) (ABTS) (ε405 = 36 800 M–1 cm–1),26 and one unit is defined as the amount that decomposes 1 μmol ABTS in a solution of 0.4 mM ABTS and 2.5 mM hydrogen peroxide at pH 4.5 and 25 °C.27
For spectroscopic analysis, the compound I* ferryl intermediate mixture of KatG was generated by mixing in 1 mL final volume 3.6 nmol of subunit in 50 mM potassium phosphate buffer, pH 7.0, with 36 nmol of peracetic acid also in 50 mM potassium phosphate buffer, pH 7.0, and let sit at 20 °C for 1 min when 300 nmol of kanamycin was added. The change in absorbance in the 350–700 nm range was monitored.
Mass Spectrometric Analysis
Samples were prepared in 50 mM ammonium acetate, pH 7, using the same concentrations and incubation times as used for the spectroscopic analysis. Protein, if present during incubation, was removed by filtration through Amicon Ultra 50 K molecular weight cut-off filters and concentrated to dryness. Reconstituted samples at 1 mg/mL in water were cocrystallized with an equal volume of 2,5-dihydroxybenzoic acid prepared in water/acetonitrile 3:1 with 0.2% formic acid on a metal target. Spectra were acquired on a Bruker UltraflexExtreme MALDI TOF–TOF instrument.
Antibiotic Susceptibility Assays
To determine antibiotic susceptibility, cultures were grown in LB medium to an absorbance of 1.0–1.5 at 600 nm and diluted to a final absorbance of 1.0 at 600 nm followed by serial dilutions from 1 to 10–5. Aliquots of 2 μL were spotted on LB plates supplemented with antibiotics at 0.5, 1.0, and 1.5 μg/mL.
Crystallization of the KatG Variants
The catalase-peroxidase from Burkholderia pseudomallei, BpKatG, was expressed and purified as described.28 Crystals of BpKatG grown in the absence or presence of 100 mM maltose were obtained at room temperature by the vapor diffusion hanging drop method at 20 °C over a reservoir solution containing 15–17.5% poly(ethylene glycol) 4000 (w/v), 20% 2-methyl-2,4-pentanediol, 100 mM maltose, 70 mM NaCl, and 0.1 M sodium citrate, pH 5.6.8,12 Crystals were primitive orthorhombic space group P212121 with two subunits in the crystal asymmetric unit. For treatment with H2O2, the crystal was soaked for 1 min in mother liquor saturated with CO and containing 5 mM H2O2. Diffraction data were collected using synchrotron beam line CMCF 08ID-1 at the Canadian Light Source in Saskatoon, SK, from crystals flash-cooled in reservoir buffer and cooled with a nitrogen cryostream. Data were processed and scaled using XDS29 and SCALA30 (Table S3). The structure refinements starting with the native BpKatG structure (1MWV) were completed using the REFMAC31 program and manual modeling with the molecular graphics COOT32 program. Figures were generated using PYMOL (the PYMOL Molecular Graphics System, Schrodinger, LLC).
Acknowledgments
This work was supported by Discovery Grants 2012-9600 (to P.C.L.) and 2015-05550 (to A.K.) from the Natural Sciences and Engineering Research Council (NSERC) of Canada, and by the Canada Research Chair Program (to P.C.L.). The Canadian Light Source is supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council Canada, the Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan. P.M.D.S. is supported by the University of Manitoba Graduate Fellowship.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.8b00356.
Effect of CO on catalase and peroxidase reactions of BpKatG (Figure S1); heme and surrounding residues of WT BpKatG after soaking of the crystal in CO-saturated mother liquor containing 5 mM H2O2 (Figure S2); mass spectrometry analysis of the oxidation products of maltose (Figure S3); schemes for the oxidation of kanamycin and maltose (Figure S4); bacterial strains and plasmids used (Table S1); oligonucleotides used in this study (Table S2); refinement statistics of BpKatG with maltose bound (6B9B) and after treatment with H2O2 in CO-containing buffer (5KT9) (Table S3) (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Claiborne A.; Fridovich I. Purification of the o-dianisidine peroxidase from Escherichia coli B. Physicochemical characterization and analysis of its dual catalatic and peroxidatic activities. J. Biol. Chem. 1979, 254, 4245–4252. [PubMed] [Google Scholar]
- Loewen P. C.; Triggs B. L.; George C. S.; Hrabarchuk B. E. Genetic mapping of katG, a locus that affects synthesis of the bifunctional catalase-peroxidase hydroperoxidase I in Escherichia coli. J. Bacteriol. 1985, 162, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triggs-Raine B. L.; Doble B. W.; Mulvey M. R.; Sorby P. A.; Loewen P. C. Nucleotide sequence of katG, encoding catalase HPI of Escherichia coli. J. Bacteriol. 1988, 170, 4415–4419. 10.1128/jb.170.9.4415-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.; Heym B.; Allen B.; Young D.; Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 1992, 358, 591–593. 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- Shoeb H. A.; Bowman B. U. Jr.; Ottolenghi A. C.; Merola A. J. Enzymatic and nonenzymatic superoxide-generating reactions of isoniazid. Antimicrob. Agents Chemother. 1985, 27, 408–412. 10.1128/AAC.27.3.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.; Wiseman B.; Deemagarn T.; Donald L. J.; Duckworth H. W.; Carpena X.; Fita I.; Loewen P. C. Catalase-peroxidases (KatG) exhibit NADH oxidase activity. J. Biol. Chem. 2004, 279, 43098–43106. 10.1074/jbc.M406374200. [DOI] [PubMed] [Google Scholar]
- Yamada Y.; Fujiwara T.; Sato T.; Igarashi N.; Tanaka N. The 2.0 A crystal structure of catalase-peroxidase from Haloarcula marismortui. Nat. Struct. Biol. 2002, 9, 691–695. 10.1038/nsb834. [DOI] [PubMed] [Google Scholar]
- Carpena X.; Loprasert S.; Mongkolsuk S.; Switala J.; Loewen P. C.; Fita I. Catalase-peroxidase KatG of Burkholderia pseudomallei at 1.7A resolution. J. Mol. Biol. 2003, 327, 475–489. 10.1016/S0022-2836(03)00122-0. [DOI] [PubMed] [Google Scholar]
- Carpena X.; Wiseman B.; Deemagarn T.; Herguedas B.; Ivancich A.; Singh R.; Loewen P. C.; Fita I. Roles for Arg426 and Trp111 in the modulation of NADH oxidase activity of the catalase-peroxidase KatG from Burkholderia pseudomallei inferred from pH-induced structural changes. Biochemistry 2006, 45, 5171–5179. 10.1021/bi060017f. [DOI] [PubMed] [Google Scholar]
- Regelsberger G.; Jakopitsch C.; Engleder M.; Ruker F.; Peschek G. A.; Obinger C. Spectral and kinetic studies of the oxidation of monosubstituted phenols and anilines by recombinant Synechocystis catalase-peroxidase compound I. Biochemistry 1999, 38, 10480–10488. 10.1021/bi990886n. [DOI] [PubMed] [Google Scholar]
- Wengenack N. L.; Jensen M. P.; Rusnak F.; Stern M. K. Mycobacterium tuberculosis KatG is a peroxynitritase. Biochem. Biophys. Res. Commun. 1999, 256, 485–487. 10.1006/bbrc.1999.0358. [DOI] [PubMed] [Google Scholar]
- Wiseman B.; Carpena X.; Feliz M.; Donald L. J.; Pons M.; Fita I.; Loewen P. C. Isonicotinic acid hydrazide conversion to Isonicotinyl-NAD by catalase-peroxidases. J. Biol. Chem. 2010, 285, 26662–26673. 10.1074/jbc.M110.139428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski M. A.; Dwyer D. J.; Hayete B.; Lawrence C. A.; Collins J. J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Dwyer D. J.; Kohanski M. A.; Collins J. J. Role of reactive oxygen species in antibiotic action and resistance. Curr. Opin. Microbiol. 2009, 12, 482–489. 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen P. C.; Switala J.; Triggs-Raine B. L. Catalases HPI and HPII in Escherichia coli are induced independently. Arch. Biochem. Biophys. 1985, 243, 144–149. 10.1016/0003-9861(85)90782-9. [DOI] [PubMed] [Google Scholar]
- Colin J.; Wiseman B.; Switala J.; Loewen P. C.; Ivancich A. Distinct role of specific tryptophans in facilitating electron transfer or as [Fe(IV) = O Trp(*)] intermediates in the peroxidase reaction of Bulkholderia pseudomallei catalase-peroxidase: a multifrequency EPR spectroscopy investigation. J. Am. Chem. Soc. 2009, 131, 8557–8563. 10.1021/ja901402v. [DOI] [PubMed] [Google Scholar]
- Ivancich A.; Donald L. J.; Villanueva J.; Wiseman B.; Fita I.; Loewen P. C. Spectroscopic and kinetic investigation of the reactions of peroxyacetic acid with Burkholderia pseudomallei catalase-peroxidase, KatG. Biochemistry 2013, 52, 7271–7282. 10.1021/bi400963j. [DOI] [PubMed] [Google Scholar]
- Ndontsa E. N.; Moore R. L.; Goodwin D. C. Stimulation of KatG catalase activity by peroxidatic electron donors. Arch. Biochem. Biophys. 2012, 525, 215–222. 10.1016/j.abb.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Zhao X.; Khajo A.; Jarrett S.; Suarez J.; Levitsky Y.; Burger R. M.; Jarzecki A. A.; Magliozzo R. S. Specific function of the Met-Tyr-Trp adduct radical and residues Arg-418 and Asp-137 in the atypical catalase reaction of catalase-peroxidase KatG. J. Biol. Chem. 2012, 287, 37057–37065. 10.1074/jbc.M112.401208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn S. H.; Park K. M.; Moon J. H.; Lee S. H.; Kim M. S. Quantification of carbohydrates and related materials using sodium ion adducts produced by matrix-assisted laser desorption ionization. J. Am. Soc. Mass Spectrom. 2016, 27, 1887–1890. 10.1007/s13361-016-1495-9. [DOI] [PubMed] [Google Scholar]
- Cade C. E.; Dlouhy A. C.; Medzihradszky K. F.; Salas-Castillo S. P.; Ghiladi R. A. Isoniazid-resistance conferring mutations in Mycobacterium tuberculosis KatG: catalase, peroxidase, and INH-NADH adduct formation activities. Protein Sci. 2010, 19, 458–474. 10.1002/pro.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J.; Wright G. D. Bacterial resistance to aminoglycoside antibiotics. Trends Microbiol. 1997, 5, 234–240. 10.1016/S0966-842X(97)01033-0. [DOI] [PubMed] [Google Scholar]
- Amin I. M.; Richmond G. E.; Sen P.; Koh T. H.; Piddock L. J.; Chua K. L. A method for generating marker-less gene deletions in multidrug-resistant Acinetobacter baumannii. BMC Microbiol. 2013, 13, 158. 10.1186/1471-2180-13-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K. H.; Schweizer H. P. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 2005, 5, 30. 10.1186/1471-2180-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth M.; Jensen P. K. Determination of catalase activity by means of the Clark oxygen electrode. Biochim. Biophys. Acta, Enzymol. 1967, 139, 171–173. 10.1016/0005-2744(67)90124-6. [DOI] [PubMed] [Google Scholar]
- Childs R. E.; Bardsley W. G. The steady-state kinetics of peroxidase with 2,2′-azino-di-(3-ethyl-benzthiazoline-6-sulphonic acid) as chromogen. Biochem. J. 1975, 145, 93–103. 10.1042/bj1450093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R.; Wiseman B.; Deemagarn T.; Jha V.; Switala J.; Loewen P. C. Comparative study of catalase-peroxidases (KatGs). Arch. Biochem. Biophys. 2008, 471, 207–214. 10.1016/j.abb.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Carpena X.; Switala J.; Loprasert S.; Mongkolsuk S.; Fita I.; Loewen P. C. Crystallization and preliminary X-ray analysis of the catalase-peroxidase KatG from Burkholderia pseudomallei. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002, 58, 2184–2186. 10.1107/S0907444902017869. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M. D.; Ballard C. C.; Cowtan K. D.; Dodson E. J.; Emsley P.; Evans P. R.; Keegan R. M.; Krissinel E. B.; Leslie A. G.; McCoy A.; McNicholas S. J.; Murshudov G. N.; Pannu N. S.; Potterton E. A.; Powell H. R.; Read R. J.; Vagin A.; Wilson K. S. Overview of the CCP4 suite and current developments. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2011, 67, 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N.; Vagin A. A.; Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1997, 53, 240–255. 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2009, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.